Abstract
The ability of the telomeric DNA-binding protein, TRF2, to stimulate t-loop formation while preventing t-loop deletion is believed to be crucial to maintain telomere integrity in mammals. However, little is known on the molecular mechanisms behind these properties of TRF2. In this report, we show that TRF2 greatly increases the rate of Holliday junction (HJ) formation and blocks the cleavage by various types of HJ resolving activities, including the newly identified human GEN1 protein. By using potassium permanganate probing and differential scanning calorimetry, we reveal that the basic domain of TRF2 induces structural changes to the junction. We propose that TRF2 contributes to t-loop stabilisation by stimulating HJ formation and by preventing resolvase cleavage. These findings provide novel insights into the interplay between telomere protection and homologous recombination and suggest a general model in which TRF2 maintains telomere integrity by controlling the turnover of HJ at t-loops and at regressed replication forks.
Keywords: recombination, resolvase, telomere, TRF2
Introduction
The termini of eukaryotic chromosomes are composed of specialised nucleoprotein structures called telomeres that are essential for the protection of chromosome ends against degradation and illicit repair. In vertebrates, telomeric DNA contains several kilobases of tandemly repeated 5′ TTAGGG motifs, terminated by a 3′ oriented single-stranded G-rich tail. In human telomeres, two proteins, TRF1 and TRF2 (TTAGGG repeat factors 1 and 2), specifically recognise the telomeric double-stranded sequence, whereas POT1 binds to the 3′ overhang (Liu et al, 2004a; de Lange, 2005).
Although TRF1, TRF2 and POT1 (together with TIN2, TPP1 and Rap1) form a complex called shelterin or telosome (Liu et al, 2004a; de Lange, 2005), they can also have an independent role(s) at telomeres. For instance, TRF2 protects telomeres against checkpoint recognition and recombination (van Steensel et al, 1998). These protective functions of TRF2 are thought to result from both a TRF2-dependent folding of telomeres into a lasso-like structure called the t-loop (Griffith et al, 1999; Stansel et al, 2001) and the ability of TRF2 to regulate various DNA transactions and enzymatic activities (Bae and Baumann, 2007; Gilson and Geli, 2007).
Four-way DNA structures such as Holliday junctions (HJs) or chickenfeet are structural intermediates in several processes (recombination, repair or replication). Telomeres present a special challenge for managing four-way DNA structures because of (i) the presence of resident telomeric proteins; (ii) inherent difficulties during their replication, which might lead to the formation of a high number of chickenfeet (Fouché et al, 2006b; Gilson and Geli, 2007; Verdun and Karlseder, 2007); (iii) the telomerase-independent mechanisms of telomere maintenance (alternative lengthening of telomeres) that involve recombination (Dunham et al, 2000) and (iv) the hypothetical presence of an HJ at the foot of the t-loop formed by migration of the single-strand 3′ tail inside the D-loop (Stansel et al, 2001). The deletion of t-loops was proposed to be a key event for the creation of extrachromosomal telomeric DNA circles, and for telomere shortening in cells overexpressing TRF2 (Ancelin et al, 2002; Karlseder et al, 2002; Wang et al, 2004). This process, named t-loop homologous recombination (t-loop HR, Wang et al, 2004) depends upon the RAD51 paralog XRCC3 and two TRF2-interacting factors, the XPF protein and the Werner helicase (Zhu et al, 2003; Li et al, 2008) and is inhibited by the N-terminal basic domain of TRF2 (named B) (Wang et al, 2004). In agreement with a key role played by B in the regulation of recombination, Griffith and coworkers showed recently by electron microscopy that this domain binds specifically to a plasmid-based HJ containing telomeric repeats (Fouché et al, 2006a). These data suggest a model of t-loop HR regulation based on a dual role of TRF2, both as an activator by its B-independent capacity to mediate t-loop formation and as an inhibitor through the binding of B on the HJ structure present at the foot of the t-loop.
The mechanism by which TRF2 controls four-way DNA junctions is expected to be of paramount importance for telomere protection, as aberrant recombination of t-loop or stalled replication forks can lead to a severe loss of telomeric DNA and ultimately to cell growth arrest and genome instability. In this report, we demonstrate that the basic domain of TRF2 not only binds to telomeric HJs but also opens their centre, favours their migration and prevents their resolution by resolvases from different sources. We discuss models for the role of TRF2 in the formation, stabilisation and resolution of telomeric HJ and the implications for chromosome end protection.
Results
TRF2 reduces the resolution of the telomeric HJ by human, yeast and bacterial resolvases
To investigate the effect of TRF2 on HJ resolution, we constructed a synthetic semimobile telomeric junction tHJ by incubation of four 54 nt oligonucleotides, each containing two human-type telomeric 5′TTAGGG repeats surrounded by two heterologous 21 nt long nontelomeric sequences (Supplementary Figure 1). Thus, tHJ harbours a 12 nt homologous region at its centre, allowing the junction branch point to migrate through 13 distinct positions (from −6 to +6, Supplementary Figure 2A). This junction is readily recognised by TRF2, to give several distinct complexes that can be visualised by EMSA (top gel, Supplementary Figure 2C). These results indicate that TRF2 forms stable complexes with HJ DNA, as observed previously with double-strand DNA. Deletions of the C-terminal Myb-like telobox domain (TRF2ΔM, Supplementary Figure 2B) or the N-terminal B domain (TRF2ΔB, Supplementary Figure 2B) do not seem to modify the nature of the complexes seen by EMSA, but binding affinity is substantially reduced for the TRF2ΔB mutant (TRF2 concentration at half binding increases from 20 to 75 nM). Deletion of the Myb-like domain only marginally affects junction binding (half binding occurs at 30 nM for TRF2ΔM). Competition experiments performed with TRF2, TRF2ΔB and TRF2ΔM show that the B domain is responsible for the binding preference of TRF2 for the structure of the junction but does not exhibit sequence specificity (Supplementary Figure 3). Thus, efficient binding of TRF2 to tHJ requires its N-terminal B domain, in agreement with the results obtained by Fouché et al (2006a).
