Abstract
An unusual deubiquitinating (DUB) activity exists in HeLa cell extracts that is highly specific for cleaving K63-linked but not K48-linked polyubiquitin chains. The activity is insensitive to both N-ethyl-maleimide and ubiquitin aldehyde, indicating that it lacks an active site cysteine residue, and gel filtration experiments show that it resides in a high molecular weight (∼600 kDa) complex. Using a biochemical approach, we found that the K63-specific DUB activity co-fractionated through seven chromatographic steps with three multisubunit complexes: the 19S (PA700) portion of the 26S proteasome, the COP9 signalosome (CSN) and a novel complex that includes the JAMM/MPN+ domain-containing protein Brcc36. When we analysed the individual complexes, we found that the activity was intrinsic to PA700 and the Brcc36 isopeptidase complex (BRISC), but that the CSN-associated activity was due entirely to an interaction with Brcc36. None of the complexes cleave K6, K11, K29, K48 or α-linked polyubiquitin, but they do cleave K63 linkages within mixed-linkage chains. Our results suggest that specificity for K63-linked polyubiquitin is a common property of the JAMM/MPN+ family of DUBs.
Keywords: Brcc36, deubiquitinating enzyme, isopeptidase, JAMM, Poh1
Introduction
Diversity in ubiquitin (Ub) signalling is generated both by the numerous receptors that recognise mono- or polyubiquitinated substrates (Hicke et al, 2005), and Ub's ability to be linked into different types of polymeric chains depending on which of its seven lysine residues connect the individual Ub units. Seminal experiments with yeast expressing individual lysine-to-arginine Ub mutants first indicated that different polyUb linkages could generate unique functional outcomes (Spence et al, 1995). Subsequently, all seven possible Ub–Ub linkages were found in vivo (Peng et al, 2003). Among these, the best understood are linkages to Ub K48 and K63. Whereas K48-linked polyUb generally targets modified substrates to the 26S proteasome for degradation (Pickart, 2004), K63-linked chains serve nonproteolytic roles in intracellular protein trafficking (Clague and Urbe, 2006), transcription (Weake and Workman, 2008), DNA repair (Huang and D'Andrea, 2006) and kinase signalling (Chen, 2005). Structural studies indicate that polyUb chains assembled through K48 and K63 adopt different conformations (Varadan et al, 2002, 2004), which may explain how these chains serve as distinct signalling elements that have diverse effects in the cell.
In addition to the numerous enzymes dedicated to the construction of Ub chains, many enzymes are also dedicated to their disassembly. Humans encode nearly 100 deubiquitinating enzymes (DUBs), including several recently shown to act on K63-linked chains. These include CYLD (Kovalenko et al, 2003), which is mutated in an inherited cancer syndrome called Familial cylindromatosis (Bignell et al, 2000). Like most DUBs, CYLD is a thiol protease that contains an active site cysteine residue.
AMSH, another K63-selective DUB (McCullough et al, 2004), participates in multivesicular body sorting (Clague and Urbe, 2006) and unlike CYLD, is a member of a small family of DUBs called JAMM/MPN+ proteins (Verma et al, 2002; Yao and Cohen, 2002; Guterman and Glickman, 2004), which lack active site cysteines. Instead, these proteins are metalloproteases that coordinate a catalytically essential zinc ion within their active sites. Of the five other human JAMM/MPN+ proteins, four are also known to possess isopeptidase activity. Poh1 is a stoichiometric subunit of the 19S (PA700) proteasome regulatory complex that releases K48-linked polyUb chains from substrates targeted to the proteasome for degradation (Yao and Cohen, 2002). Jab1 is a component of the COP9 signalosome (CSN), a multisubunit complex highly similar to the lid subcomplex of the 19S particle (Wei and Deng, 2003), which cleaves the Ub-like protein Nedd8 from cullin subunits of the SCF family of E3 ligases (Cope et al, 2002). Brcc36 is part of a nuclear complex that includes the BRCA1 protein and is targeted to DNA damage foci after irradiation (Sobhian et al, 2007; Wang et al, 2007). The last two JAMM/MPN+ proteins are KIAA1915, which regulates chromatin structure by removing Ub from histone H2A (Zhu et al, 2007), and FLJ14981, which has no known function, but its sequence suggests that it is an isopeptidase as well.
During an investigation of polyUb-binding factors, we observed a DUB activity in HeLa cell extracts that specifically cleaved K63-linked polyUb and had properties similar to JAMM/MPN+ proteins. Here, we describe a new DUB that was responsible for a portion of this activity that we call BRISC (Brcc36 isopeptidase complex). We also show that specific disassembly of K63-linked chains is a property of the Poh1 DUB within the 19S regulatory complex of the proteasome. Our results suggest that several of the JAMM/MPN+ DUBs have an unsuspected, shared specificity for K63-linked polyUb.
Results
K63-linked polyUb disassembly in HeLa cell extracts
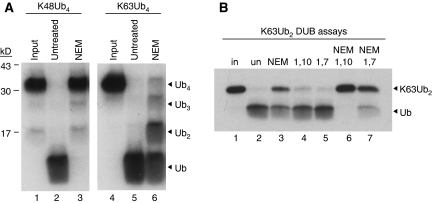
We originally attempted to purify proteins that bind specifically to K63-linked polyUb by passing HeLa cell extracts through a column containing immobilised K63-linked polyUb tetramers. Unexpectedly, a potent DUB activity in the extract cleaved the K63 linkages and disassembled the polyUb affinity resin. The activity was resistant to N-ethyl-maleimide (NEM), which alkylates the active site cysteine present in most DUBs, and specifically cleaved K63-linked but not K48-linked polyUb chains (Figure 1A, compare lanes 3 and 6).
Figure 1.
HeLa cells possess an NEM-resistant, 1,10-o-phenanthroline-sensitive, K63-specific DUB activity. (A) HeLa cell extracts (1% NP40) were treated with NEM (lanes 3 and 6) or left untreated (lanes 2 and 5), and then incubated with 125I-K48Ub4 (lanes 2–3) or 125I-K63Ub4 (lanes 5–6). Input tetramers are shown (lanes 1 and 4). Products were visualised by SDS–PAGE autoradiography. (B) Cell extracts were left untreated (lane 2) or incubated with the indicated phenanthroline compounds +/− NEM (lanes 3–7), then mixed with 125I-K63Ub2. Products were visualised as in (A).
