Abstract
Objective
Virus infections are the most common causes of encephalitis, a syndrome characterized by acute inflammation of the brain. Over 150 different viruses have been implicated in the pathogenesis of encephalitis, however due to limitations with diagnostic testing, etiologies of over half of the cases remain unknown.
Methods
To investigate whether HHV-6 is an etiological agent of encephalitis, we examined for evidence of virus infection by determining the presence of viral sequence using PCR and assessed HHV-6 antibody reactivity in the cerebrospinal fluids (CSF) of encephalitis patients with unknown etiology. In a cohort study, we compared virus specific antibody levels in CSF samples of patients with encephalitis, relapsing-remitting MS and other neurologic diseases (OND).
Results
Our results demonstrated elevated levels of HHV-6 IgG as well as IgM levels in a subset of encephalitis patients compared with OND. Moreover, cell-free viral DNA that is indicative of active infection was detected in 40% (14/35) of encephalitis patients, while no amplifiable viral sequence was found in either relapsing-remitting MS or OND patients. Additionally, a significant correlation between PCR detection and anti-HHV-6 antibody response was also demonstrated.
Interpretation
Collectively, these results suggested HHV-6 as a possible pathogen in a subset of encephalitis cases.
INTRODUCTION
Encephalitis is a rare and potentially life-threatening illness affecting the CNS characterized by an acute inflammatory process of brain. Clinical manifestations of encephalitis include a diverse and complex set of symptoms such as fever, headache, and in some cases altered mental status accompanied with seizures. Infectious agents that are bacterial, rickettsial, fungal, and parasitic are capable of inducing encephalitis; however, viral etiologies still constitute majority of the cases, as over 150 different viruses have been implicated.1 Among numerous viral agents that are capable of eliciting CNS inflammation, the most common pathogens clinicians encounter are enteroviruses, arboviruses, as well as members of the herpesvirus family. A major challenge in treating patients with symptoms of encephalitis is the prompt identification of the specific disease-inducing agents, as symptoms are not always specific for a particular pathogen and multiple organisms can share similar clinical manifestations. Currently, despite extensive neurological evaluations and laboratory testing, the etiology of majority of the cases still routinely cannot be clearly defined.1, 2
To better characterize the disease pathogenesis and identify potential infectious agents, the California Encephalitis Project (CEP) was initiated in 19981, 3 in collaboration with Emerging Infections Program, Centers for Disease Control. As reported recently, between 1998 through 2005, a total of 1570 patients with encephalitis were enrolled in the CEP based on specific defined criteria.1 Confirmed, probable or possible infectious etiologies were identified for only 29% of the cases with majority of them linked to viral infections. While a wide range of viruses are capable of playing a role in the pathogenesis, members of the herpesviruses including herpes simplex viruses, varicella zoster virus (VZV), Epstein-Barr virus (EBV), and human herpesvirus -6 (HHV-6), were identified as some of the most important pathogens involved.1, 2 Various studies have demonstrated immunoevasive mechanisms exploited by herpesviruses that allow their long latency periods within the host.4–6 Moreover, their tendency to reactivate following primary infection has also been linked to a number of disorders.7–12 In particular, HHV-6, a neurotropic virus of the β-herpesvirus family has been implicated in the development of neurologic complications such as multiple sclerosis, mesial temporal sclerosis, epilepsy, and more recently encephalitis.9, 13–19
Given the associations of HHV-6 with neurologic disorders, it was of interest to examine whether there was a relationship with HHV-6 infection or reactivation in patients with unknown origin of encephalitis. HHV-6 has been associated with a wide spectrum of neurological disorders including febrile seizure, epilepsy, multiple sclerosis, and encephalitis.8, 9, 13, 17–22 A member of the β-herpesvirus family, HHV-6 is most closely related to the human cytomegalovirus based on sequence homology and also possesses the characteristic property of becoming latent in the host following primary infection and reactivating later in life. Although HHV-6 was initially isolated from lymphocytes,23 recent studies have demonstrated that it can infect various cell types both in vitro and in vivo, including central nervous system (CNS) glial cells, peripheral monocytes and macrophages, thus has the potential to alter a number of normal metabolic or cellular processes.24 Two variants of HHV-6 that share over 95% genomic sequence homology have been described,25 the HHV-6 A and B variants. While the epidemiology and clinical significance of HHV-6 variant A still remains elusive, it has been associated more often with neurologic disorders26 such as multiple sclerosis27–29 and more recently rhomboencephalitis.14 The HHV-6B variant has been determined to be the etiological agent of exanthem subitum7, a self-limiting childhood disorder. Its reactivation has been observed in recipients of bone marrow, hematopoietic stem-cell, and solid organ transplants post-procedure,30 presumably due to reduced immune status. In addition, a number of reports have hypothesized that reactivation of HHV-6B may be associated with development of neurologic complications in a subset of transplant recipients.8, 15 Moreover, recent reports have also demonstrated evidence of HHV-6 reactivation in immunocompetent encephalitis/meningitis patients. In some patients, high viral load as well as antibodies against HHV-6 could be demonstrated in the CSF of these patients. Collectively, these studies highlight the role of HHV-6 as a pathogenic agent in CNS diseases.17–19, 31
In the current study, CSF from 35 patients referred to the CEP over a 6-month period between February-August 2006 with clinical encephalitis but of unknown origin was examined for evidence of HHV-6. Using a highly sensitive nested PCR assay against the HHV-6 U57 gene region, we amplified cell-free viral sequences from CSF of a subset of encephalitis patients. In addition, we also assessed the levels HHV-6 antibody in the CSF using a novel electrochemiluminescent assay.32 Our results demonstrated a statistical significant (p<0.05) increase in the frequency of HHV-6 DNA sequence detection in the CSF of encephalitis patients compared with patients with other neurological diseases (OND). This observation was further supported with elevated levels of HHV-6 antibody response. Collectively, these results suggest that HHV-6 is an underappreciated pathogen associated with a subset (40%, 14/35) of patients with encephalitis.
METHODS
DNA extraction
DNA was isolated using commercially available extraction kits according to manufacturers' instructions. The QIAamp Viral RNA kit was used for DNA extraction from serum (1.2 ml) and CSF liquer (1.2 ml) and the DNeasy tissue kit (all kits from Qiagen, Valencia, CA) was used for DNA extraction from fresh/frozen and formalin-fixed brain tissue. Elution volumes of purified DNA in diethopyrocarbonate water were 100 μl for serum and CSF, and 200 μl for brain tissue.
HHV-6 DNA detection by PCR
PCR detection of HHV-6 DNA in CSF was performed with HHV-6 specific primers against the U57 and U12 gene regions. Briefly, 10 μl of extracted DNA from cell-free specimen was amplified with PCR Mastermix (Fermentas, cat. K0171) at a final primer concentration of 0.5 μM in 50 μl reaction volume for 35 cycles using the GeneAmp PCR System 9700. Nested PCR for U57 was performed by amplifying 5 μl of the primary PCR product using a set of internal primers specific for HHV-6 U57. To avoid PCR contamination, extraction, PCR mastermix preparation, and sample loading were performed at three separate locations in the lab. Primer sequences for both U57 and U12, expected size of amplicons, and PCR conditions are shown in supplemental table 3. Validation of assay sensitivity for U12 and U57 using CSF samples is demonstrated in supplemental figure 1. HHV-6 viral load was measured using TaqMan quantitative PCR with 9700HT Fast Real Time PCR System (Applied Biosystems) as previously described. 8, 33 Briefly, for cell-free specimen such as CSF, 5 μl of extracted CSF DNA was amplified with a final concentration of 0.5 μM HHV-6 variant A or B specific primers and probes in a 20 μl reaction using TaqMan Universal Mastermix (Applied Biosystems ). Based on the HHV-6 plasmid standard curve34 spanning from 107 to 101 copies, the viral load was calculated and expressed as HHV-6 copies/ml. For brain biopsy samples, 50 ng of DNA was used in each 20 μl TaqMan PCR reaction and viral load per million cells was calculated by normalizing HHV-6 copy numbers to the house-keeping gene β-actin using the following formula [HHV-6 copy number/(β-actin copy number/2)] x 106.
DNA Sequencing
PCR products were purified using the Qiagen MinElute PCR Purification Kit (Qiagen, CA). After purification, sample concentration was determined using a spectrophotometer at 260/280 nm (Eppendorf Biophotometer) wavelengths. For direct sequencing of PCR products with Dye-Terminators (Perkin Elmer), 200 nanograms of purified PCR product and 3.5 picomoles of forward or reverse PCR primers were added in nuclease-free water up to 12 ml of total volume. Sequences obtained were submitted to NCBI MegaBlast for analysis
Anti-HHV-6 IgG and IgM Immunoassays
We established an assay to detect antibodies against HHV-6 proteins using an established electrochemiluminescence technology (MSD, Gaithersburg, MD). 96-well high-bind plates (L13XB) were spotted with 1 mg of HHV-6B or mock-infected cell lysate in 5 ml Tris-buffered saline (TBS) and allowed to dry at room temperature. 200 ml of Blocker A solution was added to each well and plates were blocked on a shaker for 1 hr at room temperature and then washed twice with 300 ml PBS. Serum/CSF samples were diluted in MSD Antibody Diluent and added to plates in 25 μl per well. Samples were tested in duplicate. Plates were placed on a shaker at room temperature for 1hr, then washed twice with 300 μl PBS. 25 μl Sulfo-Tag™ labeled goat anti-human IgG (diluted to 1 mg/ml in Antibody Diluent) was added to each well and plates were placed on a shaker at room temperature for 1hr, then washed twice with 300 μl PBS. Lastly, each 96-well received 150 μl MSD Read Buffer T and plates were immediately read on a MSD PR400 plate reader.
Cell culture and lysate preparation
The susceptible T cell line (SupT1) was infected with HHV-6A (U1102 strain) or HHV-6B (Z29 strain) cell-free inocula at a ratio of 100 virus particles per cell for 3 hours. Cells were subsequently washed 3 times with 5% FBS in complete RPMI containing 1% L-glutamine and 1% Pen/Strep cells/ml and kept in a 37°C cell culture incubator. Cells were observed for cytopathic effect (CPE) daily and harvested when the CPE was greater than 80%. Cells were washed with 1X TBS 3 times and cell lysate was prepared using a Polytron homogenizer following 3 rounds of freeze-thaw cycle. Viral load/ml of cell lysate was determined using HHV-6 strain specific TaqMan Quantitative PCR.34 Control lysates were prepared from uninfected SupT1 cells. Total protein concentration of all lysates were quantitated by Lowry protein assay (BioRad Laboratories) and lysates were diluted to 1 mg/ml in TBS.
Statistical Analysis
Because of the relatively small sample sizes of our cohorts, non-parametric tests were used for statistical comparison due the reliance on fewer assumptions. Fischer Exact Test for significance between different cohorts was performed using 2-way contingency table analysis and Spearman correlation was performed correlation between HHV-6 IgG and IgM antibody titers.
RESULTS
Study subjects
The California Encephalitis Project (CEP) was established in 1998 to better understand the disease pathogenesis of encephalitis and study the clinical epidemiological characteristics in California. As previously described,1 specimens from patients who met a set of specifically defined criteria for encephalitis between February-August 2006 of unknown etiology were referred to the CEP by their treating physicians for diagnostic testing (Supplemental Table 1). A “case patient” was defined as a patient hospitalized with encephalopathy exhibiting a depressed or altered level of consciousness lasting > 24 h, lethargy, or a personality change with one of the following characteristics: fever, seizure, focal neurological findings, pleocytosis, or electroencephalography or neuroimaging findings consistent with encephalitis. Summary of CEP demographics and clinical information are shown in supplemental table 2.
Control CSF samples from relapsing-remitting multiple sclerosis (RRMS) and post-bone marrow patients (post-BMT)8 were collected at the Neurology clinic at NIH. RRMS diagnosis was made according to the accepted McDonald criteria.35 The NIH RRMS group (n=18; median age=37.0, range 27–54; 11 females/7 males) had a median disease duration of 4.9 years with a range from 2 months to 28 years. The median time since last relapse from when the sample was taken was 0.42 years with a range from 0–2.75 years. The mean number of gadolinium-enhancing lesion load was 3(± 6) with a range from 0–29. Eight CSF from allogeneic post-BMT patients with neurologic complications associated with HHV-6 were included as PCR positive controls (n=8; median age=37.0, range 16–62; 5 females/3 males). In addition, we also obtained CSF samples from other neurological diseases (OND) from both the CEP and the Neurology clinic at NIH. OND cohort consisted of patients (n=14; median age=20, range 4–57; 4 females/10 males) with West Nile Virus encephalitis, subactue sclerosing panencephalitis, severe combined immunodeficiency syndrome, epilepsy, lymphoma, HTLV-1 associated myelopathy, anoxic encephalitis, arsenic poisoning, neuroleptic malignant syndrome, probable stroke, sagittal sinus venous thrombosis, and schizophrenia.
The current study was approved by the respective Institutional Review Boards at all sites. For the post-BMT and NIH patients informed consent was obtained on all subjects.
Detection of Cell-free HHV-6 DNA in cerebrospinal fluids of Encephalitis patietns
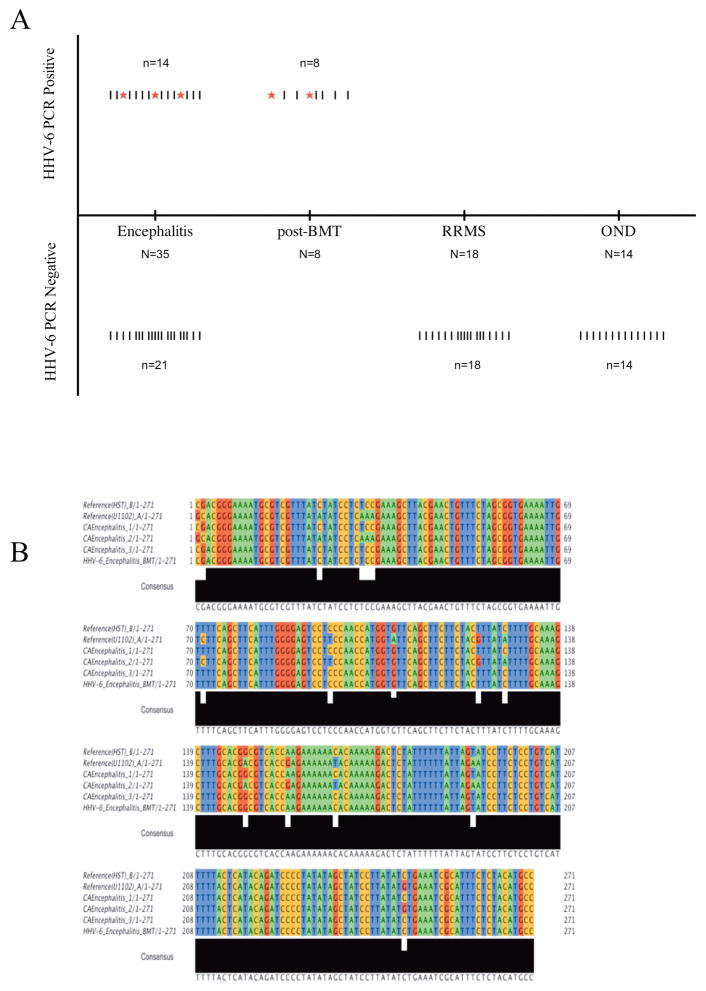
CSF from patients diagnosed with encephalitis but of unknown etiology (Supplemental Table 1) were tested for presence of HHV-6 DNA by nested polymerase chain reaction (PCR).36 Using primers sets specific for the major capsid protein (U57) gene region of HHV-6, cell-free viral DNA was amplified from CSF of encephalitic patients (40%, 14/35). In contrast, none (0/14) of the patients with other neurological diseases (OND), such as stroke, schizophrenia, and HTLV-1 associated myelopathy patients demonstrated detectable level of HHV-6 DNA sequence. Similarly, HHV-6 sequence was not detected in CSF obtained from 18 relapsing-remitting multiple sclerosis (RRMS) patients. As positive controls, CSF from 8 patients who exhibited HHV-6 associated neurological complications post-bone marrow transplant procedure8 demonstrated amplifiable HHV-6 DNA sequence in CSF samples (Figure 1A). To ensure specificity of PCR amplification, a previously determined known negative CSF sample was run as a negative control for every 3 patient CSF samples tested.