To examine whether the binding of TRF2 could affect the processing of HJs, we analysed the effect of TRF2 on HJ cleavage by the human GEN1 HJ resolving activity. GEN1 has been recently identified as the protein responsible for the resolving activity formerly known as ResA (Ip et al, 2008). It was shown that GEN1 cleaves HJ specifically and with perfect symmetry, as observed with other resolving activities such as T4 endonuclease VII, T7 endonuclease I, RuvC and CceI (Ip et al, 2008). When tHJs were treated with a catalytically active N-terminal fragment of GEN1 purified from Escherichia coli (GEN11–527, which resolves HJs in a manner that is identical to that catalysed by the full-length protein), we observed efficient cleavage (Supplementary Figure 4). Denaturing PAGE analysis revealed that GEN11–527 cleaves the HJ by introduction of symmetrically related nicks in pairs of opposite strands. In strands 1 and 3, cleavage occurs in G-rich regions, whereas in strands 2 and 4, the nicks are observed in C-rich regions. The efficiency of resolution of the tHJ by GEN11–527 is reduced by preincubation with TRF2, reaching an inhibition of >80% for the highest concentration of TRF2 (Figures 1 and 2, Supplementary Figure 5). This inhibition is found to depend on the N-terminal B domain of TRF2, as deletion of this domain totally removes the inhibition of cleavage (Figures 1 and 2, Supplementary Figure 5). All cleavage sites are affected in an equal manner for sites on the G-rich strands and on the 3′ side of the C-rich strands. However, sites located on the 5′ side of the latter strands seem more resistant to TRF2-inhibition (asterisks in Figure 2 and Supplementary Figure 5). This behaviour may depend on the Myb-like domain of TRF2, as its deletion harmonises the cleavage profiles. Overall, these data show that the B-dependent binding of TRF2 on a telomeric HJ has a marked effect on its cleavage by this human HJ resolving enzyme and might provide important insight into how TRF2 could control the processing of HJs. To determine whether this inhibition is a general feature of TRF2 or is specific to GEN1, we investigated the effect of TRF2 on three archetypal enzymes representing the three major groups of resolving enzymes (Rafferty et al, 2003; Déclais and Lilley, 2008): Endonuclease I of phage T7 (abbreviated as T7 Endo I) of the nuclease family, the yeast mitochondrial CceI enzyme of the integrase family and the unclassified RusA resolving enzyme. T7 Endo I cleaves HJs 1 nt 5′ to the junction centre (Déclais et al, 2006) and cleaves tHJ on all strands at 12 different positions corresponding to 12 of the 13 possible positions of the branch point (Supplementary Figures 6 and 7). The absence of cleavage for the most 5′ terminal positions as well as the presence of favoured cleavage sites could be explained by some sequence selectivity of the enzyme (Picksley et al, 1990; Déclais et al, 2006). CceI exhibits sequence specificity for cleavage (White and Lilley, 1996), and its main site (3′ side of a CT dinucleotide) can be found in the two C-rich strands of the telomeric junction (Supplementary Figure 8 and data not shown). The same strands will be cleaved by RusA, which also presents sequence specificity (5′ side of a CC dinucleotide) (Giraud-Panis and Lilley, 1998), although, in this case, one putative site seems to be less prone to cleavage (Supplementary Figure 8).
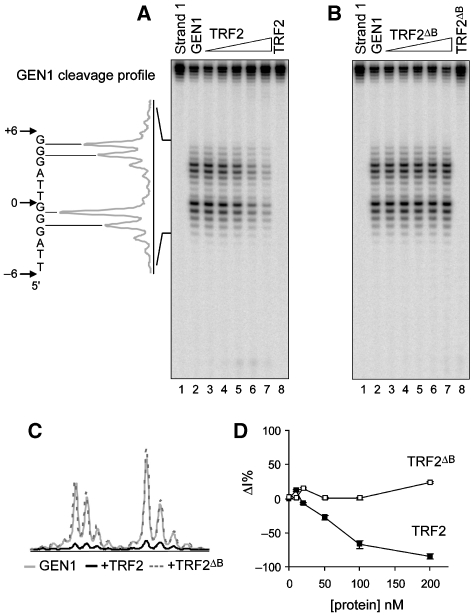
Figure 1.
TRF2 protects tHJ from cleavage by GEN1 in a B domain-dependent manner. (A) A quantity of 5 nM of tHJ labelled on strand 1 was incubated with increasing amounts of TRF2 before cleavage with GEN11–527. Concentrations of TRF2 used were 10, 20, 50, 100 and 200 nM. Lane 1 shows the uncleaved junction, and in lane 8 only 200 nM of TRF2 was added. Positions of GEN1 major cleavage sites (assigned by comparison with the result of a Maxam and Gilbert A+G sequencing reaction, Supplementary Figure 4) and the cleavage profile are represented on the left side. (B) Same experiment as in (A) with TRF2ΔB. (C) Comparison of cleavage profiles for GEN1 alone (grey line) or in the presence of 200 nM of TRF2 (black line) or TRF2ΔB (dotted line). (D) Graph representing the variations in percentage of the intensities of each cleavage band (ΔI%) as a function of TRF2 concentration (closed squares) or TRF2ΔB (open squares). The values correspond to average ΔI were calculated using the four major cleavage bands. Error bars correspond to standard errors in these values.
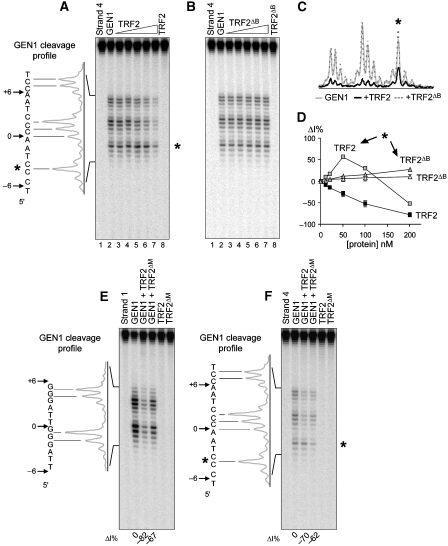
Figure 2.
TRF2 protects tHJ from cleavage by GEN1 in a B domain-dependent manner. (A) A quantity of 5 nM of tHJ labelled on strand 4 was incubated with increasing amounts of TRF2 (10, 20, 50, 100 and 200 nM) before cleavage with GEN11–527. Lane 1 shows the uncleaved junction, and in lane 8 only 200 nM of TRF2 was added. Positions of GEN11–527 major cleavage sites and the cleavage profile are represented on the left side. Note the distinct behaviour obtained for the 5′ terminal cleavage site (*). (B) Same experiment as in (A) with TRF2ΔB. (C) Comparison of cleavage profiles for GEN11–527 alone (grey line) or in the presence of 200 nM of TRF2 (black line) or of TRF2ΔB (dotted line). (D) Graph representing the variations in percentage of cleavage intensity (ΔI%) as a function of the concentration of TRF2 (closed and grey squares) or TRF2ΔB (open and grey triangles). Grey symbols correspond to the behaviour of the 5′ terminal cleavage site (*). Other symbols with their corresponding error bars represent the average behaviour and the corresponding standard errors of all other major cleavage sites. (E) A quantity of 5 nM of tHJ labelled on strand 1 was incubated with 200 nM of TRF2 or of TRF2ΔM before cleavage with GEN11–527. The first lane shows the uncleaved junction, and in the last two lanes only telomeric proteins were added. Numbers below the gel represent the variations in cleavage intensity (ΔI%) in each sample containing the telomeric protein compared with the sample only containing the enzyme. Negative numbers show protection. (F) Same experiment as in (E) with strand 4. *Marks the position of the 5′ terminal cleavage site.
