Abstract
The protein kinases ataxia-telangiectasia mutated (ATM) and ATM-Rad3 related (ATR) are activated in response to DNA damage, genotoxic stress and virus infections. Here we show that during infection with wild-type adenovirus, ATR and its cofactors RPA32, ATRIP and TopBP1 accumulate at viral replication centres, but there is minimal ATR activation. We show that the Mre11/Rad50/Nbs1 (MRN) complex is recruited to viral centres only during infection with adenoviruses lacking the early region E4 and ATR signaling is activated. This suggests a novel requirement for the MRN complex in ATR activation during virus infection, which is independent of Mre11 nuclease activity and recruitment of RPA/ATR/ATRIP/TopBP1. Unlike other damage scenarios, we found that ATM and ATR signaling are not dependent on each other during infection. We identify a region of the viral E4orf3 protein responsible for immobilization of the MRN complex and show that this prevents ATR signaling during adenovirus infection. We propose that immobilization of the MRN damage sensor by E4orf3 protein prevents recognition of viral genomes and blocks detrimental aspects of checkpoint signaling during virus infection.
Keywords: adenovirus, ATR kinase, DNA damage response, MRN complex
Introduction
The cellular DNA damage sensing and repair machinery orchestrates cell-cycle checkpoints and DNA repair pathways (Kastan and Bartek, 2004; Harper and Elledge, 2007). At the heart of the cellular response to DNA damage are the PI3 kinase-like kinases, ataxia-telangiectasia mutated (ATM) and ATM-Rad3 related (ATR), which phosphorylate multiple protein targets. Although these kinases have many overlapping substrates (Matsuoka et al, 2007; Stokes et al, 2007), they respond differently to distinct types of damage. Recent studies have begun to shed light on how these kinases are activated after DNA damage and replication stress (Lavin, 2007; Lee and Paull, 2007; Zou, 2007; Burrows and Elledge, 2008). In response to DNA double-strand breaks (DSBs), ATM is autophosphorylated on S1981 and subsequent dimer dissociation yields active monomers (Bakkenist and Kastan, 2003). ATR responds to a broader spectrum of DNA damage substrates and is crucial for maintaining genomic stability during the cell-cycle S phase (Cimprich and Cortez, 2008). ATR exists in a stable complex with an ATR-interacting protein ATRIP (Cortez et al, 2001) and is recruited to sites of DNA damage by replication protein A (RPA) associated with single-stranded DNA (ssDNA) (Zou and Elledge, 2003). It has been suggested that RPA-coated ssDNA is the substrate that initiates ATR-mediated checkpoint signaling (Zou, 2007). TopBP1 is a mediator protein that binds and activates ATR/ATRIP complexes (Kumagai et al, 2006; Mordes et al, 2008). Although ATM and ATR respond to different types of stimuli, they are integrated into a molecular circuit that links cellular DNA replication machinery with DNA damage response pathways (Hurley and Bunz, 2007).
The Mre11, Rad50 and Nbs1 proteins form the MRN complex, which is involved in the cellular DNA damage response (Stracker et al, 2004; Lavin, 2007). The MRN complex is a sensor of DSBs (Petrini and Stracker, 2003) and is required for full activation of ATM in response to DSBs (Carson et al, 2003; Uziel et al, 2003; Horejsi et al, 2004; Lee and Paull, 2005, 2007). MRN binds directly to ATM and stimulates kinase activity to phosphorylate substrates (Lee and Paull, 2004; Falck et al, 2005; You et al, 2005). Although MRN complex function has been convincingly linked to ATM activation in response to DSBs, its role in ATR activation and downstream signaling in response to different types of damage is less clear. The MRN complex prevents DSBs during chromosomal replication (Costanzo et al, 2001) and is required for the intra S-phase checkpoint in response to replication stress (Zhong et al, 2005; Olson et al, 2007b). DNA-damaging agents that stall replication induce hyperphosphorylation of the middle subunit of RPA (RPA32) that modulates its activity (Binz et al, 2004; Liu et al, 2006). Nbs1 is required for ATR-induced hyperphosphorylation of RPA32 in response to hydroxyurea (HU) (Manthey et al, 2007; Olson et al, 2007a). MRN has also been implicated in ATR activation and checkpoint signaling in response to UV treatment but may not be universally required for phosphorylation of all substrates at low doses (Difilippantonio et al, 2005; Zhong et al, 2005; Jazayeri et al, 2006; Myers and Cortez, 2006; Olson et al, 2007a).
Many viruses interact with cellular DNA damage sensing and repair pathways (Lilley et al, 2007). In the case of human adenovirus (Ad), the cellular DNA repair machinery presents an obstacle to productive infection (Stracker et al, 2002). Infection with mutant Ad deleted of the E4 region results in formation of virus genome concatemers by a cellular DNA repair pathway (Boyer et al, 1999; Stracker et al, 2002). The MRN complex is required for concatemer formation, and wild-type Ad serotype 5 (Ad5) has evolved two ways to prevent concatemers by targeting the MRN complex (Stracker et al, 2002). The viral E1b55K and E4orf6 proteins induce proteasome-mediated degradation of MRN proteins (Stracker et al, 2002; Carson et al, 2003), whereas expression of the Ad5-E4orf3 protein mislocalizes MRN proteins into intranuclear track-like structures and cytoplasmic aggregates (Stracker et al, 2002, 2005; Araujo et al, 2005; Evans and Hearing, 2005).
