Abstract
In the Escherichia coli system catalysing oxidative protein folding, disulphide bonds are generated by the cooperation of DsbB and ubiquinone and transferred to substrate proteins through DsbA. The structures solved so far for different forms of DsbB lack the Cys104–Cys130 initial-state disulphide that is directly donated to DsbA. Here, we report the 3.4 Å crystal structure of a DsbB–Fab complex, in which DsbB has this principal disulphide. Its comparison with the updated structure of the DsbB–DsbA complex as well as with the recently reported NMR structure of a DsbB variant having the rearranged Cys41–Cys130 disulphide illuminated conformational transitions of DsbB induced by the binding and release of DsbA. Mutational studies revealed that the membrane-parallel short α-helix of DsbB has a key function in physiological electron flow, presumably by controlling the positioning of the Cys130-containing loop. These findings demonstrate that DsbB has developed the elaborate conformational dynamism to oxidize DsbA for continuous protein disulphide bond formation in the cell.
Keywords: crystal structure analysis, disulphide bond, DsbA, DsbB, membrane protein
Introduction
The formation of disulphide bonds is crucial for the folding and maturation of many secreted proteins. Intracellular events of oxidative protein folding are efficient and accurate due to dedicated catalytic systems that have evolved in the biological kingdoms (Sevier and Kaiser, 2002; Heras et al, 2007). Escherichia coli has the best characterized disulphide bond formation system, composed of a series of Dsb enzymes (Kadokura et al, 2003; Ito and Inaba, 2008), in which disulphide-introducing (oxidative) and disulphide-isomerizing (reductive) pathways cooperate without futile cross-talk in the periplasm. A cytoplasmic membrane protein, DsbB, has a pivotal function in the disulphide-introducing pathway by generating disulphide bonds de novo in conjunction with bound ubiquinone (UQ) (or menaquinone under anaerobiosis) and transferring them to DsbA, the direct disulphide bond donor to substrate proteins (Bardwell et al, 1993; Kobayashi et al, 1997; Bader et al, 2000; Inaba et al, 2006b). DsbA has an extremely oxidizing (reduction-prone) active-site cysteines (Cys30 and Cys33) among thioredoxin-related proteins and therefore functions as an excellent disulphide donor (Grauschopf et al, 1995).
The central issues of this oxidative system include (i) how DsbB rapidly converts the oxidizing equivalents of quinone species into a protein disulphide bond and (ii) how DsbB efficiently and exclusively oxidizes DsbA despite DsbA's strong tendency to remain reduced. Our biochemical and biophysical characterization of DsbB led to proposals of the mechanisms for quinone-coupled disulphide generation and DsbB-mediated oxidation of DsbA as described in our previous publications (Inaba et al, 2004, 2005, 2006b). Notably, the crystal structure of a mutationally stabilized DsbB–DsbA binary complex provided essential insights into the above central issues of the system (Inaba et al, 2006a; Inaba and Ito, 2008). DsbB has a four-helix bundle scaffold composed of its transmembrane regions (TM1–TM4) and two periplasmic loops (P1 and P2) containing the respective essential cysteine pairs, Cys41–Cys44 and Cys104–Cys130 (Jander et al, 1994) (Figure 1). The head group of UQ is located around the N-terminal end of TM2, near which the side chains of Cys41, Cys44 and Arg48 line up. This structural feature, referred to as the reaction centre of disulphide bond manufacture, agrees excellently with our model of disulphide bond generation (Inaba et al, 2004, 2006b; Takahashi et al, 2004). In this model, a charge transfer (CT) complex and a covalent adduct are formed between Cys44 and UQ in a manner stabilized electrostatically by the guanidinium group of Arg48; the covalent Cys44–UQ bond then induces a nucleophilic attack by Cys41, leading to de novo formation of the Cys41–Cys44 bond.
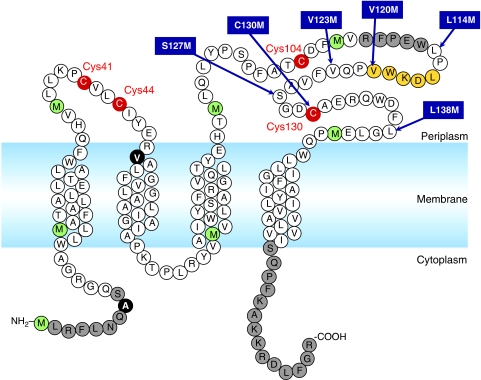
Figure 1.
Primary sequence of DsbB showing characteristic features. The essential cysteines are shown in red and methionines are in green. The two positions shown by solid black circles are cysteines in the wild-type sequence. The segment constituting the horizontal α-helix is shown in orange. Regions that lack corresponding electron density in the solved crystal structure of DsbB(Cys41Ser) are shown in grey. The sites where SeMet was introduced to model the horizontal helix and the following loop are indicated by arrows that specify the substitutions.
The binary complex structure also revealed that the Pro100–Phe106 segment of DsbB is accommodated in the deep hydrophobic groove of DsbA, where Cys104, now forming an intermolecular disulphide bond with Cys30 of DsbA, is sequestered from its original partner, Cys130. We argued that this physical separation prevents Cys130 from a counterproductive, backward attack against the Cys104(DsbB)–Cys30(DsbA) intermolecular disulphide, thus channelling the reaction forward to either the rapid pathway or the slow pathway (Inaba et al, 2005) (see also Discussion and Supplementary Figure S1). In either case, Cys130, which is reduced upon formation of the binary complex, is likely to approach the Cys41–Cys44 pair in P1 to deliver electrons to the quinone-coupled reaction centre (Kadokura and Beckwith, 2002). Thus, DsbB appears to undergo well-designed conformational changes in oxidizing DsbA.
To define the conformational changes unequivocally, however, structural information on isolated DsbB is required. Recently, the NMR structure has been solved for DsbB[CSSC] with Cys44Ser and Cys104Ser alterations (Zhou et al, 2008). Although this structure sheds light on the DsbB state with an inter-loop disulphide bond between Cys41 and Cys130, we still lack the structure of DsbB in its initial state with the Cys104–Cys130 disulphide bond. To delineate the dynamism of DsbB in the catalytic pathways, we here determined the crystal structure of the DsbB(Cys41Ser) variant at 3.4-Å resolution. The technical difficulty inherent in crystallization of this membrane protein (Inaba et al, 2006a) was overcome by co-crystallization with a monoclonal Fab antibody fragment. Although the DsbB(Cys41Ser) variant constitutively forms a CT complex between UQ and Cys44, it mimics the initial state of DsbB in that it contains the Cys104–Cys130 disulphide that is ready to interact with reduced DsbA and receive a nucleophilic attack by its Cys30 residue.
Conformational differences among DsbB(Cys41Ser)–Fab, DsbB(Cys130Ser)–DsbA(Cys33Ala) and DsbB[CSSC] illuminated the sequential relocations of the DsbB cysteines that would take place during catalysis. In addition, our systematic mutation analyses suggested that the peripherally membrane-associated α-helix contained in the P2 loop of DsbB (called ‘horizontal helix') has a key function in efficient physiological electron flow in the DsbB catalysis of DsbA oxidation. On the basis of these structural and biochemical results, we present a detailed molecular view on how the DsbA–DsbB oxidative system operates to facilitate protein disulphide bond formation in the cell.