Remarkably, TRF2 impairs the action of all three enzymes (Supplementary Figures 6–8), similarly to that observed with GEN1. In the case of CceI, cleavage of both strands is inhibited and all sites are equally affected (Supplementary Figure 8 and data not shown). For RusA, inhibition is stronger on strand 4 but, as for CceI, all cleavages are equally impaired within one strand. The effect of TRF2 on T7 Endo I is more complex and more resembles that observed on GEN1 cleavage. Although the inhibition can be observed on four centrally located positions (black symbols in graph and profile in Supplementary Figures 6 and 7), other positions show an increase in T7 Endo I activity (again 5′ located sites, grey symbols) or even the absence of an effect (open symbols). It is worth noting that, as for GEN1, removing the Myb-like domain results in the loss of the 5′ cleavage activation.
Inhibition of T7 Endo I, RusA and CceI cleavage, like that of GEN1, is mainly dependent on the B domain of TRF2 (Supplementary Figures 6–8). Therefore, TRF2 binding to tHJ through its Myb-like domain is not sufficient per se to mask the cleavage sites of these enzymes. In further agreement with a specific role of B, TRF1 (which does not contain an N-terminal basic domain) does not inhibit the cleavage by T7 Endo I, CceI or RusA (Supplementary Figure 9). As expected, because TRF1 and TRF2 share a nearly identical Myb-like domain, activation of Endo I cleavage at 5′ positions is visible for the TRF1-bound junction (Supplementary Figure 9A).
Overall, we conclude that TRF2 prevents cleavage by the human GEN1 enzyme and three archetypal resolvases that recognise and cleave HJ in different ways and represent the three major types of known resolving enzymes. This is achieved through the recognition of the junction centre by the B domain.
An assay to measure the rate of formation and migration of a telomeric HJ
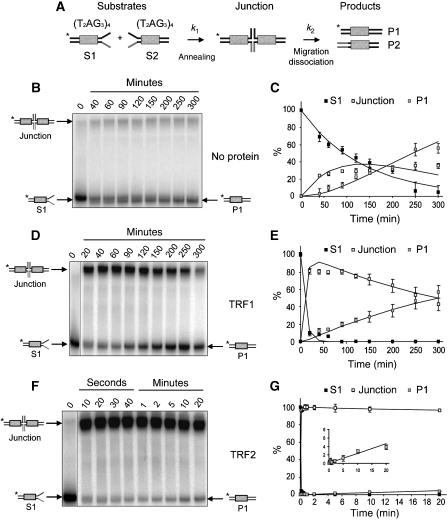
Next, we analysed the influence of TRF2 on the rate of branch migration through a telomeric sequence. For this purpose, we adapted an in vitro assay originally designed by Panyutin and Hsieh (1994). A HJ is obtained through the annealing of two homologous double-stranded DNA (S1 and S2) containing four TTAGGG repeats located 22 bp away from the 3′ terminus of the homologous region. They both end by single-stranded tails presenting complementary sequences between each other (Figure 3A, Supplementary Figure 10). Once formed, this junction can spontaneously migrate by random walking, with a rate that will depend on the assay conditions and DNA sequence (Panyutin and Hsieh, 1994; McKinney et al, 2005). Finally, an irreversible dissociation step leads to fully hybridised duplex products (P1 and P2). Disappearance of the labelled S1 substrate and appearance of the junction and of the P1 product can be followed by gel electrophoresis of the sample (Supplementary Figure 10B), allowing quantification of all species. The presence of the single-stranded tails in S1 and the hybridisation state of P1 were both verified by cleavage with the HindIII and BamHI restriction enzymes (H and B, respectively, Supplementary Figure 10C). In an additional control, we noted that when using an S2 substrate containing a 6-bp heterologous sequence (hS2, Supplementary Figure 10D), the junction was still formed but migration prevented, and thus no product was formed (Supplementary Figure 10E).
Figure 3.
TRF2 greatly increases the junction formation but slows down migration. (A) Reaction scheme of the migration assay and corresponding rates k1 and k2. (B) Migration assay performed in the absence of telomeric proteins. Numbers above indicate the time points corresponding to each sample. (C) Variations of the percentage of the S1, junction and P1 species through time corresponding to the experiment shown in (B). The lines represent the fitting curves obtained with the rates calculated using the experimental data. (D) Migration assay performed in the presence of 300 nM of TRF1. (E) Same as (C) for TRF1. (F) Migration assay performed in the presence of 100 nM of TRF2. (G) Same as (C) for TRF2. The insert shows an enlarged part of the graph corresponding to the early time points.
A kinetic analysis for junction formation and migration was performed. To simplify the analysis, the migration and final dissociation steps were associated in a single step described by k2 (Figure 3A), as performed previously by Panyutin and Hsieh (1994). Both k1 and k2 were obtained by fitting the experimental data to the three equations presented in Materials and methods.
In the absence of telomeric proteins (Figure 3B and C), the different steps are slow (Table I). Although the slowness of the annealing process can be explained by the low concentration of S1 used (2 nM), the migration/dissociation step is far slower than what could be expected from previous results on random DNA. As a comparison, Panyutin et al measured a half time of 20 min for the substrate of a strand-exchange reaction on a 956-bp DNA fragment (30°C, 100 mM NaCl). In the case of our 88 bp substrate, 40% of the junction still remains 50 min after disappearance of the substrate (30°C, 50 mM NaCl). This slowness suggest that the junction spends a significantly larger amount of time in the telomeric sequence compared with random DNA, a phenomenon already observed by Fouché et al (2006b).
Table 1.