JAMM/MPN+ domain proteins are a small class of (putative) isopeptidases that likely comprised the NEM-resistant, K63-specific activity we observed in the HeLa extracts. Instead of a catalytic cysteine residue, JAMM/MPN+ proteins coordinate a zinc ion within their active sites, which is necessary for their activity (Tran et al, 2003; Ambroggio et al, 2004; Sato et al, 2008). This property should render JAMM/MPN+ proteins both resistant to NEM and sensitive to the zinc-chelating agent 1,10-o-phenanthroline. To test this, we incubated HeLa extracts with a combination of NEM and either 1,10-o-phenanthroline (Figure 1B, lane 6) or the related, non-chelating compound 1,7-o-phenanthroline (Figure 1B, lane 7), and assessed the ability of the extract to deconjugate K63-linked polyUb dimers. The NEM-resistant, K63-directed DUB activity was completely inhibited by 1,10-o-phenathroline (Figure 1B, lane 6), but not by 1,7-o-phenathroline, which had only a very minor effect (Figure 1B, lane 7), suggesting that the activity was likely to be a zinc metalloprotease such as a JAMM/MPN+ protein.
Three distinct JAMM/MPN+ protein complexes co-purify with K63-directed DUB activity
We first considered that AMSH, a JAMM/MPN+ protein involved in endosomal and multivesicular body trafficking (Clague and Urbe, 2006), comprised the activity in the HeLa extract as AMSH cleaves K63-linked, but not K48-linked, polyUb chains (McCullough et al, 2004). AMSH, however, did not fractionate with our activity (data not shown).
To purify the K63-specific DUB, we fractionated HeLa cell extracts and followed the activity through its ability to cleave 125I-labelled K63-linked tetraUb in the presence of NEM. Although the activity partitioned into both the S100 and nuclear extracts, the nuclear activity disappeared after the first anion exchange column (Figure 2A and data not shown); attempts to recover it by combining the elutions from this column were unsuccessful. The activity in the S100 extract persisted and was followed through six additional chromatographic steps (Figure 2A; Supplementary Figure S1).
Figure 2.
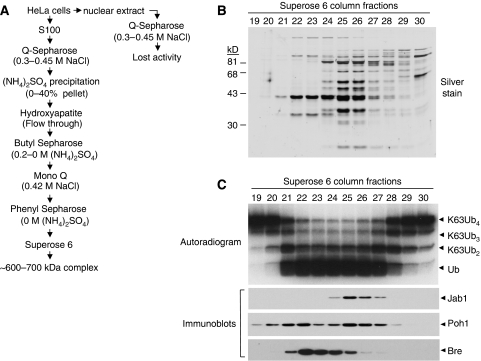
Three JAMM/MPN+ domain-containing complexes fractionate with the K63-specific DUB activity. (A) Scheme for purifying NEM-resistant, K63-specific DUB activity; see Materials and methods for details. (B) Silver-stained gel showing proteins in Superose 6 column fractions that overlap with the peak of K63-specific, NEM-resistant DUB activity. (C) (Upper panel) Fractions from the Superose 6 column were tested for their ability to disassemble 125I-K63Ub4. (Lower panels) An aliquot of each fraction was immunoblotted with antibodies to Poh1, Jab1 and Bre/Brcc45.
The activity eluted from a Superose 6 gel filtration column at a position corresponding to approximately 600–700 kDa. Fractions from this column were examined by SDS–PAGE (Figure 2B) and assessed for NEM-resistant, K63-specific DUB activity (Figure 2C, upper panel). We pooled fractions 24–26, ran them on SDS–PAGE and cut the gel into 12 sections that were then treated with trypsin and analysed by mass spectroscopy. Surprisingly, components of three JAMM/MPN+containing multisubunit complexes were identified in the final fractions. These were the 19S proteasome, including the JAMM/MPN+ protein Poh1 (Verma et al, 2002; Yao and Cohen, 2002), the CSN, including the JAMM/MPN+ protein Jab1 (Cope et al, 2002), and an uncharacterised complex, including the JAMM/MPN+ protein Brcc36 (Sobhian et al, 2007; Wang et al, 2007) (Supplementary Figure S2).
We confirmed the presence of these complexes by immunoblotting aliquots of the Superose 6 fractions with antibodies to Poh1, Jab1 or a known Brcc36-binding protein called Bre/Brcc45 (Dong et al, 2003) (Figure 2C, lower panels). We probed for Bre/Brcc45 both because Brcc36 antibodies were not available at the time, and because Bre/Brcc45 was one of the proteins identified by mass spectroscopy in the activity-containing fractions (Supplementary Figure S2). Immunoblotting confirmed that all three proteins overlapped with the active fractions from the column, although none coincided perfectly. This suggested that the DUB activity we observed was a composite of multiple complexes. Poh1 appeared in two peaks, most likely due to heterogeneity of the 19S proteasome particle.
Poh1 in the 19S proteasome complex and a novel Brcc36-containing complex each can disassemble K63-linked polyUb
Because three different JAMM/MPN+ protein-containing complexes co-fractionated with the NEM-resistant K63-specific activity, we were unsure whether a subset of the complexes was responsible for the activity or if all of them contributed. To distinguish between these possibilities, we tested whether more highly purified complexes possessed K63-specific DUB activity.
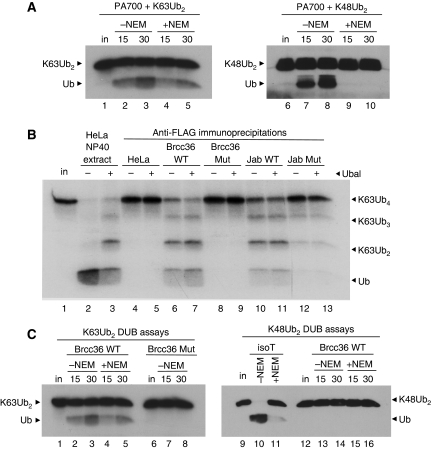
We obtained highly purified bovine PA700, the proteasome 19S subcomplex that contains Poh1 (DeMartino, 2005). Like the HeLa extract activity, PA700 efficiently cleaved K63-linked dimers (Figure 3A, lanes 2–5), even in the presence of NEM (Figure 3A, lanes 4–5). Untreated PA700 also cleaved K48-linked dimers (Figure 3A, lanes 7–8), although this activity was blocked by NEM (Figure 3A, lanes 9–10). The K48-directed activity was likely due to either Uch37 (Yao et al, 2006) or Usp14 (Borodovsky et al, 2001), two PA700-associated DUBs which, unlike Poh1, are NEM-sensitive thiol proteases. Uch37 was present in the PA700 preparation (Supplementary Figure S3); Usp14 was not tested.
Figure 3.