Results
Crystallization of the DsbB–Fab complex
Our attempts to crystallize DsbB alone have been unsuccessful for several years. As a means to overcome this difficulty, we employed a strategy of co-crystallizing DsbB with its specific antibody. We thus prepared monoclonal antibodies against wild-type DsbB and screened out those forming a stable complex with DsbB; however, the complex between wild-type DsbB and the Fab fragment of the selected antibody did not produce high-quality crystals. We reasoned that the preparation of wild-type DsbB was conformationally heterogenous due to the presence of a sub-population having a rearranged Cys41–Cys130 interloop disulphide bond (Inaba et al, 2004; Li et al, 2007; Zhou et al, 2008). We then used the Cys41Ser mutant form of DsbB, in which Cys104–Cys130 disulphide was formed uniformly, whereas Cys44 engaged in CT interaction with UQ. We prepared a DsbB(Cys41Ser)–Fab complex of 1:1 stoichiometry by size-exclusion chromatography, which yielded crystals of space group C2 symmetry (Table I), with two units of the complex per asymmetric unit (Figure 2A). X-ray diffraction data sets were collected at the maximum resolution of 3.4 Å using the SPring-8 beamline BL44XU. Phases were determined by molecular replacement using a published Fab structure (PDB ID: 1IGT). After rigid-body refinement, they yielded a map with interpretable electron density in the DsbB portion. A model of DsbB(Cys41Ser) was built de novo without using any published DsbB structures, and the final model was obtained by several cycles of rebuilding and refinement (Table I).
Table 1.
Data collection and structure determination
| DsbB(Cys41Ser)–Fab | DsbB(Cys130Ser)–DsbA(Cys33Ala) | |
|---|---|---|
| Data collection | ||
| Beamline | BL44XU at SPring-8 | BL44XU at SPring-8 |
| Space group | C2 | P42212 |
| Cell dimensions (Å) | a=269.3, b=51.5, c=125.8 | a=b=165.5, c=65.9 |
| α=γ=90.0°, β=106.9° | α=β=γ=90.0° | |
| Wavelength (Å) | 0.90000 | 0.90000 |
| Resolution range (Å) | 47.51–3.40 (3.58–3.40) | 61.20–3.70 (3.90–3.70) |
| No. of total observations | 87 328 (12 552) | 70 735 (10 322) |
| No. of unique reflections | 23 294 (3334) | 10 179 (1450) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.8) |
| I/σ(I) | 12.5 (2.3) | 18.7 (3.8) |
| Multiplicity | 3.7 (3.8) | 6.9 (7.1) |
| Rmergea | 0.079 (0.658) | 0.060 (0.527) |
| Rmeasb | 0.093 (0.769) | 0.066 (0.570) |
| Refinement | ||
| Resolution range (Å) | 45.64–3.40 | 20.0–3.70 |
| Rcwork | 0.271 | 0.304 |
| Rdfree | 0.351 | 0.334 |
| RMSD | ||
| Bond length (Å) | 0.011 | 0.012 |
| Bond angle (deg) | 1.5 | 1.5 |
| Ramachandran analysise | ||
| Most favoured (%) | 69.2 | 68.7 |
| Allowed (%) | 27.3 | 27.5 |
| Generously allowed (%) | 2.8 | 2.7 |
| Disallowed (%) | 0.7 | 1.0 |
| aRmerge=ΣΣj∣〈I(h)〉−I(h)j∣/ΣΣj∣〈I(h)〉∣, where 〈I(h)〉 is the mean intensity of symmetry-equivalent reflections. | ||
| bRmeas=Σ√(n/n–1) Σj∣I(h)–I(h)j∣/ΣΣj∣〈I(h)〉∣, the multiplicity weighted Rmerge. | ||
| cRwork=Σ(∣∣Fp(obs)–Fp(calc)∣∣)/Σ∣Fp(obs)∣. | ||
| dRfree=R factor for a selected subset (5%) of the reflections that was not included in prior refinement calculations. | ||
| eFor one DsbB–Fab or DsbB–DsbA complex performed in PROCHECK. | ||
| The number in parentheses represents statistics in the highest resolution shell. | ||
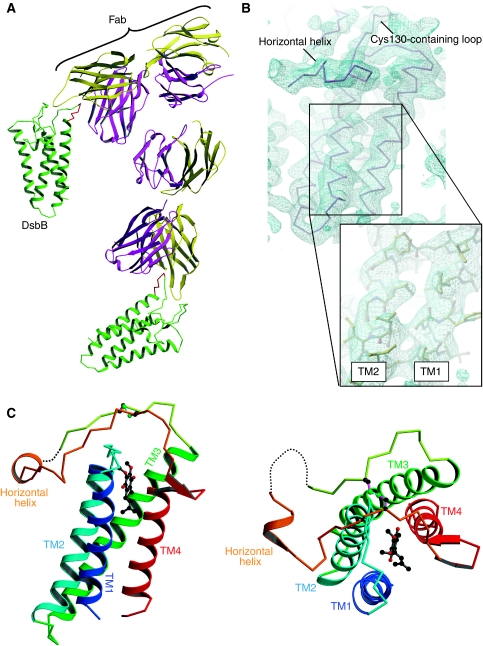
Figure 2.
Overall structure of the DsbB(Cys41Ser)–Fab complex. (A) Ribbon representation of the complex, in which DsbB molecules, heavy and light chains of Fab are shown in green, magenta and yellow, respectively. The Tyr96–Phe101 DsbB segment that is attached to Fab is shown in red. (B) Electron density map of the DsbB portion of the complex. The Cα trace of DsbB is superimposed on the electron density map drawn at the 1.0 σ contour level. The inset shows a close-up view of the side chain structure in the transmembrane region of DsbB. (C) Ribbon representation of the membrane-parallel (left) and top (right) views of DsbB(Cys41Ser). TM1 (residues 12–35), TM2 (residues 43–63), TM3 (residues 69–96), TM4 (residues 142–161) and horizontal helix (residues 116–120) of DsbB(Cys41Ser) are shown in blue, cyan, green, red and orange, respectively. UQ is represented by ball and stick (black, carbon atoms; red, oxygen atoms). The dotted line indicates the Arg109–Trp113 region, the electron density of which was invisible. For simplicity, the Fab portion is not displayed.