Annealing and migration/dissociation rates of the HJ in the presence of telomeric proteins
| Components | k1 M−1 s−1 | k2 s−1 |
|---|---|---|
| DNA alone | 6.2±0.4 × 103 | 1.2±0.1 × 10−4 |
| TRF1 | 9±2 × 104 | 4±0.5 × 10−5 |
| TRF2 | >107 | 3±1 × 10−5 |
| TRF2ΔB | 2.9±0.3 × 106 | 3±2 × 10−5 |
| TRF2ΔM | 1.1±0.2 × 106 | 4±1 × 10−4 |
| B peptide | 2.8±0.5 × 104 | 6±2 × 10−4 |
| TRF2AΔB | 1±0.2 × 105 | 6±3 × 10−5 |
TRF2 accelerates the rate of junction formation
We measured the rate of junction formation in the presence of saturating amounts of TRF1, TRF2 and truncated forms of TRF2 (Figure 3D–G, Supplementary Figure 11 for longer time points with TRF2 and Table I). When TRF1 is added, the annealing rate is increased by about 10-fold; this is probably due to the capacity of TRF1 to form paired synapses in DNA (Griffith et al, 1998), thereby helping to bring the substrates together. With TRF2, the annealing step is strikingly accelerated (k1 is more than 100 times higher in the presence of TRF2 than TRF1). TRF2ΔB and TRF2ΔM also increase annealing suggesting that both domains participate in this step. Likewise, we observed that a chemically synthesised B peptide corresponding to residues 1–45 of TRF2 can also accelerate the annealing (about 5 times), although not as efficiently as TRF2ΔM. The participation of the sequence-specific binding domain (Myb-like domain) in an event requiring a sequence 22 bp away from the telomeric tract suggests that the annealing step involves more than a simple B-dependent recognition of the junction. TRF2 is known to oligomerise on DNA, and we have reported recently that this property, missing in TRF1, is controlled by the homodimerisation domain (Amiard et al, 2007). This was deduced from data obtained using a mutant in which the B domain of TRF2 was replaced by the acidic domain of TRF1 (TRF2AΔB). This TRF2 mutant is still capable of binding telomeric DNA but creates complexes resembling those of the dimeric TRF1 protein (Supplementary Figure 12). In the migration assay, this protein behaves similarly to TRF1; annealing is fast, but not as fast as with TRF2ΔB, suggesting that oligomerisation of TRF2 has an important function in the formation of the junction. Oligomerisation has been proposed to allow TRF2 to bring together two otherwise-separated DNA regions. In the migration assay, a similar mechanism could help to bring the substrates together, therefore, greatly increasing the annealing rate.
The Myb-like domain of TRF2 slows the rate of junction migration, while the basic domain accelerates it
TRF1 and TRF2 reduce the rate of branch migration by 3–4-fold (Table I), for which the domain responsible is the Myb-like C-terminal telobox domain (compare the results obtained for TRF2 and TRF2ΔB). By contrast, an increase in the migration rate is observed with TRF2ΔM and the B peptide alone, indicating that the basic domain of TRF2 favours branch migration. Therefore, the two DNA-binding domains of TRF2 have opposite effects on branch migration; the Myb-like sequence-specific domain decreases the rate of branch migration, whereas the N-terminal junction-specific domain accelerates it. The Myb-dependent activation of T7 Endo I and GEN1 cleavages that we observed for TRF2 and TRF1 is probably due to this retardation of migration. The binding of this domain on the telomeric sequence could stabilise junctions with a branch point located at the ends of the telomeric tract. We note that these positions correspond to conformations where the telomeric sequence is only located within two of the four arms and, therefore, presents a straighter global conformation.
The basic domain induces sensitivity to attack by potassium permanganate in duplex and four-way junction DNA structures
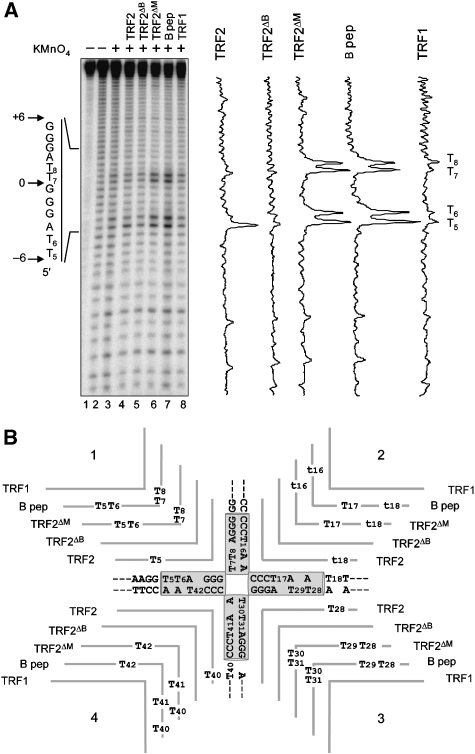
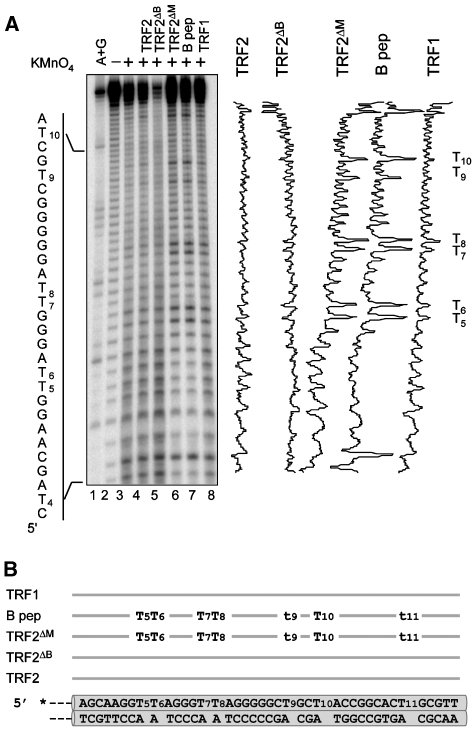
As the B domain increases junction migration (see above), B-dependent decrease in resolving enzyme cleavage cannot be explained by a stabilisation of a subset of junctions. As an alternative explanation, binding of B could protect the junction centre by competing with the enzyme, or by modifying the junction structure, or both. Therefore, we investigated whether B modifies the junction structure by potassium permanganate (KMnO4) probing, a chemical compound that allows detection of unpaired or distorted pyrimidine bases, with a preference for thymines (Buckle et al, 1991). Analysing all four strands of tHJ with KMnO4 in the absence of divalent cations (Figure 4 and Supplementary Figure 13) shows background levels of activity due in part to piperidine cleavage (Figure 4A, lane 2) and in part to the mobility of the junction branch point that leads to a certain degree of unpairing in the homologous central sequence (Figure 4A, lane3). When TRF2 is bound on the junction in saturating conditions (corresponding EMSA are shown in Supplementary Figure 13A), permanganate-hypersensitive thymines appear on all four strands (titration experiments show that this effect is dependent upon TRF2 concentration, as expected, Supplementary Figure 14). All these sites correspond to possible positions of the junction branch point (−6, −5 or +6 positions). The basic domain of TRF2 is necessary and sufficient to induce the KMnO4 reactivity: hypersensitive sites are not detected using TRF2ΔB or TRF1, and stronger and broader thymine reactivity is induced by the basic domain alone (B peptide) or by TRF2ΔM. In the latter cases, hypersensitive sites correspond to all thymines within the homologous core. Thymines located in the arms of the junction are not affected underlining the marked preference of the B domain for the junction centre where the junction branch point is located. These results indicate that binding of the basic domain results in an alteration of the structure of the central base pairs of tHJ.
