Abstract
Background
Isolation of cell colonies is an essential task in most stem cell studies. Conventional techniques for colony selection and isolation require significant time, labor, and consumption of expensive reagents. New microengineered technologies hold the promise for improving colony manipulation by reducing the required manpower and reagent consumption.
Methods
Murine embryonic stem cells were cultured on arrays composed of releasable elements termed micropallets created from a biocompatible photoresist. Micropallets containing undifferentiated colonies were released using a laser-based technique followed by cell collection and expansion in culture.
Results
The micropallet arrays provided a biocompatible substrate for maintaining undifferentiated murine stem cells in culture. A surface coating of 0.025% gelatin was shown to be optimal for cell culture and collection. Arrays composed of surface roughened micropallets provided further improvements in culture and isolation. Colonies of viable stem cells were efficiently isolated and collected. Colonies sorted in this manner were shown to remain undifferentiated even after collection and further expansion in culture.
Conclusions
Qualitative and quantitative analyses of sorting, collection efficiency and cell viability after release and expansion of stem cell colonies demonstrated that the micropallet array technology is a promising alternative to conventional sorting methods for stem cell applications.
Keywords: Embryonic stem cell, micropallet array, cell separation
The ability to genetically modify murine embryonic stem (ES) cells through gene targeting and homologous recombination has had a tremendous impact on biological and biomedical research (1). There are now widespread efforts underway to produce genetically engineered mice (GEMs) representing gene “knock outs” for each gene in the mouse genome and for “knock-in” of humanized genes in the modeling of human diseases (1,2). The selection and isolation of clonal ES colonies for generating cell lines and for producing GEMs is a time-intensive and laborious process. In conventional methods, typically 20 × 106 cells are electroporated in the presence of the transfection vector with only 10-50% of cells surviving this transfection step. The remaining cells are then grown for 2 weeks in 10-cm tissue culture dishes in media containing selective antibiotics. Typically, the media must include leukemia inhibitory factor (LIF), a cytokine necessary for maintaining the ES cells in an undifferentiated state in the absence of a feeder cell layer (3). While elimination of the feeder layer reduces complexity, purified LIF is an expensive reagent that must be used in significant quantities for the lengthy culture and volumes of media conventionally required to establish cell lines. After 2 weeks of cell expansion, clonal colonies of adequate size for manual picking are usually present. Colonies composed of undifferentiated cells are identified microscopically by their cellular and colony morphology (4). Each prospective colony is manually picked with a pipette, disaggregated, and further expanded. Samples from each colony are then prepared for genetic analysis (e.g., PCR) (5).
Large consortia have been organized to produce mouse models on a massive scale (6). These efforts have utilized robotics to automate many of the steps in GEM production, yet choosing and isolating colonies by manual colony picking remains a bottleneck in this process (3,6). It can be expected that the production of ES cell lines will be a long-term practice for modeling human disease, pharmaceutical testing, and in creating GEMs in various mouse strains that, while less robust than the 129 and C57BL/6 strains used for large-scale projects, are preferred in specific research areas (6). Implementation of new techniques can be expected to enhance the speed and decrease the costs compared with conventional methods for generating stable gene-targeted clones. Correspondingly, new technologies to support generation of these cell lines are in high demand.
In response to these needs, there has been an effort to develop microengineered devices for stem cell applications (7-12). Recently, a new cell sorting strategy using lab-on-a-chip techniques has been developed that promises a number of advantages for ES cell sorting and cloning (13-17). This technology is comprised of an array formed from transparent pedestals of micron-size dimensions, termed micropallets, on which cells are cultured. Each micropallet with its attached cell/colony can be released from the array on an individual basis using a pulsed laser and then collected in order to sort and expand cells/colonies of interest. The micropallets are formed from a biocompatible photoresist (e.g., SU-8, 1002F) which is photolithographically defined on a standard glass microscope slide to create the array (15). The array can be made in various sizes. Typically, a 1.6 × 1.6 cm array can contain more than 105 individual micropallets depending on their dimensions. By virtue of the small dimensions and ease of handling, cells can be cultured and manipulated using small volumes of media, an advantage when expensive growth factors such as LIF are required.
In this work, strain 129 murine ES cells were seeded on 1002F micropallets by allowing them to settle onto the array from a suspension. When the cells were plated at densities yielding 1 or 0 cells per micropallet and then cultured, clonal colonies were generated on the pallets initially receiving a single cell (16). Colonies composed of undifferentiated cells could easily be identified by inspecting the morphology of the cells and colonies under a microscope. A two-step procedure was used to release and collect target colonies. Individual ES colonies were removed by dislodging the micropallet with a single 5-ns laser pulse focused to a micron-sized spot at the interface of the micropallet’s base and glass substrate (14). The detached micropallets with their attached ES colonies were then collected by simply inverting the array over a collection plate (17). This entire procedure occurred while the array was immersed in media so that cell dehydration was not an issue (15). Isolated colonies were then analyzed for viability and differentiation status immediately and after continued expansion. The data demonstrated that the micropallet system provides an efficient approach for the analysis and sorting of ES cells. This technique is one of a number of new approaches that will be required for the analysis and handling of individual cells. In this same Focus Issue the working group of Andreas Schmid describes an approach for the capture of single microbes in suspension by dielectrophoresis to provide a single-cell bioreactor. These types of tools are required to manipulate and investigate single cells without resorting to manpower intensive approaches. The micropallet technique in particular holds great promise for reducing the time, cost and manpower required to isolate stable clones of cells and microbes.
Materials and Methods
Materials
Glasgow Minimum Essential Medium (G-MEM, BHK-21), ES cell-qualified fetal bovine serum (FBS), MEM non-essential amino acid, L-glutamine, MEM sodium pyruvate, penicillin/streptomycin, phosphate buffered saline (PBS) 1X, pH 7.4, 0.05% trypsin with EDTA solution and Alexa Fluor 633 protein labeling kit were obtained from Invitrogen (Carlsbad, CA). Beta-mercaptoethanol was obtained from Sigma-Aldrich (St. Louis, MO). ES cell qualified 0.1% gelatin solution and LIF at 107 units/ml were purchased from Millipore (Temecula, CA). Sylgard 184 silicone elastomer kit was purchased from Dow Corning (Midland, MI). EPON resin 1002F (phenol, 4,4′-(1-methylethylidene)bis-, polymer with 2,2′-[(1-methylethylidene) bis(4,1-phenyleneoxymethylene]bis[oxirane]) was obtained from Miller-Stephenson (Sylmar, CA). SU-8 photoresist and SU-8 developer (1-methoxy-2-propyl acetate, also used for 1002F) were obtained from MicroChem Corp. (Newton, MA). (Heptadecafluoro-1,1,2,2-tetrahydrodecyl)trichlorosilane was from Gelest Inc. (Morrisville, PA). The alkaline phosphatase Stem TAG™ CBA-300 kit was obtained from Cell Biolabs, Inc. (San Diego, CA). Slurry of 1.0 μm alumina particles in water was purchased from MTI Corp. (Richmond, CA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Murine ES Cell Lines and their Medium
Feeder-independent murine embryonic stem cells (129 strain) were obtained from the University of North Carolina-Animal Models Core Facility (UNC-AMC, Chapel Hill, NC). FBS was heat inactivated in a water bath at 56°C for 30 min. LIF-supplemented media (G-MEM with L-glutamine, beta-mercaptoethanol, sodium pyruvate and penicillin/streptomycin, 15% FBS, and 533 U/mL LIF) was used for ES cell culture. The pH of the cell culture medium was adjusted to 7.4 using cell culture tested and sterile 1 N NaOH. All solutions involved with cell culture were filtered using 0.22 μm vacuum filters (Best Lab Deals, Garner, NC) before use.
Fabrication of Films and Pallets from SU-8 and 1002F Photoresists
1002F-50 photoresist was made by dissolving EPON resin 1002F and triarylsulfonium hexafluoroantimonate salts in gamma-butyrolactone (GBL) at a ratio of 61% w/w 1002F resin, 6.1% w/w photoinitiator and 32.9% w/w solvent. SU-8 and 1002F films and 1002F pallets were fabricated on ∼76 × 25 × 1 mm3 pre-cleaned microscope glass slides (Corning Glass Works, Corning, NY). Films (50 μm thickness) and pallets (100 × 100 μm squares, 50 μm tall, 30 μm inter-pallet space) were fabricated in a manner similar to that described previously (13).
Roughening Pallet Surfaces
For trials involving roughened 1002F pallets and films, an in-situ procedure of roughening the miniaturized structures using aqueous slurries containing homogeneous size alumina particles was used. Briefly, a 1:1 ratio composed of diluted slurries of 1.0 μm alumina particles in distilled water were used in a built-in-house assembled magnetic force-based roughening system. The optimal roughed 1002F surface was obtained after a 30 s polishing procedure as confirmed by atomic force microscopy. The roughened 1002F micropallet arrays and films were rinsed with distilled water and then ethanol 5X and dried with N2 gas immediately after roughening. The roughened arrays were then silanized (see below).
Surface Coatings for Virtual Air Walls
After fabrication of 1002F pallets on glass substrates (and after roughening where applicable), the micropallet arrays were baked at least 30 min on a hot plate at 65°C to remove any solvent trapped on the surface. The formation of a hydrophobic perfluoroalkylsilane layer on the silicone oxide surface was then carried out in a low-pressure reactor as described previously (14).
Fabrication of PDMS Chambers for ES Cell Culture and Collection
Polydimethylsiloxane (PDMS) O-rings (I.D./O.D.∼18/35 mm) were constructed from Sylgard 184 cured in a plastic mold as described previously (18). The mold was made simply by putting a Teflon ring (O.D.∼18 mm) inside a 35-mm diameter polystyrene tissue culture dish. After pouring the PDMS into the mold, the assembly was heated in a 65°C oven for at least 30 min, after which time the assembly was cooled and the solid O-ring was extracted from the mold. The chamber was constructed by using a thin layer of Sylgard 184 to glue the fabricated O-ring to a 1002F microarray (cell sorting, release experiments) or glass slides covered with SU-8 or 1002F films (cell growth study experiments). To facilitate attachment, the entire assembly was heated on a 65°C hot plate for at least 20 min. Immediately before use, the arrays with O-rings were sterilized by rinsing with 95% ethanol, and then dried in a tissue culture hood. The entire assembly was placed into a 60 × 15 mm polystyrene tissue culture dish (BD Bioscience, San Jose, CA) for culturing cells and colonies.
An O-ring identical to that constructed above was attached to a 60 × 15 mm polystyrene tissue culture dish (BD Bioscience) using a similar protocol. This O-ring/tissue culture dish constructed from polystyrene was used to collect released pallets and their attached ES colonies. Before use, the chamber was rinsed with 95% ethanol and PBS three times. The chamber was then coated with 1 ml of 0.1% gelatin in PBS for at least 1 h under sterile conditions. The extra gelatin was then removed and the chamber rinsed with ES cell media prior to use.
Coating of Pallets and Films with Gelatin for ES Cell Culture
To coat gelatin on the micropallets top surface for ES cell growth, the arrays were treated by 0.025% sterile gelatin in PBS for at least 1 h at room temperature in a tissue culture hood. The extra gelatin was removed and the array rinsed once each with PBS and ES media. A similar coating protocol was used for the PDMS collection chambers constructed on regular polystyrene tissue culture dishes, glass slides covered with SU-8 films, and glass slides covered with 1002F films with either 0.025% or 0.1% gelatin.
For experiments involving fluorescence analysis of gelatin-coated 1002F pallets, a 0.025% gelatin solution was labeled and purified using an Alexa Fluor 633 protein labeling kit following the manufacturer’s instructions. Briefly, 2 ml of 0.025% gelatin in PBS was mixed with 25 μl Alexa Fluor 633 reactive dye and incubated for 1 h at room temperature in the dark while mixing using a magnetic stirrer. The free dye was then removed from the conjugated gelatin using a purification column containing gel filtration media. In imaging experiments, 1 ml of Alexa Fluor 633 conjugated gelatin was added to regular and roughened 1002F micropallet arrays and the fluorescence intensity of the arrays was measured by fluorescence microscopy using an inverted epi-fluorescence microscope (TE 300, Nikon, Melville, NY) and a cooled CCD camera (Photometrix Coolsnap FX, Tucson, AZ). Fluorescence data were collected using Metafluor software (Universal Imaging Corporation, Downingtown, PA). ImageJ software (National Institute of Health, Bethesda, MD) was used for quantitative analysis of the fluorescence micrographs.
ES Cell Culture on Micropallet Arrays and Films
All cells were maintained at 37°C in a humidified 5% CO2 atmosphere in culture medium. After the micropallet arrays were silanized and coated with gelatin, the murine ES cells were seeded onto arrays containing approximately 9,000 pallets. In each experiment, ES cells were plated by placing 1 ml of cell suspension in the PDMS culture chamber housing the microarray. In order to minimize clumping of ES cells, before counting cells and transferring them into the PDMS chamber, the cell suspension was gradually aspirated using a 1 ml pipette and then filtered through a 40 μm Nylon filter (BD Biosciences). The cell density of the suspension (∼3,000 cells/ml) was determined empirically to produce ≤1 cell per pallet. Cells cultured on the arrays were maintained in the incubator and imaged under transillumination mode with an inverted microscope (TE 300, Nikon). In all experiments, the culture media was replaced every 1-2 days.
For the ES growth rate study on gelatin-coated polystyrene, SU-8 and 1002F films (both regular and roughened surfaces), 1 ml of an ES cell suspension (2,000 cells/ml) in culture media was placed into the PDMS chamber after initial aspiration and filtration as described above. The cell growth rates were determined by microscopic observation of ES colony areas after one week in LIF-supplemented culture using a 60X (NA = 0.7) objective. For each data point, the average ES colony area was determined from the region of interest corresponding to the colony circumference on several collected optical micrographs (n > 10) using the ImageJ software.
Laser-Based Pallet Release and Collection of Released Colony-Pallets
Release of pallets by a single pulse (5 ns, 532 nm) from a focused Nd:YAG laser (Minilite™ II, Continuum, Santa Clara, CA) was performed as described previously (14). Prior to laser release, the micropallet array was rinsed with fresh ES culture medium three times to remove any nonadherent or dead cells. Fresh medium (1 ml) was then added to the chamber housing the array. Prior to use, the collection chamber pre-coated with gelatin (see above) was rinsed with fresh culture medium and placed directly above the micropallet chamber in a sterile environment to mate the two PDMS O-rings of the collection chamber and the micropallet array. The assembly was placed on the microscope stage and selected ES colony-pallets were released with the pulsed laser. The assembly was then inverted to transfer the media and released pallets into the collection chamber. The collection chamber and micropallet array components were separated in a sterile environment. The collection chamber containing the released colonies and ES growth media was placed into a 60 × 15 mm tissue culture dish (BD Bioscience) and transferred to an incubator. The growth of the collected ES colonies was observed over time by transmitted light microscopy with an inverted microscope.
Alkaline Phosphatase Differentiation Test of Expanded ES Colonies
The alkaline phosphatase differentiation test of ES cells was performed using the Stem TAG™ CBA-300 kit following the manufacturer’s instructions. Briefly, the cultured ES colonies in collection chambers were rinsed with PBS containing 0.05% Tween-20 (PBST) after aspirating the ES culture media. For each chamber after adding 0.4 ml fixing solution, mixtures were incubated at room temperature for ∼2 min. The fixative was removed and the cells were washed twice with 1 ml of PBST. After aspirating the final wash, 0.4 ml of freshly prepared staining solution was added to each chamber containing fixed cells and incubated at room temperature for approximately 30 min in the dark. Finally, the staining solution was removed and the stained colonies washed twice with 1 ml PBST. After staining, cell colonies were imaged using an inverted microscope equipped with a high resolution digital camera (EOS Rebel XTi, Canon, Japan).
Results and Disscusion
Growth Rate of Murine ES Cells on Various Culture Surfaces
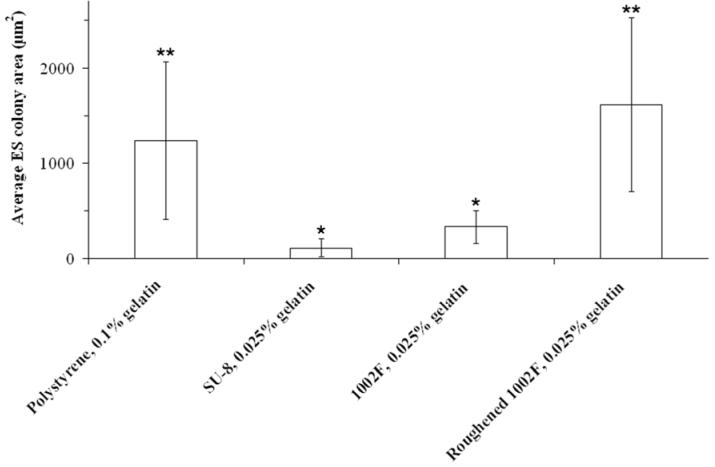
ES cells are significantly influenced by their environment as they propagate, and must be cultured on specialized surfaces in the presence of differentiation inhibitory factors (e.g., LIF) to maintain pluripotency (19-26). SU-8 and 1002F photoresists have been demonstrated in prior work to be appropriate materials on which to culture standard tumor cell lines including HeLa, RBL, A172 and 208F (13). The adherence and viability of cells cultured on 1002F were better than for SU-8 even in the absence of surface coatings (13). Since ES cells have more stringent growth requirements, it was necessary to study ES cell growth characteristics on these recently developed surfaces. In these studies, murine strain 129 ES cells were grown on micropallet arrays or thin films of the photoresists coated with gelatin. ES cell growth was compared to the standard ES culture substrate of polystyrene coated with 0.1% gelatin (27). It was found that the use of 0.1% gelatin as a coating for the array resulted in the formation of a continuous film of gelatin covering the array. This film prevented the virtual walls from localizing plated cells to individual micropallets and led to difficulties in the release and collection of released micropallets (data not shown). To prevent these problems, the percentage of gelatin used to coat the photoresists was reduced in a stepwise manner to 0.025%. To assess cell growth characteristics on this lower concentration of gelatin, ES cells were grown on thin films of SU-8, 1002F, and roughened 1002F photoresists, and their growth was compared to that on polystyrene coated with 0.1% gelatin (Fig. 1). The average colony area on 0.025% gelatin-coated SU-8 (110 ± 96 μm2) and 1002F (332 ± 172 μm2) surfaces was significantly lower than that on 0.1% gelatin-coated polystyrene (1,237 ± 826 μm2). In contrast, average colony growth on the 0.025% gelatin-coated and roughened 1002F surface (1,612 ± 909 μm2) showed a dramatic improvement over all other photoresist surfaces and was equivalent to the control polystyrene. These studies demonstrated that 0.025% gelatin-coated, roughened 1002F provided an optimal substrate for ES cell culture on the micropallet arrays. The increase in the ES cell growth rate for the roughened 1002F surface relative to an unroughened surface may have been due to enhanced adhesion of the gelatin to the surface of the micropallets as a result of the nanometer scale surface roughness generated by the roughening procedure (Shadpour & Allbritton, unpublished data).
Fig. 1.
Growth rates of murine ES colonies on various surfaces after one week in culture. The histograms represent the mean colony area (n > 10 colonies for each experimental condition) with error bars representing the standard deviations. Statistically significant differences were determined using the Student’s t-test (α = 0.05). Colony areas on both unroughened arrays were less than either the control or the roughened arrays (P < 0.001). Asterisks represent paired statistical differences as shown (*P = 0.005, **P = 0.3).
Fluorescence Study of Gelatin-Coated Micropallet Arrays
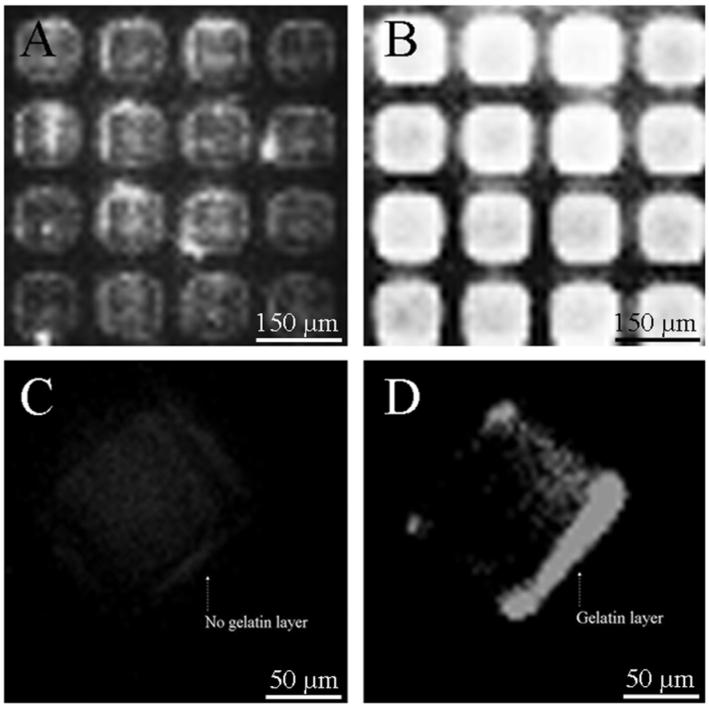
To confirm the presence and assess the homogeneity of the 0.025% gelatin surface layer on regular and roughened 1002F surfaces, unroughened and roughened 1002F micropallet arrays were coated using Alexa-Fluor-633-conjugated gelatin (Fig. 2). Under fluorescence microscopy, the unroughened 1002F array demonstrated patchy fluorescence of individual micropallets. In contrast, micropallets on the roughened array were seen to stain homogeneously and with higher fluorescence intensity. A quantitative comparison of the fluorescence intensity of individual micropallets (n = 30) revealed an average signal-to-background ratio (SBR) of 7.8 ± 2.4 vs. 11.7 ± 0.6 for the unroughened and roughened micropallets, respectively. The consistency and quality of the gelatin coating on micropallets viewed in cross-section after their release and collection were observed to be improved on the roughened pallet surface as well (Fig. 2 C&D).
Fig. 2.
Fluorescence micrographs of 1002F micropallet arrays and released pallets after incubation with Alexa Fluor 633 conjugated gelatin. (A&B) Unroughened “A” and roughened “B” 1002F micropallet arrays. (C&D) Released 1002F micropallets from unroughened (C) and roughened (D) arrays shown in “A” and “B”, respectively.
Isolation of Murine ES Colonies Grown on Micropallet Arrays
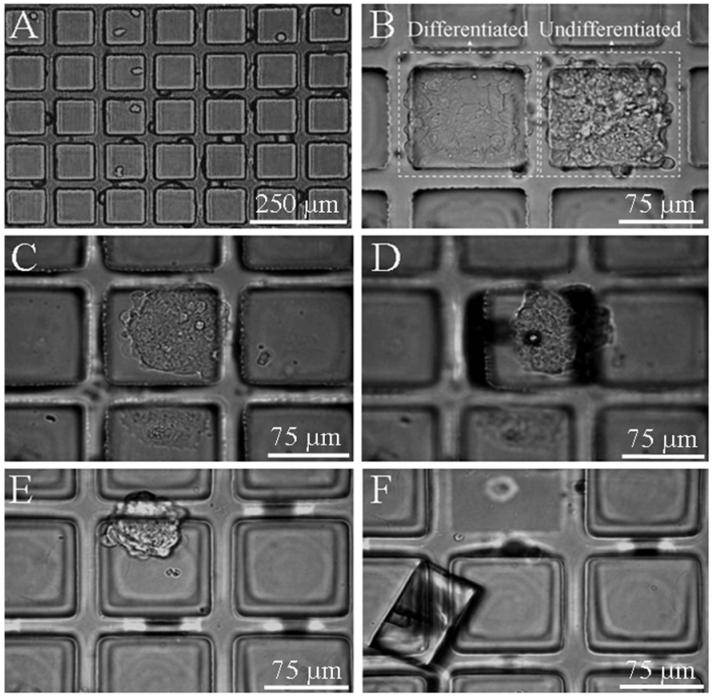
To evaluate the use of micropallet arrays for culturing and isolating clonal colonies of undifferentiated ES cells, a single cell suspension of murine strain 129 ES cells was plated on unroughened and roughened 1002F micropallet arrays coated with 0.025% gelatin at a density to yield ≤1 cell per micropallet. After 24 hours the media was exchanged and the arrays were examined. Single ES cells were seen to be attached to micropallets on both types of arrays (Fig. 3A). The cells were maintained for 5 days in culture and re-examined for colony formation. As expected, two morphologies of ES colonies were found (Fig. 3B). Some colonies were composed of larger and flattened cells with well-defined membranes indicating the cells making up the colony were differentiated (4). Colonies were considered to be composed of undifferentiated ES cells by the presence of a dome-shaped colony of rapidly growing cells with tight borders and close packing (4). For the unroughened array, 44% ± 12 (n = 50) of the colonies inspected were judged to be undifferentiated. For the roughened array, 70% ± 15 (n = 43) of the colonies inspected remained undifferentiated. The standard culture surface of 0.1% gelatin-coated polystyrene revealed that 73% ± 6 (n = 58) of the colonies remained in an undifferentiated state supporting that the roughened 1002F surface coated with 0.025% gelatin was an equivalent culture substrate.
Fig. 3.
Micrographs of murine ES cell growth on 1002F micropallet arrays coated with 0.025% gelatin at 24 h “A” and 5 days “B-F” after plating cells on the arrays. (A) Micrograph showing murine strain 129 ES cells plated on the array at a density ≤1 cell per micropallet. (B) Micrograph showing a morphology-based identification of undifferentiated murine ES colonies on pallets. (C&D) Micrographs of a micropallet containing an undifferentiated ES cell colony on a roughened 1002F micropallet array before “C” and after “D” release with a pulsed laser. In “D” the colony is seen to remain adherent to the released micropallet. (E&F) Micrographs of an undifferentiated ES cell colony on an unroughened array before “E” and after “F” release with a pulsed laser. In “F” the colony has become dislodged from the released micropallet.
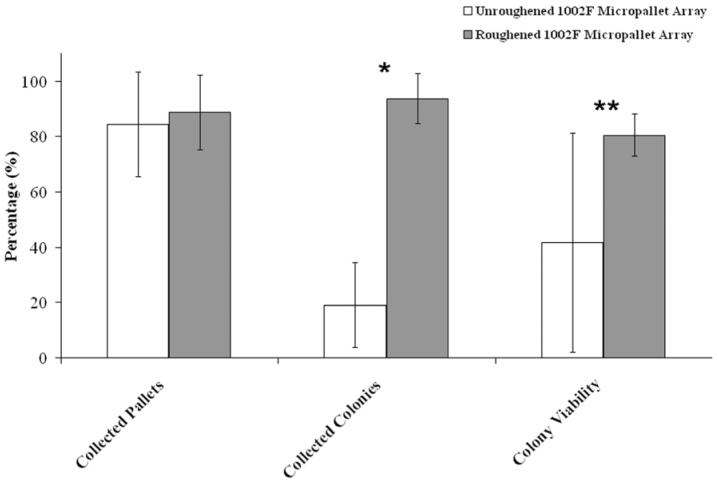
Based on the morphology of the cells and colonies, micropallets containing colonies of undifferentiated ES cells were released using a pulsed laser as previously described (Fig. 3 C&D) (14). After release, a significant number of colonies on the unroughened arrays dislodged from the released micropallets (Fig. 3 E&F). In contrast, the majority of colonies cultured on the roughened 1002F micropallets remained adherent to the released micropallets (Fig. 3C&D). These findings support that the colonies adhered more readily to the roughened 1002F surface, thus facilitating subsequent collection of the selected cells (see below). To quantify the collection efficiency of the micropallet array for sorting ES cell colonies, additional experiments were undertaken. In these experiments, ten 1002F micropallet arrays (unroughened and roughened) coated with 0.025% gelatin were used to compare the efficiency of collection and the outgrowth of cells (Fig. 4). Micropallets carrying undifferentiated ES colonies were released with the laser and transferred into a collection vessel. For each array, a minimum of 7 micropallets were released with a total of 66 micropallets released from six unroughened arrays and 53 micropallets from four roughened arrays. For the unroughened arrays, the median percentage of collected micropallets was 84% (50% confidence interval [CI] ± 7, 90% CI ± 19). This was similar (P = 0.6) for micropallets collected from the roughened array at 89% (50% CI ± 4, 90% CI ± 14). However, due to the improved adhesion of colonies, the median percentage of micropallets containing adherent cells after isolation was 94% (50% CI ± 3, 90% CI ± 9) (n = 47 collected micropallets) for roughened 1002F arrays, but only 19% (50% CI ± 6, 90% CI ± 15) (n = 55 collected micropallets) for the unroughened arrays. These data demonstrate that the roughened 1002F micropallet arrays provided an efficient method for isolating ES colonies.
Fig. 4.
Quantitative summary of collection and viability of released colonies. Each histogram and error bar represents the median value and 90% confidence interval of the results of experiments (n ≥ 4) conducted on different days. Asterisks represent paired statistical differences as shown (*P < 0.0001, **P = 0.2).
Collection and Clonal Expansion of Isolated ES Cell Colonies in an Undifferentiated State
After release and collection of the micropallets containing colonies of undifferentiated cells in the above experiment, the colonies were maintained in culture for clonal expansion as described previously (Figs. 4 and 5) (17). The expansion was carried out in a culture chamber composed of a polystyrene base coated with 0.1% gelatin which closely mimicked a conventional Petri dish as is typically used for ES cell culture (27). Viability of collected ES colonies, as defined by the ability of an isolated colony to continue to expand after 4 days in cell culture, was determined. For those micropallets retaining their colonies (n = 42) that were collected from the four roughened 1002F arrays, the median percentage of colonies that continued to expand was 80% (50% CI ± 2, 90% CI ± 7). For the six unroughened arrays, the median colony viability was 42% (50% CI ± 13, 90% CI ± 40) (n = 11 collected micropallets with retained colonies). These differences in viability did not reach statistical significance (P = 0.2). The results show that the degree of viability compares very favorably with routine culture techniques for ES cells which have a reported viability of 60% upon plating of undifferentiated murine stem cells (28).
Fig. 5.
Micrographs of selectively released 1002F pallets with undifferentiated murine ES colonies in collection chamber as obtained from gelatin coated and roughened 1002F micropallet arrays (see Fig. 3, C-D). Shown in pictures are cell colonies (A) 1 day, (B) 2 days, (C) 3 days, and (D) 4 days culture in collection chamber after being released from micropallet arrays (see text for details).
Expansion of the collected ES colonies was undertaken by continued culture in LIF supplemented media. After 4 days in culture, 60% of colonies from the unroughened array remained undifferentiated based on their morphology. For those colonies isolated from the roughened array, 91% remained undifferentiated. This enhanced maintenance of the undifferentiated state of the cells may have been the result of the improved gelatin coating on the roughened micropallets during the initial period after colony isolation. It is known that the presence of a gelatin layer is necessary for ES cells to remain undifferentiated during culture (29). While the bulk of each expanding colony obtained from the roughened array remained undifferentiated after 4 days in culture, it should be noted that differentiation of some cells at the periphery of expanding colonies could be observed (Fig. 5D). This phenomenon is typical of ES colonies as they expand in culture and can be minimized by daily replenishment of culture media (30).
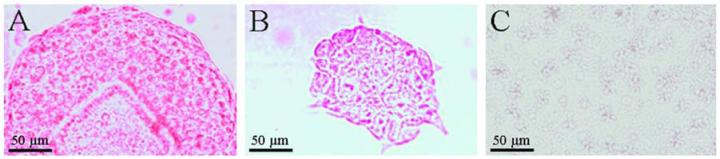
To further substantiate that the expanded colonies remained undifferentiated, alkaline phosphatase staining was performed on ES cell colonies isolated from roughened 1002F arrays. After release, the collected colonies were placed in culture for 3 days in LIF-supplemented media before alkaline phosphatase staining (Fig. 6A). As controls, undifferentiated murine ES cells were plated in standard tissue culture dishes coated with 0.1% gelatin, and were cultured for 3 days in the presence (Fig. 6B) or absence (Fig. 6C) of LIF. As seen above, the majority of colonies collected from the micropallet arrays, and the control cells cultured in the presence of LIF remained undifferentiated as evidenced by positive staining for alkaline phosphatase (21). In contrast, ES cells cultured in the absence of LIF did not stain for alkaline phosphatase indicating that they had become differentiated. These results confirmed that ES cells grown on the roughened 1002F micropallet arrays could be selected based on their morphology, and subsequently isolated and expanded in an undifferentiated state.
Fig. 6.
Alkaline phosphatase staining of murine ES cell colonies (A) after release and 3 days expansion in collection chamber and (B) positive and (C) negative controls (see text for details).
Conclusion
A microengineered cell array is reported for the isolation and expansion of murine ES cell colonies. Selected colonies of undifferentiated ES cells are shown to be released in a viable manner using a pulsed laser and efficiently collected. The collected colonies can be further expanded in culture with a high rate of viability. Moreover, morphologic parameters and staining for alkaline phosphatase demonstrate that the majority of isolated colonies remain in an undifferentiated state. The technique provides a flexible approach to perform colony sorting of a few or a large number of colonies. As reported, the method requires only a small number of cells (i.e., 3,000) to generate clonal colonies; however, the number of colonies to be sorted can be increased simply by enlarging the array. The process of establishing clonal colonies followed by isolation of desired colonies can be performed in less than one week. Furthermore, after an initial isolation step, cells remaining on an array can be put back into culture and the process repeated if desired. An additional attribute is that the compact size and large density of the micropallet arrays enable cells to be cultured in small volumes of media; an important issue if expensive supplements are contemplated, e.g., LIF. One envisioned application of this platform is for selection of gene-targeted ES colonies very soon after the transfection procedure. In the area of microbial applications, the method could be utilized for the morphologic analysis of microbes adherent to a surface followed by sorting based on their size, shape, or growth pattern. The technique may also be used to sample an adherent biofilm displaying a unique visible characteristic. The method will considerably reduce the time, manpower and reagent costs imposed by conventional approaches to accomplish these various tasks.
ACKNOWLEDGMENT
This research was supported by National Institute of Health (EB007612). Stephen Knight from Department of Genetics (UNC, Chapel Hill) is greatly acknowledged for initial cell preparations and technical support with culture procedures. NLA and CES disclose a financial interest in Intellego, Ltd.
Grant sponsor: National Institutes of Health (EB007612)
LITERATURE CITED
- 1.Downing GJ, Battey JF. Technical assessment of the first 20 years of research using mouse embryonic stem cell lines. Stem Cells. 2004;22:1168–1180. doi: 10.1634/stemcells.2004-0101. [DOI] [PubMed] [Google Scholar]
- 2.Beckers J, de Angelis MH. Large-scale mutational analysis for the annotation of the mouse genome. Current Opinion in Chemical Biology. 2001;6:17–23. doi: 10.1016/s1367-5931(01)00277-0. [DOI] [PubMed] [Google Scholar]
- 3.Draper JS, Nagy A. Improved embryonic stem cell technologies. Handbook of Experimental Pharmacology. 2007;178:107–128. doi: 10.1007/978-3-540-35109-2_5. [DOI] [PubMed] [Google Scholar]
- 4.Heo J, Lee J-S, Chu I-S, Takahama Y, Thorgeirsson SS. Spontaneous differentiation of mouse embryonic stem cells in vitro: characterization by global gene expression profiles. Biochemical and Biophysical Research Communications. 2005;332:1061–1069. doi: 10.1016/j.bbrc.2005.04.173. [DOI] [PubMed] [Google Scholar]
- 5.Lodish H, Berk A, Kaiser CA, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira P. Molecular Cell Biology. W.H. Freedman; New York: 2007. p. 1150. [Google Scholar]
- 6.Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab on a Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 8.Dutton G. Biotech firms make strides in advancing genetransfer methods - Novel approaches significantly exhance levels of expression. Genetic Engineering News. 1999;19:1–5. [Google Scholar]
- 9.Kim L, Vahey MD, Lee HY, Voldman J. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab on a Chip. 2006;6:394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 10.Lee JW, Na DS, Kang JY, Lee SH, Ju BK. Differentiation of mouse P19 embryonic carcinoma stem cells injected into an empty zebrafish egg chorion in a microfluidic device. Bioscience Biotechnology and Biochemistry. 2006;70:1325–1330. doi: 10.1271/bbb.50609. [DOI] [PubMed] [Google Scholar]
- 11.Valero A, Post JN, van Nieuwkasteele JW, ter Braak PM, Kruijer W, van den Berg A. Gene transfer and protein dynamics in stem cells using single cell electroporation in a microfluidic device. Lab on a Chip. 2008;8:62–67. doi: 10.1039/b713420g. [DOI] [PubMed] [Google Scholar]
- 12.Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, Weiner LP. A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab on a Chip. 2008;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pai J-H, Wang Y, Salazar GTA, Sims CE, Bachman M, Li GP, Allbritton NL. Photoresist with Low Fluorescence for Bioanalytical Applications. Analytical Chemistry. 2007;79:8774–8780. doi: 10.1021/ac071528q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar GTa, Wang Y, Young G, Bachman M, Sims CE, Li GP, Allbritton NL. Micropallet Arrays for the Separation of Single, Adherent Cells. Analytical Chemistry. 2007;79:682–687. doi: 10.1021/ac0615706. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Sims CE, Marc P, Bachman M, Li GP, Allbritton NL. Micropatterning of Living Cells on a Heterogeneously Wetted Surface. Langmuir. 2006;22:8257–8262. doi: 10.1021/la061602k. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Young G, Aoto Phillip C, Pai J-H, Bachman M, Li GP, Sims Christopher E, Allbritton Nancy L. Broadening cell selection criteria with micropallet arrays of adherent cells. Cytometry. Part A: the journal of the International Society for Analytical Cytology. 2007;71:866–74. doi: 10.1002/cyto.a.20424. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Young G, Bachman M, Sims CE, Li GP, Allbritton NL. Collection and Expansion of Single Cells and Colonies Released from a Micropallet Array. Analytical Chemistry. 2007;79:2359–2366. doi: 10.1021/ac062180m. [DOI] [PubMed] [Google Scholar]
- 18.Shadpour H, Sims CE, Allbritton NL. Enrichment and expansion of cells using antibody-coated micropallet arrays. Analytical Chemistry. 2008 doi: 10.1002/cyto.a.20741. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mummery CL, van den Eijnden-van Raaij AJ. Growth factors and their receptors in differentiation and early murine development. Cell Differ Dev. 1990;30:1–18. doi: 10.1016/0922-3371(90)90069-9. [DOI] [PubMed] [Google Scholar]
- 20.Whetton AD, Dexter TM. Influence of growth factors and substrates on differentiation of haemopoietic stem cells. Curr Opin Cell Biol. 1993;5:1044–1049. doi: 10.1016/0955-0674(93)90090-d. [DOI] [PubMed] [Google Scholar]
- 21.Reubinoff BE, Pera MF, Fong C-Y, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nature Biotechnology. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 22.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 23.Murray P, Edgar D. The regulation of embryonic stem cell differentiation by leukemia inhibitory factor (LIF) Differentiation. 2001;68:227–234. doi: 10.1046/j.1432-0436.2001.680410.x. [DOI] [PubMed] [Google Scholar]
- 24.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 25.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 26.Hatada S, Arnold LW, Hatada T, Cowhig JEJ, Ciavatta D, Smithies O. Isolating gene-corrected stem cells without drug selection. PNAS. 2005;102:16357–16361. doi: 10.1073/pnas.0508263102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenlee AR, Kronenwetter-Koepel TA, Kaiser SJ, Ellis TM, Liu K. Combined effects of Matrigel and growth factors on maintaining undifferentiated murine embryonic stem cells for embryotoxicity testing. Toxicology in Vitro. 2004;18:543–553. doi: 10.1016/j.tiv.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 28. http://ourworld.compuserve.com/homepages/TheBroons/tnp5.htm.
- 29.Greenlee AR, Kronenwetter-Koepel TA, Kaiser SJ, Liu K. Comparison of Matrigel and gelatin substrata for feeder-free culture of undifferentiated mouse embryonic stem cells for toxicity testing. Toxicology in Vitro. 2005;19:389–397. doi: 10.1016/j.tiv.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Nagy A, Gertsensstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2003. p. 764. [Google Scholar]