Coincident with concatemer formation, infection with E4-deleted Ad also elicits a cellular DNA damage response (Stracker et al, 2002; Carson et al, 2003). The ATM and ATR kinase signaling pathways are activated, as detected by autophosphorylation of ATM and phosphorylation of many known ATM and ATR substrates (Carson et al, 2003). The response is also characterized by recruitment of the MRN complex and other DNA damage response proteins to viral replication centres. In contrast, this cellular DNA damage response is not observed during infection with wild-type Ad5 that expresses E4 proteins (Carson et al, 2003). Ad infection provides a novel system to analyze the mechanism and consequences of ATR signaling. Here, we examine the effect of E4orf3-mediated MRN redistribution on ATM and ATR signaling. We show that activation of ATR signaling during viral infection is dependent on the MRN complex but unlike other types of damage is independent of ATM. The MRN complex is not required for accumulation of RPA, ATRIP, ATR and TopBP1 at viral replication centres. We examine MRN dynamics in living cells and show that E4orf3-induced immobilization prevents MRN from responding to the virus and other types of DNA. These results show a novel role for MRN in activation of ATR signaling independent of ATM and downstream of recruitment of RPA/ATR/ATRIP/TopBP1.
Results
The MRN complex is required for robust ATR signaling during infection
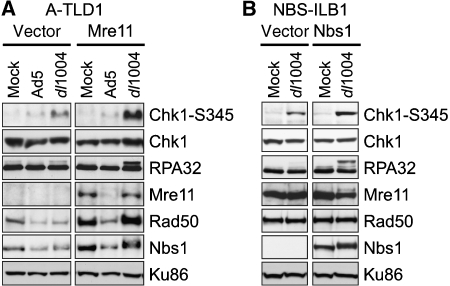
We have showed earlier the phosphorylation of ATM and ATR kinase substrates in response to E4-deleted Ad infection (Carson et al, 2003). Some sites were modified by either ATM or ATR (e.g., Chk2-T68 and Nbs1-S343), whereas others were ATR specific (e.g., Chk1-S345 and RPA32). We showed that MRN is required for ATM activation (Carson et al, 2003), but the requirement for MRN in ATR signaling during virus infection has not been clarified. To investigate the role of MRN specifically in ATR-dependent signaling responses to virus, we examined phosphorylation events during infections of cells with hypomorphic mutations in Mre11 and Nbs1. Infections were performed in mutant and complemented versions of the A-TLD1 cell line that expresses mutant Mre11 (Carson et al, 2003), and the NBS-ILB1 cell line that harbours an Nbs1 mutation (Cerosaletti et al, 2000). Lysates from cells infected with the E4-deleted virus dl1004 were analyzed by immunoblotting with a phospho-specific antibody to Chk1-S345, and with an antibody to RPA32 (Figure 1). Phosphorylation of Chk1 and RPA32 was detected in cell lines complemented with wild-type cDNAs. In contrast, infection with E4-deleted Ad in the A-TLD1 (Figure 1A) and NBS (Figure 1B) mutant lines produced significantly reduced signaling. Wild-type Ad infection did not generate these phosphorylation events due to degradation of MRN proteins. These data show that the MRN complex is required for robust ATR signaling in response to Ad infection.
Figure 1.
The Mre11 and Nbs1 proteins are required for robust ATR signaling in response to mutant Ad infection. (A) Infections were performed in a transformed cell line deficient for Mre11 (A-TLD1) that was transduced with an empty retrovirus vector (vector) or a vector expressing the wild-type Mre11 cDNA (Mre11). Cells were uninfected (Mock) or infected with wild-type Ad5 (Ad5) and the E4-deletion mutant (dl1004). Immunoblotting of lysates prepared at 30 hpi used antibodies for total protein (Chk1, RPA32, Mre11, Rad50 and Nbs1) or phospho-specific sites (Chk1-S345). Ku86 served as a loading control. (B) Infections were performed in the transformed NBS cell line (NBS-ILB1) transduced with an empty retrovirus vector (vector) or a vector expressing the wild-type Nbs1 cDNA.
The presence of Ad5-E4orf3 reduces ATR signaling but not ATM
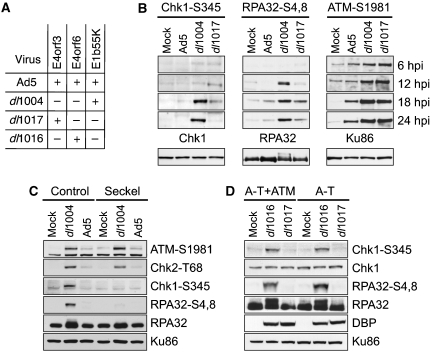
As the Ad5-E4orf3 protein affects MRN complex localization, we investigated whether E4orf3 alters ATM and ATR signaling during infection. Because the viral E1b55K and E4orf6 proteins downregulate damage signaling through degradation of the MRN complex (Carson et al, 2003), we tested a mutant that is deleted of E4orf6 and E1b55K but retains E4orf3 (dl1017) (Bridge and Ketner, 1990), and compared it with wild-type Ad5 and the large E4-deleted mutant (see Figure 2A). The E4-deleted virus induced phosphorylation of ATR substrates Chk1 and RPA32, as detected by a shift in the mobility of RPA32 and by recognition with phospho-specific antibodies (Chk1-S345 and RPA32-S4,8) (Figure 2B). These phosphorylation events were greatly reduced during infection with dl1017 that expresses E4orf3, indicating that ATR signaling is reduced in the presence of Ad5-E4orf3.
Figure 2.
Phosphorylation of ATR substrates is reduced in the presence of Ad5-E4orf3 and is independent of ATM. (A) Genotypes of mutant viruses used for infection (Bridge and Ketner, 1990). (B) HeLa cells were mock infected or infected with indicated viruses. Cells were harvested at 6, 12, 18 and 24 hpi and prepared for analysis by immunoblotting using antibodies to phospho-specific residues (Chk1-S345, RPA32-S4,8 and ATM-S1981). Below are lysates at 24 hpi probed with antibodies against total protein (Chk1, RPA32 and Ku86) to serve as loading controls. (C) Immunoblots of infections in control cells (48BR) or Seckel cells with mutant ATR (GM18366). (D) Immunoblots of infections in A-T cells with mutant ATM (AT221JE-T) or a complemented line (A-T+ATM). DBP was a marker of virus infection and Ku86 served as a loading control.