Figure 4.
TRF2 creates potassium permanganate hypersensitive sites on tHJ. (A) Permanganate probing experiment performed with 5 nM of double-stranded tHJ labelled on strand 1 in the presence of 500 nM of TRF1, TRF2, TRF2ΔB or TRF2ΔM and 5 μM of the B peptide. Lane 1 shows the undigested strand. Lanes 2 and 3 show the background cleavage obtained through piperidine cleavage alone and with KMnO4, respectively. Sequence of the telomeric tract is given on the left side. On the right side are presented the relative intensity profiles obtained after quantification of the samples lanes. (B) Summary of the probing assay obtained for all proteins on all junction strands. Each line above the junction corresponds to the action of the indicated protein, capital ‘Ts' represent major hypersensitive sites, lower case ‘ts' correspond to minor sites.
Analysis of the activity of TRF2ΔM and of the B peptide on a double-stranded substrate shows all the thymines of the sequence to be reactive (Figure 5). The B domain is expected to bind nonspecifically at every possible position on a simple duplex, lacking a branch point. The appearance of hypersensitive sites at all thymines, therefore, indicates that the B domain has the intrinsic ability to open or distort any double-stranded DNA sequence to which it binds.
Figure 5.
The B domain of TRF2 creates potassium permanganate hypersensitive sites on the double stranded dsT2 probe. (A) Permanganate probing experiment performed with 5 nM of double stranded dsT2 labelled on the top strand in the presence of 500 nM of TRF1, TRF2, TRF2ΔB or TRF2ΔM and 5 μM of the B peptide. Lane 1 shows the result of a Maxam and Gilbert A+G sequencing reaction performed on the same strand. Lanes 2 and 3 show the background cleavage obtained through piperidine cleavage alone and with KMnO4, respectively. Sequence of the DNA substrate is given on the left side. On the right side are presented the relative intensity profiles obtained after quantification of the samples lanes. (B) Summary of the probing assay, major sites are indicated by capital Ts and minor sites by lower case ts.
The B domain creates or stabilises opened intermediates in DNA
To study base-pair opening in DNA induced by the B domain, we used differential scanning calorimetry (DSC). We analysed the thermal melting of a small double-stranded duplex containing two telomeric repeats (18dsT2), and the influence of the B peptide on this process. DSC is sensitive to the melting of all folded components and their complexes, giving rise to peaks in the DSC curves which maximum corresponds to their melting temperature (Tmax). By good fortune, the B peptide alone gave no thermal signal in the DSC (Figure 6C), suggesting that this peptide does not undergo folding. However, to interpret the DSC data correctly, we needed to know whether the peptide is induced to fold when it binds the DNA probe.
Figure 6.
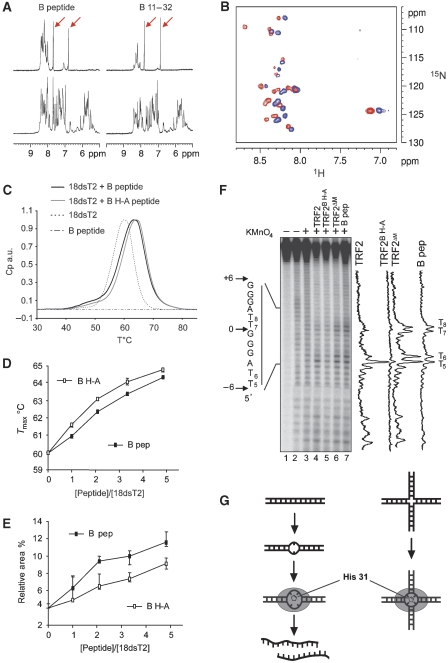
The B domain causes base-pair opening. (A) Spectral region from 5 to 10 ppm of the one-dimensional 1H NMR spectra of a 0.1-mM solution of the B and B11–32 peptides alone (top), and in the presence of 0.1 mM of a the 18dsT2 probe (bottom). The arrows mark resonances assigned to the histidine side chain protons. (B) Superposition of the 15N-HSQC spectra of a 1-mM solution of the B11–32 peptide in the absence of DNA (blue) and in the presence of an equal concentration of the 18dsT2 probe (red). (C) Normalised differential scanning calorimetry curves for DNA, peptide and DNA–peptide complexes (103.7 μM of peptides). (D) Variations of the Tmax as a function of the molar ratio between the B (closed rectangles) or B H-A (open rectangles) peptides and the 18dsT2 probe. (E) Variations of the relative area of the peak corresponding to the intermediate species as a function of the molar ratio between the B (closed rectangles) or B H-A (open rectangles) peptides and the 18dsT2 probe. Error bars in (D) and (E) were drawn using the minima and maxima obtained for each value. (F) Permanganate probing experiment performed with 5 nM of the junction tHJ labelled on strand 1 in the presence of 500 nM of TRF2, TRF2DM, TRF2B H-A or 5 μM of B peptide. Lane 1 shows the undigested strand. Lanes 2 and 3 show the background cleavage obtained through piperidine cleavage alone and with KMnO4, respectively. Sequence of the telomeric tract is given on the left side. On the right side are presented the relative intensity profiles obtained after quantification of the samples lanes. (G) The binding of the B domain on DNA creates or stabilises DNA opened intermediates both in a duplex (left side) and in a four-way DNA junction (right side). This melting effect requires His31 of the B domain.
For this purpose, we performed nuclear magnetic resonance (NMR) experiments on the full-length B peptide, and a shorter version of this peptide comprising residues 11–32, and the 18dsT2 probe (Figure 6A and B). In the absence of DNA, the amide protons of the peptides resonate between 7 and 8.7 ppm, indicating that the peptides do not possess secondary structure, thereby corroborating the absence of structure revealed by DSC. This was also confirmed by 1H-15N HSQC experiments on the B11–32 peptide (Figure 6B). The presence of DNA induces the appearance of new resonances corresponding to DNA protons, and the shifting of some resonances of the peptides confirms the interaction between both species (Figure 6A). Importantly, binding does not increase the amplitude of the range we observed for the chemical shifts of the peptide, which argues against a folding of this peptide in the presence of DNA. This absence of secondary structure was also confirmed by 1H-15N HSQC experiments on the bound form of the B11–32 peptide (Figure 6B). Thus, the signal observed in the DSC scan of the DNA–peptide complex will only correspond to the melting behaviour of the DNA.