Figure 1. PCR amplication of HHV-6 sequence from CSF of encephalitis patients.
A) Results of HHV-6 U57 nested PCR from various neurologic cohorts. “|” denotes each patient sample tested. “*” denotes samples that were both positive by primary and nested PCR amplication. B) Alignment of HHV-6 U12 ORF amplified by primary PCR from CSF of encephalitis patients. PCR amplicons from patients CEP1, 2 and 3 were aligned against HHV-6 A (U1102) and -6B (HST) RefSeq sequences from Genbank. CEP1 and 3 demonstrated homology to the HST strain of HHV-6 while CEP 2 showed homology to the U1102 strain.
Within this cohort, 3 patients with encephalitis were found to have high levels of HHV-6 DNA amplifiable by primary PCR with the U57 primers set (Figure 1A). To confirm this observation, we performed additional PCR amplifications with primers specific for another HHV-6 gene region, namely U12. As shown in figure 1B, DNA sequence analysis of U12 PCR product from these 3 patients demonstrated homology to the published HHV-6 sequences in Genbank. Moreover, alignment of the sequenced PCR products with refseq for variants A (U1102) and B (HST) revealed the presence of HHV-6B in the CSF of 2 patients and HHV-6 variant A (U1102) in 1 encephalitic patient (Figure 1B). The U57 nested PCR products for HHV-6 positive CSF were also subtyped by sequence analysis. Of the 14 encephalitis CSF positive for HHV-6 U57 by nested PCR (Figure 1A), 2 specimens demonstrated homology to the HHV-6 variant A (U1102) and 12 demonstrated homology to the B (HST) variant (Supplemental Table 1).
Characterization of HHV-6 electrochemiluminescent antibody assay
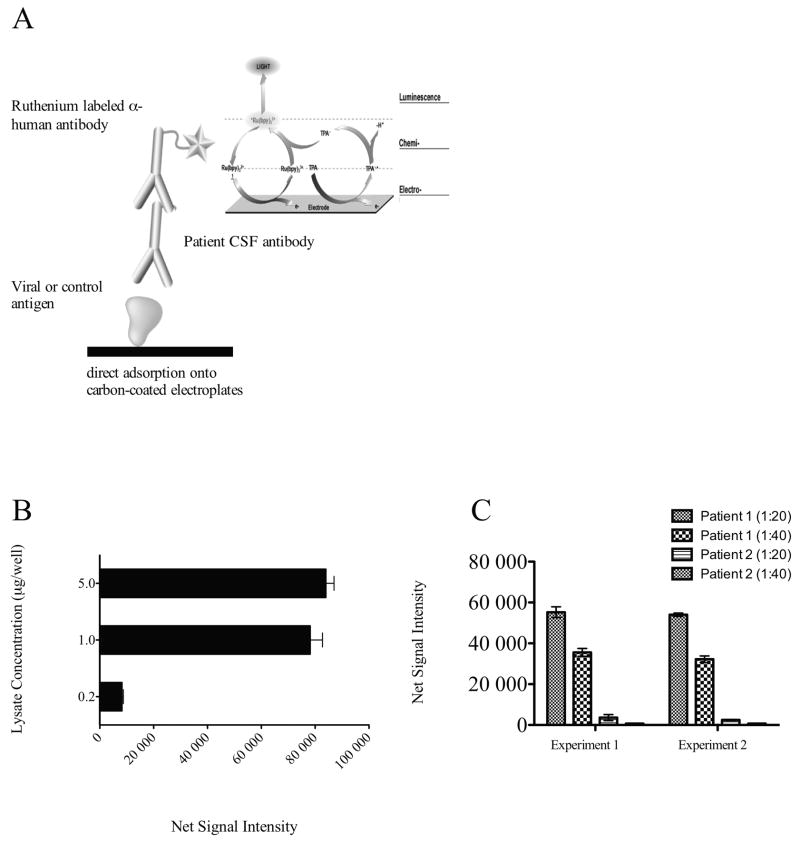
The high frequency of HHV-6 DNA detection in the CSF of encephalitis patients is consistent with a number of recent studies14, 17–20, 31 that suggest a possible role of HHV-6 in neurological complications. To extend the significance of this observation, we asked whether host immune response against HHV-6 could also be detected in the CSF of encephalitis patients with unknown etiology. Using a sensitive electrochemiluminescent (ECL) antibody assay, we measured levels of HHV-6 specific antibody in the CSF. A schematic of this assay is depicted in figure 2A. The HHV-6 antigens used in this assay were derived from lysates of HHV-6B infected T cell lines (SupT1) and control background lysates were obtained from matched uninfected SupT1 cells. We first validated this assay by testing the ability of a well-characterized monoclonal antibody against the HHV-6 P41 protein to bind to the lysates of infected as well as uninfected cells. A specific binding (net signal intensity) was defined as the difference between signal intensity obtained with HHV-6 infected lysate and uninfected lysate. As shown in figure 2B, HHV-6 anti-p41 monoclonal antibody demonstrated specific binding to the viral protein in a dose dependent manner. Moreover, the optimal amount of lysate concentration to be used for subsequent experiments was determined to be 1ug/well as maximal signal was obtained using this concentration (Figure 2B). Next, we assessed the reproducibility of this assay by examining values obtained from two separate experiments. HHV-6 antibody reactivity in the CSF of two representative patients with encephalitis in replicate experiments demonstrated high reproducibility as shown in figure 2C.
Figure 2. Validation of HHV-6 Antibody ECL assay.
A) Schematic diagram representing HHV-6 antibody ECL assay. Briefly, lysate from either HHV-6 infected cells or control non-infected parental cell was deposited onto the carbon-coated electroplates. Patient CSF at 1:20 or 1:40 dilution was then added and allowed to incubate for 1 hour. For detection, a ruthenium labeled anti-human IgG was added. Electrical charged is applied to the electroplate for excitation of bound ruthenium molecules. B) Validation of the ECL assay using a specific monoclonal antibody against HHV-6 P41 protein. Ruthenium labeled anti-P41 antibody was added at 1ug/mL concentration to increasing amount of lysate as represented and reactivity was measured. Net signal intensity was calculated by [Value of HHV-6 lysate - value of SupT1 lysate]. C) Representative replicate experiments demonstrated high reproducibility. HHV-6 IgG in CSF samples from two patients were tested in separate experiments at 1:20 and 1:40 dilutions. Comparable results were obtained in these independent experiments from the same samples.
Anti-HHV-6 immunoreactivty in CSF of patients with unknown origin of encephalitis
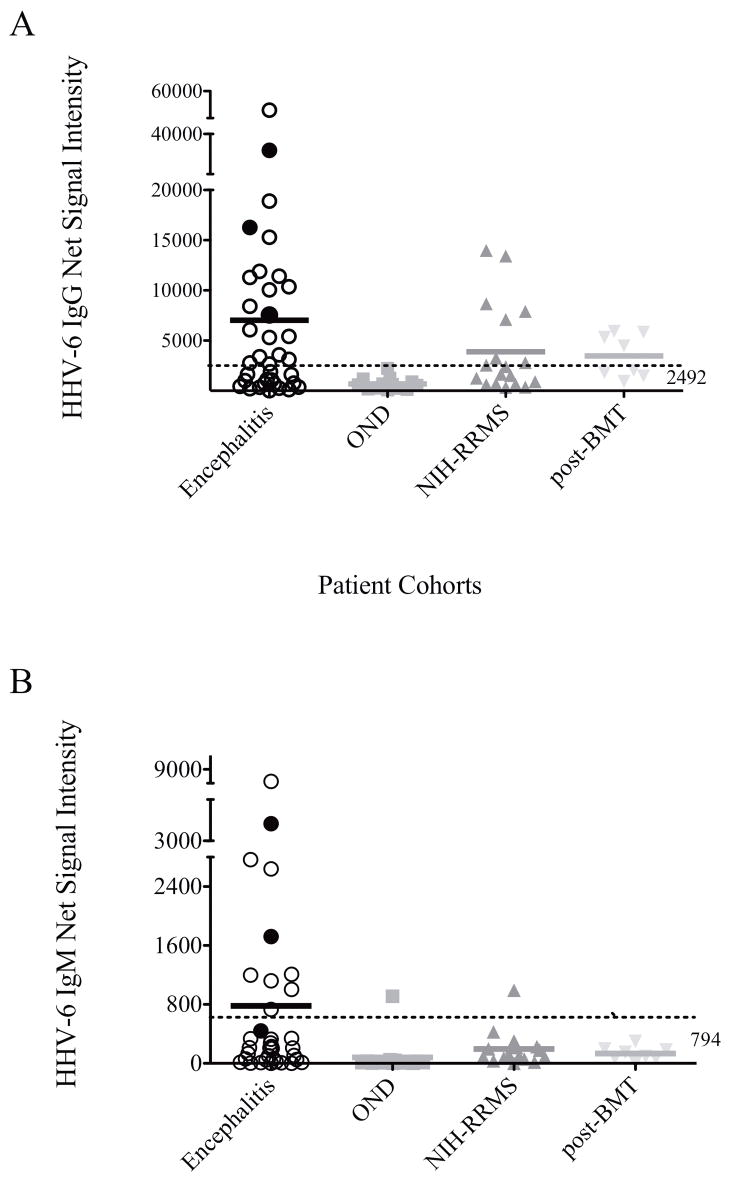
Using this ECL antibody assay, we investigated the presence of HHV-6 antibody in the CSF of encephalitis patients enrolled in the CEP. In a cohort of 35 patients, our result demonstrated elevated HHV-6 IgG antibody in a subset of individuals (37%, 13/35) with unknown origin of encephalitis compared with OND controls (Figure 3A, Table 1). Similarly, we were able to show that RRMS patients and post-BMT recipients with neurological complications also had elevated IgG reactivity against HHV-6 in their CSF. This finding is consistent with previous observations that suggested an association of HHV-6 with neurological disorders.8, 9, 13, 17–22 To better evaluate the percentage of patients with elevated CSF HHV-6 IgG reactivity, we empirically defined the background HHV-6 IgG level in the CSF as 3 standard deviations above the mean HHV-6 IgG value of OND patients. As shown in table 1, 37% of the encephalitis patients demonstrated significantly elevated HHV-6 IgG reactivity in the CSF above background whether we use OND as controls (p=0.018) or all non-encephalitis cases (p=0.03). Collectively, these findings are consistent with exposure of HHV-6 in subsets of patients with encephalitis of unknown origin.
Figure 3. Elevated HHV-6 IgG and IgM in CSF of patients with encephalitis.
A) Antibody against HHV-6 in CSF of patient with encephalitis was significantly elevated compared with patients with other neurological diseases (OND). Elevated IgG was observed in NIH-RRMS patients and post-BMT recipients. The “normal range” was determined to be 2942 as defined by 3 standard deviations above the mean of OND patients. Antibody levels of CEP1,2, and 3 who demonstrated amplifiable HHV-6 sequence by primary PCR are indicated by filled circles. B) HHV-6 IgM in CSF of patient with encephalitis was specifically elevated compared with other neurological diseases cohorts. The “normal range” was determined to be 794 as defined by 3 standard deviations above the mean of OND patients. Antibody levels of CEP1,2, and 3 who demonstrated amplifiable HHV-6 sequence by primary PCR are indicated by filled circles.
Table 1.
Summary of HHV-6 Antibody Reactivity and Statistical Analysis
| CSF IgG | ||||||
|---|---|---|---|---|---|---|
| N | Mean signal intensity | Standard Deviation | % Positive | Fischer exact p-value (Encephalitis vs. Controls) | Fischer exact p-value (Encephalitis vs. OND) | |
| Encephalitis | 35 | 7024 | 10584 | 13/35 (37%) | 0.03 | 0.018 |
| Non-Encephalitis Controls | ||||||
| RRMS | 18 | 3891 | 4411 | 5/18 (27%) | - | - |
| pBMT | 8 | 3474 | 2113 | 1/8 (12.5%) | - | - |
| OND | 14 | 686 | 602 | 0/14 (0%) | - | - |
| CSF IgM | ||||||
| N | Mean signal intensity | Standard Deviation | % Positive | Fischer exact p-value (Encephalitis vs. Controls) | Fischer exact p-value (Encephalitis vs. OND) | |
| Encephalitis | 35 | 781 | 1540 | 10/35 (28%) | 0.0098 | N.S. |
| Non-Encephalitis Controls | - | |||||
| RRMS | 18 | 195 | 234 | 1/18 (6%) | - | - |
| pBMT | 8 | 133 | 92 | 0/8 (0%) | - | - |
| OND | 14 | 80.3 | 239 | 1/13 (7%) | - | - |
Increased HHV-6 IgM in encephalitis patients and correlation with IgG in CSF
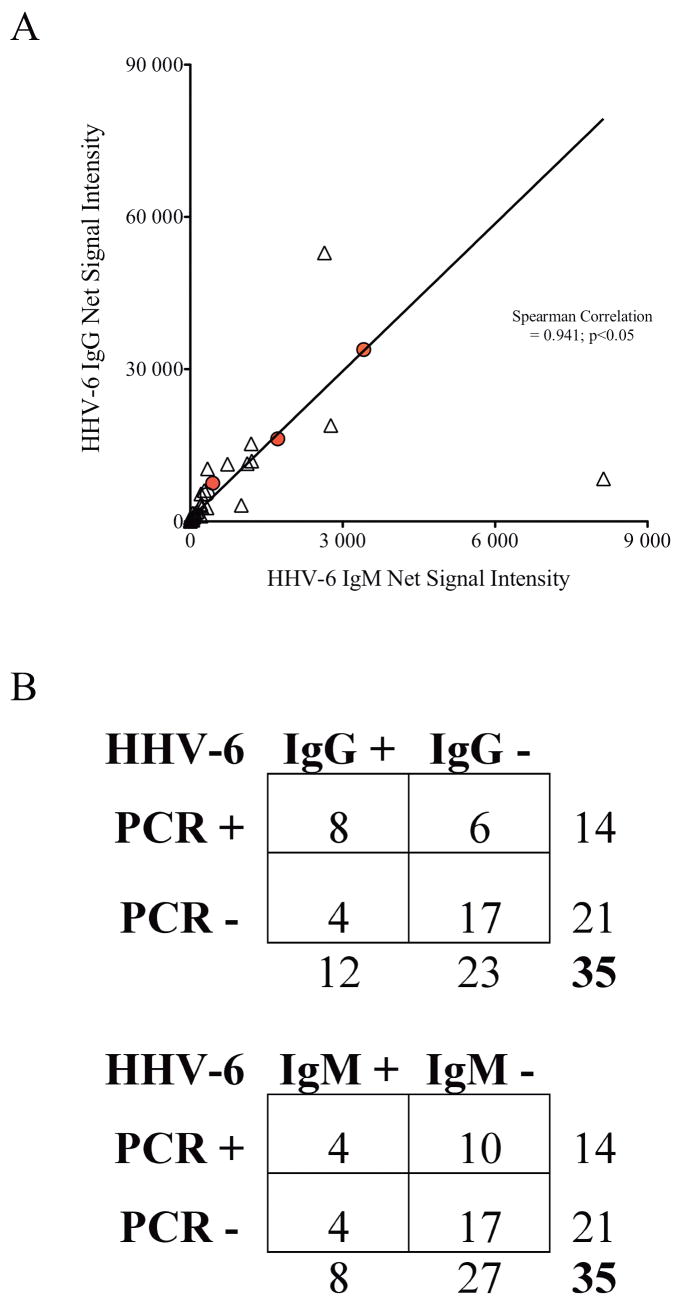
While IgG antibody reactivity may suggest history of exposure to HHV-6, IgM responses are more reflective of recent viral encounter such as primary infection or reactivation. Therefore, we also investigated the levels of HHV-6 IgM in CSF of the various cohorts of neurologic patients. In the encephalitis cohort, consistent with findings of enhanced HHV-6 IgG in CSF, a subset of patients demonstrated elevated HHV-6 IgM reactivity (Figure 3B). Moreover, levels of HHV-6 IgG antibody strongly correlated with HHV-6 IgM reactivity (Spearman correlation=0.941, p<0.05, Figure 4A), suggesting recent exposure to HHV-6 in a subset of patients with unknown origin of encephalitis with elevated levels of HHV-6 antibody. As shown in table 1, 28% of the encephalitis patients had HHV-6 IgM reactivity that was significantly elevated (p=.0098) when compared with controls CSF from all non-encephalitis cases. Correlation between PCR detection and antibody reactivity was also performed (Figure 4B). A statistical significant correlation was observed between HHV-6 DNA and HHV-IgG titer (Fischer’ exact p=0.03) and no correlation was detected with HHV-6 IgM titer (Fischer’s exact p=0.69). These observations of enhanced HHV-6 IgG and IgM reactivity coupled with increased frequency of cell-free HHV-6 DNA detection in the CSF supports the hypothesis of recent HHV-6 exposure in at least a subset of patients with encephalitis.
Figure 4. Correlation of HHV-6 antibody titers and PCR detection.
A) Significant correlation was demonstrated between levels of HHV-6 IgG and IgM in patients with encephalitis (spearman correlation=0.941; p<0.05). Filled circles represent HHV-6 IgG and IgM of CEP1,2, and 3. B) Statistically significant correlation was observed between HHV-6 PCR detection and HHV-6 IgG (Fischer’s exact test p=0.03) but not with HHV-6 IgM (Fischer’s exact test p=0.69).
Detection of HHV-6 in brain biopsy from a patient with encephalitis
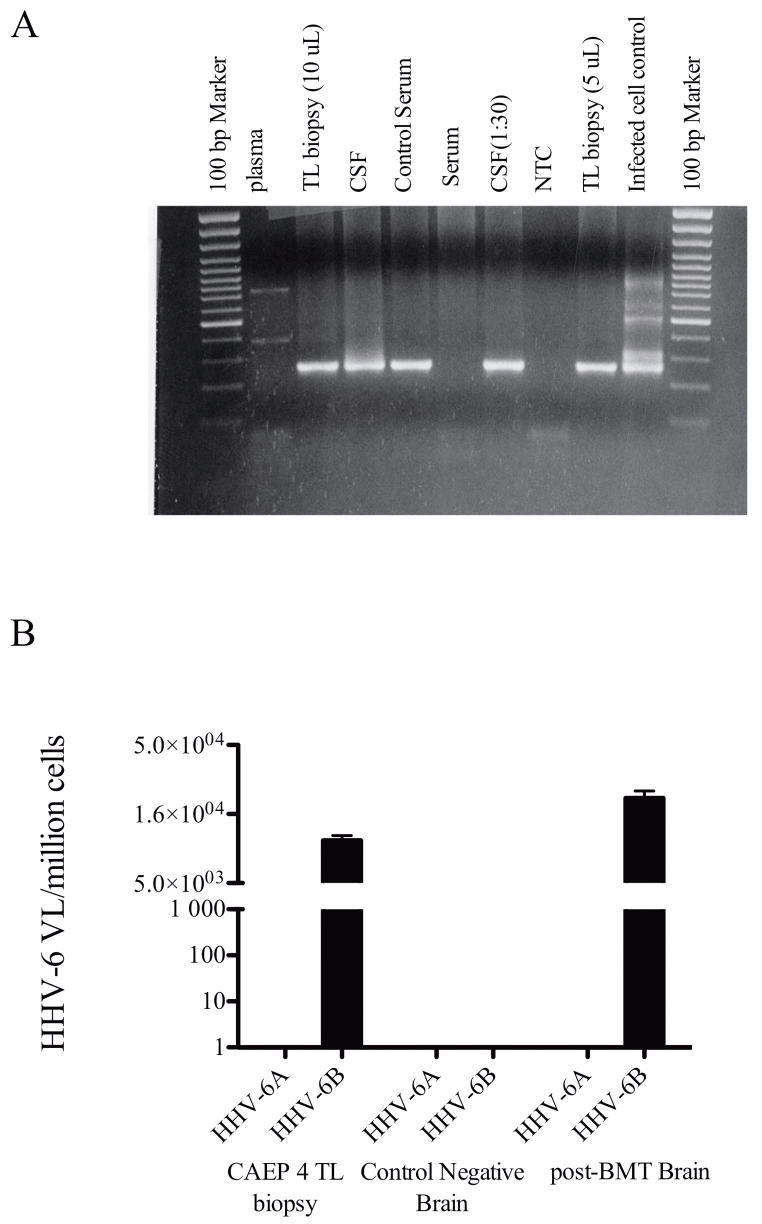
While detection of viral DNA and antibody reactivity in the CSF are suggestive of CNS pathology associated with HHV-6 in a portion of encephalitic cases (40%), increased lymphocytes infiltrating into CNS during disease progression could result in positive viral detection as peripheral lymphocytes have been suggested to be an in vivo reservoir of HHV-6.23, 37 We had the unique opportunity to assess the relationship of HHV-6 virus load of the CNS compared to CSF from one encephalitis patient (CEP4, case ID #16 in supplemental table 1) exhibiting seizure, weakness, dysarthria and diplopia who underwent a brain biopsy. As shown in figure 5A, nested PCR of HHV-6 MCP gene region demonstrated presence of HHV-6 in the CSF and brain biopsy specimen. Surprisingly, no detectable HHV-6 sequence could be amplified from plasma or serum of this patient at the time of CSF sampling. Furthermore, we determined the viral load of this brain biopsy by using a variant-specific TaqMan quantitative real-time PCR assay.34 Shown in figure 5B, the viral burden in the brain biopsy was equivalent to approximately 1 x 104 copies of HHV-6 variant B/million cells. Subsequent sequence analysis of the U57 PCR products confirmed the presence of HST strain of HHV-6 variant B. Interestingly, detection of HHV-6 was confined to CNS compartments as no amplifiable HHV-6 sequence could be found in either plasma or serum in this patient (Figure 5A). Together, these findings strongly support that detection of HHV-6 in CSF of encephalitis patients may reflect events in the CNS.