We also examined the effect of E4orf3 on ATM signaling. In contrast to ATR, ATM signaling was intact during infection with dl1017, as evidenced by ATM autophosphorylation on S1981 (Figure 2B). Analysis of substrates that are phosphorylated by both ATM and ATR (Chk2-T68, Nbs1-S434 and BRCA1) revealed that their phosphorylation was decreased but not abolished by E4orf3 expression (Supplementary Figure S1A), showing that E4orf3 inhibits ATR but not ATM. Immunofluorescence during infection with dl1017 showed that MRN mislocalization by E4orf3 did not prevent ATM activation, and that phosphorylated Nbs1 colocalized with Rad50 in nuclear tracks (Supplementary Figure S1B). Together, these data suggest that MRN mislocalization by E4orf3 is not sufficient to prevent ATM activation, indicating that ATM and ATR signaling pathways can be independently regulated during Ad infection.
To determine the interdependence of ATR and ATM, we examined signaling in cells with kinase mutations. Cells derived from a Seckel syndrome patient with mutation in ATR (O'Driscoll et al, 2003) and from an A-T patient with mutation in ATM were infected with viruses that express E4orf3 (Ad5 or dl1017) and those that lack E4orf3 (dl1004 or dl1016). Phosphorylation of Chk1-S345 and RPA32-S4,8 was abolished in the Seckel cells (Figure 2B). ATM activation and signaling were intact in Seckel cells, as detected by autophosphorylation on S1981 (Figure 2C) and phosphorylation of downstream substrates (Supplementary Figure S1C). Infections of A-T cells or a matched complemented line with the mutant virus dl1016 that lacks both E1b55K and E4orfs 1-3 (ΔE1b55K/ΔE4orf1-3) induced ATR signaling (Figure 2C). However, signaling to Chk1 and RPA32 was abrogated in the presence of E4orf3 (dl1017). These results show that ATR activation in response to Ad infection, and its inhibition by E4orf3, are independent of ATM.
Accumulation of ATR at viral replication centres is independent of MRN
Viral replication centres can be detected by staining with antibodies to the viral DNA-binding protein (DBP) that coats ssDNA accumulated at viral replication sites (Pombo et al, 1994). The cellular RPA32 protein also accumulates at these sites (Stracker et al, 2005), with both wild-type and mutant viruses (Figure 3A). The E4orf3 protein is excluded from viral replication centres and appears in intranuclear tracks and cytoplasmic aggregates (Araujo et al, 2005; Evans and Hearing, 2005), with both wild-type Ad5 or the dl1017 mutant (Figure 3A). The Ad5-E4orf3 protein is sufficient to redistribute the MRN complex, as detected by staining for Nbs1 (Figure 3A). In contrast, infection with dl1004 leads to accumulation of MRN at viral centres. We, therefore, investigated whether MRN mislocalization by E4orf3 correlates with inhibition of ATR signaling (Figure 3B). Phosphorylation of RPA32 at viral centres, as detected by staining with the RPA32-S4,8 antibody, was only observed when the MRN complex was associated with viral replication centres (i.e., in the absence of E4orf3 during infection with dl1004). When the virus expressed E4orf3, RPA32 phosphorylation was not detected. This shows that E4orf3 mislocalization of the MRN complex prevents its accumulation at viral replication centres, and that this correlates with the absence of ATR signaling to RPA32.
Figure 3.
Signaling by ATR at viral replication centres. (A) E4orf3 sequesters Nbs1 into tracks away from viral replication centres. HeLa cells were mock infected or infected with indicated viruses. Cells were fixed at 16 hpi, and immunofluorescence was performed. Staining shows that Nbs1 but not RPA32 is excluded from viral replication centres in the presence of E4orf3. (B) Colocalization of the MRN complex with RPA32 at replication centres correlates with RPA32 phosphorylation. Immunofluorescence with a phospho-specific antibody to RPA32-S4,8 shows colocalization with viral replication centres stained with an antibody to DBP only in the absence of E4orf3. (C) Accumulation of RPA32, ATR and ATRIP at viral centres is independent of E4. (D) TopBP1 accumulates at viral centres independently of E4. (E) Activation is observed for ATM, but not ATR, in Seckel cells. (F) ATR signaling is restored in A-TLD cells by expression of Mre11 or the nuclease-defective mutant Mre11-3. In all images, the nuclei were located by co-staining cellular DNA with DAPI (blue).
We also investigated whether E4orf3 affects localization of other proteins involved in ATR signaling. We observed that ATR, RPA and ATRIP accumulated at viral replication centres with all viruses tested, irrespective of E4orf3 expression (Figure 3C) (Carson et al, 2003; data not shown). These data suggest that E4orf3 does not abrogate ATR signaling by mislocalizing ATR, RPA32 or ATRIP. It also shows that the presence of ATR at viral centres is not sufficient to initiate or maintain the DNA damage signaling during infection. The TopBP1 protein is a regulator of ATR that stimulates kinase activity through an interaction with ATRIP (Kumagai et al, 2006; Mordes et al, 2008). We examined localization of TopBP1 during infection and found it localized in viral DBP centres with wild-type and E4 mutant viruses (Figure 3D). Immunofluorescence of infected Seckel cells confirmed that phosphorylation of RPA32-S4,8 was not observed in the absence of functional ATR, although RPA32 and ATRIP accumulated at viral centres (Figure 3E). Lack of RPA32 phosphorylation was confirmed in Seckel cells from multiple origins and could also be restored by introduction of the wild-type ATR gene (Supplementary Figure S2). ATM activation was detectable in Seckel cells as revealed by staining for ATM-S1981 and is therefore independent of ATR in response to Ad infection (Figure 3E). Although RPA and ATR were detected at viral centres in the absence of functional MRN complex, phosphorylation of RPA32 was not observed during infections of NBS and A-TLD1 cells (Figure 3F, Supplementary Figure S2C and D). ATR signaling was restored when infections were performed in A-TLD1 cells transduced with retroviruses that express either wild-type Mre11 or the nuclease-defective Mre11-3 mutant (Figure 3F). Together, these data show that the MRN complex is not required for localization of ATR with ssDNA at viral replication centres but is required for ATR signaling.