The 18dsT2-melting curve (Figure 6C) shows a maximum at 60°C, and closer analysis reveals the presence of a small shoulder between 40 and 47°C that corresponds to 4% of the total area of the transition peak (Figure 6E). This shoulder could indicate that the melting of our telomeric probe is not a simple transition between a closed and an open state but rather involves opened conformational intermediates (Spink, 2008). This behaviour was expected to some extent, as it has been shown in several reports that telomeric DNA has the tendency to open more easily than random DNA (Amiard et al, 2007 and references within). Adding the B peptide to the probe significantly modifies the melting curve. First the curve is shifted to a higher temperature, giving a rise in the Tmax that depends on the concentration of peptide (Figure 6D). This, again, supports the interaction between the peptide and the DNA probe and indicates that the B peptide leads to an overall stabilisation of the probe. More importantly, the area of the low-temperature shoulder that was observed for the DNA sample increases (Figure 6C). This indicates that the binding of the peptide stabilises transition intermediates or stimulates their appearance in the telomeric DNA. In support of this, competition experiments performed by EMSA using a nontelomeric 54 bp duplex and the TRF2ΔM protein show a slight but significant preference for a DNA containing a two base-pairs T bubble (Supplementary Figure 15). Furthermore, when the duplex contains a two base-pairs T bulge, which creates a local kinking of the helix axis, the competition is even more effective. This is consistent with preferred binding of the B domain of TRF2 to branched DNA molecules, and suggests that opening is also favoured. Overall, these data indicate that the B domain of TRF2 both stabilises the telomeric DNA and creates or stabilises opened intermediates (a model is shown on the left part of Figure 6G).
Histidine 31 participates in DNA binding, base-pair opening and protection against resolving enzyme cleavage
Poor resolution of the 1H resonances of the 1D spectra does not allow the assignment of all resonances. However, two narrow resonances at 6.9 and 7.8 ppm corresponding to protons of the histidine 31 imidazole group can be distinguished. These resonances are shifted in the presence of the DNA indicating that H31 participates in the interaction (Figure 6A). DSC experiments on a mutated peptide containing alanine in place of this histidine (B H-A peptide) also confirm the involvement of this residue (Figure 6C–E). The mutation causes a significant decrease in the fraction of intermediate species, suggesting that it is less efficient in base-pair opening (lower relative area Figure 6E). Furthermore, this mutant also causes an increase in the stability of the DNA (exemplified by a higher Tmax; Figure 6D). Taken together, these data indicate that His31 both participates in the binding of the peptide and in the opening of the double-stranded DNA. This suggests that His31 is also involved in the opening of the junction centre. Indeed, in saturating-binding conditions, the TRF2 mutant with alanine replacing His31 in the full-length protein (TRF2B H-A) causes a marked decrease in the KMnO4 hypersensitivity of the junction in the presence of TRF2 (Figure 6F on strand 1, data not shown for binding and for the other strands).
Moreover, TRF2B H-A does not inhibit cleavage by GEN11–527, T7 Endo I and CCE1, even under condition of saturating binding (Supplementary Figure 16), consistent with a role of the His31-dependent melting in junction-resolving enzyme inhibition.
Discussion
We demonstrated in this work that TRF2 is able to affect all the major biochemical aspects of an HJ: its formation, its migration and its resolution.
TRF2 promotes Holliday junction formation
We have shown that formation of HJ is greatly enhanced by the combined action of the TRF2 Myb-like domain and of the B domain aided by the oligomerisation of the protein, which increases substrate assembly. Although the B domain participates in this annealing step, it appears dispensable (deleting this domain only causes a three-fold decrease in the annealing rate; Table I). Taken together with the fact that both TRF2 and TRF2ΔB were shown to stimulate single-strand invasion (Amiard et al, 2007), these findings suggest that TRF2 could assist in the first steps of homologous recombination between telomeric repeats in a basic domain-independent manner. Similarly, TRF2 is expected to stimulate junction formation at the basis of a t-loop. In view of the marked preference of TRF2 for branched structure, TRF2 could also readily promote the formation of a chickenfoot in the context of a stalled replication fork on telomeres. Such a junction-promoting activity could also explain the inhibition of gene conversion observed in a TRF2-depleted background using a reporter model (Mao et al, 2007).
Further analysis reveals that both full-length TRF1 and TRF2 decrease but do not block junction migration. This effect is mainly driven by the Myb-like domain, which explains why both proteins behave in the same manner, their C-terminal telobox domains being almost identical. Migration of DNA strands and junction branch point have been hypothesised for the formation of the t-loop HJ and of a potential double HJ at the base of the loop (de Lange, 2005). That neither TRF1 nor TRF2 prevents such migration in vitro supports this model. Telomeric junction migration could also be enhanced by other proteins such as the Werner protein or the Bloom protein in conjunction with Topoisomerase IIIα (Constantinou et al, 2000; Opresko et al, 2002; Plank et al, 2006; Temime-Smaali et al, 2008). Another possible candidate is the RAD54 protein that can also promote branch migration (Bugreev et al, 2006) and is involved in telomere length maintenance (Jaco et al, 2003).
The basic domain of TRF2 creates or stabilises opened intermediates of telomeric DNA
The N-terminal basic domain of TRF2 induces potassium permanganate reactivity, and opens DNA as observed by DSC, suggesting that this domain has the ability to melt duplex telomeric DNA and the centre of an HJ. In agreement with these melting properties, the basic domain increases branch-point migration. Moreover, through NMR, DSC and permanganate probing experiments, we showed that H31 participates in DNA binding and helix opening. To our knowledge, participation of a histidine in DNA melting has not been commonly reported with the exception of the T4 Endonuclease VII resolving enzyme, which uses two histidines to widen the junction centre (Biertumpfel et al, 2007). Such a mechanism could be used by TRF2, although, in that case, we observe base-pair opening rather than widening. As the B domain of TRF2 does not contain aromatic residues, the molecular mechanism of this opening seems to differ from a classical intercalation but alternatively it could involve the telomeric DNA itself. Indeed, telomeric DNA has been shown to be able to open more easily than random DNA (this work and Amiard et al, 2007 and references within).