Figure 5. HHV-6 detection in temporal lobe biopsy of a patient with unknown origin of encephalitis.
A) HHV-6 U57 nested PCR amplification of plasma, temporal lobe biopsy, CSF, and serum samples from a patient with unknown origin of encephalitis. The results showed specific amplification of HHV-6 sequence in the CNS compartments and not in the periphery. B) HHV-6 virus load in the temporal lobe biopsy was measured by HHV-6 strain specific quantitative TaqMan real-time PCR. As shown, temporal lobe biopsy from CEP4 demonstrated 1x104 copies of HHV-6B/million cells while no appreciable HHV-6A virus was detected. As controls, a specimen from a non-HHV-6 related tumor biopsy and a specimen from a post-BMT patient with neurologic complications were used as negative and positive controls, respectively.
DISCUSSION
Based on a series of recent investigations, HHV-6 has become increasingly recognized as an emerging CNS pathogen. HHV-6 has been demonstrated to be neurotropic 38 and has been linked a number of neurologic disorders including to multiple sclerosis, post-transplant limbic encephalitis, mesial temporal sclerosis and encephalitis/meningitis in immunocompetend individuals.8, 9, 13, 17–22 A hallmark of HHV-6 is its long-term survival in a latent or persistent form in the host. Theses studies suggest that the CNS is a site of latency for this agent where periodic reactivation of HHV-6 may be linked to the association of HHV-6 with a variety of neurologic diseases.8, 9, 13
Since HHV-6 has been implicated as an etiological agent in encephalitis17–19, here we investigated for the prevalence of HHV-6 in CSF samples from a cohort of 35 patients who were submitted to the CEP with clinical encephalitis but tested negative for other commonly associated pathogenic agents.1, 3 Our results demonstrated that a significant percentage of these patients (14/35, 40%) had detectable HHV-6 DNA sequence in their CSF, while control patients with RRMS and other neurologic diseases (OND) were negative for HHV-6 DNA sequences (Figure 1A). This finding is consistent with the recent report of detection of HHV-6 in CSF of a cohort of encephalitis patients with unknown origin from New York State.19 As had been previously suggested, detection of HHV-6 in cell-free compartments such as saliva, serum and CSF maybe indicative of active viral replication.27, 29 Therefore, the increased frequency of HHV-6 detection in encephalitis patients compared to controls (RRMS and OND) may suggest a possible association of HHV-6 in the pathogenesis of the disease. However, it is worth noting that the prevalence of HHV-6 detection reported by Tovakali et al. 19 was significantly lower compared to our study (1.75 vs. 40%). Factors such as number of patients tested, sensitivity of the virus-specific primers used, time of CSF sampling relative to onset of clinical symptoms and PCR methods are possible variations that may have contributed to the overall differences. A highly sensitive nested PCR technique with a detection limit as low as 10 viral copies per reaction (Supplemental figure 1) was used in this study. The observed increase in the percentage of HHV-6 positive detection using nested PCR amplification suggest that only low levels of cell-free virus may be present in the CSF for majority of the patients.
To substantiate our PCR findings, immune reactivity against HHV-6 as determined by antibody levels in the CSF of patients with encephalitis was examined. Using a sensitive electro-chemilluminescence assay (Figure 2A) we demonstrated that 37% of the encephalitis patients also had elevated levels of HHV-6 IgG antibodies in the CSF compared with OND patients (Figure 3A). Importantly, anti-HHV-6 IgG reactivity was strongly correlated with the PCR detection of HHV-6 DNA in CSF of encephalitis patients (Fischer exact p=0.03, Figure 4B). We further explored the HHV-6 IgM response in our cohorts of patients and found that elevated IgM response was specifically observed in patients with encephalitis (28%), but not RRMS (6%), post-BMT (0%) or OND (7%) patients (Figure 3B), although this did not reach statistical significance based on our empirically defined cutoff for HHV-6 IgM antibody level and is likely due to the small sample size of OND and also the one outlier in the OND cohort who was diagnosed with lymphoma. However, a clear statistically significant increase in HHV-6 IgM could be demonstrated in encephalitis patients when compared with neurologic controls inclusive of RRMS, post-BMT and OND patients suggesting that elevated IgM titers in the CSF of encephalitis patients may indicate a recent exposure or infection of HHV-6 (Table 1). While there was a statistical significant correlation between HHV-6 DNA detection and IgG antibody titer, their positivity were not always concordant, highlighting that either PCR or antibody testing alone may not be sufficient for diagnosis (Figure 4B).
Since encephalitis is characterized as inflammation of the CNS where peripheral lymphocytes harboring latent HHV-6 virus may be recruited to the brain, detection of HHV-6 in the CNS has been suggested to be a secondary event related to peripheral lymphocyte recruitment. While we could not definitely exclude the possible contribution of infiltrating lymphocytes resulting in the high frequency of HHV-6 DNA detection in the CNS, several lines of evidence suggest otherwise. First, there was no correlation between the number of white blood cells in the CSF and HHV-6 DNA detection (Supplemental Table 1, Fischer’s exact test p=0.55). Second, RRMS patients who may have compromised blood brain barriers with pleocytosis were negative for HHV-6 DNA in their CSF (Figure 1A). Lastly, the unique opportunity to examine a brain biopsy sample from one encephalitis patient (CEP4) revealed that while high levels (1 x 104 copies/million cells) of HHV-6 were present in the CNS, only low levels of viral DNA was detected in the CSF. Furthermore, plasma and serum samples from this individual were free of evidence of HHV-6 viral sequences (Figures 5A and B). Collectively, these observations are more consistent with HHV-6 reactivation in CNS reservoirs.
HHV-6 has been characterized as an opportunistic virus and immunocompromised individuals are more susceptible to virus reactivation leading to development of associated neurologic complications such as encephalitis.8 The precise mechanism of how or whether HHV-6 infection can result in encephalitis still remains to be defined. Interactions between host immunity and CNS glial cells following exposure to herpesviruses could contribute to neuropathology of encephalitis.39–41 In addition, proinflammatory cytokines such as TNF-a, IL-1, and IL-6 produced by inflammatory virus infected cells may also play a role in activating CNS glial cells.42–45 Therefore, CNS tissue destruction may be a consequence of activated astrocytes or scavenger microglial cells. In support of this hypothesis, reactive astrocytes have been demonstrated from brain specimens of patients with post-bone marrow transplant neurologic complications and mesial temporal sclerosis associated with HHV-6 infection.8, 13 Moreover, it has been reported recently that HHV-6 infection in astrocytes may alter normal cellular function. As a model for viral induced glutamate excitotoxicity in mesial temporal lobe epilepsy, expression of a high affinity glutamate transporter, EAAT2, was found to be decreased following infection of HHV-6 in purified human primary astrocytes.46 Together, these studies support an association of HHV-6 in neurologic complications through various mechanisms.
Collectively, our study demonstrated an association of HHV-6 in a surprising high percentage of immunocompetent patients with encephalitis, consistent with a number of studies suggesting a CNS pathogenic role for HHV-6. Further investigations with larger cohorts of patients with encephalitis will help elucidate the contribution of HHV-6 in CNS disorders.
Supplementary Material
Validation of U12 and U57 PCR assays on CSF. HHV-6 positive CSF of 1.3 x 105 copies/ml determined by real-time TaqMan quantitative was amplified using primers specific for U57 (MCP) and U12 gene regions of HHV-6. Ten microliters (1.3 x102 copies) of the CSF was amplified in lane 1 and lanes 2–6 represented subsequent 10 fold serial dilutions. Lane 7 was a negative CSF control and lane 8 represented HHV-6 infected supernatant. With primary PCR using U57 (MCP) external primers, the detection limit approximately 100 copies/reaction (lane 1, top panel). Nested PCR with U57 (MCP) internal primers had a sensitive of 10 copies/reaction (lane 2, middle panel). For U12, the detection limit was as low as 1 copy/reaction (lane 3, bottom panel). Low range molecular weight DNA ladder (Fermentas) was as molecular weight marker.
California Encephalitis Project Patient Demographics and Clinical Information
Summary of California Encephalitis Patients
U12 and U57 Primers Sequences and PCR conditions
Acknowledgments
We would like to thank Dr. Susan Gagnon Ph.D. for her technical assistance with the HHV-6 ECL assay. We thank Dr. Unsong Oh for providing the relapsing-remitting MS and OND CSF samples. And we would like to thank Dr. John Crawford for providing clinical insights and critical discussions on the manuscript. This research was supported in part by the Intramural Research Program of the NIH, NINDS.
References
- 1.Glaser CA, Honarmand S, Anderson LJ, et al. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis. 2006;43:1565–1577. doi: 10.1086/509330. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy PG. Viral encephalitis: causes, differential diagnosis, and management. J Neurol Neurosurg Psychiatry. 2004;75(Suppl 1):i10–15. doi: 10.1136/jnnp.2003.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaser CA, Gilliam S, Schnurr D, et al. In search of encephalitis etiologies: diagnostic challenges in the California Encephalitis Project, 1998–2000. Clin Infect Dis. 2003;36:731–742. doi: 10.1086/367841. [DOI] [PubMed] [Google Scholar]
- 4.Lusso P. HHV-6 and the immune system: mechanisms of immunomodulation and viral escape. J Clin Virol. 2006;37(Suppl 1):S4–10. doi: 10.1016/S1386-6532(06)70004-X. [DOI] [PubMed] [Google Scholar]
- 5.Froberg MK. Review: CMV escapes! Ann Clin Lab Sci. 2004;34:123–130. [PubMed] [Google Scholar]
- 6.Koelle DM, Corey L. Herpes simplex: insights on pathogenesis and possible vaccines. Annu Rev Med. 2008;59:381–395. doi: 10.1146/annurev.med.59.061606.095540. [DOI] [PubMed] [Google Scholar]
- 7.Yamanishi K, Okuno T, Shiraki K, et al. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 8.Fotheringham J, Akhyani N, Vortmeyer A, et al. Detection of active human herpesvirus-6 infection in the brain: correlation with polymerase chain reaction detection in cerebrospinal fluid. J Infect Dis. 2007;195:450–454. doi: 10.1086/510757. [DOI] [PubMed] [Google Scholar]
- 9.Soldan SS, Berti R, Salem N, et al. Association of human herpes virus 6 (HHV-6) with multiple sclerosis: increased IgM response to HHV-6 early antigen and detection of serum HHV-6 DNA. Nat Med. 1997;3:1394–1397. doi: 10.1038/nm1297-1394. [DOI] [PubMed] [Google Scholar]
- 10.Opsahl ML, Kennedy PG. Early and late HHV-6 gene transcripts in multiple sclerosis lesions and normal appearing white matter. Brain. 2005;128:516–527. doi: 10.1093/brain/awh390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prober C. Sixth disease and the ubiquity of human herpesviruses. N Engl J Med. 2005;352:753–755. doi: 10.1056/NEJMp048302. [DOI] [PubMed] [Google Scholar]
- 12.Sotelo J, Martinez-Palomo A, Ordonez G, Pineda B. Varicella-zoster virus in cerebrospinal fluid at relapses of multiple sclerosis. Ann Neurol. 2008 doi: 10.1002/ana.21316. [DOI] [PubMed] [Google Scholar]
- 13.Donati D, Akhyani N, Fogdell-Hahn A, et al. Detection of human herpesvirus-6 in mesial temporal lobe epilepsy surgical brain resections. Neurology. 2003;61:1405–1411. doi: 10.1212/01.wnl.0000094357.10782.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford JR, Kadom N, Santi MR, et al. Human herpesvirus 6 rhombencephalitis in immunocompetent children. J Child Neurol. 2007;22:1260–1268. doi: 10.1177/0883073807307086. [DOI] [PubMed] [Google Scholar]
- 15.Seeley WW, Marty FM, Holmes TM, et al. Post-transplant acute limbic encephalitis: clinical features and relationship to HHV6. Neurology. 2007;69:156–165. doi: 10.1212/01.wnl.0000265591.10200.d7. [DOI] [PubMed] [Google Scholar]
- 16.Challoner PB, Smith KT, Parker JD, et al. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc Natl Acad Sci U S A. 1995;92:7440–7444. doi: 10.1073/pnas.92.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birnbaum T, Padovan CS, Sporer B, et al. Severe meningoencephalitis caused by human herpesvirus 6 type B in an immunocompetent woman treated with ganciclovir. Clin Infect Dis. 2005;40:887–889. doi: 10.1086/427943. [DOI] [PubMed] [Google Scholar]
- 18.Isaacson E, Glaser CA, Forghani B, et al. Evidence of human herpesvirus 6 infection in 4 immunocompetent patients with encephalitis. Clin Infect Dis. 2005;40:890–893. doi: 10.1086/427944. [DOI] [PubMed] [Google Scholar]
- 19.Tavakoli NP, Nattanmai S, Hull R, et al. Detection and typing of human herpesvirus 6 by molecular methods in specimens from patients diagnosed with encephalitis or meningitis. J Clin Microbiol. 2007;45:3972–3978. doi: 10.1128/JCM.01692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vianello F, Barbaro F, Cogo P, et al. Co-infection with Mycoplasma pneumoniae and human herpesvirus 6 (HHV-6) in an immunocompetent child with meningoencephalitis: a random association? Infection. 2008;36:174–176. doi: 10.1007/s15010-007-6249-y. [DOI] [PubMed] [Google Scholar]
- 21.Schvoerer E, Frechin V, Fritsch S, et al. Atypical symptoms in patients with herpesvirus DNA detected by PCR in cerebrospinal fluid. J Clin Virol. 2006;35:458–462. doi: 10.1016/j.jcv.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Caserta MT, Hall CB, Schnabel K, et al. Neuroinvasion and persistence of human herpesvirus 6 in children. J Infect Dis. 1994;170:1586–1589. doi: 10.1093/infdis/170.6.1586. [DOI] [PubMed] [Google Scholar]
- 23.Ablashi DV, Salahuddin SZ, Josephs SF, et al. HBLV (or HHV-6) in human cell lines. Nature. 1987;329:207. doi: 10.1038/329207a0. [DOI] [PubMed] [Google Scholar]
- 24.Fotheringham J, Jacobson S. Human herpesvirus 6 and multiple sclerosis: potential mechanisms for virus-induced disease. Herpes. 2005;12:4–9. [PubMed] [Google Scholar]
- 25.Dominguez G, Dambaugh TR, Stamey FR, et al. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall CB, Caserta MT, Schnabel KC, et al. Persistence of human herpesvirus 6 according to site and variant: possible greater neurotropism of variant A. Clin Infect Dis. 1998;26:132–137. doi: 10.1086/516280. [DOI] [PubMed] [Google Scholar]
- 27.Alvarez-Lafuente R, De Las Heras V, Bartolome M, et al. Human herpesvirus 6 and multiple sclerosis: a one-year follow-up study. Brain Pathol. 2006;16:20–27. doi: 10.1111/j.1750-3639.2006.tb00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cermelli C, Berti R, Soldan SS, et al. High frequency of human herpesvirus 6 DNA in multiple sclerosis plaques isolated by laser microdissection. J Infect Dis. 2003;187:1377–1387. doi: 10.1086/368166. [DOI] [PubMed] [Google Scholar]
- 29.Akhyani N, Berti R, Brennan MB, et al. Tissue distribution and variant characterization of human herpesvirus (HHV)-6: increased prevalence of HHV-6A in patients with multiple sclerosis. J Infect Dis. 2000;182:1321–1325. doi: 10.1086/315893. [DOI] [PubMed] [Google Scholar]
- 30.Zerr DM. Human herpesvirus 6 and central nervous system disease in hematopoietic cell transplantation. J Clin Virol. 2006;37(Suppl 1):S52–56. doi: 10.1016/S1386-6532(06)70012-9. [DOI] [PubMed] [Google Scholar]
- 31.Sawada J, Nakatani-Enomoto S, Aizawa H, et al. An adult case of relapsing human herpesvirus-6 encephalitis. Intern Med. 2007;46:1617–1620. doi: 10.2169/internalmedicine.46.0239. [DOI] [PubMed] [Google Scholar]
- 32.Yao K, Gagnon S, Akhyani N, et al. Reactivation of human herpesvirus-6 in natalizumab treated multiple sclerosis patients. PLoS ONE. 2008;3:e2028. doi: 10.1371/journal.pone.0002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fotheringham J, Donati D, Akhyani N, et al. Association of human herpesvirus-6B with mesial temporal lobe epilepsy. PLoS Med. 2007;4:e180. doi: 10.1371/journal.pmed.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nitsche A, Muller CW, Radonic A, et al. Human herpesvirus 6A DNA Is detected frequently in plasma but rarely in peripheral blood leukocytes of patients after bone marrow transplantation. J Infect Dis. 2001;183:130–133. doi: 10.1086/317651. [DOI] [PubMed] [Google Scholar]
- 35.McDonald WI, Compston A, Edan G, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 36.Secchiero P, Carrigan DR, Asano Y, et al. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J Infect Dis. 1995;171:273–280. doi: 10.1093/infdis/171.2.273. [DOI] [PubMed] [Google Scholar]
- 37.Lusso P, Markham PD, Tschachler E, et al. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6) The Journal of experimental medicine. 1988;167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campadelli-Fiume G, Mirandola P, Menotti L. Human herpesvirus 6: An emerging pathogen. Emerg Infect Dis. 1999;5:353–366. doi: 10.3201/eid0503.990306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardell JL, Dazin P, Islar J, et al. Apoptotic effects of Human Herpesvirus-6A on glia and neurons as potential triggers for central nervous system autoimmunity. J Clin Virol. 2006;37(Suppl 1):S11–16. doi: 10.1016/S1386-6532(06)70005-1. [DOI] [PubMed] [Google Scholar]
- 40.Meeuwsen S, Persoon-Deen C, Bsibsi M, et al. Modulation of the cytokine network in human adult astrocytes by human herpesvirus-6A. J Neuroimmunol. 2005;164:37–47. doi: 10.1016/j.jneuroim.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Kong H, Baerbig Q, Duncan L, et al. Human herpesvirus type 6 indirectly enhances oligodendrocyte cell death. J Neurovirol. 2003;9:539–550. doi: 10.1080/13550280390241241. [DOI] [PubMed] [Google Scholar]
- 42.Enoki H, Takeda S, Matsubayashi R, Matsubayashi T. Steroid therapy in an infant with human herpesvirus 6 encephalopathy. Brain Dev. 2006;28:597–599. doi: 10.1016/j.braindev.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Arena A, Stassi G, Speranza A, et al. Modulatory effect of HHV-6 on MCP-1 production by human monocytes. New Microbiol. 2002;25:335–340. [PubMed] [Google Scholar]
- 44.Caruso A, Rotola A, Comar M, et al. HHV-6 infects human aortic and heart microvascular endothelial cells, increasing their ability to secrete proinflammatory chemokines. J Med Virol. 2002;67:528–533. doi: 10.1002/jmv.10133. [DOI] [PubMed] [Google Scholar]
- 45.Mayne M, Cheadle C, Soldan SS, et al. Gene expression profile of herpesvirus-infected T cells obtained using immunomicroarrays: induction of proinflammatory mechanisms. J Virol. 2001;75:11641–11650. doi: 10.1128/JVI.75.23.11641-11650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fotheringham J, Williams EL, Akhyani N, Jacobson S. Human herpesvirus 6 (HHV-6) induces dysregulation of glutamate uptake and transporter expression in astrocytes. J Neuroimmune Pharmacol. 2008;3:105–116. doi: 10.1007/s11481-007-9084-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of U12 and U57 PCR assays on CSF. HHV-6 positive CSF of 1.3 x 105 copies/ml determined by real-time TaqMan quantitative was amplified using primers specific for U57 (MCP) and U12 gene regions of HHV-6. Ten microliters (1.3 x102 copies) of the CSF was amplified in lane 1 and lanes 2–6 represented subsequent 10 fold serial dilutions. Lane 7 was a negative CSF control and lane 8 represented HHV-6 infected supernatant. With primary PCR using U57 (MCP) external primers, the detection limit approximately 100 copies/reaction (lane 1, top panel). Nested PCR with U57 (MCP) internal primers had a sensitive of 10 copies/reaction (lane 2, middle panel). For U12, the detection limit was as low as 1 copy/reaction (lane 3, bottom panel). Low range molecular weight DNA ladder (Fermentas) was as molecular weight marker.
California Encephalitis Project Patient Demographics and Clinical Information
Summary of California Encephalitis Patients
U12 and U57 Primers Sequences and PCR conditions