Three JAMM-MPN+-containing protein complexes possess K63-specific, NEM-resistant DUB activity. (A) Untreated (lanes 2–3 and 7–8) or NEM-treated (lanes 4–5 and 9–10) PA700 was incubated with 125I-K63Ub2 (lanes 2–5) or unlabelled K48Ub2 (lanes 7–10) for the indicated times. Products were visualised by autoradiography (lanes 1–5) or by immunoblotting with anti-Ub (lanes 6–10). (B) Anti-Flag immunoprecipitations were performed on lysates from HeLa cells that were untransduced (lanes 4–5), or stably expressing C-terminally Flag and HA-tagged WT (lanes 6–7) or mutant (H122Q) Brcc36 (lanes 8–9); or WT (lanes 10–11) or mutant (H138Q) Jab1 (lanes 12–13). Immunoprecipitates were incubated with 125I-K63Ub4 in the absence (even lanes) or presence (odd lanes) of Ubal. Input (in) 125I-K63Ub4 is shown (lane 1). (C) Brcc36 wild-type (WT) (lanes 2–5 and 13–16), H122Q mutant (Mut) complexes (lanes 7–8), or Isopeptidase T (isoT) (lanes 10–11) were left untreated (lanes 2–3, 7–8, 10 and 13–14) or incubated with NEM (lanes 4–5, 11 and 15–16), and then mixed with either 25I-K63Ub2 (lanes 2–8) or unlabelled K48Ub2 (lanes 10–11 and 13–16) for the indicated times. Reaction products were visualised either by autoradiography (lanes 1–8) or by immunoblotting with anti-Ub (lanes 9–16).
Next, we tested whether Brcc36 and the Jab1-containing CSN also displayed K63-specific activity. As sources of these proteins, we generated HeLa cell lines that stably express C-terminally Flag- and HA-tagged versions of Brcc36 or Jab1. We immunoprecipitated Brcc36–Flag–HA or Jab1–Flag–HA with anti-Flag-conjugated beads and eluted bound proteins through competition with Flag peptide. The Brcc36 complex, which we will call BRISC, contained three additional, and apparently stoichiometric, subunits identified by mass spectroscopy as Bre/Brcc45 (Dong et al, 2003), Abro1 (Wang et al, 2007) and HSPC142 (Supplementary Figure S4A).
Immunoprecipitated Brcc36–Flag–HA and Jab1–Flag–HA possessed potent K63-directed DUB activity (Figure 3B, lanes 6–7 and 10–11). We did not detect any K63-directed DUB activity when we subjected non-transduced HeLa cells to the identical procedure (Figure 3B, lanes 4–5). We also generated cell lines stably expressing mutant versions of each protein, Brcc36 H122Q and Jab1 H138Q, in which one of the zinc-binding histidines in the JAMM/MPN+ motif of each protein was changed to glutamine. The zinc-binding mutations in Brcc36 and Jab1 did not affect their assembly into multisubunit complexes (Supplementary Figure S5). The Brcc36 H122Q immunoprecipitate lacked any detectable DUB activity (Figure 3B, lanes 8–9), indicating that the intact, zinc-bound JAMM/MPN+ domain was necessary for its activity. The K63-linkage specificity of both this complex and PA700 was evident from assays that used either western blotting or radioiodination for detection (see Supplementary Figure S6A; Figure 6).
The Jab1 H138Q mutant CSN, although less active than its wild-type (WT) counterpart, still retained some activity (Figure 3B, lanes 12–13). As we will show later, Brcc36 interacts with the CSN and is responsible for the CSN-associated, K63-specific DUB activity. Interestingly, more Brcc36 co-precipitated with the WT Jab1 CSN than with the mutant Jab1 H138Q CSN (see below). This difference accounted for the increased DUB activity contributed by the WT Jab1 CSN in Figure 3B (compare lanes 10–11 with lanes 12–13).
The JAMM/MPN+ DUBs have ‘endo-protease' activity specific for polyUb K63-linkages
To confirm the specificity of BRISC, we assessed its ability to cleave K63- versus K48-linked dimers. BRISC showed K63-directed, NEM-resistant cleavage activity (Figure 3C, lanes 4–5) but lacked the ability to cleave K48-linked (Figure 3C, lanes 13–16) or α-linked polyubiquitin (Supplementary Figure S6A). Unlike PA700, untreated BRISC did not cleave K48 linkages (Figure 3C, lanes 13–14), suggesting that the Brcc36 subunit is the only DUB associated with this complex.
We probed the specificity of the DUB complexes further by testing their ability to cleave other types of polyUb linkages. We used the E3 ligase KIAA10 to assemble mixed polyUb tetramers linked through K29 (You and Pickart, 2001) and K6 (Pickart and Peng, unpublished data). Unlike the DUB Isopeptidase T (Hadari et al, 1992) (Figure 4, lane 2), neither PA700 (Supplementary Figure S6B, lanes 5–8) nor BRISC (Supplementary Figure S6B, lanes 10–13) cleaved these types of linkages. In addition, neither of these complexes cleaved K11-linked dimers (Supplementary Figure S6C), which were generated with the Ub conjugating enzyme E2-EPF (Baboshina and Haas, 1996).
Figure 4.
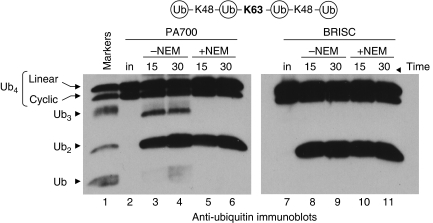
PA700 and BRISC specifically cleave K63 linkages within a mixed polyUb polymer and act as ‘endo' Ub isopeptidases. PA700 and BRISC were left untreated (lanes 3–4 and 8–9, respectively) or reacted with NEM (lanes 5–6 and 10–11), then incubated with K48–K63–K48-linked Ub4 for the indicated times. Markers (lane 1) consist of substrate partially digested by Isopeptidase T (isoT). Reactions were immunoblotted with anti-Ub. Note that input substrate (lanes 2 and 7) consists of a doublet representing linear (upper band) and cyclic (lower band) Ub4 (see Supplementary Figure S3).
Finally, we investigated whether these complexes could cleave K63 linkages within the context of a mixed-linkage chain. We obtained a commercially available polyUb tetramer consisting of a K63 linkage flanked by two K48-linkages. Specific cleavage of the K63 linkage in this substrate should generate only two K48-linked dimers. The mixed-linkage substrate migrated as a doublet (Figure 4, lane 2), which we determined to consist of the expected linear chain (upper band) and a cyclic form (lower band) containing an extra K63 linkage (Supplementary Figure S7). In the absence of NEM, PA700 slowly cleaved the K48 linkages, yielding both tri- and mono-Ub (Figure 4, lanes 3–4), and rapidly cleaved the K63 linkages, yielding dimers as the predominant products. The NEM-sensitive, K48-directed activity was likely due to Uch37 or USP14 (Figure 4, lanes 5–6).
BRISC specifically generated dimers from this substrate (Figure 4, lanes 8–11), indicating that it only cleaved the K63 linkage at the centre of the chain. The Jab1 immunoprecipitate, like PA700, possessed an NEM-sensitive, K48-directed activity that generated monomers, likely due to associated USP15 (Hetfeld et al, 2005) (Supplementary Figure S8). However, its most potent activity was the NEM-resistant, K63-directed activity (Supplementary Figure S8) contributed by co-purified BRISC (see below). Together, the data show the exquisite specificity of these JAMM/MPN+ isopeptidases for cleaving K63-linked polyUb, and their abilities to cut in the middle of a polyUb chain.