TRF2 inhibits the cleavage of HJ by the human GEN1 and three archetypal junction-resolving enzymes
Analysis of the effect of TRF2 on HJ resolution reveals the ability of this protein to impair enzymatic cleavage by four different enzymes including the last addition of the family, the human GEN1 HJ-resolving enzyme, through a process that involves the B domain. Interestingly, a point mutation at His31, which abolishes the B-dependent DNA melting at the junction centre, precludes the capacity of TRF2 to inhibit GEN1, T7 Endo I and CCE1, suggesting that one of the mechanisms by which B inhibits the cleavage could be to open the junction centre. The B-dependent inhibition of the human resolvase GEN1 could provide an elegant explanation for the inhibition of t-loop HR due to TRF2 and its activation in the context of the overexpression of TRF2ΔB. Indeed, t-loop HR has been shown to involve XRCC3, a protein that forms a complex with RAD51C and associates with the HJ resolvase ResA, which is now known to be the GEN1 endonuclease (Liu et al, 2004b; Wang et al, 2004; Ip et al, 2008). In this context, it is possible that inhibition of t-loop HR could be attributed to the B-dependent inhibition of the GEN1 resolvase we observe, therefore explaining the biological effect of the overexpression of TRF2ΔB—that is, loss of t-loop protection and resolution.
The ‘Janus effect' of TRF2 on telomere protection and recombination
This work unveils two basic properties of TRF2 with respect to HJs: it stimulates their formation and prevents their resolution. As TRF2 is a major telomere capping protein, this suggests that HJs are involved in telomere protection. Indeed, homologous recombination is critical for telomere replication and the formation of HJ is likely to be involved in t-loop formation or stabilisation (Stansel et al, 2001; Verdun and Karlseder, 2006). We propose that part of the capping functions of TRF2 relies on its intrinsic ability to control HJ turnover: the capacity to oligomerise, which appears to enhance HJ formation and the ability of the B domain to bind and to open the junction centre, which inhibits the action of different types of resolvase. Therefore, TRF2, similarly to Janus, is a double-faced telomere caretaker: its ability to favour the presence of HJ at telomere may contribute to telomere capping, but it also increases the susceptibility of telomere for deletion by homologous recombination. The balance between the pro- and anti-recombinogenic intrinsic properties of TRF2 is likely to be modulated in the cell by its interaction with other factors such as the Werner, topoisomerase IIIα, Apollo or Rap1 proteins. Future studies will probably shed further light on this.
Materials and methods
DNA probes, proteins, peptides and enzymes
Sequences and purifications of DNA probes can be found in Supplementary data. TRF2 proteins and mutants were as follows TRF2 (3–500), TRF2ΔB (45–500), TRF2ΔM (3–437), TRF1 (2–439), TRF2AΔB contains residues 2 to 67 from TRF1 followed by residues 47–500 from TRF2, TRF2B H-A contains residues 3–500 of TRF2 and bears a His to Ala mutation at position 31. All telomeric proteins were fused to a N-terminal tag containing six histidines and were purified using the procedure as described in Amiard et al (2007).
T7 Endonuclease I, MBP-CceI, MBP-RusA and GEN11–527 were purified as already published (White and Lilley, 1996; Giraud-Panis and Lilley, 1998; Déclais et al, 2006; Ip et al, 2008). The TRF2 B (1–45), B 11–32 (11–32) and the B H-A (1–45 H31 has been replaced by alanine) peptides were synthesised by Invitrogen. Their sequence is presented in Supplementary data.
EMSA and competition assays
Proteins were incubated 15 min at 20°C with 5 nM DNA in 10 μl of 20 mM Tris–Acetate pH 8, 0.1 mg ml−1 BSA, 1 mM dTT, 50 mM KAc, 30 mM NaCl, 5% (v/v) glycerol (binding buffer). For competition experiments, various concentrations of unlabelled competitor were added to the mixture before the addition of the proteins. Gel electrophoresis was performed as published (Amiard et al, 2007) and gels were analysed using a Phosphorimager FLA5100 (Fuji) and the Multigauge software (Fuji).
Resolution assays
A quantity of 5 nM of tHJ labelled on one strand was incubated with different concentrations of telomeric proteins in 10 μl of binding buffer containing 10 mM MgAc2 and digested for 60 min using 2 nM of T7 Endonuclease I, MBP-CceI and MBP-RusA or 50 nM of GEN11–527. Reactions were performed at 4°C for Endo I or 37°C for MBP-CceI, MBP-RusA and GEN11–527.
The reaction was stopped by addition of 6 μg of proteinase K followed by 20 min incubation at 4°C or 15 min at 37°C. Formamide was added to a final concentration of 60% (v/v) and the samples loaded on a 10% denaturing acrylamide (19:1, bis:mono) 1 × TBE gel. After migration in 1 × TBE, gels were treated as above. Each lane on the gel was quantified giving profiles that were corrected for loading differences. Intensities of each band were quantified and variations in percentage from the initial protein-free values were calculated.
Migration assay
A quantity of 2 nM of labelled S1 substrate was incubated in 90 μl of binding buffer at 30°C in the absence or presence of the appropriate protein for 15 min. The migration assay started by addition of 10 μl of S2 substrate at a final concentration of 20 nM. Aliquots (10 μl) were taken at different time points, and the reaction was stopped by addition of 2 μl of 60 mM MgCl2, 6 μg/ml EthBr, 0.65% SDS and 20 μg of Proteinase K and incubated during 5 min at 30°C. Samples were kept on dry ice before loading on a 8% native acrylamide (19:1, bis:mono) 1 × TB gel with 3 mM MgCl2 and 0.5 μg/ml EthBr and migrated at 15 V/cm for 4 h. After migration, gels were dried and analysed as above.
Experimental data obtained for the concentration of each species were fitted using:
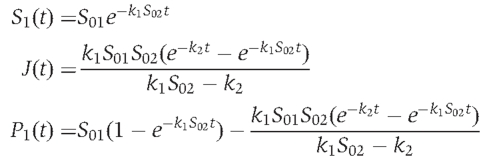 |
where S01 is the initial concentration of S1 substrate; S02 is the initial concentration of S2 substrate; t corresponds to the time in sec; k1 is the annealing rate constant (M−1 s−1); k2 is the migration/dissociation rate constant (s−1).
Permanganate probing
A quantity of 5 nM of tHJ labelled on either strand was incubated for 15 min in the presence or absence of various concentrations of the appropriate protein in 10 μl of binding buffer at 20°C. KMnO4 was added to a final concentration of 6 mM. After 5 min, 3 μl of stop solution was added (6 M β-mercaptoethanol, 1.8 M NaOAc) and the DNA was ethanol precipitated. DNA was cleaved by addition of piperidine (10%) and incubation at 95°C during 30 min. The samples were lyophilised three times resuspended in 5 μl of 60% formamide with dyes and loaded on a 10% 1 × TBE acrylamide (19:1, bis:mono) denaturing gel. After electrophoresis in 1 × TBE, the gel was dried and exposed as above. Each lane on the gel was quantified giving profiles that were corrected for loading differences. The relative intensity profiles presented in the figures were obtained by subtracting the control profile without protein to each sample profile.