Mislocalization of the MRN complex by Ad5-E4orf3 is required for disruption of ATR signaling
Although E4orf3 proteins from all serotypes tested form nuclear tracks, rearrange the promyelocytic leukemia protein PML and redistribute components of the PML bodies, only Ad5-E4orf3 mislocalizes the MRN complex (Stracker et al, 2005). Sequence analysis for the highly conserved E4orf3 proteins revealed residues specific to the subgroup C viruses (Ad1, Ad2 and Ad5) that uniquely mislocalize MRN. On the basis of these alignments, we converted isoleucine at position 104 of Ad5 to arginine, which is present in all other serotypes (Figure 4A). The Ad5 E4orf3-I104R mutant retained the ability to form nuclear tracks and disrupt PML but was defective for redistribution of the MRN complex (Figure 4B). This identified a region of E4orf3 involved in targeting the MRN complex and provided a separation-of-function mutant.
Figure 4.
A region in Ad5-E4orf3 important for targeting the MRN complex. (A) Sequence alignment of E4orf3 proteins. E4orf3 sequences from different human (subgroup noted in brackets) and simian adenoviruses were aligned using the CLUSTAL algorithm and the region around residue I104 is shown. Conserved residues are shown boxed, with CLUSTAL colour scheme reflecting amino acids of similar chemical nature. The I104 residue is highlighted, showing that this site differs between subgroup C and all other sequenced E4orf3 genes. (B) The Ad5 E4orf3-I104R mutant does not redistribute the MRN complex. Plasmids for wild-type and mutant E4orf3 were transfected into HeLa cells. Immunofluorescence shows that the E4orf3 mutant protein still forms tracks and disrupts PML structures but is unable to redistribute members of the MRN complex, which remain diffusely nuclear. Representative images are shown and nuclei are located by co-staining cellular DNA with DAPI.
We next examined the ability of Ad5-E4orf3, Ad12-E4orf3 and E4orf3-I104R proteins to prevent ATR signaling during infection with E4-deleted virus (Figure 5). The cells were transfected with E4orf3 expression vectors, and then infected with dl1004. Immunoblotting revealed abrogated ATR signaling in the presence of Ad5-E4orf3 (reduced Chk1-S345 and RPA32-S4,8 phosphorylation and RPA32 mobility shift), although ATM autophosphorylation was not significantly altered (Figure 5A). In contrast, the Ad12-E4orf3 and E4orf3-I104R proteins, which failed to mislocalize the MRN complex, did not inhibit ATR signaling (Figure 5A and B). In this experiment, there was a slight decrease in ATM signaling (Figure 5B), but this was not reproducible and did not correlate with MRN mislocalization. Cells from the same experiment were also examined by immunofluorescence for RPA32-S4,8 phosphorylation, as a marker of ATR signaling (Figure 5C). We observed that Ad5-E4orf3, but not Ad12-E4orf3 or E4orf3-I104R, prevented RPA phosphorylation at viral replication centres. Cotransfection of a plasmid expressing GFP served as a marker for transfected cells. Control staining showed that viral DBP centres were formed in all cells, and that expression of E4orf3 proteins resulted in PML disruption (data not shown). Together, these data support the hypothesis that mislocalization of the MRN complex by E4orf3 leads to abrogation of ATR signaling.
Figure 5.
ATR signaling in response to virus infection is abrogated by E4orf3 proteins that mislocalize the MRN complex. HeLa cells were transfected with an empty plasmid (vector) or with vectors expressing E4orf3 proteins. After 24 h, cells were either mock infected, infected with the E1b55K/E4orf6 mutant (dl1017) (positive control), or infected with the E4-deletion mutant (dl1004). (A, B) Lysates from cells at 24 hpi were immunoblotted with antibodies to Chk1-S345, RPA32-S4,8, ATM-1981 and RPA32. (C) Cells were fixed at 18 hpi, and immunofluorescence was performed with an antibody to RPA32-S4,8. Images are shown merged with DAPI staining. A plasmid expressing GFP was cotransfected with that for E4orf3 at a ratio of 1:10 and GFP staining is shown in the lower left insert panel as a positive control for transfection.
The MRN complex is immobilized by Ad5-E4orf3
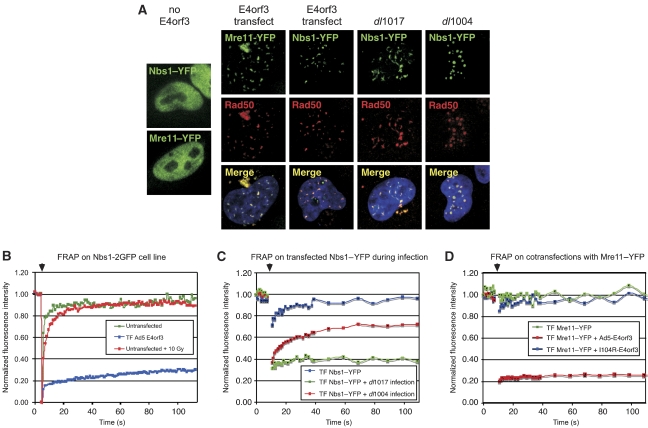
We reported earlier that Ad5-E4orf3 expression alters MRN complex solubility (Araujo et al, 2005). To examine the effect of E4orf3 on protein dynamics in live cells, we used fluorescently tagged fusion proteins of Mre11 and Nbs1. We expressed Mre11–YFP and Nbs1–YFP by cell transfection either in the presence of E4orf3 alone or in virus-infected cells (Figure 6). The fusion proteins were distributed diffusely in the nucleoplasm in untreated cells (Lukas et al, 2003). In the presence of E4orf3, provided by virus infection or plasmid transfection, the fusion proteins localized into track-like structures and accumulated in cytoplasmic aggregates (Figure 6A). Both Mre11–YFP and Nbs1–YFP fusion protein colocalized with endogenous Rad50 protein. Similar results were obtained using a stable cell line expressing Nbs1–2GFP (Lukas et al, 2003) (data not shown). These results show that the fusion proteins are correctly incorporated into E4orf3-induced structures. During infection with E4-deleted virus, Nbs1–YFP formed foci at viral replication centres (Figure 6A), as reported earlier for endogenous MRN (Stracker et al, 2002).
Figure 6.
E4orf3 abrogates MRN function by immobilizing the MRN complex. (A) HeLa cells were transfected with Nbs1–YFP or Mre11–YFP alone or together with Ad5-E4orf3 plasmid vector. After 24 h, cells were infected with dl1004 or dl1017. Cells were fixed 18 hpi, and immunofluorescence was performed with a Rad50 antibody. Images shown are merged with DAPI staining. (B–D) FRAP analysis of Nbs1 and Mre11 in cells expressing E4orf3 by infection or transfection. The unbleached portion of the cell served to normalize the overall fluorescence decay during the repeated image collection. Arrows above indicate the time of bleaching. (B) Stable U2OS cells expressing Nbs1–2GFP were either untreated, transfected with E4orf3, or treated with 10 Gy gamma-irradiation. (C) HeLa cells were transfected with Nbs1–YFP and then mock treated or infected with dl1004 and dl1017. Cells were analyzed by FRAP at 18 hpi. (D) HeLa cells transfected with Mre11–YFP together with either empty vector or vectors expressing Ad5-E4orf3 and E4orf3-I104R were analyzed by FRAP.
We then examined the dynamics of MRN in E4orf3 tracks using fluorescence recovery after photobleaching (FRAP) to determine the rate at which a bleached area is repopulated with new fluorescent protein. As reported earlier, we observed that the Mre11 and Nbs1 proteins are highly mobile in the absence of E4orf3 (Figure 6B). FRAP analysis of Nbs1 foci at viral replication centres during infection with E4-deleted virus dl1004 showed that the signal recovered with kinetics slower than that of an undamaged area (Figure 6C). The increased residence time at virus centres resembles that observed for recruitment of the MRN complex at DSBs (Lukas et al, 2003). In contrast, the kinetics of fluorescence recovery was much more significantly affected by the presence of E4orf3, where the fluorescence signal did not recover after photo-bleaching of MRN in tracks induced by E4orf3 during transfection (Figure 6B) or infection (Figure 6C). We also compared MRN mobility in the presence of wild-type and I104R mutant E4orf3 proteins (Figure 6D). Although wild-type Ad5-E4orf3 expression led to immobilization of Mre11–YFP, the I104R mutant barely affected Mre11 dynamics. This supports our observations from fixed images that show I104R does not alter Mre11 localization. Together, these data show that MRN in tracks induced by Ad5-E4orf3 represent immobilized proteins.
E4orf3 prevents ATR-dependent signaling in response to nonviral damage
To assess the impact of MRN immobilization on the ability of cells to respond to DNA damage, we generated spatially restricted DSBs through laser microirradiation (Lukas et al, 2003). Local DNA damage was generated in subnuclear regions by a focussed laser beam programmed to move once across individual cell nuclei. Previous studies using visualization with specific antibodies and fluorescently tagged proteins have shown that DNA damage proteins are redistributed to these DSBs (Lukas et al, 2003, 2004). We examined recruitment of endogenous cellular repair proteins to laser-induced DSBs in cells expressing E4orf3. The sites of microirradiation can be visualized by recruitment of the mediator protein Mdc1 (Lukas et al, 2004), which is unaffected by E4orf3 (Figure 7A). Cells expressing Ad5-E4orf3 localized endogenous Nbs1 into the characteristic nuclear tracks and displayed minimal accumulation of Nbs1 at microirradiation sites compared with neighbouring untransfected cells. This result shows that Nbs1 immobilization in E4orf3-induced tracks prevents its accumulation at DSBs.
Figure 7.
E4orf3 prevents ATR-dependent damage signaling induced by nonviral sources. (A) U2OS cells transfected with a plasmid vector expressing Ad5-E4orf3 were laser microirradiated. Cells were fixed and stained for endogenous Nbs1 and Mdc1. Arrows indicate cells with E4orf3-induced tracks of Nbs1. (B) Stable U2OS cell lines expressing Nbs1–2GFP were transfected with a plasmid expressing Ad5-E4orf3, together with prRFP-C1, which was used as a marker for transfected cells. Cells were laser microirradiated (as indicated by dashed line) and images show the recruitment of Nbs1 to sites of damage. (C) HeLa cells were mock infected, or infected with E1-deleted recombinant Ads expressing GFP (rAd-GFP) or E4orf3 (rAd-E4orf3). At 24 hpi, the cells were mock treated or treated with 2 mM hydroxyurea (HU) for 2 h, and the cells were harvested for immunoblotting. Cellular proteins were detected with antibodies to RPA32 and specific phosphorylated sites at RPA-S4,8 and Nbs1-S343. Ku86 served as a loading control.
To examine the effect of E4orf3 expression on kinetics of the DNA damage response, we combined laser microirradiation with live cell imaging (Figure 7B). The Nbs1–2GFP cell line was cotransfected with the Ad5-E4orf3 expression vector and a plasmid expressing RFP to serve as a transfection marker. Generation of subnuclear restricted DSBs resulted in rapid recruitment of Nbs1–2GFP. In contrast, there was barely detectable recruitment of Nbs1 in cells expressing Ad5-E4orf3, even at 15 min after treatment. These data indicate that the MRN complex immobilized in E4orf3-induced tracks is unable to respond efficiently to exogenous DNA damage. As MRN has been shown to be required for ATR signaling in response to HU treatment (Manthey et al, 2007), we used recombinant Ad vectors to express Ad5-E4orf3 or GFP in cells and then exposed them to HU (Figure 7C). Signaling was abrogated by E4orf3 as shown by decreased hyperphosphorylation of RPA32 and staining with phospho-specific antibodies to RPA32-S4,8, Nbs1-S343 (Figure 7C), and Chk1-S345 (data not shown). Together, these data show that immobilization of MRN by E4orf3 prevents the ATR-mediated response to replication stress.
Discussion
The role of MRN in ATR activation
The MRN complex has an important function in the cellular DNA repair response to Ad infection. We showed earlier that during infection with E4-deleted Ad, MRN is required for concatemerization of the viral genome and activation of ATM signaling (Stracker et al, 2002; Carson et al, 2003). In this report, we show that MRN also has an important function in ATR signaling in response to E4-deleted Ad. Substrates known to contribute to ATR kinase activation include ssDNA coated with RPA, and junctions of single- and double-stranded DNA (MacDougall et al, 2007; Zou, 2007). Our data show that ssDNA at viral replication centres (Pombo et al, 1994) is sufficient for recruitment of ATR and ATRIP, but that ATR signaling to Chk1 and RPA32 is only detected when the MRN complex also accumulated at viral centres. Although we have focussed on MRN, we cannot exclude additional roles for other proteins implicated in ATR activation and checkpoint signaling, such as the 9-1-1 complex, the Rad17 complex and Claspin (Zou, 2007). The cellular E1b55K-associated protein E1B-AP5 was recently implicated in ATR signaling during Ad infection (Blackford et al, 2008), but this factor localizes to wild-type Ad5 centres and, therefore, does not explain the induction of ATR signaling in the absence of E4.
MRN has recently been implicated in facilitating ATR activation and signaling in response to some types of damage. Processing of DSBs in an MRN-dependent manner results in the formation of ssDNA and ATR activation (Adams et al, 2006; Jazayeri et al, 2006; Myers and Cortez, 2006). MRN is involved in ATR-mediated phosphorylation events in response to replication stress, although signaling events may also be MRN independent, depending on the substrate and dose of damaging agent (Pichierri and Rosselli, 2004; Stiff et al, 2005; Zhong et al, 2005; Olson et al, 2007b). During infection with E4-deleted Ad, it is not clear which DNA structures serve as the trigger for ATR signaling, and there may be multiple ways that the MRN complex contributes to ATR activation. Although the nuclease activity of Mre11 is required for joining Ad genomes into concatemers (Stracker et al, 2002), we found that it was not required for ATR signaling. In contrast to DSBs caused by IR or replication stress, where the nuclease activity of Mre11 is required for resection (Buis et al, 2008), we found that Mre11 nuclease activity is not required for generation of ssDNA and recruitment of RPA/ATR/ATRIP at viral centres. However, signaling may be triggered by further processing of the viral genome, for example, by removal of the terminal protein from the 5′ end of the genome by other nucleases to generate free DNA ends. MRN-dependent processing of DSBs has been suggested to generate small oligonucleotides that stimulate ATM activity (Jazayeri et al, 2008). It will be interesting to determine whether these are generated during virus infection and whether they play a role in ATR activation. The MRN complex could have a direct role in stimulating ATR kinase activity, as has been shown in vitro for ATM (Lee and Paull, 2004). The MRN complex may also facilitate phosphorylation of downstream substrates through recruitment or retention of proteins to viral centres, as has been proposed for stalled replication forks (Stiff et al, 2005). MRN associates with RPA at sites of DNA damage to mediate the intra-S-phase checkpoint (Robison et al, 2004; Olson et al, 2007b) and also interacts with ATR/ATRIP (Olson et al, 2007a). Either of these two interactions could contribute to ATR activation by MRN at viral centres. The ATM and ATR kinases may be coordinated and interdependent in response to some types of damage (Hurley and Bunz, 2007). In response to IR, activation of ATR is ATM dependent (Jazayeri et al, 2006; Myers and Cortez, 2006), whereas in response to HU and UV activation of ATM is ATR dependent (Liu et al, 2005; Stiff et al, 2006). In the case of virus infection, we have found that although both rely upon the MRN complex, ATM and ATR signaling are independent of each other. This may reflect the fact that during infection there are numerous substrates for kinase activation, including replication intermediates and double-strand ends. In addition to the viral genome, infection may also induce chromosomal damage to the host genome. Early Ad genes alter cell-cycle progression, which could lead to collapse of replication forks, and also cause genomic instability and chromosomal aberrations (Caporossi and Bacchetti, 1990; Lavia et al, 2003). Therefore, although the phosphorylated ATR substrates predominantly accumulate at viral replication centres, it is possible that chromosomal damage also contributes to induction of ATR signaling during infection.
The function of viral E4 proteins
There is functional redundancy between the E4orf3 and E4orf6 products from Ad, and either is sufficient to promote viral replication, prevent concatemerization of the viral genome, and enable viral late protein production. Both E4 proteins target the MRN complex to prevent concatemer formation (Stracker et al, 2002; Evans and Hearing, 2003). Together with previous observations of MRN degradation by E1b55K/E4orf6 (Carson et al, 2003), our data show that both E4 proteins target MRN to prevent damage signaling. Inactivation of MRN is also likely to be responsible for the ability of E4 proteins to promote viral DNA replication. We and others have found that MRN inhibits replication of E4-deleted mutant Ad, although the mechanism is unclear (Evans and Hearing, 2005; Mathew and Bridge, 2007, 2008; Lakdawala et al, 2008). Replication of cellular DNA is tightly regulated to ensure that the genome is replicated only once per cell cycle (Arias and Walter, 2007). MRN is recruited to cellular replication origins and can inhibit firing of new origins of DNA replication upon damage (Olson et al, 2007b) and suppress rereplication (Wu et al, 2004; Lee et al, 2007). Mre11 has recently been suggested to bind the Ad genome (Mathew and Bridge, 2008), but it is unclear how this inhibits replication. Checkpoint signaling by ATM and ATR is not responsible for the defective replication of E4-deleted Ad (Lakdawala et al, 2008). The virus uses its own protein-priming mechanism and polymerase, which could be affected by MRN binding to the origin or its participation in removal of the terminal protein from the viral genome.
Our work links the E4orf3-induced redistribution of proteins associated with PML nuclear bodies to their role in sensing DNA damage (Everett, 2006). We show that relocalization of the MRN complex dramatically reduces its dynamics, essentially immobilizing the proteins in E4orf3-induced intranuclear tracks. Similar observations were made with FRAP analysis of other fluorescently tagged components of the PML bodies (unpublished observations). Experiments with laser microirradiation and live cell imaging showed that recruitment of MRN to damage sites was severely abrogated in cells expressing Ad5 E4orf3. This correlated with decreased recruitment of RPA and ATR, as well as abrogated damage signaling, and was not seen with the I104R mutant (data not shown). Together, these results show that immobilization by E4orf3 prevents the MRN damage sensor from responding to new damage sites. This supports the hypothesis that sequestering MRN (and other host factors) into E4orf3-induced tracks will prevent these proteins from sensing and accumulating at virus centres and will thus thwart host antiviral responses. It has also been suggested that sequestration of MRN in cytoplasmic aggresomes by the adenoviral E1b55K inactivates the complex and protects the viral genome (Liu et al, 2005). As the E4orf3 and E4orf6 proteins both block damage signaling and also promote production of late viral proteins, it will be interesting to determine whether these activities are linked.
Inactivation of ATR may be a general approach used by viruses to neutralize aspects of the host defense. In addition to our observations with Ad infection, it has been suggested that ATR signaling is manipulated by other DNA viruses. For example, in the case of herpes-simplex virus type 1 (HSV-1), infection activates ATM signaling pathways and results in accumulation of cellular repair proteins at viral centres (Lilley et al, 2007). Although RPA is found at HSV-1 viral replication compartments, ATR-dependent signaling is not activated, and it has been suggested that it is prevented because of spatial uncoupling of the ATR-ATRIP complex and sequestering of phosphorylated RPA in viral-induced nuclear domains (Wilkinson and Weller, 2006). Induction of the Epstein–Barr virus lytic program also elicits a cellular DNA damage response and ATM activation, but ATR signaling is minimal (Kudoh et al, 2005). Therefore, viruses appear to use multiple strategies to inactivate ATR and downstream signaling events during viral infection, to prevent negative impacts of the cellular response to replication stress on virus production. Understanding how viruses such as Ad manipulate signaling pathways will provide insights into the regulation of DNA damage responses in mammalian cells.
Materials and methods
Cell lines
HeLa and 293 cells were purchased from the American Tissue Culture Collection. W162 cells for growth of E4-deleted viruses were from G Ketner, A-T cells (AT221JET and complemented version) were from Y Shiloh, and Seckel cells were from Coriell Institute (GM18366) and A D'Andrea (F02-98). Immortalized A-TLD1 and NBS (NBS-ILB1) and matched cells reconstituted with wild-type Mre11 and NBS1 were described (Cerosaletti et al, 2000; Carson et al, 2003). The retrovirus expression plasmid for the Mre11-3 mutant (HD129/130LV) was generated by site-directed mutagenesis and A-TLD1 cells were transduced by the retrovirus as described previously (Carson et al, 2003). The U2OS-derived stable cell line with Nbs1–2GFP has been described (Lukas et al, 2003). Cells were maintained as monolayers in either Dulbecco modified Eagle's medium (DMEM) or MEM plus Earle's salts (Seckel cells) supplemented with 10 or 20% fetal bovine serum (FBS), at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids and transfections
Expression vectors for Ad5-E4orf3 and Ad12-E4orf3 proteins were described (Stracker et al, 2005). Site-directed mutagenesis of E4orf3 was performed using QuikChange (Stratagene). Cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol.
Viruses and infections
The mutant viruses dl1004 (ΔE4), dl1016 (ΔE4orf1-3/ΔE1b55K) and dl1017 (ΔE4orf6/ΔE1b55K) have been described (Bridge and Ketner, 1990) and were obtained from G. Ketner. Wild-type Ad5 and dl1017 were propagated in 293 cells. The dl1004 and dl1016 viruses were propagated on W162 cells (Weinberg and Ketner, 1983). All viruses were purified by two sequential rounds of ultra-centrifugation in cesium chloride gradients and stored in 40% glycerol at −20°C. Infections were performed in DMEM supplemented with 2% FBS. After 2 h at 37°C additional serum was added to a total of 10%.
Antibodies
Primary antibodies were purchased from Novus Biologicals Inc. (Nbs1), Genetex (Mre11-12D7, Rad50-13B3), Cell Signaling (Chk1-S345), Rockland (ATM S1981-P), Santa Cruz (ATR, PML, Chk1, Ku86), Upstate Biotechnology (ATRIP), BD Bioscience (TopBP1) and Bethyl (RPA32-S4,8). The antibody to RPA32 was from T. Melendy, the monoclonal B6 antibody to DBP was from A. Levine, polyclonal rabbit antisera to DBP was from P. van der Vliet, and the E4orf3 antibody was from T. Dobner. Secondary antibodies were from Jackson Laboratories, Eurogentec and Invitrogen Molecular Probes.
Immunoblotting and immunofluorescence
Immunoblotting and immunofluorescence were performed as described previously (Carson et al, 2003). Novex (Invitrogen) 3 to 8% gradient gels were used for the resolution of ATM. For immunofluorescence, cells grown on glass coverslips were infected at an MOI of 25–100 pfu/cell. After 16–24 h, the cells were washed, fixed, stained and counter-stained with 4′,6-diamidino-2-phenylindol (DAPI). Immunoreactivity was visualized using a Nikon microscope in conjunction with a CCD camera (Cooke Sensicam) or a Leica confocal microscope.
FRAP analysis
FRAP analysis was performed in the Nbs1–2GFP U2OS cells as reported previously (Lukas et al, 2003). Further details are provided in Supplementary data. Image collection and FRAP data were processed on a Leica confocal microscope.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Information
Acknowledgments
We thank P Concannon, A D'Andrea, T Dobner, P Jeggo, G Ketner, A Levine, T Melendy, J Petrini and P van der Vliet for generous gifts of reagents. We thank members of the Weitzman lab, past and present, for discussions and critical reading of the manuscript. We thank D Ornelles for discussions and sequence analysis for E4orf3. We acknowledge the James B Pendleton Charitable Trust for providing the Pendleton Microscopy Facility. This work was supported by NIH grant CA97093 (MDW), and by gifts from the Joe W & Dorothy Dorsett Brown Foundation and the Lebensfeld Foundation to MDW. Additional support came from The Danish National Research Foundation, Danish Cancer Society, European Commission (DNA Repair) and the John and Birthe Meyer Foundation. CTC was supported by the Timken-Sturgis Foundation and a scholarship from the ARCS Foundation. CTC and NIO were supported in part by an NIH Training Grant to the Salk Institute.
References
- Adams KE, Medhurst AL, Dart DA, Lakin ND (2006) Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 25: 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD (2005) Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J Virol 79: 11382–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 [DOI] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS (2004) Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair 3: 1015–1024 [DOI] [PubMed] [Google Scholar]
- Blackford AN, Bruton RK, Dirlik O, Stewart GS, Taylor AM, Dobner T, Grand RJ, Turnell AS (2008) A role for E1B-AP5 in ATR signaling pathways during adenovirus infection. J Virol 82: 7640–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J, Rohleder K, Ketner G (1999) Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263: 307–312 [DOI] [PubMed] [Google Scholar]
- Bridge E, Ketner G (1990) Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174: 345–353 [DOI] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ (2008) How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev 22: 1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporossi D, Bacchetti S (1990) Definition of adenovirus type 5 functions involved in the induction of chromosomal aberrations in human cells. J Gen Virol 71(Part 4): 801–808 [DOI] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD (2003) The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J 22: 6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerosaletti KM, Desai-Mehta A, Yeo TC, Kraakman-Van Der Zwet M, Zdzienicka MZ, Concannon P (2000) Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis 15: 281–286 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J (2001) Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, Manova K, Kruhlak M, Camerini-Otero RD, Sharan S, Nussenzweig M, Nussenzweig A (2005) Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol 7: 675–685 [DOI] [PubMed] [Google Scholar]
- Evans JD, Hearing P (2003) Distinct roles of the Adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J Virol 77: 5295–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Hearing P (2005) Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J Virol 79: 6207–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD (2006) Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol 8: 365–374 [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Horejsi Z, Falck J, Bakkenist CJ, Kastan MB, Lukas J, Bartek J (2004) Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene 23: 3122–3127 [DOI] [PubMed] [Google Scholar]
- Hurley PJ, Bunz F (2007) ATM and ATR: components of an integrated circuit. Cell Cycle 6: 414–417 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V (2008) Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J 27: 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T (2005) Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem 280: 8156–8163 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Lakdawala SS, Schwartz RA, Ferenchak K, Carson CT, McSharry BP, Wilkinson GW, Weitzman MD (2008) Differential requirements of the C-terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J Virol 82: 8362–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavia P, Mileo AM, Giordano A, Paggi MG (2003) Emerging roles of DNA tumor viruses in cell proliferation: new insights into genomic instability. Oncogene 22: 6508–6516 [DOI] [PubMed] [Google Scholar]
- Lavin MF (2007) ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26: 7749–7758 [DOI] [PubMed] [Google Scholar]
- Lee AY, Liu E, Wu X (2007) The Mre11/Rad50/Nbs1 complex plays an important role in the prevention of DNA rereplication in mammalian cells. J Biol Chem 282: 32243–32255 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT (2004) Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304: 93–96 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT (2007) Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26: 7741–7748 [DOI] [PubMed] [Google Scholar]
- Lilley CE, Schwartz RA, Weitzman MD (2007) Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol 15: 119–126 [DOI] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, Melendy T (2006) Phosphorylation of replication protein A by S-phase checkpoint kinases. DNA Repair (Amst) 5: 369–380 [DOI] [PubMed] [Google Scholar]
- Liu Y, Shevchenko A, Shevchenko A, Berk AJ (2005) Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol 79: 14004–14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J (2004) Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 23: 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA (2007) The structural determinants of checkpoint activation. Genes Dev 21: 898–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey KC, Opiyo S, Glanzer JG, Dimitrova D, Elliott J, Oakley GG (2007) NBS1 mediates ATR-dependent RPA hyperphosphorylation following replication-fork stall and collapse. J Cell Sci 120 (Part 23): 4221–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew SS, Bridge E (2007) The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology 365: 346–355 [DOI] [PubMed] [Google Scholar]
- Mathew SS, Bridge E (2008) Nbs1-dependent binding of Mre11 to adenovirus E4 mutant viral DNA is important for inhibiting DNA replication. Virology 374: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Cortez D (2006) Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 281: 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA (2003) A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501 [DOI] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Lee AY, Chen L, Wu X (2007a) The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem 282: 22939–22952 [DOI] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X (2007b) The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol 27: 6053–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH, Stracker TH (2003) The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol 13: 458–462 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F (2004) The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J 23: 1178–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M (1994) Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J 13: 5075–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG (2004) Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem 279: 34802–34810 [DOI] [PubMed] [Google Scholar]
- Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA (2005) Nbs1 is required for ATR-dependent phosphorylation events. EMBO J 24: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O'Driscoll M, Jeggo PA (2006) ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J 25: 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ (2007) Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci USA 104: 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Carson CT, Weitzman MD (2002) Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418: 348–352 [DOI] [PubMed] [Google Scholar]
- Stracker TH, Lee DV, Carson CT, Araujo FD, Ornelles DA, Weitzman MD (2005) Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J Virol 79: 6664–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH (2004) The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair 3: 845–854 [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y (2003) Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22: 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DH, Ketner G (1983) A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc Natl Acad Sci USA 80: 5383–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK (2006) Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J Cell Sci 119(Part 13): 2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Avni D, Chiba T, Yan F, Zhao Q, Lin Y, Heng H, Livingston D (2004) SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev 18: 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P (2005) ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25: 5363–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Bryson A, Eckersdorff M, Ferguson DO (2005) Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum Mol Genet 14: 2685–2693 [DOI] [PubMed] [Google Scholar]
- Zou L (2007) Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev 21: 879–885 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Information