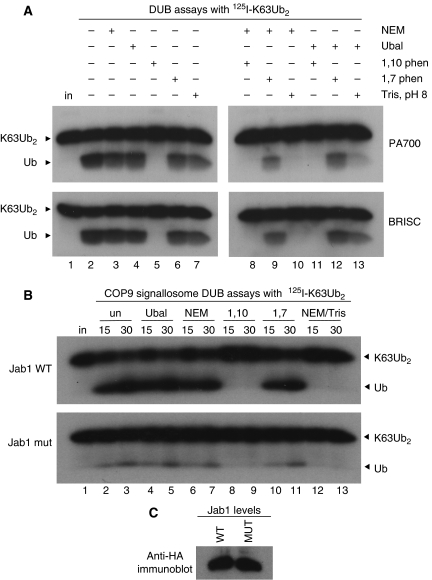
Finally, we tested whether the DUB complexes were sensitive to 1,10-o-phenanthroline (Figure 5A). Both PA700 (upper panels) and BRISC (lower panels) retained K63-directed DUB activity in the presence of NEM (lane 3), Ub aldehyde (Ubal) (lane 4), and the non-chelating compound 1,7-o-phenanthroline (lane 6). Each complex, however, was inhibited by 1,10-o-phenanthroline (lane 5). During the course of these experiments, we unexpectedly found that the NEM-resistant DUB activity in HeLa extracts was also inhibited by the combination of pH 8 Tris and NEM (Supplementary Figure S9). We used this as an additional assay to confirm that the purified complexes possessed the same properties as the original HeLa extract activity. PA700 and BRISC were both inhibited by the combination of NEM and 200 mM Tris, pH 8 (lane 10). Treatment with pH 8 Tris in the absence of NEM had little effect on the K63-DUB activities of PA700 or BRISC (lane 7), suggesting that alkylation with NEM made the JAMM/MPN+ proteins' active sites accessible to Tris (see Discussion).
Figure 5.
Purified PA700 and BRISC are sensitive to 1,10-o-phenanthroline and unprotonated Tris. (A) PA700 (upper panel) or BRISC (lower panel) were left untreated (lane 2) or incubated with the indicated compounds (lanes 3–13) and then mixed with 125I-K63Ub2. Reactions lacking Tris contained 10 mM Hepes, pH 8 (lanes 2–6, 8–9 and 11–12) in addition to the indicated compounds. Products were visualised by SDS–PAGE and autoradiography. Input (in) 125I-K63Ub2 is shown. (B) Same as (A) except Jab1 WT (upper panel) or Jab1 H138Q-containing (lower panel) CSN complexes were used. (C) Anti-HA immunoblot indicates levels of Jab1 WT and H138Q subunits in the CSN preparations used in (B).
A Brcc36 complex associates with the CSN
To test whether Jab1 was responsible for the K63-directed DUB activity associated with the CSN, we examined whether the Jab1 H138Q mutant CSN could cleave K63-linked dimers (Figure 5B). Relative to the Jab1 WT CSN, K63-DUB activity of the Jab1 mutant CSN was severely compromised (Figure 5B, compare upper and lower panels) under conditions where the levels of the Jab1 WT and Jab1 H138Q subunits were similar (Figure 5C). The mutant CSN's residual DUB activity was Ubal and NEM-resistant (Figure 5B, lanes 4–7) but was inhibited by 1,10-o-phenanthroline or NEM/Tris, pH 8 (Figure 5B, lanes 8–9 and 12–13). Although these results initially suggested that the intact Jab1 subunit possessed K63-directed DUB activity, two observations confounded this interpretation. First, the Jab1 mutant CSN completely lacked deneddylation activity (Figure 7A, lanes 6–9) yet retained some DUB activity (Figure 5B, lower panel). Second, NEM inhibited the Jab1 WT CSN's deneddylation activity (Supplementary Figure S10, lane 14) but did not affect its K63-DUB activity (Figure 5B, lanes 6–7). How could NEM treatment inhibit one activity and not the other?
We considered that another JAMM/MPN+ protein, perhaps Brcc36, had co-precipitated with the CSN and provided the residual K63 DUB activity. A BRISC-CSN interaction could also explain how three separate complexes containing JAMM/MPN+ subunits co-eluted during our original fractionation (Figure 2). Because the CSN and proteasome lid are highly homologous and expected to possess similar physical properties, it was not surprising that they co-eluted through this purification scheme. Co-elution of the dissimilar BRISC, however, was unexpected. BRISC consists of four subunits predicted to elute, if a tetramer, at less than 200 kDa. Therefore, it seemed likely that BRISC either formed higher-order oligomers or associated with additional proteins that caused it to elute as an apparent 600–700 kDa complex (Figure 2).
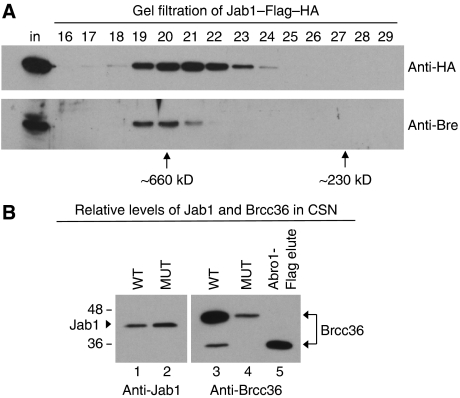
To look for an interaction between the BRISC and the CSN, we performed an anti-Flag immunoprecipitation from HeLa cells stably expressing Jab1 WT–Flag–HA and eluted bound proteins with Flag peptide. We then passed the eluate over a gel filtration column and immunoblotted with antibodies to both the HA-tag on the CSN-associated Jab1 subunit and the BRISC-associated Bre/Brcc45 subunit. Indeed, Bre/Brcc45 co-eluted with the CSN after both purification steps (Figure 6A), indicating that the complexes interacted. The Bre peak was shifted to a slightly earlier elution position relative to the Jab1 peak, suggesting that the higher molecular weight BRISC-associated portion of the CSN represented a small fraction of the total. In the original purification from HeLa extracts, there was some overlap between the Bre/Brcc45 and Jab1 peaks from the final size exclusion column, although the former eluted earlier from this column as well (Figure 2C).
Figure 6.
CSN and BRISC interact, and Jab1 WT CSN co-precipitates more Brcc36 than Jab1 mutant CSN. (A) Jab1 WT–Flag–HA immunoprecipitates were passed over a Superdex 200 size exclusion column. Column fractions were immunoblotted with anti-HA (to visualise the tagged Jab1 subunit) and anti-Bre (a BRISC subunit). The input Flag eluate before size exclusion (in) and the running positions of size standards are shown. (B) Equivalent amounts of Jab1 WT (lanes 1 and 3) or H138Q mutant (lanes 2 and 5) CSNs were run on SDS–PAGE and immunoblotted with anti-Jab1 (left panel) or anti-Brcc36 (right panel). Lane 5 contains the anti-Flag immunoprecipitate from Abro1–Flag–HA stable HeLa cells to indicate the migration position of endogenous Brcc36 in non-CSN-associated BRISC.
If BRISC provided the NEM/Ubal-resistant DUB activity associated with the CSN, why would the Jab1 H138Q mutant immunoprecipitate have so much less DUB activity than its WT counterpart (Figure 5B)? By immunoblotting, we found that WT and mutant CSNs normalised to the levels of their Jab1 subunits (Figure 6B, lanes 1–2) differed dramatically in their levels of associated Brcc36 (Figure 6B, lanes 3–4). The Jab1 mutant CSN co-precipitated approximately 10-fold less Brcc36 than the Jab1 WT CSN, a difference that could account for its decreased K63 DUB activity (Figure 5B).
Interestingly, most of the CSN-associated Brcc36 migrated slower by SDS–PAGE than the endogenous Brcc36 co-precipitated with epitope-tagged Abro1 (another BRISC subunit) (Figure 6B, lane 5; Supplementary Figure S11). The slower migrating form was not Nedd8-, Ub- or SUMO-ylated (data not shown) but may be otherwise modified or represent a novel splice form. We confirmed that the slower migrating polypeptide was indeed Brcc36 by immunoblotting with two different antibodies that were generated with N- or C-terminal Brcc36 peptides (Figure 7D; Supplementary Figure S12).
Figure 7.
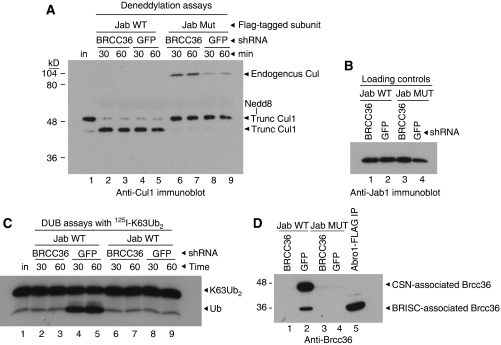
Brcc36 accounts for all of the Ubal-resistant K63 DUB activity associated with the CSN. (A) Jab1 WT or H138Q mutant (MUT) complexes were purified from HeLa cells stably expressing either a Brcc36- or GFP-directed shRNA, as indicated. Complexes were then incubated with neddylated cullin-1 (Cul1) for the indicated times and reactions were immunoblotted with anti-Cul1. Input substrate (lane 1) and co-precipitated, endogenous Cul1 are indicated. Note that the substrate was a truncated form of Cul1. (B) CSN preparations were run on SDS–PAGE and immunoblotted with anti-Jab1 to show equivalent loading. (C) Jab1 WT or H138Q mutant (MUT) complexes were purified from HeLa cells stably expressing either a Brcc36- or GFP-directed shRNA, as indicated. Complexes were then treated with Ubal and incubated with 125I-K63Ub2 for the indicated times. Products were visualised by autoradiography. (D) CSN preparations were run on SDS–PAGE and immunoblotted with anti-Brcc36.
To determine whether Brcc36 provided all the NEM/Ubal-resistant, K63 DUB activity associated with the CSN, we first performed assays using Jab1 WT and mutant immunoprecipitates normalised by Brcc36 content (Supplementary Figure S13). Under these conditions, approximately 10-fold more Jab1 H138Q mutant CSN was used per reaction than Jab1 WT CSN, yet their DUB activities were comparable (Supplementary Figure S13B). Next, we tested the activities of Jab1 immunoprecipitates purified from Brcc36 knockdown cells. We stably expressed shRNAs directed against Brcc36 or GFP (as a negative control) in our Jab1 WT–Flag–HA and Jab1 H138Q–Flag–HA cell lines (Supplementary Figure S14) and then purified CSN complexes by anti-Flag immunoprecipitation. As expected, Jab1 WT complexes purified from Brcc36 or GFP knockdown cells deneddylated a truncated Cul1-Nedd8 conjugate (Huang et al, 2004) (Figure 7A, lanes 2–5), but Jab1 H138Q mutant complexes did not (Figure 7A, lanes 6–9). A high molecular weight (∼90 kD) polypeptide recognised by the anti-Cul1 antibody was also present in the lanes containing Jab1 mutant CSNs (Figure 7A, lanes 6–9). This was neddylated, endogenous Cul1, which bound and co-precipitated with the Jab1 mutant complex during the anti-Flag affinity purification (Supplementary Figure S15).
We then examined the Ubal-resistant K63 DUB activity associated with the CSN complexes. Jab1 WT CSN purified from Brcc36 knockdown cells lacked associated K63 DUB activity (Figure 7C, lanes 2–3), whereas Jab1 WT complex purified from GFP knockdown cells retained activity (Figure 7, lanes 4–5). Consistent with this result, both the high and low molecular weight forms of Brcc36 co-precipitated with the CSN complex purified from control cells (Figure 7D, lane 2), but both forms were absent from CSN purified from Brcc36 knockdown cells (Figure 7D, lane 1). Neither of the Jab1 mutant complexes, which bound to significantly less Brcc36 than the Jab1 WT complexes (Figure 7D, lanes 3–4), had DUB activity in these assays (Figure 7C, lanes 6–9). Together, this indicated that Brcc36 provided all the NEM/Ubal-resistant, K63 DUB activity associated with the CSN.
Discussion
We used a biochemical approach to purify an NEM-resistant, K63-specific DUB activity from HeLa extracts and found that three multisubunit complexes, each possessing a JAMM/MPN+ subunit, eluted with the activity. These included the 19S (PA700) portion of the 26S proteasome, the CSN, and a novel complex containing Brcc36. Given that AMSH, another JAMM/MPN+ protein, also deconjugates K63- but not K48-linked chains (McCullough et al, 2004), we considered that such specificity may be a general property of this family of enzymes. When we analysed the individual complexes to explore this possibility, we found that although NEM-resistant, K63-specific DUB activity was associated with all three, it was intrinsic to only the PA700 and BRISC complexes; the CSN-associated activity was due entirely to co-precipitating Brcc36.
Our evidence that Poh1 is the source of the K63-specific DUB activity in PA700 is as follows: the activity is sensitive to 1,10-o-phenanthroline or NEM/Tris base, insensitive to Ubal and is robust in highly purified preparations of PA700 in which we are unable to detect Brcc36. Thus, if this proteasome-associated K63-DUB activity is not Poh1, it must be a zinc-metalloprotease DUB present in substoichiometric amounts and with very high specific activity.
The inability of PA700 or BRISC to cleave K48, K6, K11 or K29 Ub–Ub isopeptide linkages, or α-linked linear polyUb, underscores the remarkable specificity of these enzymes. In addition, they cleave K63 linkages within the context of a mixed-linkage polymer and in the middle of chains. Such endo-proteolytic activity is somewhat unusual for DUBs but is also a characteristic of CYLD (Komander, 2008), another K63-specific DUB, and Otu1 (Messick et al, 2008), a K48-specific DUB.
Several mechanisms could account for the specificity of these enzymes. One possibility is that they bind preferentially to K63-linked polyUb relative to other types of chains, possibly through recognition of a unique surface near the substrate's K63–G76 isopeptide bond. Alternatively, cleavage specificity may derive from how the isopeptide linkage of a bound polyUb chain is positioned within the enzyme's active site. PolyUb chains linked through lysines other than K63 may bind to these enzymes but not be oriented such that isopeptide bond cleavage could occur. This would allow DUBs to scrutinise different types of linkages, perhaps even within the same chain but cleave only a subset. This model is consistent with the crystal structure of the JAMM/MPN+ protein AMSH complexed with K63 diUb (Sato et al, 2008), which was published while this manuscript was under review.
The four BRISC subunits possess no obvious Ub-binding domains. This does not exclude the possibility that one or more of the subunits of this complex has a novel Ub interaction site(s), or that the Brcc36 JAMM/MPN+ motif alone is sufficient for binding to K63-linked chains. We also do not know whether the BRISC (or PA700) complex binds to one or multiple Ubs in a chain although either mechanism could, in principle, suffice to orient the K63-linkage properly within the JAMM/MPN+ active site.
Unlike the Brcc36 complex we purified, another Brcc36 complex has been reported that localises to radiation-induced DNA damage foci (Sobhian et al, 2007; Wang et al, 2007) and includes the proteins BRCA1, Abraxas and RAP80. The RAP80 subunit selectively binds to K63-linked polyUb through its two Ub-interacting motifs (Sobhian et al, 2007). RAP80 is necessary for recruiting Brcc36 to DNA damage foci (Sobhian et al, 2007, Wang and Elledge, 2007) and, in doing so, may target it to K63-polyubiquitinated substrate(s), such as histones H2A or H2AX (Huen et al, 2007, Mailand et al, 2007). Brcc36 may function to release the BRCA1 machinery from these foci once the necessary repairs have been made. The N-terminal two-thirds of Abraxas and Abro1 are 39% identical (Wang et al, 2007) and contain a conserved coiled-coil domain necessary for binding to Brcc36 (Wang and Elledge, 2007; unpublished data). This suggests that Abraxas and Abro1 act as adaptor proteins that recruit the highly selective Brcc36 DUB to substrates following certain stimuli.
Although it appears not to be a stoichiometric component of the CSN, BRISC co-precipitates with the CSN and provides all of its Ubal-resistant, K63 DUB activity. This is most clearly shown by the lack of such activity in CSN complexes purified from Brcc36 knockdown cells (Figure 7C, lanes 2–3). We also found that much more Brcc36 co-precipitated with Jab1 WT as compared with Jab1 mutant CSN complexes. This was not due to differences in the levels of expression of the Jab1 subunits (Supplementary Figure S16). Instead, we believe that the stable interaction between the Jab1 mutant CSN and neddylated cullins (Figure 7A, lanes 6–9) may ‘block' the mutant CSN from associating with BRISC, perhaps through competition for a common interaction surface. It is also possible that the Jab1 H138Q mutation directly affects the CSN interaction with BRISC; however, because the mutant CSN complex assembles and binds to Cul1-Nedd8 conjugates, we consider such a direct effect to be unlikely.
We can only speculate about the function of the 19S (PA700) proteasome-associated, K63-specific DUB activity. This activity is not unique to free PA700, because we also detect it in intact 26S proteasomes (not shown). If K48-linked polyUb chains are considered the primary degradation signal for proteasomal degradation (Pickart, 2004), why would the 26S proteasome possess a highly specific activity for cleaving K63-linked polyUb chains, which are thought to serve non-proteolytic roles? One possibility is that it could help control access or residence time of potential degradation substrates at the proteasome. This general idea has been proposed previously for the chain-trimming activities of Uch37 and Usp14/UBP6, the other major proteasome-associated DUBs (Lam et al, 1997; Guterman and Glickman, 2004; Pickart and Cohen, 2004). For Poh1 within 26S proteasomes, K63-directed chain cleavage may be particularly important if a mixed-linkage substrate is bound, or if a substrate's polyUb chain has been modified by proteasome-associated E3 ligase activities such as Hul5 (Crosas et al, 2006). Alternatively, Poh1 might spare K63-linked substrates from proteasome-mediated degradation. PA700 cleaves a mixed-linkage K48–K63–K48 polymer largely into dimers, indicating that the chain's K63 linkage is cleaved much more rapidly than its K48 linkages. This may promote release of K63-linked substrates from the proteasome before they are committed to degradation. K48-linked substrates, however, may remain bound long enough to allow further processing by the proteasome, which includes Poh1-mediated cleavage of the isopeptide bond directly linking the substrate and proximal Ub (Verma et al, 2002; Yao and Cohen, 2002). How Poh1 cleaves this bond, given its K63-linkage selectivity, may relate both to the ATP-dependence of this reaction when Poh1 is incorporated into 26S proteasomes (Verma et al, 2002; Yao and Cohen, 2002) and to the ‘coupling' of substrate deubiquitination and degradation by the 26S proteasome (Liu et al, 2006). Perhaps ATP hydrolysis is required to ‘position' the proximal Ub-substrate isopeptide bond toward the Poh1 active site to allow cleavage to occur.
While characterising the K63-specific DUB activities, we serendipitously found that the combination of NEM and unprotonated Tris inhibited the activities of JAMM/MPN+ proteins. Two crystal structures of an archaeal JAMM/MPN+ protein called AF2198 (Tran et al, 2003; Ambroggio et al, 2004) offer a mechanism for the Tris-mediated effect. One group grew crystals in the absence of Tris; this structure shows a catalytically essential water molecule positioned adjacent to the protein's active site zinc atom (Ambroggio et al, 2004). The other group grew crystals in 100 mM Tris, pH 7.8 (Tran et al, 2003), which would have abundant unprotonated Tris (the pKa of Tris is approximately 8.1). In this structure, a Tris molecule has displaced the water, thus removing the nucleophile from the active site.
Poh1 and Brcc36, however, were only inhibited by Tris if first alkylated by NEM. These proteins are larger than AF2198 and are incorporated into multisubunit complexes. Possibly, modification of one or more cysteine residues by NEM altered their conformations to facilitate access by a Tris molecule. Whatever the mechanism, using NEM +/− Tris at pH ∼8 can be a convenient diagnostic and effective inhibitor of JAMM/MPN+ DUBs.
In short, we found that the PA700 portion of the proteasome and a novel Brcc36-containing complex are highly specific, K63-directed DUBs that also possess endo-isopeptidase activity. Our data suggest that K63-selectivity is a general property of the JAMM/MPN+ family of DUBs. Also, our data indicate that a single DUB, Brcc36, is associated with three different complexes—BRISC, the BRCA1 DNA-damage-targeted complex and the CSN.
Materials and methods
Purification of NEM-resistant, K63-specific DUB activity
All steps were done at 4°C or on ice. We prepared cytosolic and nuclear extracts from a 20-l HeLa cell culture pellet (obtained from the National Cell Culture Center, Minneapolis, MN) by published methods (Dignam et al, 1983). Briefly, cells were washed with PBS, then with hypotonic lysis buffer (10 mM Hepes, pH 7.3, 10 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 0.1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride (PMSF)) and resuspended in the same buffer. Cells were lysed by Dounce homogenisation and examined visually to assure complete lysis. Lysate was spun at 2700 g for 10 min to pellet the nuclei. The post-nuclear supernatant was then spun at 100 000 g for 60 min in a Ti.70 rotor to generate the S100 cytosol fraction.
The nuclei were stirred in 10 mM Hepes, pH 7.9, 420 mM NaCl, 1 mM DTT, 1 mM PMSF for 30 min, and then spun for 30 min at 20 000 g to pellet insoluble material. The resulting supernatant comprised the nuclear extract, which was dialysed into HDE (10 mM Hepes, pH 7.3, 1 mM DTT, and 0.1 mM EDTA). The S100 fraction (0.4 g protein) and nuclear extract (0.1 g protein) were incubated for 30 min with 50 ml Q Sepharose (Pharmacia) equilibrated in HDE. The resin was transferred to a column, washed with four column volumes of HDE and then eluted stepwise with four column volumes of HDE containing 0.1, 0.3, 0.45 or 0.8 M NaCl. The S100 K63-DUB activity (see Deubiquitination assays below) eluted in the 0.3–0.45 M fraction, but we were unable to detect the nuclear DUB activity in any of the column elutions; therefore, only the S100 activity was subjected to further purification.
We added (NH4)2SO4 to the 0.3–0.45 M NaCl Q Sepharose elution to a final concentration of 40% saturation. Precipitated material was pelleted (7600 g for 10 min), resuspended in 20 ml of 50 mM potassium phosphate buffer, pH 7.4, containing 1 mM DTT, and applied to a 4-ml hydroxyapatite column (Bio-Rad) equilibrated with the same buffer. The activity was recovered in the flow-through fraction, which was then adjusted to 0.45 M (NH4)2SO4 and loaded onto a 2-ml Butyl Sepharose column (Pharmacia) equilibrated with 50 mM potassium phosphate buffer, pH 7.4, containing 0.45 M (NH4)2SO4. The column was eluted with a 40-ml linear gradient from 0.45 M to 0 M (NH4)2SO4 and then with 10 ml of 50 mM potassium phosphate buffer, pH 7.4, 0.5% Triton X-100, 1 mM DTT. The active fractions (14 ml) were pooled, dialysed into pH 7.3 HDE and bound to a Mono Q column (Pharmacia) equilibrated with pH 7.3 HDE. Proteins were eluted with a 30-ml linear gradient of 0.2–0.6 M NaCl in HDE. Active fractions were pooled and to a final concentration of 0.5 M (NH4)2SO4. The solution was then applied to a 0.4-ml Phenyl Sepharose column (Pharmacia) equilibrated with 20 mM Hepes, pH 7.3, 0.5 M (NH4)2SO4, and 1 mM DTT. The column was washed with this buffer and eluted stepwise with 1.6 ml of the same buffer containing 0.25, 0.1 and 0 M (NH4)2SO4, and finally with 1.6 ml 20 mM Hepes, 7.3, 0.5% Triton X-100 and 1 mM DTT. The 0 M (NH4)2SO4 fraction was concentrated to 200 μl in an Amicon Ultra-4 device (Millipore) and run on a Superose 6 gel filtration column (Pharmacia) equilibrated with 150 mM NaCl in HDE. Fractions (0.5 ml) were assayed for K63-specific, NEM-resistant DUB activity, and 15 μl aliquots were run on two 10% SDS–PAGE gels; one was silver stained and the other transferred to PVDF and immunoblotted with anti-Poh1, anti-Jab1 and anti-Bre antibodies (all from Zymed). Fractions 24–26 were pooled, concentrated, run on a 4–20% gradient gel (Bio-Rad) and stained with Colloidal Coomassie (Novagen). The gel was sliced into 12 sections and proteins from each were eluted, trypsinised and analysed by mass spectrometry (see supplement).
Assembly of polyUb chains
PolyUb dimers and tetramers were made as described (Pickart and Raasi, 2005) using E2-25K or Ubc13/Mms2 to generate K48 or K63-linked chains, respectively, E2-EPF (Baboshina and Haas, 1996) to generate K11-linked chains, and the combination of UbcH5a and the catalytic domain of KIAA10 (You and Pickart, 2001; unpublished results) to generate K29/6 chains. Mixed-linkage K48–K63–K48 tetraUb was purchased from Boston Biochem.
HeLa cell culture and stable cell lines
Stable cell lines were generated according to published methods (Nakatani and Ogryzko, 2003). Briefly, cDNAs were subcloned into pOZ-C, a retroviral vector that encodes C-terminal Flag and HA tags and has an internal ribosomal entry sequence preceding the cytosolic domain of the IL-2 receptor. Transduced cells were purified by physical separation using IgG-coated magnetic beads (Dynal) bound to anti-IL-2 antibodies (Pharmingen). The shRNA (TRCN0000073970) directed against Brcc36 was in the pLKO.1 vector (Open Biosystems). It was packaged into lentiviruses, which were used to infect HeLa cells according to the online protocol from the Broad Institute TRC. We picked individual clones from stable shRNA-expressing cells selected in 1 μg/ml puromycin. HeLa S3 cells were maintained in DME supplemented with 6% FBS and Penicillin/Streptomycin (Invitrogen).
Individual DUB complexes
Bovine red blood cell PA700 (DeMartino, 2005) was a gift from Dr George DeMartino (UT Southwestern Medical Center, Dallas, TX). Wild-type and mutant Jab1–Flag–HA and Brcc36–Flag–HA complexes were purified from stable HeLa cell lines (see above). HeLa cells (20 × 15-cm dishes) were washed with PBS and lysed in 10 ml of 20 mM Hepes, pH 7.3, 1% Triton X-100, 150 mM NaCl, 5 mM β-mercaptoethanol, 0.1 mM EDTA, 1 mM PMSF, 2 μg/ml aprotinin, 5 μg/ml leupeptin and 5 μg/ml soybean trypsin inhibitor. Lysates were spun for 30 min at 20 000 g in an SS34 rotor to remove insoluble material. Anti-Flag M2 antibody-coupled beads (100 μl; Sigma) were equilibrated in the above buffer, added to the clarified lysate, and rotated at 4°C for 1 h. The beads were washed with the above buffer, then with buffer lacking Triton X-100, and finally eluted with 3 × 100 μl of 0.2 mg/ml Flag peptide (Sigma) in 20 mM Hepes, pH 8, 150 mM NaCl, 5 mM β-mercaptoethanol.