NMR
One-dimensional 1H-NMR spectra were acquired at 27°C using a Bruker Avance III DRX 500 spectrometer with a 5-mm QXI probe. NMR experiments were performed on samples containing 0.1 mM of the free and bound peptide in 90% H2O/10% D2O, 50 mM NaCl, 20 mM Hepes, pH 7.0. 1D spectra were collected with 16 K points and 64 scans. 2D heteronuclear spectra (1H-15N, HSQC recorded with 15N at natural abundance, 0.37%) were recorded for the free and bound B11–32 peptide (1 mM) in 10 mM phosphate buffer, pH 6.0, on a 600-MHz spectrometer (Bruker DRX600 AvanceIII) equipped with a TCI cryoprobe. About 128 increments were collected, with spectral widths of 8400 Hz (1H) and 6000 Hz (15N), and 512 scans per t1 increment were made.
Differential scanning calorimetry
DSC experiments were performed using the 6300 nano-DSC III (Calorimetry Sciences Corporation). In a 1-ml volume of calorimetry buffer (Tris 20 mM, pH=8, NaCl 50 mM, previously heated and degased), 32 μM of the 18dsT2 probe was incubated with either peptide at various concentrations (0, 32, 66, 103.7 and 147 μM) at room temperature for 15 min then scanned against the same buffer in the reference capillary cell from 20 to 90°C (1°C/min). Five heating and cooling cycles were recorded. Data were analysed with the software provided by the manufacturer (CpCalc) to correct for baseline effects, and to calculate the excess heat capacity (Cp). The five heating curves were averaged and spline interpolated using Matlab. Average curves were normalised and only heating curves are shown on Figure 6C, cooling curves giving similar results. For calculation of the relative area of the intermediate species transition, the data were fitted by a Gaussian mixture using an optimisation Matlab script based on the EM algorithm (Expectation-Maximization) and the means, variances and relative proportions of the two Gaussian distributions as fitting parameters.
Supplementary Material
Supplementary Figures
Acknowledgments
This work was supported by grants from the Ligue Nationale contre le Cancer (Eric Gilson ‘équipe labellisée'), Cancer Research UK and from the Institut National du Cancer (INCa, program TELINCA). AP is supported by a fellowship from the Ligue Contre le Cancer de Haute Savoie, SA was supported by a fellowship from the Association pour la Recherche sur le Cancer.
References
- Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, Paoletti J, Gilson E, Giraud-Panis MJ (2007) A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E (2002) Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol 22: 3474–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae NS, Baumann P (2007) A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 26: 323–334 [DOI] [PubMed] [Google Scholar]
- Biertumpfel C, Yang W, Suck D (2007) Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 449: 616–620 [DOI] [PubMed] [Google Scholar]
- Buckle M, Fritsch A, Roux P, Geiselmann J, Buc H (1991) Kinetic studies on promoter-RNA polymerase complexes. Methods Enzymol 208: 236–258 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV (2006) Rad54 protein promotes branch migration of Holliday junctions. Nature 442: 590–593 [DOI] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC (2000) Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep 1: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Déclais AC, Lilley DM (2008) New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr Opin Struct Biol 18: 86–95 [DOI] [PubMed] [Google Scholar]
- Déclais AC, Liu J, Freeman AD, Lilley DM (2006) Structural recognition between a four-way DNA junction and a resolving enzyme. J Mol Biol 359: 1261–1276 [DOI] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR (2000) Telomere maintenance by recombination in human cells. Nat Genet 26: 447–450 [DOI] [PubMed] [Google Scholar]
- Fouché N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD (2006a) The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem 281: 37486–37495 [DOI] [PubMed] [Google Scholar]
- Fouché N, Ozgur S, Roy D, Griffith JD (2006b) Replication fork regression in repetitive DNAs. Nucleic Acids Res 34: 6044–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Lilley DM (1998) Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J Mol Biol 278: 117–133 [DOI] [PubMed] [Google Scholar]
- Griffith J, Bianchi A, de Lange T (1998) TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol 278: 79–88 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361 [DOI] [PubMed] [Google Scholar]
- Jaco I, Munoz P, Goytisolo F, Wesoly J, Bailey S, Taccioli G, Blasco MA (2003) Role of mammalian Rad54 in telomere length maintenance. Mol Cell Biol 23: 5572–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, de Lange T (2002) Senescence induced by altered telomere state, not telomere loss. Science 295: 2446–2449 [DOI] [PubMed] [Google Scholar]
- Li B, Jog SP, Reddy S, Comai L (2008) WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol Cell Biol 28: 1892–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z (2004a) Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O'Regan P, West SC (2004b) RAD51C is required for Holliday junction processing in mammalian cells. Science 303: 243–246 [DOI] [PubMed] [Google Scholar]
- Mao Z, Seluanov A, Jiang Y, Gorbunova V (2007) TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc Natl Acad Sci USA 104: 13068–13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Freeman AD, Lilley DM, Ha T (2005) Observing spontaneous branch migration of Holliday junctions one step at a time. Proc Natl Acad Sci USA 102: 5715–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA (2002) Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem 277: 41110–41119 [DOI] [PubMed] [Google Scholar]
- Panyutin IG, Hsieh P (1994) The kinetics of spontaneous DNA branch migration. Proc Natl Acad Sci USA 91: 2021–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley SM, Parsons CA, Kemper B, West SC (1990) Cleavage specificity of bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I on synthetic branch migratable Holliday junctions. J Mol Biol 212: 723–735 [DOI] [PubMed] [Google Scholar]
- Plank JL, Wu J, Hsieh TS (2006) Topoisomerase IIIalpha and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci USA 103: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty JB, Bolt EL, Muranova TA, Sedelnikova SE, Leonard P, Pasquo A, Baker PJ, Rice DW, Sharples GJ, Lloyd RG (2003) The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold. Structure 11: 1557–1567 [DOI] [PubMed] [Google Scholar]
- Spink CH (2008) Differential scanning calorimetry. Methods Cell Biol 84: 115–141 [DOI] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD (2001) T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J 20: 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C, Gomez D, Giraud-Panis MJ, Londono-Vallejo A, Gilson E, Amor-Gueret M, Riou JF (2008) Topoisomerase IIIalpha is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J 27: 1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2006) The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127: 709–720 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2007) Replication and protection of telomeres. Nature 447: 924–931 [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- White MF, Lilley DM (1996) The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J Mol Biol 257: 330–341 [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12: 1489–1498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures