Abstract
Interferon (IFN)-alpha is an innate immune cytokine that induces significant depressive symptoms in clinical populations. A number of mechanisms have been considered regarding the relationship between IFN-alpha and depression, including the effects of IFN-alpha on the hypothalamic-pituitary-adrenal (HPA) axis. Here, we examined the impact of mouse interferon (mIFN)-alpha and its signaling pathways on the functioning of the glucocorticoid receptor (GR), which plays a key role in HPA axis regulation. mIFN-alpha treatment (100–1000 IU/ml) of HT22 mouse hippocampal cells for 24 hours was found to significantly inhibit dexamethasone (DEX)-induced GR-mediated MMTV-luciferase activity and significantly decrease DEX-induced GR-binding to its DNA response element. Of note, mIFN-alpha treatment for 24 hours had no effect on DEX-induced GR translocation or GR protein expression. Inhibition of DEX-induced GR function by mIFN-alpha was significantly reversed by pharmacological inhibition of janus kinase/signal transducer and activator of transcription (Jak-STAT) signaling pathways, but not by inhibition of p38 mitogen-activated protein kinase. Moreover, pretreatment of cells with siRNA targeted to STAT5, but not STAT1 or STAT2, significantly attenuated IFN-alpha inhibition of DEX-induced MMTV-luciferase activity. Immunoprecipitation experiments revealed nuclear co-immunoprecipitation of activated STAT5 and GR following IFN-alpha plus DEX treatment. Taken together, these results indicate that negative regulation of GR function by IFN-alpha in hippocampal HT22 cells is mediated by activation of Jak/STAT signaling pathways leading to nuclear STAT5-GR protein-protein interactions. Given the role of GR in depressive disorders, IFN-alpha effects on GR function in cells of hippocampal origin may contribute to HPA axis alterations and depressive symptoms in IFN-alpha-treated patients.
Keywords: Jak-STAT, interferon-alpha, STAT5, glucocorticoid receptor, major depression, cytokine
INTRODUCTION
Interferon (IFN)-alpha is a cytokine of the innate immune response that has both antiviral and antiproliferative activities (Abbas and Lichtman, 2003). Accordingly, IFN-alpha has become a mainstay in the treatment of certain cancers such as malignant melanoma and viral infections including hepatitis C (Heathcote, 2007). Despite its therapeutic efficacy, IFN-alpha is well-known to induce the development of depressive symptoms in 20–50% of patients, depending on the dose (Raison et al., 2005).
A number of mechanisms have been considered regarding the relationship between IFN-alpha and depression including the effects of IFN-alpha on the hypothalamic-pituitary adrenal (HPA) axis. For example, one study found that patients who developed major depression during IFN-alpha therapy were more likely to exhibit exaggerated HPA axis responses to the first IFN-alpha injection (Capuron et al., 2003). Given the role of corticotropin releasing hormone (CRH) in the regulation of HPA axis outflow, these results suggest that increased sensitivity of CRH pathways to IFN-alpha may represent a vulnerability factor for the development of IFN-alpha-induced depression. Of note, hypersecretion of CRH is believed to play a central role in the development of depression, and IFN-alpha has been shown to stimulate CRH expression in the hypothalamus as well as the amygdala of laboratory animals (Raber et al., 1997). IFN-alpha has also been shown to lead to flattening of the diurnal cortisol slope and increased evening cortisol concentrations, both of which were correlated with increased IFN-alpha-induced depressive symptoms (Raison et al., 2008).
CRH hypersecretion in major depression and flattening of the cortisol slope is believed to be related in part to impaired negative feedback regulation of the HPA axis by glucocorticoids, an effect that may be mediated by decreased glucocorticoid receptor (GR) function (Nemeroff, 1996; Raison and Miller, 2003). Indeed, depressed patients have been shown to exhibit decreased GR function both in vivo and in vitro as manifested by failure of the synthetic glucocorticoid dexamethasone (DEX) to suppress cortisol secretion during the DEX suppression test (DST) and DEX-CRH test (Holsboer, 2000; Ising et al., 2005) and reduced sensitivity of peripheral blood mononuclear cells to the in vitro inhibitory effects of DEX on mitogen-induced lymphocyte proliferation (Pariante, 2004). In addition, flattening of the cortisol slope has been associated with non-suppression of cortisol during the DST in patients with metastatic breast cancer (Spiegel et al., 2006). Of relevance to IFN-alpha, a recent report demonstrated that IFN-alpha treatment of several cell lines for 72 hours was associated with decreased GR mRNA and protein expression (Cai et al., 2005). In addition, plasma levels of IFN-alpha have been associated with decreased GR binding affinity and decreased sensitivity to cortisol in peripheral blood monocytes of patients with acquired immunodeficiency syndrome (AIDS), although no direct effect of IFN-alpha on GR function was observed in this study (Norbiato et al., 1996). Finally, IFN-alpha is known to stimulate p38 mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription (STAT) pathways (Platanias, 2005), and both p38 MAPK and STAT induction have been shown to inhibit GR function (Biola et al., 2001; Stocklin et al., 1996; Wang et al., 2004). Taken together, these data suggest that one mechanism by which IFN-alpha may contribute to depression is through disruption of GR function and its role in the regulation of HPA axis responses.
The current study was designed to further explore the effects of IFN-alpha on GR function as well as the signaling pathways involved. For these studies, mouse HT22 cells, a hippocampus-derived cell line, and mouse IFN-alpha (mIFN-alpha) were used. Hippocampal GR have been shown to be essential for maintenance of normal HPA axis function and have been implicated in the underlying HPA axis pathophysiology of major depression (De Kloet et al., 1998). Moreover, administration of mIFN-alpha to mice has been shown to activate IFN-alpha signaling molecules in multiple brain regions including neurons in the hippocampus (Wang et al., 2008).
MATERIALS AND METHODS
Cells and Reagents
Mouse hippocampal HT22 cells, kindly provided by Dr. Y. Sagara (University of California, San Diego, CA), were grown at 37 °C and 5% CO2 in DMEM supplemented with 10% heat-inactivated (56 °C, 30 minutes) fetal bovine serum (Hyclone, Logan, UT), 50 U/ml penicillin, and 50 mg/ml streptomycin. GR-green fluorescent protein (GFP) chimera was a gift from Dr. Carmine Pariante, Institute of Psychiatry, London, UK. Pharmacologic reagents included recombinant type-I mouse IFN-alpha (mIFN-alpha) (PBL Biomedical Laboratories, Piscataway, NJ), DEX (Sigma Aldrich, St Louis, MO), janus kinase (Jak) Inhibitor I (Calbiochem, San Diego, CA) and SB203580 (Calbiochem, San Diego, CA).
Transfection
HT22 cells were either transiently or stably transfected with the mouse mammary tumor virus (MMTV)-luciferase reporter gene construct containing multiple glucocorticoid receptor binding sites [(glucocorticoid response elements (GREs)] upstream of the promoter region. Transfections were accomplished using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA). Stably transfected HT22 cells were produced by treatment for 2 weeks with G418 and cloned by limiting dilution. Clones were then screened for DEX-induced MMTV-luciferase activity. Transient transfections with the MMTV-luciferase reporter or GR-GFP were performed in serum-free medium. Stripped fetal bovine serum (FBS) was added to wells 5 hours after transfection.
Luciferase assay
HT22 cells were seeded into 12-well plates and grown for 20–24 hours until 70–80% confluent. After drug treatments, HT22 cells were washed once with cold 1X phosphate buffered saline (PBS), and lysed using a passive lysis buffer (see below). Cells were then centrifuged at 10,000 rpm for 15 seconds at room temperature (RT) to remove cellular debris. Luciferase activity was measured using a microplate luminometer (Luminoscan Ascent, Thermo Labsystems, Helsinki, Finland) and luciferase substrate (Promega, Madison, WI). All activity values reported are the means of treatments from three independent experiments represented as fold change relative to the vehicle control.
Nuclear, cytosolic, and whole cell extracts
Cell monolayers were rinsed with 1× PBS and then harvested in a nuclear homogenization buffer (NHB) containing 20 mM Tris (pH 7.4), 10 mM NaCl, 3 mM MgC2, 5 uM dithiothreitol, 1 uM phenylmethylsulfonyl fluoride, 1 uM pepstatin, 50 trypsin-inhibitory mU of aprotinin, 10 uM leupeptin, and 2 mM sodium vanadate. Igepal CA-630 (Nonidet P-40) was added to a final concentration of 0.15%, and cells were homogenized with 16 strokes in a Dounce homogenizer. The homogenates were centrifuged at 3500 rpm for 5 minutes. Supernatants were saved as cytosolic extract, and the nuclear pellets were resuspended in 0.5 volumes of NHB and were centrifuged as before. The pellet of intact nuclei was resuspended again in one-half of the original volume of NHB and centrifuged again. The majority of the pellet (intact nuclei) was resuspended in an extraction buffer containing 20 mM HEPES (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM ethylenediaminetetraacetic acid (EDTA), 5 uM dithiothreitol, 1 uM phenylmethylsulfonyl fluoride, 1 uM pepstatin, 50 trypsin-inhibitory mU of aprotinin, 10 uM leupeptin, 2 mM sodium vanadate, and 25% glycerol or immunoprecipitation (IP) buffer containing 10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM ethylene glycol tetraacetic acid, 1 mM EDTA, 1% Triton X-100, 1 uM phenylmethylsulfonyl fluoride, 1 uM pepstatin, 50 trypsininhibitory mU of aprotinin, 10 uM leupeptin, and 2 mM sodium vanadate. Nuclei were extracted for 30 minutes on ice. The samples were then subjected to centrifugation at 10,000 rpm at 4°C for 10 minutes. These supernatants contained nuclear protein. To generate whole cell extracts, cells were harvested in 1X PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, and 1% NP-40 containing protease inhibitors (1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml pepstatin, and 1 μg/ml leupeptin]. After 30 minutes incubation on ice, cell lysates were centrifuged at 13,000 × g for 10 minutes at 4°C, and the supernatant was collected. For normalization of sample loading for relevant assays, protein concentrations were determined using a commercial bicinchoninic acid (BCA) assay (Pierce, Rockford, IL) with bovine serum albumin as the protein standard.
Western blot analysis
50 μg cytosolic protein or whole cell protein were mixed with sodium dodecyl sulfate (SDS) buffer and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). For Western blot analysis of nuclear extracts, 25 μg total protein was mixed with SDS buffer before SDS-PAGE. Separated proteins were then electrophoretically transferred onto a nitrocellulose membrane. The membrane was blocked for 1 hour in a 5% milk/tris-buffer saline tween 20 solution, and then incubated overnight in the presence of the primary antibody (1:1000 dilution) raised against GR (M-20; Santa Cruz Biotechnology, Santa Cruz, CA) or phospho (p-)STAT5a/b (Tyr694; Cell Signaling, Danvers, MA). The washed membrane was subsequently incubated with the secondary antibody (1:2000 dilution) for 1 hour. The membrane was washed again and visualized using a commercially available chemoluminescence kit from Amersham Biosciences Corp. (Piscataway, NJ).
Localization of Transiently Transfected GR-GFP
HT22 cells were grown on coverslips before transfection with 2 μg of an expression vector for a GR-GFP chimera using Lipofectamine 2000 transfection reagent, and allowed to recover for 16 hr. Cells were then treated with vehicle, IFN-alpha and/or DEX as described. Cells were fixed/permeabilized with methanol at −20° C for 10 minutes. 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) was then added at 0.5 mg/ml in 1X PBS, followed by incubation at RT for 10 minutes. Unbound dye was removed by washing 3 times with 1X PBS. Coverslips were then mounted using VECTASHIELD® mounting medium for fluorescence microscopy with DAPI (Vector Laboratories, Burlingame, CA) or ToPro (Invitrogen Life Technologies). GR-GFP was visualized using a Nikon E800 fluorescence microscope. Following transient transfection of HT22 cells with either the parental GFP construct or the chimeric GFP–GR construct, fluorescence was observed in ~20–30% of the cells, reflecting the level of transfection efficiency.
Electrophoretic mobility shift assay
A synthetic oligonucleotide containing a GRE sequence (5′-AAG ATT CAG GTC ATG ACC TGA GGA GA) was obtained from Invitrogen Life Technologies. The oligonucleotide was annealed and then labeled at the 5′ end using T4-polynucleotide kinase and [γ-32P] ATP according to manufacturer instructions. In total, 5–10 μg of nuclear extracts from various treatments were incubated with 1 μg poly d(I-C) for 15 minutes at RT to bind nonspecific DNA binding proteins. [γ-32P] DNA (1 μl, final concentration 1 nM) was then added and incubated for 15 minutes at RT. A 100-fold excess of unlabeled DNA was added 5 minutes before the addition of [γ-32P] DNA to compete with the specific DNA protein binding. Reaction mixtures were loaded onto a 5% nondenaturing polyacrylamide gel (acrylamide : bisacrylamide, 30 : 0.8) and run at 150V in 1X TBE buffer (0.09M Tris, 0.09M borate, and 2mM EDTA, pH 8.3). Gels were dried, and protein-DNA binding was visualized by autoradiography.
siRNA procedures
The predesigned STAT1 siRNA (5′-TCCTATTATTATTTAATATAA-3′), STAT2 siRNA (5′-TTGGGTGTTACTACCAGGAAA-3′) and non-targeting siRNA (5′-AAGGAGGCTGAACATTCCGTC-3′) were purchased from Qiagen (Qiagen Inc., Valencia, CA). In order to insure that both STAT5 isoforms were reduced, Stat5 specific siRNA (5′-CUACAGUCCUGGUGUGAGA-3′) targeting a region homologous to both STAT5a and STAT5b isoforms were obtained from the siDesign center (Dharmacon, Lafayette, CO). All siRNA duplexes were used at 100nM, and were transfected for 48 hours using 2μL Lipofectamine 2000 reagent/20pmol of siRNA. After 48 hours, cells were co-transfected with the MMTV-luciferase construct and renilla plasmid. After 4 hours, the transfection complex was replaced with stripped serum media containing the indicated treatments. Cells lysates were harvested following treatment and analyzed for luciferase and renilla activities using the Dual-Luciferase reporter assay system (Promega, Madison, WI). Luciferase values were corrected for transfection efficiency using renilla activity.
Immunoprecipitation
Nuclear protein (300 μg) was preincubated with protein A agarose, and the resulting supernatant was then incubated with 5 ug of the polyclonal anti-GR antibody (M-20; Santa Cruz Biotechnology) for 2 hours at 4°C. Protein-A agarose (Santa Cruz, San Diego, CA) was added to the mixture, and the sample was rotated for an additional 1 hour. Bound GR and any associated proteins were isolated by pelleting this mixture. The pellets were rinsed twice with 1X PBS, and bound proteins were eluted from the agarose by incubation at 100°C for 10 minutes after the addition of Laemmli sample buffer. These samples were separated by SDS-PAGE and analyzed by Western blotting with either STAT5 pY or GR antibodies.
Data analysis and statistics
Descriptive statistics [including the mean and standard error of the mean (SEM)] were used to characterize dependent measures in all experiments. Two-way analysis of variance (ANOVA) was used to explore main effects of mIFN-alpha and DEX treatments as well as their interaction in studies with MMTV-luciferase activity, GR protein levels, or GR-DNA binding as dependent outcomes. Results of experiments exploring the effects of pharmacological inhibitors on mIFN-alpha inhibition of DEX-induced MMTV-luciferase activity were analyzed in a two-step process. Main effects of DEX and mIFN-alpha, as well as their interaction, were first analyzed in pharmacological inhibitor control conditions (i.e. vehicle treated cells) with a two-way ANOVA in order to first confirm mIFN-alpha effects on DEX-induced MMTV-luciferase activity. Separate one-way ANOVAs were then used to determine the overall main effect of Jak-STAT or p38 MAPK inhibition on mIFN-alpha’s effects on DEX-induced MMTV-luciferase activity. For the study involving siRNA treatments targeting relevant STAT pathways, a one-way ANOVA was used to identify the main effect of siRNA in cells treated with mIFN-alpha and DEX following two-way ANOVA analysis of the main effects of DEX and mIFN-alpha and their interaction. Results of the study employing siRNA targeting only STAT5 were first analyzed with a two-way ANOVA analysis of the main effects of DEX and mIFN-alpha and their interaction, followed by a one-way ANOVA comparing all treatment conditions. Significant main effects were followed by Fisher’s Least Significant Difference Tests to explore relevant post hoc comparisons. The level of significance was set at p<0.05, and all tests of significance were two-tailed.
RESULTS
mIFN-alpha attenuates DEX-induced MMTV-luciferase activity and DEX-induced GR-GRE binding
RT-PCR analysis of nuclear extracts revealed that HT22 cells express mRNA coding for the mouse IFN-alpha receptor 1 (see Supplementary Figure 1). To determine the effect of IFN-alpha treatment on GR function in HT22 cells, cells stably transfected with the MMTV-luciferase reporter gene construct were treated with vehicle for 24 hours, mIFN-alpha (100 or 1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50nM) for 2 hours, or mIFN-alpha (100 or 1000 U/ml) for 22 hours followed by mIFN-alpha plus DEX for 2 hours. There was a significant main effect of DEX (F[1,12]=249.98, p<0.01) and mIFN-alpha (F[2,12]=30.92, p<0.01) treatment on MMTV-luciferase activity as well as a significant interaction between these treatment conditions (F[2,12]=26.63, p<0.001). mIFN-alpha at both 100 and 1000 U/ml significantly decreased the effect of DEX alone on DEX-induced MMTV-luciferase activity, with the higher dose of mIFN-alpha showing a trend toward greater inhibition (p=0.06) (Figure 1). Of note, mIFN-alpha alone (100 U/ml) significantly decreased MMTV-luciferase activity relative to the vehicle control condition (p<0.05).
Figure 1. Dexamethasone-induced luciferase activity in HT22 cells is inhibited by mIFN-alpha.
HT22 cells were grown in 12-well culture plates until 80% confluent. Transient transfections were performed using Lipofectin reagent and pAH-Luc plasmid in serum-free medium. Cells were then treated with vehicle for 24 hours, mIFN-alpha (100 or 1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50nM) for 2 hours, or mIFN-alpha (100 or 1000 U/ml) for 22 hours followed by mIFN-alpha plus DEX for 2 hours. All samples were assayed in triplicate, and values shown represent the mean (± SEM) of observations obtained from three independent experiments. Luciferase activity was measured as described. * p<0.05 vs. medium group in same DEX treatment group, FLSD.
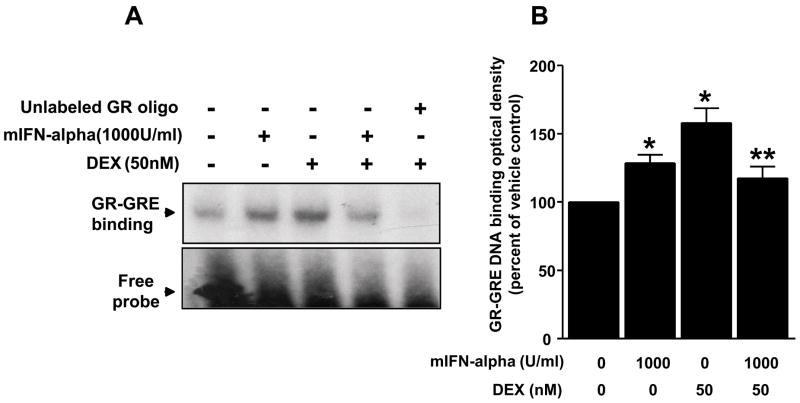
To examine the effect of mIFN-alpha (1000 U/ml) on DEX-induced GR-GRE binding, nuclear extracts were obtained from HT22 cells treated as indicated above. A representative electrophoretic mobility shift assay (EMSA) from 3 independent experiments is shown in Figure 2. Densitometric analysis of the pooled experiments revealed a main effect of DEX on GR-GRE binding (F[1,8]=9.78, p<0.02), as well as a significant interaction between DEX and IFN-alpha treatment (F[1,8] = 21.18, p<0.01). Post hoc analysis revealed that treatment of cells with mIFN-alpha plus DEX resulted in significantly less GR-GRE binding than DEX alone (p<0.05) (Figure 2). In addition, mIFN-alpha alone increased GR-GRE binding relative to vehicle-treated control cells (p<0.05).
Figure 2. mIFN-alpha decreases dexamethasone-induced GR-GRE binding in HT22 cells.
Panel A: Cells were grown in 100-mm culture dishes until 100% confluency. Cells were treated with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50nM) for 2 hours, or mIFN-alpha (1000 U/ml) for 22 hours followed by mIFN-alpha plus DEX for 2 hours. Nuclear extracts were then isolated, and 10 μg nuclear protein from each treatment condition was used in gel mobility shift assay using 32P-labeled synthetic double strand GRE oligonucleotide. A representative gel is shown. Panel B: Data shown represent mean ± SEM optical density obtained from three independent experiments expressed as percent of control (vehicle condition). * p<0.05 vs. vehicle control, ** p<0.05 vs. DEX alone.
mIFN-alpha treatment does not affect GR protein expression or GR nuclear translocation
To determine whether the effects of mIFN-alpha on MMTV-luciferase activity and GR-GRE binding were due to reduced GR protein expression, HT22 cells were treated with vehicle for 24 hours or mIFN-alpha (1000 U/ml) for 1, 5, 12, or 24 hours. Whole cell extracts were probed for GR protein, and a representative Western blot from 3 independent experiments is shown in Figure 3. Using data from the pooled experiments, densitometric analysis revealed no main effect of mIFN-alpha treatment on GR protein expression over the time points examined (F[4,10]=0.41, p>0.05)(Figure 3).
Figure 3. Treatment with mIFN-alpha for 1, 5, 12, and 24 hours does not decrease whole cell expression of GR protein in HT22 cells.
Cells were grown in 6-well culture plates until 80% confluent, and then treated with vehicle for 24 hours or mIFN-alpha (1000 U/ml) for the lengths of time indicated. Whole cell GR was determined via western blot. Panel A: Values shown represent the mean (± SEM) optical density of GR protein from three independent experiments expressed as percent of control (vehicle condition). Panel B: A representative blot of GR protein expression following the indicated mIFN-alpha treatments is shown.
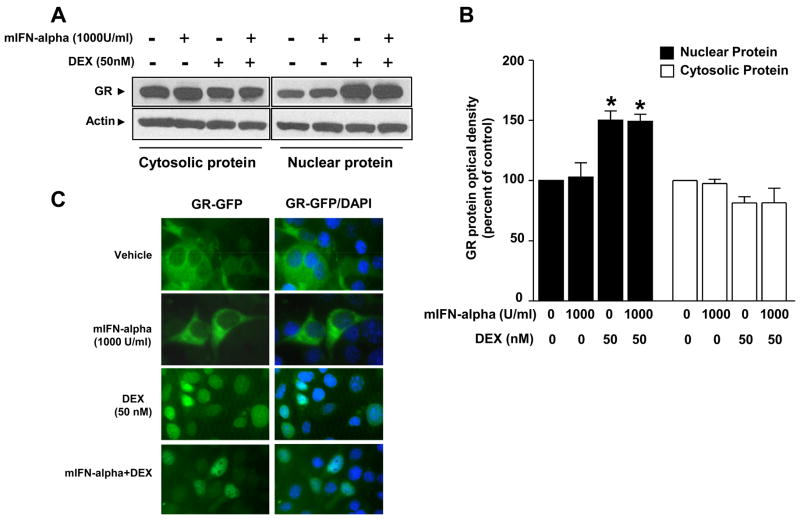
The extent to which mIFN-alpha impairs DEX-induced GR translocation from cytoplasm to the nucleus was examined using Western blot (a representative blot of 3 independent experiments is shown in Figure 4). Cells were treated as indicated in Figure 1 and described above. A significant main effect of DEX treatment (50 nM) was found for both cytosolic (F[1, 8]=6.45, p<0.05) and nuclear (F[1,8]=39.49, p<0.01) GR protein. However, mIFN-alpha treatment (1000 U/ml) had no main effect on GR protein levels in either cytosolic or nuclear fraction (cytosolic: F[1,8]=0.03, p>0.05; nuclear: F[1,8]=0.015, p>0.05) (Figure 4). In addition, there was no interaction between mIFN-alpha and DEX treatment (cytosolic: F[1,8]=0.04, p>0.05; nuclear: F[1,8]=0.06, p>0.05). Post hoc analyses indicated that nuclear extracts obtained from cells treated with DEX in the presence or absence of mIFN-alpha exhibited similar amounts of nuclear GR protein that was significantly increased relative to the control condition in each case (p<0.05) (Figure 4). Increases in nuclear protein were accompanied by decreased cytosolic GR protein in each of the two treatment conditions.
Figure 4. Nuclear translocation of GR in HT22 cells is not attenuated by mIFN-alpha.
Panel A: Cells were grown in DMEM supplemented with 10% FBS to confluency. Cells were treated with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50nM) for 2 hours, or mIFN-alpha (1000 U/ml) for 22 hours followed by mIFN-alpha plus DEX for 2 hours. Western blot analyses were conducted on nuclear and cytosolic extracts using an antibody raised against GR. Representative blots of cytosolic and nuclear protein extracts are shown. Panel B: Values shown represent the mean (± SEM) optical density of GR protein from three independent experiments expressed as percent of control (vehicle condition). * p<0.05 vs. vehicle control group within the same compartment. Panel C: Cells transfected with a GR-green fluorescent protein (GFP) chimera were grown in DMEM containing charcoal-stripped serum for 16 hours before experimental treatments began. Cells were then treated with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 23 hours followed by vehicle plus DEX (50nM) for 1 hour, or mIFN-alpha (1000 U/ml) for 23 hours followed by mIFN-alpha plus DEX for 1 hour. GR-GFP fluorescent images depict representative cells.
The lack of an effect of mIFN-alpha on DEX-induced GR translocation was confirmed by evaluation of nuclear translocation using a GR-GFP construct and fluorescence microscopy. Treatment of cells with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 23 hours and vehicle plus DEX (50nM) for 1 hour or mIFN-alpha (1000 U/ml) for 23 hours followed by mIFN-alpha plus DEX for 1 hour resulted in comparable amounts of nuclear GR-GFP (see Figure 4). One hour of DEX treatment was used in these experiments based on preliminary studies demonstrating maximal GR translocation under these conditions. No effects of any of the treatments were found on the parental GFP construct (Supplementary Figure 2).
Inhibition of Jak-STAT signaling, but not p38 MAPK signaling, reverses mIFN-alpha-induced changes in DEX-induced MMTV luciferase activity
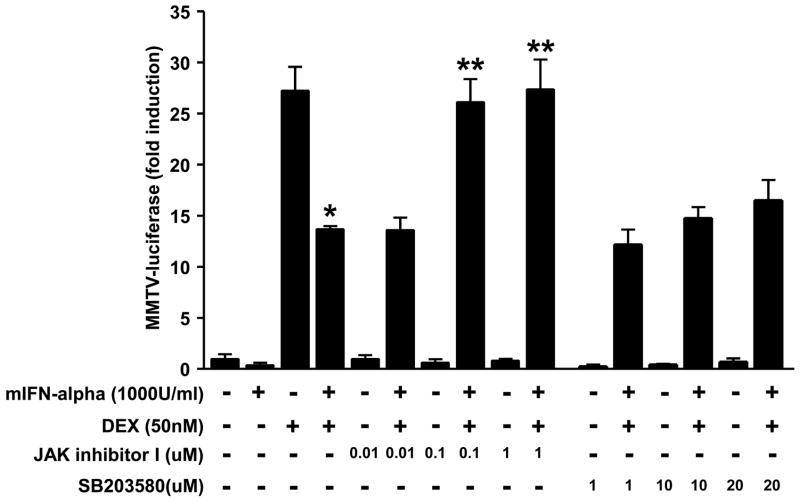
Prior work has shown that IFN-alpha exerts its effects via Jak-STAT signaling pathways as well as p38 MAPK (Platanias, 2005). Consistent with these data, mIFN-alpha (1000 U/ml) was found to potently induce phosphorylated (activated) STAT1, STAT 2, STAT5 and p38 in a time dependent fashion (see Supplementary Figure 3). We therefore investigated the extent to which pharmacological blockade of Jak-STAT and p38 MAPK would reverse inhibition of DEX-induced GR function by mIFN-alpha. MMTV-luciferase activity was examined in stably transfected HT22 cells treated with vehicle for 24 hours, mIFN-alpha for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50nM) for 2 hours or mIFN-alpha (1000 U/ml) for 22 hours followed by IFN-alpha plus DEX for 2 hours in the presence or absence of increasing concentrations of the non-specific Jak inhibitor, Jak Inhibitor I (0.1–10.0 μM) or the p38 MAPK inhibitor, SB203580 (1–20μM), applied 2 hours prior to vehicle or mIFN-alpha treatment (and continued until cell harvest). In separate experiments, Jak Inhibitor I and SB203580 were shown to inhibit activation (phosphorylation) of STAT5 and p38, respectively (see Supplementary Figures 4 and 5). In agreement with observations from earlier experiments, significant main effects of DEX (F[1,8]=279.62, p<0.001) and mIFN-alpha (F[1,8]=35.96, p<0.01) treatment on MMTV-luciferase activity were noted. A significant interaction between DEX and mIFN-alpha treatments was also found (F[1,8]=29.90, p<0.01). Post hoc analysis indicated that treatment with mIFN-alpha plus DEX resulted in significantly lower levels of MMTV-luciferase activity than treatment with DEX alone (Figure 5). When statistical analyses were restricted to cells treated with mIFN-alpha plus DEX in the presence or absence of Jak Inhibitor 1 or SB203580, a main effect of Jak Inhibitor 1 on MMTV-luciferase activity was noted (F[3, 28]=17.01, p<0.01), while no effect was found for SB203580 (F[3, 28]=1.18, p>0.05). Post hoc analysis revealed that cells treated with both mIFN-alpha and DEX plus 1 or 0.1 μM Jak 1 Inhibitor displayed higher levels of MMTV-luciferase activity relative to cells treated with mIFN-alpha plus DEX alone (Figure 5).
Figure 5. Inhibition of Jak-STAT signaling, but not p38 MAPK signaling, reverses mIFN-alpha-induced changes in DEX-induced MMTV luciferase activity.
Panel A: HT22 cells stably transfected with the MMTV-luciferase reporter gene construct were grown in 12-well culture plates until 80% confluent. Cells were then treated with vehicle for 24 hours, mIFN-alpha for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50nM) for 2 hours or mIFN-alpha (1000 U/ml) for 22 h followed by IFN-alpha plus DEX for 2 hours in the presence or absence of increasing concentrations of Jak Inhibitor I (0.01–1.0 μM) or SB203580 (1–20μM). All samples were assayed in triplicate, and values shown represent the mean (± SEM) of observations obtained from three independent experiments. Luciferase activity was measured as described. * p<0.05 vs. cells treated with DEX alone; ** p<0.05 vs. cells treated mIFN-alpha plus DEX.
Inhibition of STAT5 protein expression reverses the effect of mIFN-alpha on DEX-induced MMTV luciferase activity
The role of the Jak-STAT signaling pathway in mIFN-alpha-mediated GR inhibition was further explored using siRNAs targeted against STAT1, STAT2, or STAT5 mRNA (see Supplementary Figure 6 for demonstration of successful targeting of STAT protein expression using siRNA). Cells were transfected with siRNA followed by transfection with the MMTV-luciferase reporter construct as described in the Methods section. Cells were then treated with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50 nM) for 2 hours, or mIFN-alpha for 22 hours followed by mIFN-alpha plus DEX for 2 hours. Confirming observations in earlier experiments, main effects of DEX (F[1,8]=50.69, p<0.01) and mIFN-alpha (F[1,8]=32.35, p<0.01) on MMTV-luciferase activity were observed, as was a significant interaction between the two treatments (F[1,8]=31.10, p<0.01). Post hoc analysis indicated that mIFN-alpha significantly reduced DEX-induced GR luciferase activity (Figure 6). Analysis of the effects of siRNAs directed against specific STAT proteins on MMTV-luciferase activity restricted to cells treated with mIFN-alpha plus DEX revealed a main effect of siRNA treatment (F[4, 10]=51.93, p<0.01). Post hoc analysis revealed that transfection of cells with siRNA directed against STAT5 significantly reversed the inhibitory effects of mIFN-alpha on DEX-induced GR-mediated gene transcription. However, transfection of cells with siRNA directed against STAT1 or STAT2 had no effect.
Figure 6. siRNA directed against STAT5, but not STAT1 or STAT2, reverses mIFN-alpha inhibition of dexamethasone-induced MMTV-luciferase activity in HT22 cells.
Panel A. Cells were grown in 60-mm culture dishes until 80% confluent. Cells were transiently transfected for 24 hours with 100 nM of either target siRNA directed against STAT1, STAT2, or STAT5, or were transfected with a non-target siRNA, followed by transfection with the MMTV-luciferase reporter construct. Following transfection, cells were then treated with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50 nM) for 2 hours, and mIFN-alpha for 22 hours followed by mIFN-alpha plus DEX for 2 hours. Values shown represent the mean (± SEM) of observations obtained from three independent experiments. * p<0.05 vs. cells treated with DEX alone; ** p<0.05 vs. cells treated with mIFN-alpha plus DEX.
Inhibition of STAT5 protein expression does not alter DEX-induced MMTV-luciferase activity in the absence of mIFN-alpha
To determine whether STAT5 plays a role in DEX-induced MMTV luciferase activity in the absence of IFN-alpha, DEX-induced MMTV-luciferase responses were compared in cells transfected with siRNA targeted against STAT5 versus cells transfected with non-targeted siRNA. Other relevant control conditions were also incorporated (STAT5 siRNA + mIFN-alpha, STAT5 siRNA alone). HT22 cells were transfected with or without STAT5 siRNA, or non-target siRNA, followed by transfection with the MMTV-luciferase reporter (described above). Cells were then treated with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50 nM) for 2 hours, or mIFN-alpha for 22 hours followed by mIFN-alpha plus DEX for 2 hours. As in prior experiments, main effects of DEX (F[1,8]=41.59, p<0.01) and mIFN-alpha (F[1,8]=32.58, p<0.01), and a significant interaction between the two treatment conditions (F[1,8]=31.54, p<0.01) on MMTV-luciferase activity were observed. Post hoc analysis indicated that mIFN-alpha significantly reduced DEX-induced GR luciferase activity (Figure 7) in cells not transfected with siRNA targeted against STAT5. Analysis of MMTV-luciferase activity across all treatment conditions revealed a significant main effect of treatment condition (F[8, 18]=22.40, p<0.01). Post hoc analysis revealed that DEX-induced GR-mediated gene transcription did not significantly differ in cells transfected with siRNA targeted against STAT5 versus cells not transfected with siRNA (Figure 7).
Figure 7. siRNA inhibition of STAT5 protein expression does not alter DEX-induced MMTV-luciferase activity in the absence of mIFN-alpha treatment.
Panel A: HT22 cells were grown in 12-well culture plates until 80% confluent. Cells were then transiently transfected for 24 hours with 100 nM of siRNA directed against STAT5, or were transfected with a non-target siRNA, followed by transfection with the MMTV-luciferase reporter construct. Following transfection, cells were then treated with vehicle for 24 hours, mIFN-alpha (1000 U/ml) for 24 hours, vehicle for 22 hours followed by vehicle plus DEX (50 nM) for 2 hours, and mIFN-alpha for 22 hours followed by mIFN-alpha plus DEX for 2 hours. Values shown represent the mean (± SEM) of observations obtained from three independent experiments. * p<0.05 vs. cells treated with DEX alone; ** p<0.05 vs. cells treated with mIFN-alpha plus DEX.
STAT5 and GR are co-immunoprecipitated in HT22 cells treated with mIFN-alpha and dexamethasone
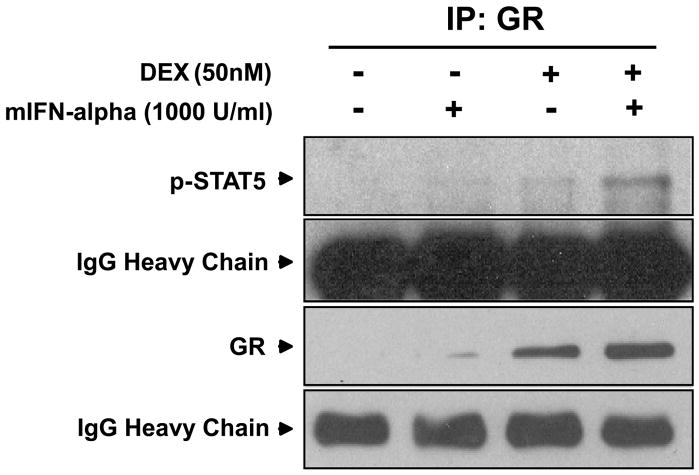
Direct protein-protein interactions between GR and STAT5 were examined using immunoprecipitation. Nuclear protein was extracted from cells treated with mIFN-alpha (1000 U/ml), mIFN-alpha plus DEX (50 nM), or vehicle for 30 minutes and incubated with anti-GR antibodies for immunoprecipitation. Western blot analysis was then performed using a specific anti-phospho-STAT5 antiserum. The 30 minute treatment time in these experiments was derived from a previous study that examined the effect of IL-2 on GR-STAT5 protein-protein interactions (Biola et al., 2001). As shown in Figure 8, antibodies to GR precipitated phospho-STAT5 in the mIFN-alpha plus DEX treatment condition, indicating that STAT5 and GR engage in nuclear protein-protein interactions after treatment with both DEX and mIFN-alpha.
Figure 8. STAT5 and GR engage in protein-protein interactions after treatment with IFN-alpha and dexamethasone in HT22 cells.
Images are representative of three separate experiments. Nuclear extracts were generated from cells treated with vehicle, mIFN-alpha (1000 U/ml), or mIFN-alpha plus DEX (50 nM) for 30 minutes and immunoprecipitated using protein A-or protein-G-agarose beads with an anti-GR antibody. Proteins were then resolved using 10% Tris-HCl gels, electrophoretically transferred to nitrocellulose membranes, and blotted with anti-p-STAT5 or anti-GR antibodies.
DISCUSSION
The data demonstrate that treatment of mouse HT22 cells with mIFN-alpha inhibits GR function as manifested by decreased DEX-induced GR-mediated gene transcription and decreased GR-GRE binding. Under similar treatment conditions, mIFN-alpha had no effects on whole cell GR protein expression or GR nuclear translocation. Pharmacologic inhibition of Jak-STAT signaling pathways, but not p38 MAPK, reversed mIFN-alpha effects on GR function. Disrupting expression of the gene encoding for STAT5 with siRNA also reversed mIFN-alpha effects on GR function, while treatment of cells with siRNA directed against STAT1 and STAT2 failed to show significant effects. Of note, inhibition of STAT5 using siRNA had no effect on DEX-induced MMTV-luciferase activity in the absence of IFN-alpha. Co-immunoprecipitation of phospho-STAT5 and GR following treatment with mIFN-alpha plus DEX demonstrated nuclear phospho-STAT5-GR protein-protein interactions. Taken together, the data suggest that IFN-alpha inhibits GR function by activating STAT5, which then interacts with GR in the nucleus to prevent GR-GRE binding and reporter gene activation.
In the absence of ligand, GR exists in the cytoplasm associated with an assembly of chaperone molecules, notably heat shock proteins. Upon binding to hormone, GR dissociates from the chaperone complex allowing a conformational change that includes exposure of a nuclear localization signal. GR then associates with the cytoskeleton (Galigniana et al., 1998) and translocates to the nucleus through nuclear pores. Once inside the nucleus, GR forms homodimers and binds to GREs on relevant glucocorticoid-sensitive genes or associates with other transcription factors through direct protein-protein interactions (Guiochon-Mantel et al., 1996). Given the many steps in GR signaling, there are numerous points at which cytokines such as IFN-alpha could disrupt GR function, including effects on GR protein expression, hormone binding, nuclear translocation and GR interactions with its DNA response element as well as nuclear transcription factors (Pace et al., 2007).
A number of studies have found that cytokines can alter GR expression (Miller et al., 1999; Pace et al., 2007). Relevant to IFN-alpha, treatment of hepatoblastoma (Huh7) and T cell leukemia-derived (Jurkat) cell lines with IFN-alpha for 72 hours was found to decrease GR mRNA and protein (Cai et al., 2005). However, consistent with the observations reported herein, changes in GR expression were not apparent after 24 hours of IFN-alpha treatment (Cai et al., 2005), suggesting that inhibition of GR function as a result of mIFN-alpha treatment for relatively short periods of time (e.g. 24 hours) does not involve decreased GR protein expression. Regarding hormone binding, plasma levels of IFN-alpha have been found to be inversely associated with monocyte GR binding affinity in AIDS patients (Norbiato et al., 1996). Nevertheless, although binding affinity was not directly measured in the current study, data demonstrating intact GR nuclear translocation following IFN-alpha plus DEX suggest that ligand-dependent conformational changes and interaction with relevant cytoskeletal elements following hormone binding (indirect reflections of affinity for hormone) were not disrupted (Galigniana et al., 1998).
Several studies have examined changes in nuclear translocation of GR as a result of cytokine exposure. For example, both IL-1 and IL-2 have been shown to inhibit DEX-induced GR translocation in vitro (Goleva et al., 2002; Pariante et al., 1999; Wang et al., 2004). In addition, IL-1 receptor deficient mice fail to develop stress-induced glucocorticoid resistance, which has been shown to be secondary to decreased GR translocation in mouse splenocytes and is associated with increased susceptibility to endotoxic shock (Engler et al., 2008; Quan et al., 2001). Inhibition of GR translocation by IL-1 has been found to involve p38 MAPK signaling pathways (Wang et al., 2004). Indeed, treatment with the pharmacological p38 inhibitor, SB203580, as well as antisense oligonucleotides targeting p38 reversed IL-1-induced inhibition of GR-mediated gene transcription in LMCAT cells (Wang et al., 2004). Interestingly, p38 has also been implicated in the effects of IL-2 and IL-4 on GR binding affinity in peripheral blood mononuclear cells (Irusen et al., 2002). Although IFN-alpha has been shown to activate p38 MAPK signaling pathways (Platanias, 2005), IFN-alpha had no effects on GR nuclear translocation, and inhibition of p38 MAPK did not significantly attenuate the effects of IFN-alpha on GR function, indicating that Jak-STAT signaling pathways may be more relevant mediators of mIFN-alpha’s effect on GR.
Many of the effects of IFN-alpha are mediated by Jak-STAT signaling, including primary viral defense and immunosurveillance for malignant cells (Caraglia et al., 2005; Platanias, 2005; Platanias and Fish, 1999). Upon activation, type I IFN-alpha receptors dimerize within the lipid bilayer, permitting transphosphorylation of these receptors. IFN-alpha receptors, in turn, phosphorylate key Jak tyrosine residues. Phosphorylated Jak tyrosine moieties then recruit inactive STAT monomers from the cytoplasm such as STAT1, STAT2, and STAT5. These STATs are phosphorylated by activated Jaks, leading to STAT dimerization and subsequent nuclear translocation where these complexes function as transcription factors (Platanias, 2005; Rogatsky and Ivashkiv, 2006). With respect to GR, the data indicate that IFN-alpha-induced activation of Jak-STAT pathways led to inhibition of GR function through STAT5-GR protein-protein interactions. These data are consistent with similar results obtained following treatment of cells with IL-2 (Biola et al., 2001; Goleva et al., 2002). Unlike IFN-alpha however, inhibition of GR function by IL-2 (as noted above) appears to involve both impairment of GR translocation as well as direct nuclear protein-protein interactions between GR and STAT5 (Goleva et al., 2002). Of note, prolactin has also been shown to disrupt DEX-induced MMTV-luciferase activity through STAT5-GR protein-protein interactions. Indeed, co-immunoprecipitation studies have demonstrated that GR and STAT5 interact within nuclear extracts obtained from cells treated with both prolactin and DEX (Stocklin et al., 1996). Interestingly, while STAT5 inhibits DEX-induced GR-mediated gene transcription, activation of GR may not, in turn, inhibit STAT-5-mediated gene transcription. Such is the case for the STAT5-regulated gene beta-casein. For example, treatment of HC11 mammary epithelial cell with both prolactin and glucocorticoids results in increased expression of beta-casein mRNA compared to treatment with prolactin alone (Stocklin et al., 1996). This synergism between GR and STAT5, leading to increased STAT5-mediated gene transcription, has also been described in liver cells in response to growth hormone (Engblom et al., 2007). Finally, it should be noted that IFN-alpha led to increased nuclear localization of the GR (Figure 8), which paradoxically was associated with reduced DEX-induced MMTV-luciferase activity (Figure 1). These data suggest that while STAT5-GR interactions may increase GR protein in the nucleus, these interactions inhibit GR transcriptional activity, possibly through the disruption of binding of steroid receptor co-factors relevant to GR transcription.
Interestingly, while STAT5 appears to inhibit GR function, there were no effects of STAT1 or STAT2 on the GR in the current study. Of note, STAT1 has been shown to play a critical role in the induction of IFN-alpha responsive genes in the brain of mice following IFN-alpha administration (Wang et al., 2008). Nevertheless, activation of Jak-STAT pathways by IFN-alpha appears to lead to both genomic and non-genomic effects, including the interaction of STAT proteins with other transcription factors including the GR. Such effects, including specifically the interaction of STAT5 with GR, may in turn influence downstream expression of relevant glucocorticoid responsive genes.
Although it remains to be determined whether the effects of IFN-alpha on GR function generalize to primary hippocampal neurons or neurons located in other brain regions as well as other cell types throughout the body, inhibition of GR by STAT5 in a cell line of hippocampal origin is of interest, given the role that hippocampal GR play in the regulation of HPA axis function (Herman et al., 2005). Alterations in hippocampal morphology and function are associated with major depression (Duman et al., 1997), and these changes are believed to contribute to the HPA axis abnormalities seen in depressed patients. Of note, our group has previously reported that robust HPA axis responses to the initial injection of IFN-alpha are associated with vulnerability to IFN-alpha-induced depression (Capuron et al., 2003). Moreover, chronic IFN-alpha administration has been associated with flattening of the diurnal cortisol curve and increased evening plasma cortisol concentrations, both of which were correlated with depressive symptoms (Raison et al., 2008). To the extent that impaired hippocampal GR-mediated feedback inhibition of CRH and HPA axis function may be involved in these effects (as reflected by altered responses to DEX challenge tests in both patients with major depression and patients with flattened diurnal cortisol curves) (Pariante, 2004; Raison et al., 2008; Spiegel et al., 2006), the findings presented herein may shed light on possible molecular mechanisms behind IFN-alpha-induced neuroendocrine changes and depression in relevant clinical populations. Because Jak-STAT signaling may underlie such effects, inhibition of STAT proteins including STAT5 and/or Jak-STAT signaling pathways may represent a possible therapeutic strategy for normalizing GR function and mood alterations in patients chronically exposed to IFN-alpha or other innate immune cytokines.
In conclusion, treatment of a mouse hippocampal cell line with mIFN-alpha disrupts DEX-induced activity of GR. Because IFN-alpha is widely used as immunotherapy for various diseases, and because studies have suggested that IFN-alpha-induced depression is associated with altered HPA axis function, the data provide potentially important insights into the pathophysiological mechanism(s) that mediate these effects. Disruption of Jak-STAT signaling upstream of STAT5, as well as disruption of STAT5 gene expression abrogated the effects of mIFN-alpha on GR function. Thus, STAT5, like p38 MAPK (Wang et al., 2004) and Jun N-terminal kinase (Wang et al., 2005), may be an important contributor to impaired GR signaling (and its relationship to mood disorders)(Raison and Miller, 2003), and therefore may be a relevant factor in the pathophysiology of major depression that occurs in patients with increased inflammation.
Supplementary Material
Supplementary Figure 1. HT22 cells express mouse IFN-alpha receptor 1.
To confirm IFN-alpha receptor expression in mouse HT22 cells, HT22 cells were grown in DMEM supplemented with 10% FBS in 60-mm culture dishes to confluency. Total RNA was isolated using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to manufacturer instructions. After extraction, RNA samples were dissolved in RNase-free water, and their concentrations and the A260/280 ratio were determined using the MBA 2000 System (Perkin-Elmer, Shelton, CT, USA). 1 μg total RNA was then reverse-transcribed using the SuperScript First-strand Synthesis System for real time (RT)-polymerase chain reaction (PCR) (Invitrogen Life Technologies) with a random primer. The primers used for the amplification of the mouse IFN-alpha receptor 1 mRNA were 5′-CGAGGCGAAGTGGTTAAAAG-3′ (forward) and 5′-CCTGGCACAGGCATTTTATT-3′ (reverse). PCR was carried out using SuperMix (Invitrogen) after optimization of primer concentrations, template concentrations, and PCR cycles. The PCR was held at 94°C for 2 minutes, followed by thermal cycling for 35 cycles at 94°C for 15 seconds, 60°C for 15 seconds, and then 72°C for 30 seconds. RT-PCR analysis of nuclear extracts revealed that HT22 cells express mRNA coding for the mouse IFN-alpha receptor 1.
Supplementary Figure 2.
HT22 cells were grown on coverslips before transfection with the parental GFP construct and then allowed to recover for 16 hours. Cells were then treated with vehicle, IFN-alpha and/or DEX as described for Figure 4. Cells were then fixed/permeabilized with methanol at −20 °C for 10 minutes. 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) was then added at 0.5 mg/ml in 1X PBS, followed by incubation at RT for 10 minutes. Unbound dye was removed by washing 3 times with 1X PBS. Coverslips were then mounted using VECTASHIELD® mounting medium for fluorescence microscopy with DAPI (Vector Laboratories, Burlingame, CA) or ToPro (Invitrogen Life Technologies). GFP was visualized using a Nikon E800 fluorescence microscope.
Supplementary Figure 3. Treatment of HT22 cells with mIFN-alpha induces p-STAT1, p-STAT2, p-STAT5, and p-p38 expression.
HT22 cells were treated with mIFN-alpha (100 U/ml) for 0, 5, 15, 30, or 60 min, after which time STAT1, p-STAT1, STAT2, p-STAT2, STAT5, p-STAT5, p38, and p-p38 proteins were measured in whole cell extracts by Western blot.
Supplementary Figure 4. Treatment of HT22 cells with Jak inhibitor I blocks IFN-alpha-induced expression of whole cell p-STAT5.
HT22 cells were treated with mIFN-alpha (1000 U/ml) for 60 min in the presence of increasing concentrations of Jak Inhibitor I (0.01–1.0 μM) before STAT5 and p-STAT5 proteins were measured in whole cell extracts by Western blot. Lanes shown were all run on the same blot.
Supplementary Figure 5. Treatment of HT22 cells with SB203580 blocks mIFN-alpha-induced expression of whole cell p-p38.
HT22 cells were treated with mIFN-alpha (1000 U/ml) for 60 min in the presence of increasing concentrations of SB203580 (10–20μM) before p38 and p-p38 proteins were measured in whole cell extracts by Western blot.
Supplementary Figure 6. Treatment of HT22 cells with siRNA directed against different STATs selectively decreases mIFN-alpha-induced expression of STAT1, STAT2, and STAT5 protein.
HT22 cells were transiently transfected with 100 nM of either target or non-target siRNA for 24 hours, followed by treatment with mIFN-alpha for 24 hours. At the end of this 48 hour period, STAT proteins were measured in nuclear extracts by Western blot to confirm inhibition of STAT protein expression (representative blots of three independent experiments for each siRNA are shown). The optical density of the STAT proteins is expressed as the percent of the vehicle control condition. There was a main effect of siRNA treatment in each case (STAT1 (F[3,8]=6.68, p<0.05), STAT2 (F[3,8]=11.31, p<0.01), and STAT5 (F[3,8]=19.11, p<0.01) with target siRNA treatment leading to significant decreases in the respective STAT protein compared to non-target siRNA and control conditions (p<0.05).
Acknowledgments
This research was funded by grants from the NIMH (MH069124 and MH075102).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas AK, Lichtman AH. Cellular and Molecular Immunology. 5. Saunders; Philadelphia, PA: 2003. [Google Scholar]
- Biola A, Lefebvre P, Perrin-Wolff M, Sturm M, Bertoglio J, Pallardy M. Interleukin-2 inhibits glucocorticoid receptor transcriptional activity through a mechanism involving STAT5 (signal transducer and activator of transcription 5) but not AP-1. Molecular Endocrinology. 2001;15:1062–1076. doi: 10.1210/mend.15.7.0657. [DOI] [PubMed] [Google Scholar]
- Cai W, Khaoustov VI, Xie Q, Pan T, Le W, Yoffe B. Interferon-alpha-induced modulation of glucocorticoid and serotonin receptors as a mechanism of depression.[see comment] Journal of Hepatology. 2005;42:880–887. doi: 10.1016/j.jhep.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Capuron L, Raison CL, Musselman DL, Lawson DH, Nemeroff CB, Miller AH. Association of exaggerated HPA axis response to the initial injection of interferon-alpha with development of depression during interferon-alpha therapy. Am J Psychiatry. 2003;160:1342–1345. doi: 10.1176/appi.ajp.160.7.1342. [DOI] [PubMed] [Google Scholar]
- Caraglia M, Marra M, Pelaia G, Maselli R, Caputi M, Marsico SA, Abbruzzese A. Alpha-interferon and its effects on signal transduction pathways. Journal of Cellular Physiology. 2005;202:323–335. doi: 10.1002/jcp.20137. [DOI] [PubMed] [Google Scholar]
- De Kloet ER, Vreugdenhil E, Oitzl MS, Joels M. Brain corticosteroid receptor balance in health and disease. Endocrine Reviews. 1998;19:269–301. doi: 10.1210/edrv.19.3.0331. [DOI] [PubMed] [Google Scholar]
- Duman RS, Heninger GR, Nestler EJ. A molecular and cellular theory of depression.[see comment] Archives of General Psychiatry. 1997;54:597–606. doi: 10.1001/archpsyc.1997.01830190015002. [DOI] [PubMed] [Google Scholar]
- Engler H, Bailey MT, Engler A, Stiner-Jones LM, Quan N, Sheridan JF. Interleukin-1 receptor type 1-deficient mice fail to develop social stress-associated glucocorticoid resistance in the spleen. Psychoneuroendocrinology. 2008;33:108–117. doi: 10.1016/j.psyneuen.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engblom D, Kornfeld J, Schwake L, Tronche F, Reinmann A, Beug H, Hennighausen L, Moriggl R, Schutz G. Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes and Development. 2007;21:1157–1162. doi: 10.1101/gad.426007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galigniana MD, Scruggs JL, Herrington J, Welsh MJ, Carter-Su C, Housley PR, Pratt WB. Heat shock protein 90-dependent (geldanamycin-inhibited) movement of the glucocorticoid receptor through the cytoplasm to the nucleus requires intact cytoskeleton. Molecular Endocrinology. 1998;12:1903–1913. doi: 10.1210/mend.12.12.0204. [DOI] [PubMed] [Google Scholar]
- Goleva E, Kisich KO, Leung DY. A role for STAT5 in the pathogenesis of IL-2-induced glucocorticoid resistance. Journal of Immunology. 2002;169:5934–5940. doi: 10.4049/jimmunol.169.10.5934. [DOI] [PubMed] [Google Scholar]
- Guiochon-Mantel A, Delabre K, Lescop P, Milgrom E. The Ernst Schering Poster Award. Intracellular traffic of steroid hormone receptors. Journal of Steroid Biochemistry & Molecular Biology. 1996;56:3–9. doi: 10.1016/0960-0760(95)00268-5. [DOI] [PubMed] [Google Scholar]
- Heathcote EJ. Antiviral therapy: chronic hepatitis C. Journal of viral hepatitis. 2007;14:82–88. doi: 10.1111/j.1365-2893.2007.00921.x. [DOI] [PubMed] [Google Scholar]
- Herman JP, Ostrander MM, Mueller NK, Figueiredo H. Limbic system mechanisms of stress regulation: hypothalamo-pituitary-adrenocortical axis. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2005;29:1201–1213. doi: 10.1016/j.pnpbp.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Holsboer F. The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology. 2000;23:477–501. doi: 10.1016/S0893-133X(00)00159-7. [DOI] [PubMed] [Google Scholar]
- Irusen E, Matthews JG, Takahashi A, Barnes PJ, Chung KF, Adcock IM. p38 Mitogen-activated protein kinase-induced glucocorticoid receptor phosphorylation reduces its activity: role in steroid-insensitive asthma. Journal of Allergy & Clinical Immunology. 2002;109:649–657. doi: 10.1067/mai.2002.122465. [DOI] [PubMed] [Google Scholar]
- Ising M, Kunzel HE, Binder EB, Nickel T, Modell S, Holsboer F. The combined dexamethasone/CRH test as a potential surrogate marker in depression. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:1085–1093. doi: 10.1016/j.pnpbp.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Miller AH, Pariante CM, Pearce BD. Effects of cytokines on glucocorticoid receptor expression and function. Glucocorticoid resistance and relevance to depression. Adv Exp Med Biol. 1999;461:107–116. doi: 10.1007/978-0-585-37970-8_7. [DOI] [PubMed] [Google Scholar]
- Nemeroff CB. The corticotropin-releasing factor (CRF) hypothesis of depression: new findings and new directions. Mol Psychiatry. 1996;1:336–342. [PubMed] [Google Scholar]
- Norbiato G, Bevilacqua M, Vago T, Clerici M. Glucocorticoids and interferon-alpha in the acquired immunodeficiency syndrome. J Clin Endocrinol Metab. 1996;81:2601–2606. doi: 10.1210/jcem.81.7.8675584. [DOI] [PubMed] [Google Scholar]
- Pace TWW, Hu F, Miller AH. Cytokine-effects on glucocorticoid receptor function: Relevance to glucocorticoid resistance and the pathophysiology and treatment of major depression. Brain, Behavior, & Immunity. 2007;21:9 – 19. doi: 10.1016/j.bbi.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariante CM. Glucocorticoid receptor function in vitro in patients with major depression. Stress. 2004;7:209–219. doi: 10.1080/10253890500069650. [DOI] [PubMed] [Google Scholar]
- Pariante CM, Pearce BD, Pisell TL, Sanchez CI, Po C, Su C, Miller AH. The proinflammatory cytokine, interleukin-1alpha, reduces glucocorticoid receptor translocation and function. Endocrinology. 1999;140:4359–4366. doi: 10.1210/endo.140.9.6986. [DOI] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature Reviews. Immunology. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Platanias LC, Fish EN. Signaling pathways activated by interferons. Exp Hematol. 1999;27:1583–1592. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- Quan N, Avitsur R, Stark JL, He L, Shah M, Caligiuri M, Padgett DA, Marucha PT, Sheridan JF. Social stress increases the susceptibility to endotoxic shock. Journal of Neuroimmunology. 2001;115:36–45. doi: 10.1016/s0165-5728(01)00273-9. [DOI] [PubMed] [Google Scholar]
- Raber J, Koob GF, Bloom FE. Interferon-alpha and transforming growth factor-beta 1 regulate corticotropin-releasing factor release from the amygdala: comparison with the hypothalamic response. Neurochemistry International. 1997;30:455–463. doi: 10.1016/s0197-0186(96)00082-4. [DOI] [PubMed] [Google Scholar]
- Raison CL, Borisov AS, Woolwine BJ, Massung B, Vogt GJ, Miller AH. Interferon-alpha effects on diurnal hypothalamic-pituitary-adrenal axis activity: Relationship with proinflammatory cytokines and behavior. Molecular Psychiatry. 2008 doi: 10.1038/mp.2008.58. epub, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Demetrashvili M, Capuron L, Miller AH. Neuropsychiatric adverse effects of interferon-alpha: recognition and management. CNS Drugs. 2005;19:105–123. doi: 10.2165/00023210-200519020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Miller AH. When not enough is too much: the role of insufficient glucocorticoid signaling in the pathophysiology of stress-related disorders. American Journal of Psychiatry. 2003;160:1554–1565. doi: 10.1176/appi.ajp.160.9.1554. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Ivashkiv LB. Glucocoritcoid modulation of cytokine signaling. Tissue Antigens. 2006;68:1 – 12. doi: 10.1111/j.1399-0039.2006.00599.x. [DOI] [PubMed] [Google Scholar]
- Spiegel D, Giese-Davis J, Taylor CB, Kraemer H. Stress sensitivity in metastatic breast cancer: analysis of hypothalamic-pituitary-adrenal axis function. Psychoneuroendocrinology. 2006;31:1231–1244. doi: 10.1016/j.psyneuen.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- Wang J, Campell IL, Zhang H. Systemic interferon-alpha regulates interferon-stimulated genes in the central nervous system. Molecular Psychiatry. 2008;13:293–301. doi: 10.1038/sj.mp.4002013. [DOI] [PubMed] [Google Scholar]
- Wang X, Wu H, Lakdawala VS, Hu F, Hanson ND, Miller AH. Inhibition of Jun N-terminal kinase (JNK) enhances glucocorticoid receptor-mediated function in mouse hippocampal HT22 cells. Neuropsychopharmacology. 2005;30:242–249. doi: 10.1038/sj.npp.1300606. [DOI] [PubMed] [Google Scholar]
- Wang X, Wu H, Miller AH. Interleukin-1 alpha-induced activation of p38 mitogen-activated protein kinase inhibits glucocorticoid receptor function. Molecular Psychiatry. 2004;9:65–75. doi: 10.1038/sj.mp.4001339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. HT22 cells express mouse IFN-alpha receptor 1.
To confirm IFN-alpha receptor expression in mouse HT22 cells, HT22 cells were grown in DMEM supplemented with 10% FBS in 60-mm culture dishes to confluency. Total RNA was isolated using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to manufacturer instructions. After extraction, RNA samples were dissolved in RNase-free water, and their concentrations and the A260/280 ratio were determined using the MBA 2000 System (Perkin-Elmer, Shelton, CT, USA). 1 μg total RNA was then reverse-transcribed using the SuperScript First-strand Synthesis System for real time (RT)-polymerase chain reaction (PCR) (Invitrogen Life Technologies) with a random primer. The primers used for the amplification of the mouse IFN-alpha receptor 1 mRNA were 5′-CGAGGCGAAGTGGTTAAAAG-3′ (forward) and 5′-CCTGGCACAGGCATTTTATT-3′ (reverse). PCR was carried out using SuperMix (Invitrogen) after optimization of primer concentrations, template concentrations, and PCR cycles. The PCR was held at 94°C for 2 minutes, followed by thermal cycling for 35 cycles at 94°C for 15 seconds, 60°C for 15 seconds, and then 72°C for 30 seconds. RT-PCR analysis of nuclear extracts revealed that HT22 cells express mRNA coding for the mouse IFN-alpha receptor 1.
Supplementary Figure 2.
HT22 cells were grown on coverslips before transfection with the parental GFP construct and then allowed to recover for 16 hours. Cells were then treated with vehicle, IFN-alpha and/or DEX as described for Figure 4. Cells were then fixed/permeabilized with methanol at −20 °C for 10 minutes. 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) was then added at 0.5 mg/ml in 1X PBS, followed by incubation at RT for 10 minutes. Unbound dye was removed by washing 3 times with 1X PBS. Coverslips were then mounted using VECTASHIELD® mounting medium for fluorescence microscopy with DAPI (Vector Laboratories, Burlingame, CA) or ToPro (Invitrogen Life Technologies). GFP was visualized using a Nikon E800 fluorescence microscope.
Supplementary Figure 3. Treatment of HT22 cells with mIFN-alpha induces p-STAT1, p-STAT2, p-STAT5, and p-p38 expression.
HT22 cells were treated with mIFN-alpha (100 U/ml) for 0, 5, 15, 30, or 60 min, after which time STAT1, p-STAT1, STAT2, p-STAT2, STAT5, p-STAT5, p38, and p-p38 proteins were measured in whole cell extracts by Western blot.
Supplementary Figure 4. Treatment of HT22 cells with Jak inhibitor I blocks IFN-alpha-induced expression of whole cell p-STAT5.
HT22 cells were treated with mIFN-alpha (1000 U/ml) for 60 min in the presence of increasing concentrations of Jak Inhibitor I (0.01–1.0 μM) before STAT5 and p-STAT5 proteins were measured in whole cell extracts by Western blot. Lanes shown were all run on the same blot.
Supplementary Figure 5. Treatment of HT22 cells with SB203580 blocks mIFN-alpha-induced expression of whole cell p-p38.
HT22 cells were treated with mIFN-alpha (1000 U/ml) for 60 min in the presence of increasing concentrations of SB203580 (10–20μM) before p38 and p-p38 proteins were measured in whole cell extracts by Western blot.
Supplementary Figure 6. Treatment of HT22 cells with siRNA directed against different STATs selectively decreases mIFN-alpha-induced expression of STAT1, STAT2, and STAT5 protein.
HT22 cells were transiently transfected with 100 nM of either target or non-target siRNA for 24 hours, followed by treatment with mIFN-alpha for 24 hours. At the end of this 48 hour period, STAT proteins were measured in nuclear extracts by Western blot to confirm inhibition of STAT protein expression (representative blots of three independent experiments for each siRNA are shown). The optical density of the STAT proteins is expressed as the percent of the vehicle control condition. There was a main effect of siRNA treatment in each case (STAT1 (F[3,8]=6.68, p<0.05), STAT2 (F[3,8]=11.31, p<0.01), and STAT5 (F[3,8]=19.11, p<0.01) with target siRNA treatment leading to significant decreases in the respective STAT protein compared to non-target siRNA and control conditions (p<0.05).