Deubiquitination assays
PolyUb chains were radioiodinated using Na125I and chloramine T (Ciechanover et al, 1980). Approximately 0.5 μM polyUb was used per reaction. In Figure 1, where HeLa extracts were the DUB source, 2 × 15-cm dishes of HeLa S3 cells were washed with PBS and lysed in pH 7.3 HDE containing 1% NP40 and protease inhibitors (as above). For each reaction, 8 μl of extract was treated with inhibitors (see below) for 20 min at room temperature, then incubated with 125I-labelled polyUb for 15 min at 37°C in a final volume of 12 μl. Purified enzymes (20–50 nM) were incubated for 20 min at room temperature +/− inhibitors at the following final concentrations: 4 mM NEM, 4 mM 1,10- or 1,7-o-phenanthroline, 0.5 μM Ubal, and 200 mM Tris pH 8. Final reaction volumes were 10 μl and contained 20 mM Hepes, pH 7.3 (unless noted otherwise). Positive control reactions included 1 nM Isopeptidase T. Reactions proceeded for 15 min at 37°C unless otherwise indicated and stopped with Laemmli gel loading buffer. After SDS–PAGE (13.5 or 15%), products were visualised by autoradiography or transferred to PVDF and immunoblotted with affinity-purified anti-Ub antisera.
Deneddylation assays
Neddylated cullin was made essentially as described (Huang et al, 2008). The E1 (20 nM APP-BP1) and E2 (100 nM UbcH12) enzymes were incubated with 11 μM Nedd8 and 20 nM Cul-1/Rbx1 for 2 min in 2.5 mM ATP, 50 mM Tris, pH 7.6, 0.5 mM DTT and 5 mM MgCl2. The reaction was stopped by adding EDTA to 3.3 mM. For deneddylation assays, approximately 100 nM CSN was incubated with 1 μg of the neddylated cullin in pH 7.3 HDE for the indicated times. Reaction products were run on 10% SDS–PAGE and immunoblotted with anti-Cul-1 antisera (Zymed). APP-BP1 and Cul-1/Rbx1 were generous gifts from Dr Brenda Schulman (St Jude Children's Research Hospital, Memphis, TN). UbcH12 and Nedd8 were purchased from Boston Biochem.
CSN/BRISC interaction
WT Jab1–Flag–HA complex was immunopurified as above and run over a Superose 6 column equilibrated with PBS, 1 mM DTT. A portion of each 0.5 ml fraction was run on 10% SDS–PAGE, transferred to PVDF and immunoblotted with anti-HA (Santa Cruz Biochemicals) or anti-Bre. Anti-Brcc36 (Invitrogen), which was generated with a C-terminal Brcc36 peptide, was used for all indicated blots except for in Supplementary Figure S12, where we used anti-Brcc36 (Pro Sci), which was generated against an N-terminal Brcc36 peptide.
Supplementary Material
Supplementary Figure S1–S16
Supplementary Table 1
Supplementary Information
Acknowledgments
This work was funded in part by NIH Roadmap grant RR020839 (AP, CMP and REC) and Ruth Kirschstein Postdoctoral Fellowship 5F32GM075712 (EC). We are very grateful to the following individuals for generous gifts of reagents: Dr Brenda Schulman for the APP-BP-1 and Cul-1/Rbx1, Dr George DeMartino for PA700 and Dr Sylvie Urbe for anti-AMSH antibodies. We also thank Drs Jef Boeke and Tingting Yao for critical reading of the manuscript and helpful discussions.
References
- Ambroggio XI, Rees DC, Deshaies RJ (2004) JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol 2: E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboshina OV, Haas AL (1996) Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem 271: 2823–2831 [DOI] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, Jones C, Hansen J, Blair E, Hofmann B, Siebert R, Turner G, Evans DG, Schrander-Stumpel C, Beemer FA, van Den Ouweland A et al. (2000) Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet 25: 160–165 [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL (2001) A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J 20: 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2005) Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 7: 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Heller H, Elias S, Haas AL, Hershko A (1980) ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci USA 77: 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Urbe S (2006) Endocytosis: the DUB version. Trends Cell Biol 16: 551–555 [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413 [DOI] [PubMed] [Google Scholar]
- DeMartino GN (2005) Purification of PA700, the 19S regulatory complex of the 26S proteasome. Meth Enzymol 398: 295–306 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R (2003) Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell 12: 1087–1099 [DOI] [PubMed] [Google Scholar]
- Guterman A, Glickman MH (2004) Deubiquitinating enzymes are IN/(trinsic to proteasome function). Curr Protein Pept Sci 5: 201–211 [DOI] [PubMed] [Google Scholar]
- Hadari T, Warms JV, Rose IA, Hershko A (1992) A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J Biol Chem 267: 719–727 [PubMed] [Google Scholar]
- Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, Glickman M, Schade R, Kloetzel PM, Dubiel W (2005) The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol 15: 1217–1221 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA (2004) A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein Nedd8. Nat Struct Mol Biol 11: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Zhuang M, Ayrault O, Schulman BA (2008) Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol 15: 280–287 [DOI] [PubMed] [Google Scholar]
- Huang TT, D'Andrea AD (2006) Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 7: 323–334 [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell 29: 451–464 [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G (2003) The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424: 801–805 [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385: 737–740 [DOI] [PubMed] [Google Scholar]
- Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN (2006) ATP binding and ATP hydrolysis play distinct roles in the function of the 26S proteasome. Mol Cell 24: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- McCullough J, Clague MJ, Urbe S (2004) AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol 166: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, Reyes-Turcu FE, Wilkinson KD, Marmorstein R (2008) Structural basis for ubiquitin recognition by the otu1 ovarian tumor domain protein. J Biol Chem 283: 11038–11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V (2003) Immunoaffinity purification of mammalian protein complexes. Meth Enzymol 370: 430–444 [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2004) Back to the future with ubiquitin. Cell 116: 181–190 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE (2004) Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol 5: 177–187 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Raasi S (2005) Controlled synthesis of polyubiquitin chains. Meth Enzymol 399: 21–36 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S (2008) Structural basis for specific cleavage of K63-linked polyubiquitin chains. Nature 455: 358–362 [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HJ, Allen MD, Lowe J, Bycroft M (2003) Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 42: 11460–11465 [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D (2004) Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem 279: 7055–7063 [DOI] [PubMed] [Google Scholar]
- Varadan R, Walker O, Pickart C, Fushman D (2002) Structural properties of polyubiquitin chains in solution. J Mol Biol 324: 637–647 [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR III, Koonin EV, Deshaies RJ (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 549–552 [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2008) Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419: 403–407 [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol 8: 994–1002 [DOI] [PubMed] [Google Scholar]
- You J, Pickart CM (2001) A HECT domain E3 enzyme assembles novel polyubiquitin chains. J Biol Chem 276: 19871–19878 [DOI] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG (2007) A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell 27: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S16
Supplementary Table 1
Supplementary Information