Overall structure of the DsbB–Fab complex
The refined model at 3.4-Å resolution includes most of the DsbB residues (Gln10–Ile162), to which the electron density map fits very nicely (Figure 2B). An N-terminal short α-helix (aa 2–9) reported in the NMR analysis of DsbB[CSSC] (Zhou et al, 2008) was not detected in the present analysis. The electron density map of DsbB(Cys41Ser) displays the side chain structure of bulky amino acids, especially in the transmembrane region (inset of Figure 2B). The Fab fragment is attached to the Tyr96–Phe101 segment contained in the P2 loop of DsbB (Figure 2A), which is actually very close to the site recognized by DsbA in the DsbB–DsbA complex. The overall architecture of DsbB(Cys41Ser) in complex with Fab is similar to that of DsbB(Cys130Ser) in complex with DsbA(Cys33Ala) (Inaba et al, 2006a) and to the solution structure of isolated DsbB[CSSC] (Zhou et al, 2008), in that it has a four-helix bundle scaffold in the transmembrane region and a long periplasmic loop containing a short membrane-parallel α-helix (Figure 2C); however, there are several local differences among these structures (see also the following sections and Discussion for more details). Although the electron density of the periplasmic loop connecting the horizontal helix to TM4 was invisible in the original DsbB–DsbA complex, DsbB(Cys41Ser) exhibited discrete electron density that is attributed to this region (Figure 2B). It is conceivable that the formation of Cys104–Cys130 disulphide makes this loop much less flexible. However, the segment from Arg109 to Trp113 lacked electron density in the DsbB–Fab complex (Figure 2C). In the DsbB–DsbA complex, a short intermolecular β-sheet is formed between Cys104–Phe106 of DsbB and Arg148–Val150 of DsbA, which may stabilize the DsbB loop in front of the horizontal helix (Inaba et al, 2006a).
Updated structure of the DsbB–DsbA complex
We carried out further model building of the DsbB(Cys130Ser)–DsbA(Cys33Ala) complex, in particular of the horizontal helix and the following loop segment of DsbB(Cys130Ser). Here, the methionine-marking method (Inaba et al, 2006a) was again used to determine the peptide backbone from relatively low-resolution crystallographic data. We constructed new DsbB variants with a methionine substitution for Leu114, Val123, Ser127 or Leu138. They were labelled with selenomethionine (SeMet), complexed with DsbA(Cys33Ala) and crystallized. The positional information of anomalous scattering peaks of selenium atoms from these variants as well as from previously constructed variants (Val120Met and Cys130Met) guided our coordinate remodelling (Figure 3B; Supplementary Figure S2). Our current analysis indicates that the horizontal helix is actually shorter than the previous assignment (Glu112–Ala126) and comprises the Leu116 to Val120 segment (Figure 3C). This revised assignment is in agreement with the crystal structure of the DsbB(Cys41Ser)–Fab complex as well as with the NMR structure of DsbB[CSSC].
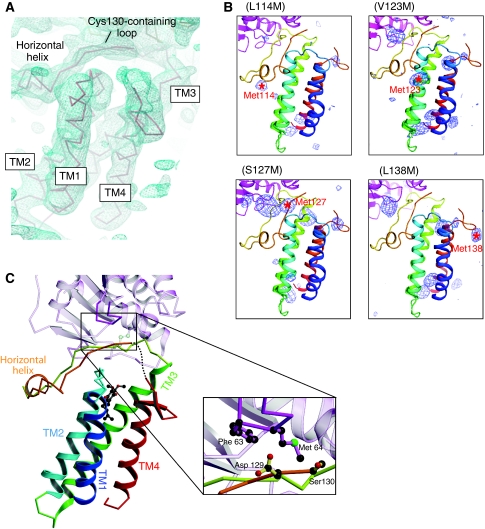
Figure 3.
Updated structure of DsbB(Cys130Ser) in disulphide-linked complex with DsbA(Cys33Ala). (A) Electron density map of the DsbB portion of the complex presented in roughly the same orientation as in (C). The Cα trace of DsbB is superposed on the electron density map drawn at the 1.0 σ contour level. Note that electron density is lacking for the Ala131–Trp135 region. (B) The Bijvoet anomalous difference-Fourier maps of new DsbB variants with a methionine substitution for Leu114, Val123, Ser127 or Leu138. The anomalous scattering peaks of selenium atoms contoured at 3.0 σ are shown in blue. The positions of selenium signals that appeared upon the indicated mutations are shown by red asterisks with residue numbers. (C) Ribbon representation of the horizontal view of the updated DsbB(Cys130Ser)–DsbA(Cys33Ala) complex structure. TM1 (residues 12–35), TM2 (residues 43–63), TM3 (residues 69–96), TM4 (residues 142–161) and horizontal helix (residues 116–120) of DsbB(Cys130Ser) are shown in blue, cyan, green, red and orange, respectively. UQ is represented by ball and stick (black, carbon atoms; red, oxygen atoms). The dotted line indicates the Ala131–Trp135 region, the electron density of which was invisible. As highlighted in the inset, the Cys130-neighbouring segment of DsbB interacts with the Phe63–Gly65 loop of DsbA (black, carbon atoms; red, oxygen atoms; green, sulphur atom). Note that Ser at position 130 is Cys in WT DsbB. For simplicity, the DsbA portion is represented semitransparently except for the Phe63–Gly65 loop.
Our rebuilding and refinement of the horizontal helix resulted in realization of the electron density that accounts for the limited region immediately after the horizontal helix. Although the electron density from this region was weak and seen only at the contour level below 1.3 σ probably because of its intrinsic flexibility (Figure 3A), we managed to define the Pro121 to Cys130 subregion. It was thus found that the Asp129–Cys130 dipeptide is adjacent to a highly conserved loop of DsbA (Phe63–Gly65) (Figure 3C), whereas this interaction contains neither hydrogen bonds nor salt bridges and would hence be weak. The NMR analysis of DsbB[CSSC] in the presence of a cysteine-less DsbA mutant also suggested that the Asp129–Cys130 dipeptide contacts DsbA even when Cys130 is disulphide-bonded to Cys41 (Zhou et al, 2008). However, this region of DsbB(Cys130Ser) seems to contact DsbA more tightly than that of DsbB[CSSC].
Similarly, a short β-hairpin just in front of TM4 was modelled based on the refined electron density map (Figure 3A) as well as from the anomalous scattering peak of SeMet138 shown in Figure 3B. The updated structure model yielded significantly lower Rwork and Rfree values than the original model (Table I), suggestive of the validity of the present model rebuilding.
DsbA-induced conformational changes in DsbB
Our crystal structure analyses of DsbA-bound and DsbA-unbound forms of DsbB established that the covalent binding of DsbA induces a significant conformational change in the second periplasmic loop of DsbB. As shown in Figure 4A, the Pro100–Cys104 segment of DsbB in the complex is shifted markedly towards the outside of the four-helix bundle, thus separating Cys104 from Cys130. Although the Cα atom of Cys104 is at a distance of 6.3 Å from that of Cys130 in DsbB(Cys41Ser), the distance is extended to 8.9 Å in the DsbB–DsbA complex. As discussed previously (Inaba et al, 2006a), attraction of this DsbB segment into the deep groove of DsbA appears to be the primary factor that induces the separation of these two cysteines. In contrast, the architecture of the transmembrane and horizontal helices of DsbB was not significantly different between the DsbA-bound and -unbound states (Figure 4A).
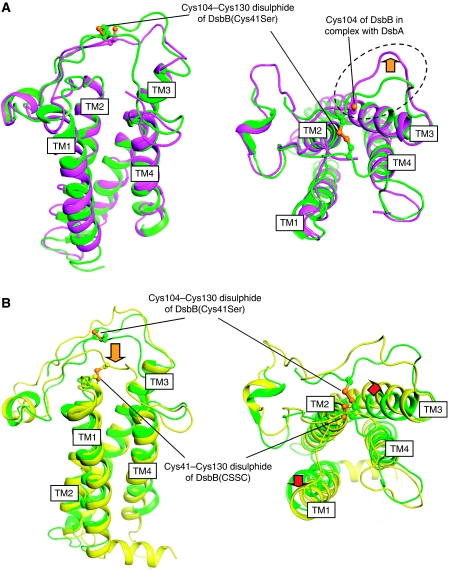
Figure 4.
Conformational transitions during the DsbB catalysis. (A) Membrane-parallel (left) and top (right) views of DsbB(Cys41Ser) in complex with Fab (shown in green) and DsbB(Cys130Ser) in complex with DsbA (shown in magenta), in which these two DsbB molecules are superimposed such that the RMSD between their Cα atoms are minimized. For simplicity, the Fab and DsbA portions are not displayed. Cys104 and Cys130 are represented by ball and stick. The dotted circle and orange arrow in the right panel highlight the marked outward movement of the Pro100–Cys104 segment upon binding to DsbA. (B) Membrane-parallel (left) and top (right) views of DsbB(Cys41Ser) in complex with Fab (shown in green) and DsbB[CSSC] (shown in yellow), in which these two DsbB molecules are superimposed such that the RMSD between their Cα atoms are minimized. For simplicity, the Fab portion is not displayed. The Cys104–Cys130 in DsbB(Cys41Ser) and the Cys41–Cys130 disulphide in DsbB[CSSC] are represented by ball and stick. The orange arrow in the left panel denotes Cys130 approach to Cys41–Cys44 disulphide upon cleavage of Cys104–Cys130 disulphide and release of DsbA from the DsbB–DsbA complex. Red arrows in the right panel denote possible movements of TM1 and TM3 induced by the formation of Cys41–Cys130 disulphide.
Mobility of Cys130-containing loop
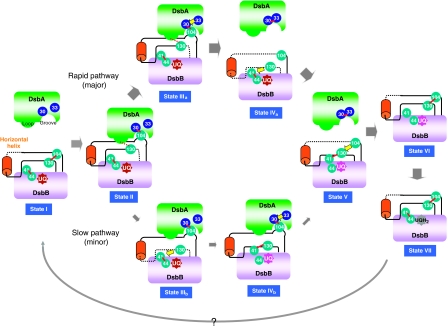
Marked structural differences are also found between DsbB(Cys41Ser) and DsbB[CSSC]. As shown in Figure 4B, the NMR structure of DsbB[CSSC] had a Cys130-containing segment that approaches Cys41 in P1 to form Cys41–Cys130 inter-loop disulphide. On the other hand, the Cys130 region of DsbB(Cys41Ser) stayed remote from P1 such that Cys130 formed a initial-state disulphide with Cys104 (Figure 4B). Notably, DsbB(Cys130Ser) in complex with DsbA (Cys33Ala) also had the residue-130 that was separated from P1, although Cys104–Cys130 disulphide was not formed in this construct (Figure 4A, left). The region around the residue-130 in the binary complex is in contact with DsbA, specifically the Phe63–Gly65 region (Figure 3C). Although some Cys130Ser mutational effect cannot be ruled out, it is possible that the release of DsbA from DsbB allows the Cys130-containing segment to access Cys41–Cys44 disulphide at significantly increased frequency to promote the rapid pathway reaction. Our results and those of Zhou et al (2008) suggest that Cys130 can reside at three different relative positions on DsbB; the Cys104-proximal location observed for DsbB(Cys41Ser), the location separated from Cys104 but still in association with the Phe63–Gly65 region of DsbA observed for the DsbB(Cys130Ser)–DsbA(Cys33Ala) complex, and the location close to the P1 Cys41 observed for the isolated DsbB[CSSC]. Unpaired but DsbA-proximal Cys130 could explain the preferred occurrence of the rapid DsbA oxidation pathway, in which the resolution of the intermolecular disulphide caused by Cys33 of DsbA precedes Cys130's approach to Cys41 (Figure 8; see Discussion for more details).
Figure 8.
Conformational transitions of DsbB for efficient DsbA oxidation. DsbB-mediated DsbA oxidation reaction is divided into seven states (I–VII). In each state, a disulphide bond and a flexible segment of DsbB are shown with a red line and a black dotted line, respectively. The present crystal structure of DsbB(Cys41Ser) represents the structure of the initial state (state I) in that it contains the Cys104–Cys130 disulphide. The DsbB–DsbA complex, the structure of which has now been updated, corresponds to state II, in which the Cys104-containing DsbB segment is pulled into the deep hydrophobic groove of DsbA to separate Cys104 from Cys130. Additionally, the Cys130-neighbouring peptide interacts with the Phe63–Gly65 loop of DsbA, as shown by a red dotted line. It is inferred that the membrane-associated horizontal helix of DsbB has a key function in properly controlling the motion of the Cys130-containing loop. In the next stage, a productive race between Cys33(DsbA) attack against the intermolecular disulphide (leading to the rapid pathway) and the Cys130(DsbB) attack against the Cys41–Cys44 disulphide (leading to the slow pathway) takes place. Cys130 approach to Cys41–Cys44 disulphide is inhibited in state II, presumably due to local contact between the Cys130-neighbouring segment and the Phe63–Gly65 loop of DsbA (red dotted line), explaining the major rapid and minor slow pathways (see Discussion for more details). In either pathway, state V with the Cys41–Cys130 interloop disulphide and the extremely flexible Cys104-containing loop is generated, as revealed by the NMR analysis of DsbB[CSSC] (Zhou et al, 2008). Cys104 then attacks Cys41–Cys130 disulphide, resulting in Cys104–Cys130 disulphide and reduced Cys41 and Cys44 (state VI). Two electrons thus moved onto the Cys41–Cys44 pair are accepted by UQ through the formation of the Cys44–UQ CT complex and covalent adduct, leading to the regeneration of fully oxidized DsbB (state VII). The reaction turns over through an unknown mechanism of UQ exchange.
It is also interesting that TM1 and TM3 in DsbB[CSSC] move slightly outward from the centre of the four-helix bundle, resulting in expansion of the space embraced by the four-helix bundle (Figure 4B, right). The backbones of TM1 and TM3 exhibit largest discrepancies between DsbB(Cys41Ser) and DsbB[CSSC] at Phe32 and Phe82, respectively; Cα atoms at Phe32 deviate by 2.86 Å and those at Phe82 deviate by 3.58 Å. Although such movements of the TMs may simply represent a difference between solution and crystal structures, it is possible that the position and orientation of the transmembrane helices of DsbB are controlled according to molecular states of the functional cysteines in the periplasmic loops (see Discussion for more details).
Structural basis of Cys–UQ CT complex on DsbB
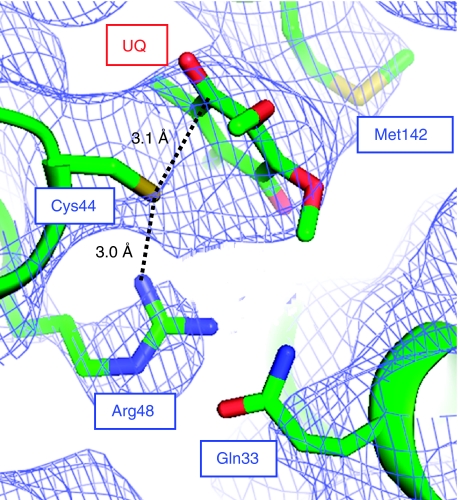
The crystal structure of DsbB(Cys41Ser) yielded direct evidence of CT complex formation between Cys44 of DsbB and UQ. Figure 5 shows the molecular geometry and electron density map of UQ and the surrounding residues, in which the S atom of Cys44 is only 3.1 Å away from the C1 atom of the UQ ring. The guanidinium moiety of Arg48 is at 3.0 Å distance from the S atom of Cys44, presumably forming a hydrogen bond with its thiolate form. These structural features are in excellent agreement with the mechanism of de novo disulphide bond generation on DsbB we proposed previously (Inaba et al, 2006b). The side chain of Gln33, which was reported to contribute to the Cys44–UQ CT complex formation (Zhou et al, 2008), could be located close to the guanidinium moiety of Arg48 although Cδ, Nɛ and Oɛ atoms of Gln33 lack any electron density (Figure 5).
Figure 5.
Formation of Cys44–UQ CT complex on DsbB. Close-up view of the UQ-neighbouring region of DsbB(Cys41Ser), in which the electron density map is shown at the 1.0σ contour level. Note that the head group of UQ and the side chains of Cys44, Arg48 and Met142, which are shown by stick representation, fit nicely onto the electron density map. Residues 38–42 of DsbB are removed for a clear view of this region.
Functional role of the horizontal helix in DsbB catalysis
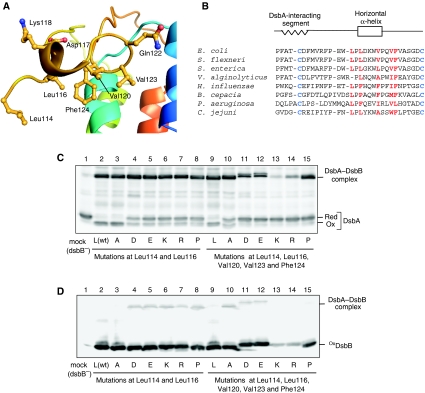
The structure of the ‘horizontal helix' revealed its clear amphiphilicity. As shown in Figure 6A, side chains of hydrophobic residues Leu114, Leu116, Val120, Val123 and Phe124 in the horizontal helix are all oriented towards the cytoplasmic membrane, whereas charged or hydrophilic residues Asp117, Lys118 and Gln122 are oriented oppositely to the periplasmic aqueous phase. The hydrophobic residues mentioned above are well conserved in DsbB orthologues (Figure 6B) (Kadokura et al, 2000; Raczko et al, 2005).
Figure 6.
Functional role of the DsbB horizontal helix in DsbA oxidation. (A) Close-up view of the horizontal helix of DsbB(Cys41Ser) in complex with Fab. Leu114, Leu116, Val120, Val123 and Phe124 located on the membrane-facing side and Asp117, Lys118 and Gln122 located on the aqueous side are represented by ball and stick. (B) Sequence alignment of DsbB orthologues for the region from Pro100 to Cys130. Conserved essential cysteines in the P2 loop are in blue. Residues in red denote membrane-facing hydrophobic residues near or inside the horizontal helix (see (A)). (C) In vivo redox state of DsbA in cells expressing each DsbB variant, in which Leu114 and Leu116 (plus Val120, Val123 and Phe124) are simultaneously replaced with alanine (A), aspartate (D), glutamate (E), lysine (K), arginine (R) or proline (P). Reduced and oxidized forms of DsbA were separated by 12.5% SDS–PAGE after cysteine alkylation with AMS and detected by western blot analysis with an anti-DsbA antibody. (D) In vivo redox states of the indicated DsbB variants, which were detected by western blot analysis (12.5%) with an anti-myc antibody.
To assess the functional importance of the horizontal helix, we introduced substitutions of charged (D, E, K and R) or helix-breaking (P) residues into the membrane-facing positions. Mutant DsbB proteins were expressed from plasmids in ΔdsbB cells and the in vivo redox states of DsbA were examined. It was found that simultaneous replacement of Leu114 and Leu116 with charged residues or with proline significantly increased the proportion of the reduced form of DsbA (Figure 6C, lanes 4–8), as compared with cells expressing wild-type DsbB (lane 2). In contrast, alanine substitutions at this position had a negligible effect (lane 3). Simultaneous mutations at Leu114, Leu116, Val120, Val123 and Phe124 resulted in more severe defects (lanes 10–15). Cellular accumulation levels of these DsbB variants, except for the five-K or five-R mutants, were similar to that of wild-type DsbB (Figure 6D, lanes 2–15). Thus, the presence of two or more charged or helix-breaking residues at these positions is incompatible with the functionality of DsbB. In vitro assays using membrane fractions of mutant and wild-type cells confirmed the mutational effects on DsbB activity (Supplementary Figure S3).
The DsbB variants examined above were mostly in the oxidized form in vivo, although many of the five-residue alterations affected band mobility in SDS–PAGE (Figure 6D). In vitro experiments indicated that all of the membrane-integrated DsbB variants were refractory to reduction with 5 mM DTT, as observed with wild-type DsbB (Supplementary Figure S4) (Kobayashi and Ito, 1999). Thus, they retain the ability to generate disulphide bonds through interaction with UQ. It is noted, however, that the activity-compromised DsbB variants exhibited a significant fraction of the disulphide-bonded DsbA–DsbB complex (Figure 6C and D). The results suggest that these mutations impaired the DsbA-oxidizing ability of DsbB without severely hampering the interaction with DsbA or UQ.
Membrane association of the horizontal helix in DsbB
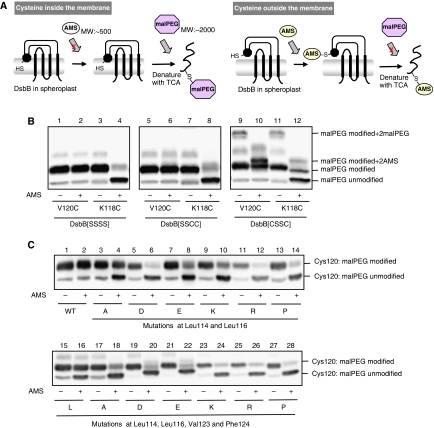
To investigate the physiological localization of the horizontal helix as well as effects of the above mutations on it, we introduced a single cysteine into the Val120 position on the membrane side of the horizontal helix and then monitored the accessibility of a membrane-impermeable alkylating reagent, AMS, to the engineered cysteine residue. In this construct, all the DsbB cysteines were eliminated by the replacement with serine to avoid any complication that could arise from alkylation of the native cysteines. Spheroplasts were prepared from cells expressing the single cysteine DsbB variant and treated with 5 mM AMS (MW: ∼500 Da). Samples were then denatured with TCA, solubilized with SDS and subjected to counter-modification with maleimide-PEG2K (malPEG) (MW: ∼2 kDa) of any cysteine that had escaped AMS modification (Figure 7A). Thus, malPEG modification that causes a marked retardation in SDS–PAGE indicates that the corresponding proportion of the protein has not been alkylated by the first reaction with AMS (Cho et al, 2007; Koide et al, 2007). Such a fraction should have a cysteine in an AMS-inaccessible environment, that is, most probably in the lipid phase (Figure 7A).
Figure 7.
Membrane association of the horizontal helix in wild-type and mutant DsbB. (A) Schematic depiction of AMS alkylation and malPEG counter-alkylation assay. Membrane-buried cysteine is not modified with AMS, a membrane-impermeable alkylating agent, in the first alkylation step. After denaturation with TCA and solubilization in SDS, however, it is exposed to the aqueous phase and modifiable with malPEG. In contrast, cysteine outside the membrane is subject to the first-step AMS alkylation and, hence, not to the second-step malPEG alkylation. The AMS (∼0.5 kDa) and the malPEG (∼2 kDa) modifications can be discriminated by the different extents of mobility retardation in SDS–PAGE. A black circle in spheroplast DsbB indicates the horizontal helix. (B) Different locations of Val120 and Lys118 relative to the membrane. [SSSS], [SSCC] and [CSSC] forms of spheroplast DsbB–myc proteins, in which either Val120Cys or Lys118Cys mutation was introduced, were subjected to the AMS alkylation and malPEG counter-alkylation assay shown in (A). MalPEG-modified species were separated from malPEG-unmodified species by SDS–PAGE (12.5%) and detected by western blot analysis with an anti-myc antibody. Extra bands observed for DsbB[CSSC] represent species having malPEG- or AMS-modified Cys41 and Cys130 as well as malPEG-modified Cys120 (or Cys118). (C) AMS alkylation and malPEG counter-alkylation assays for a series of DsbB[SSSS] derivatives having a single cysteine at position 120. Additionally, they had the indicated mutations. Detection was carried out as addressed in (B).
Cysteine at position 120 on the membrane side of the horizontal helix was malPEG-modified irrespective of AMS pretreatment (Figure 7B, lanes 1 and 2), strongly suggesting that this part of DsbB is membrane buried. In contrast, the Lys118Cys mutant with single cysteine on the other side of the helix was mostly alkylated with AMS in the first step, which interfered with malPEG counter-modification (Figure 7B, lanes 3 and 4). This result is consistent with the notion that this side of the helix is exposed to the periplasmic milieu. It is thus suggested that the horizontal helix associates ‘peripherally' with the E. coli cytoplasmic membrane. Zhou et al (2008) showed by EPR spectroscopy of nitro oxide-labelled DsbB[CSSC] that the horizontal helix was completely buried in the lipid bilayer. However, cysteine at position 118 was AMS accessible even in the constructs that were based on DsbB[SSCC] (Figure 7B, lanes 5–8) and DsbB[CSSC] (lanes 9–12), although some fraction of the latter construct was in reduced form, giving rise to extra species with additionally malPEG- (lanes 9 and 11) or AMS-modified (lanes 10 and 12) Cys41 and Cys130. Thus, it seems unlikely that the presence of Cys130–Cys41 or Cys104–Cys130 disulphide substantially affected the vertical positioning of the horizontal helix. The results of Zhou et al (2008) might have been a consequence of the reconstitution of once-solubilized DsbB proteins into artificial POPC vesicles.
Double substitutions of Ala for Leu114 and Leu116 in DsbB(V120C) somewhat increased the modifiability of Cys120 with AMS (Figure 7C, lanes 3 and 4), suggesting that the horizontal helix is now more exposed to the aqueous phase. Introduction of charged or helix-breaking residues into these two sites enhanced the AMS modifiability of Cys120 remarkably (Figure 7C, lanes 5–14), suggesting that these mutations disrupted the membrane association of the horizontal helix more markedly than Ala substitutions. Similar membrane association-interfering effects were observed with simultaneous amino-acid substitutions at positions 114, 116, 123 and 124 (Figure 7C, lanes 19–28), including Ala substitutions (lanes 17 and 18). Taken together, we conclude that the hydrophobic side of the horizontal helix is associated with the membrane and that this association is important for DsbB activity to oxidize DsbA effectively.
Discussion
Previously, crystal structures of DsbB had only been obtained as disulphide-linked complexes with DsbA (Inaba et al, 2006a, 2006b; Malojĉić et al, 2008). In this study, we succeeded in determining the crystal structure of this intractable membrane protein, with the aid of the monoclonal antibody Fab fragment and using a conformationally homogeneous Cys41Ser variant. The Fab fragment proved to bind to almost the same region of DsbB as DsbA. It seems general that Fab binds to a surface-exposed region of an antigen protein, which also serves as a docking site for a native partner protein (Tsukazaki et al, 2008).
It is interesting to note that various constructions of DsbB, designed primarily from technical demands, illuminate the different conformations that this enzyme adopts to fulfill the catalytic abilities of disulphide bond generation and DsbA oxidation. The association of DsbA and consequent formation of the intermolecular disulphide-linkage between Cys30 (DsbA) and Cys104 (DsbB) physically separate Cys104 from Cys130. As discussed previously (Inaba et al, 2006a), the sequestration of Cys130 from the intermolecular disulphide will inhibit backward electron flow from DsbB to DsbA upon formation of the binary complex.
Notably, cleavage of Cys104–Cys130 disulphide in the DsbB–DsbA complex leads to larger fluctuation of the DsbB loop immediately after Cys130, as suggested from the lack of electron density for this region in the presence of DsbA(Cys33Ala) (Figure 3A). In contrast, the NMR structure of isolated DsbB[CSSC] indicates that its Cys130-neighbouring segment is fixed in the vicinity of Cys41 and, in turn, the Cys104-containing segment becomes mobile (Zhou et al, 2008). The enhanced mobility of these cysteine-containing segments will allow the cysteines to attack their respective target disulphides with increased probability along the physiological electron flow pathway. We envisage that flexible regions in DsbB P2 (see black dotted lines in Figure 8) alternate depending on the redox states of the functional cysteines and association–dissociation status of DsbA. Such a mechanism should be well suited for promoting physiological electron movement in this oxidative system.
Although proper flexibility of active site cysteines would be essential for the catalytic activity of DsbB, excessive mobility could have deleterious effects on the catalysis. In line with this expectation, we observed that introduction of charged or helix-breaking residues into the membrane-facing side of the horizontal helix compromised DsbA oxidation activity of DsbB. Cysteine alkylation assays revealed that the horizontal helix is associated with the periplasmic leaflet of the cytoplasmic membrane through its hydrophobic side chains that align on the membrane-facing surface of the helix and that the mutations impaired this membrane association of the helix. The correlation between the DsbA oxidation activity and the membrane affinity of the horizontal helix suggested that the mobile property of the P2 loop is properly regulated by the membrane-associated horizontal helix to accomplish the effective DsbA oxidation.
By virtue of its membrane tethering, the horizontal helix divides the P2 region into two loops, which will assure that the two cysteines in P2 receive separate mobility regulation as discussed above. In addition, the horizontal helix might impose proper limitations to the mobility of these cysteines. If the Cys130 movement becomes undirected because of the mutational disruption of the membrane-horizontal helix interaction, it could attack the Cys30 (DsbA)–Cys104 (DsbB) intermolecular disulphide to cancel out the oxidation reaction. Indeed, our in vitro assay with quinone-free DsbB showed that although wild-type DsbB was capable of oxidizing ∼40% of reduced DsbA in a 1:1 stoichiometric reaction even without the aid of UQ (Inaba et al, 2005), DsbB variants having the inactivating mutations on the horizontal helix lost the ability to accept electrons from DsbA (Supplementary Figure S5). We propose that the membrane-bound horizontal helix of DsbB restricts the movement of the two catalytically essential P2 cysteines, thereby functioning as a ratchet that drives the physiological thiol-disulphide exchange reactions. The proper organization by the horizontal helix is also important for the specificity of oxidation, in which DsbB does not effectively oxidize the dimeric DsbC protein (Pan et al, 2008).
We also note that the small movement of transmembrane helices TM1 and TM3 might have functional significance. The formation of the Cys41–Cys130 disulphide in DsbB[CSSC] appears to somehow affect the local structures of the P1 loop and TM1 (Figure 4B), resulting in expansion of the UQ-accommodating space. This expansion may be involved in the movement or exchange of UQ that could accompany the catalytic reactions of DsbB. We previously observed that DsbB mutants, in which insertion or substitution mutations were introduced into the central cytosolic loop connecting TM2 and TM3, had a Cys41–Cys130 disulphide constitutively similar to DsbB[CSSC], resulting in severe inactivation (Takahashi et al, 2006). Although structural information is lacking for these inactive DsbB mutants, the altered cytosolic loop could have affected the orientation of TM3 and hence the geometry of the functional cysteines. Thus, the fine adjustment of the transmembrane helices may also be coupled with the disulphide rearrangements that occur in the periplasmic regions of DsbB.
Our current analysis and the recent reports by others on structures of DsbB in different states have established that ‘cysteine relocation' (Inaba et al, 2006a) actually occurs in DsbB to productively mediate electron transfer from DsbA to UQ through DsbB. The direct visualization of the Cys104–Cys130 proximity in the initial state of DsbB established that DsbA indeed induces the separation of these two cysteines. Although the Cys104 sequestration into the Cys130-inaccessible position prevents the backward resolution of the DsbA–DsbB intermediate (Inaba et al, 2006a), Cys130 remains associated with DsbA. In our view, this is followed by the choice between the rapid and slow DsbA oxidation pathways, namely, between the Cys33(DsbA) attacking against the intermolecular disulphide and Cys130(DsbB) attacking against the Cys41–Cys44 disulphide (see states IIIa and IIIb in Figure 8).
The transient tethering of Cys130 to a location (near the Phe63–Gly65 loop of DsbA) that is far from both Cys104 and Cys41 provides an essential basis, by which DsbB prefers the rapid pathway of DsbA oxidation; this would allow enough time for Cys33 of DsbA to attack the intermolecular disulphide before Cys130's attack against Cys41. The resulting oxidation of DsbA and its release from DsbB may enhance fluctuation of the Cys130-neighbouring segment and would allow Cys130 to quickly approach the Cys41–Cys44 disulphide to make a transient disulphide with Cys41 (state IVa in Figure 8). This is followed by the reoxidation of Cys104 and Cys130 coupled with quinone-mediated oxidation of Cys41 and Cys44 (see below).
With a certain probability, however, Cys130 that is liberated upon the DsbA–DsbB intermediate formation and intrinsically flexible can detach from the Phe63–Gly65 loop of DsbA and approach Cys41 before release of DsbA (state IIIb in Figure 8), leading to the slow pathway. This notion accounts for our previous observations that the rapid pathway predominates but the slow pathway also takes place significantly (Inaba et al, 2004, 2005). In either pathway, Cys104 liberated by the resolution of the DsbA–DsbB complex readily attacks the Cys41–Cys130 disulphide (state V in Figure 8). Indeed, Zhou et al (2008) showed that DsbB[CSSC], which we believe to mimic an intermediate of not only the slow pathway but also the rapid pathway, has increased mobility of the Cys104-containing loop to facilitate re-formation of the Cys104–Cys130 disulphide. Finally, the reduced Cys41–Cys44 pair is oxidized by UQ, and DsbB is reactivated. In summary, DsbB-mediated DsbA oxidation is ensured by the regulated dynamism of the periplasmic loops of DsbB accompanied with the programmed relocation of the essential cysteines, in which binding/release of DsbA and the membrane association of the horizontal helix have essential functions.
Materials and methods
Purification and crystallization of a DsbB–Fab complex
DsbB(Cys41Ser) was overexpressed in E. coli and purified in detergent undecyl-β-D-maltoside as described previously (Inaba et al, 2006a). A hybridoma producing the monoclonal antibody against DsbB(Cys41Ser) was prepared by MBL Co., Ltd and our in-house screenings. The Fab fragment was purified by IBL Co., Ltd and sequenced by MBL Co., Ltd. After mixing DsbB(Cys41Ser) with a stoichiometric excess of Fab, the complex was purified on a Superdex-200 column equilibrated in 20 mM HEPES pH 8.0, 150 mM NaCl and 0.1% (w/v) 1,2-diheptanoyl-sn-glycero-3-phosphocoline (DHPC; Avanti) and concentrated to ∼10 mg/ml. Crystals of space group C2 were grown by sitting-drop vapour diffusion in a mixture of 15% PEG3350, 0.1 M magnesium formate and 0.1 M MOPS (pH 7.0). Crystals grew within 10 days. They were cryoprotected in 27% PEG3350, 0.1 M magnesium formate, 0.1 M MOPS pH 7.0, 0.1% DHPC before freezing in liquid nitrogen.
Crystallographic analysis
Data for the DsbB(Cys41Ser)–Fab complex were collected on a beamline BL44XU at SPring-8 (Hyogo, Japan) with image plates/CCD hybrid detector DIP6040 (MAC Science/Bruker AXS) at cryogenic temperature (100 K). The data were integrated with HKL2000 (Otwinowski and Minor, 1997), and the crystallographic parameters are summarized in Table I. The crystal contained two complex molecules in an asymmetric unit. Phase determination was made by molecular replacement using a published Fab structure (PDB code 1IGT) as the search model. The model was refined by several cycles of manual rebuilding and refinement with Coot (Emsley and Cowtan, 2004), Program O (Jones et al, 1991) and Refmac 5 (Collaborative Computational Project, 1994).
SeMet-labelled forms of new DsbB variants (Leu114Met, Val123Met, Ser127Met and Leu138Met) were crystallized in a disulphide-linked complex with DsbA following the same sample preparation and crystallization protocols that had been worked out for the original DsbB(Cys130Ser)–DsbA(Cys33Ala) complex (Inaba et al, 2006a). Diffraction data sets were collected and processed in the same way as the DsbB(Cys41Ser)–Fab complex. Their crystallographic parameters are summarized in Supplementary Table SI. Bijvoet anomalous difference-Fourier maps were calculated using molecular replacement solution with the original DsbB–DsbA complex model (2HI7). The model was built and refined using the same softwares as above.
Detection of in vivo redox states of DsbA and DsbB variants
E. coli strain SS141 (dsbB∷kan5) was transformed with pUC119-based dsbB-his6-myc plasmids that encode a series of DsbB variants. The strains were grown aerobically at 37°C in L-broth supplemented with 50 μg/ml of ampicillin, 0.5% glucose and 0.1 mM IPTG. Whole-cell proteins were precipitated by direct treatment with final 5% of trichloroacetic. Protein precipitates were processed for AMS modification as described previously (Kobayashi and Ito, 1999). Redox states of DsbA and DsbB were visualized by western blot analysis with anti-DsbA and anti-myc antibodies, respectively.
AMS alkylation and malPEG counter-alkylation of single Cys DsbB variants
Two-step cysteine alkylation assay to investigate the location of the horizontal helix was performed by essentially the same procedures as described by Cho et al (2007), except that the concentration of alkylating reagents and the incubation time were changed appropriately. Intact spheroplasts containing a series of DsbB variants with a single cysteine, Cys120, were prepared by resuspending the cells in ice-cold buffer containing 50 mM Tris–HCl (pH 8.0), 1 mM CaCl2, 3 mM EDTA, 18% sucrose and 30 μg/ml lysozyme. Subsequently, 5 mM of AMS was added and the mixtures were incubated on ice for 1 h for the first alkylation. Samples were TCA-precipitated, washed with acetone and dissolved in 50 mM Tris, pH 7.0, 2% SDS and 5 mM malPEG2K (SUNBRIGHT ME-020MA; NOF Corporation, a kind gift from Dr Jun Hoseki, Kyoto University). This second alkylation was performed at room temperature overnight. Alkylated DsbB proteins were separated by 12.5% SDS–PAGE and visualized by western blot analysis with anti-myc antibodies.
Accession numbers
Coordinates and structure factors described herein have been deposited in the Protein Data Bank with the ID codes 2ZUQ for the DsbB–Fab complex and 2ZUP for the updated DsbB–DsbA complex.
Supplementary Material
Supplementary Figures S1–S5
Acknowledgments
We thank Tomoya Tsukazaki for useful suggestions about antibody preparation and crystallization. We also thank Eiki Yamashita and Masato Yoshimura for help with diffraction data collection. This study was supported by a grant-in-aid from the Japan Society for the Promotion of Science and MEXT (to K Inaba), by CREST from Japan Science and Technology Agency (to K Ito), by Targeted Proteins Research Program (TPRP) from MEXT, Japan (to M Suzuki) and by Kyushu University Interdisciplinary Program in Education and Projects in Research Development (to K Inaba).
References
- Bader MW, Xie T, Yu CA, Bardwell JC (2000) Disulfide bonds are generated by quinone reduction. J Biol Chem 275: 26082–26088 [DOI] [PubMed] [Google Scholar]
- Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J (1993) A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA 90: 1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Porat A, Ye J, Beckwith J (2007) Redox-active cysteines of a membrane electron transporter DsbD show dual compartment accessibility. EMBO J 26: 3509–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC (1995) Why is DsbA such an oxidizing disulfide catalyst? Cell 83: 947–955 [DOI] [PubMed] [Google Scholar]
- Heras B, Kurz M, Shouldice SR, Martin JL (2007) The name's bond…disulfide bond. Curr Opin Struct Biol 17: 691–698 [DOI] [PubMed] [Google Scholar]
- Inaba K, Ito K (2008) Structure and mechanisms of the DsbB–DsbA disulfide bond generation machine. Biochim Biophys Acta 1783: 520–529 [DOI] [PubMed] [Google Scholar]
- Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, Ito K (2006a) Crystal structure of the DsbB–DsbA complex reveals a mechanism of disulfide bond generation. Cell 127: 789–801 [DOI] [PubMed] [Google Scholar]
- Inaba K, Takahashi YH, Fujieda N, Kano K, Miyoshi H, Ito K (2004) DsbB elicits a red-shift of bound ubiquinone during the catalysis of DsbA oxidation. J Biol Chem 279: 6761–6768 [DOI] [PubMed] [Google Scholar]
- Inaba K, Takahashi YH, Ito K (2005) Reactivities of quinone-free DsbB from Escherichia coli. J Biol Chem 280: 33035–33044 [DOI] [PubMed] [Google Scholar]
- Inaba K, Takahashi YH, Ito K, Hayashi S (2006b) Critical role of a thiolate-quinone charge transfer complex and its adduct form in de novo disulfide bond generation by DsbB. Proc Natl Acad Sci USA 103: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Inaba K (2008) The disulfide bond formation (dsb) system. Curr Opin Struct Biol 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Jander G, Martin NL, Beckwith J (1994) Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J 13: 5121–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kadokura H, Bader M, Tian H, Bardwell JC, Beckwith J (2000) Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc Natl Acad Sci USA 97: 10884–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Beckwith J (2002) Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J 21: 2354–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Katzen F, Beckwith J (2003) Protein disulfide bond formation in prokaryotes. Annu Rev Biochem 72: 111–135 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ito K (1999) Respiratory chain strongly oxidizes the CXXC motif of DsbB in the Escherichia coli disulfide bond formation pathway. EMBO J 18: 1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K (1997) Respiratory chain is required to maintain oxidized states of the DsbA–DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci USA 94: 11857–11862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide K, Maegawa S, Ito K, Akiyama Y (2007) Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J Biol Chem 282: 4553–4560 [DOI] [PubMed] [Google Scholar]
- Li Y, Berthold DA, Frericks HL, Gennis RB, Rienstra CM (2007) Partial (13)C and (15)N chemical-shift assignments of the disulfide-bond-forming enzyme DsbB by 3D magic-angle spinning NMR spectroscopy. Chembiochem 8: 434–442 [DOI] [PubMed] [Google Scholar]
- Malojĉić G, Owen RL, Grimshaw JPA, Glockshuber R (2008) Preparation and structure of the charge-transfer intermediate of the transmembrane redox catalyst DsbB. FEBS Lett 582: 3301–3307 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pan JL, Sliskovic I, Bardwell JC (2008) Mutants in DsbB that appear to redirect oxidation through the disulfide isomerization pathway. J Mol Biol 377: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczko AM, Bujnicki JM, Pawlowski M, Godlewska R, Lewandowska M, Jagusztyn-Krynicka EK (2005) Characterization of new DsbB-like thiol-oxidoreductases of Campylobacter jejuni and Helicobacter pylori and classification of the DsbB family based on phylogenomic, structural and functional criteria. Microbiology 151: 219–231 [DOI] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA (2002) Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol 3: 836–847 [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Inaba K, Ito K (2004) Characterization of the menaquinone-dependent disulfide bond formation pathway of Escherichia coli. J Biol Chem 279: 47057–47065 [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Inaba K, Ito K (2006) Role of the cytosolic loop of DsbB in catalytic turnover of the ubiquinone–DsbB complex. Antioxid Redox Signal 8: 743–752 [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O (2008) Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455: 988–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cierpicki T, Jimenez RHF, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH (2008) NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell 31: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S5