Abstract
The length of the face represents an important axis of variation in mammals and especially in primates. Mice with mutations that produce variation along this axis present an opportunity to study the developmental factors that may underlie evolutionary change in facial length. The Crf4 mutant, obtained from the C57BL/6J (wt/wt) background by chemical mutagenesis by the Baylor Mouse Mutagenesis Resource, is reported to have a short-faced phenotype. As an initial step towards developing this model, we performed 3D geometric morphometric comparisons of Crf4 mice to C57BL/6J wild-type mice focusing on three stages of face development and morphology – embryonic (GD 9.5–12), neonatal, and adult. Morphometric analysis of adult Crf4 mutants revealed that in addition to a shortened face, these mice exhibit a significant reduction in brain size and basicranial length. These same features also differ at the neonatal stage. During embryonic face formation, only dimensions related to brain growth were smaller, whereas the Crf4 face actually appeared advanced relative to the wild-type at the same somite stage. These results show that aspects of the Crf4 phenotype are evident as early as embryonic face formation. Based on our anatomical findings we hypothesize that the reduction in facial growth in Crf4 mice is a secondary consequence of reduction in the growth of the brain. If correct, the Crf4 mutant will be a useful model for studying the role of epigenetic interactions between the brain and face in the evolutionary developmental biology of the mammalian craniofacial complex as well as human craniofacial dysmorphology.
Keywords: C57BL/6J, craniofacial development, Crf4, embryo, epigenetic interactions, geometric morphometrics, micro-computed tomography, mouse, neonate, skull
Introduction
Shortening of the basicranium and face are prominent and related trends in human evolution (Lieberman et al. 2002). Facial length is also an important axis of variation among species of primates and other mammals and is therefore of evolutionary interest. In humans, this trait is also of clinical significance. Abnormal growth of the face results in midfacial retrusion, prognathism and retrognathism, and malocclusions (Enlow, 1982), as well as cleft lip and palate, during embryonic development (Diewert, 1985; Diewert & Shiota, 1990; Diewert & Wang, 1992; Wang & Diewert, 1992; Diewert & Lozanoff, 1993a,b; Diewert & Lozanoff, 2002). However, the developmental genetics and regulation of variation in facial length – that is, prognathism of the jaws and midface – are largely unknown. Study of the developmental basis for variation in facial length will enhance understanding and improve treatment of clinical facial phenotypes, as well as offer testable hypotheses about the evolutionary mechanisms underlying evolutionary changes in primate – and notably human – facial length.
The midface develops from three paired epithelium-covered mesenchymal tissue buds that surround the primitive oropharynx. These paired primordia are the medial and lateral nasal prominences and maxillary prominences (Fig. 1). By gestational days 10–12, mesenchyme derived from cephalic neural crest and paraxial mesoderm (Couly et al. 1993; Creuzet et al. 2005) has already migrated into the presumptive facial primordia and epithelial-mesenchymal signaling has begun to organize cellular condensations (Ralphs, 1992). Perhaps the most single critical event of normal face formation is the contact and fusion of the three paired facial prominences. Importantly, variation in facial prominence length and shape may be the proximal cause of the failure of fusion that leads to cleft lip and palate disorders (Trasler, 1968; Wang & Diewert, 1992b; Diewert & Lozanoff, 1993b; Hallgrímsson et al. 2004; Young et al. 2007).
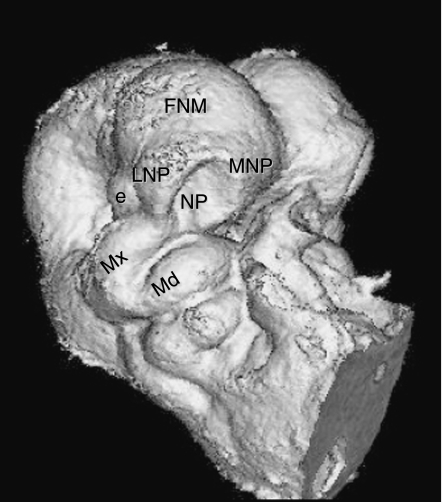
Fig. 1.
Micro-CT scan of mouse embryo, gestational day 10, right side. Six paired epithelium-covered mesenchymal tissue buds surround the primitive oropharynx, three to a side (Mx, MNP, LNP) and, after merging and fusing together, give rise to the midface. e, eye; FNM, frontonasal mass; LNP, lateral nasal prominence; Md, mandibular arch; MNP, medial nasal prominence; Mx, maxillary prominence.
There are many possible prenatal developmental mechanisms for variation in facial length, including responses to hormones (Ramirez-Yanez et al. 2005), muscle development (Ralphs, 1992), gene expression – particularly transcriptional – variation (Sears et al. 2007), size of embryonic facial primordia, and postnatal developmental pattern (Enlow, 1990). Here we focus on the Crf4 ‘short-faced’ mouse mutant (MGI:3026658) as a model for studying the developmental mechanisms underlying phenotypic variation in facial length. As an initial step to understanding this craniofacial defect, we test the hypothesis that, at the time of its formation, the Crf4 embryonic face already exhibits the mutant phenotype – shortened face – seen, but until now not quantified, in the adult mouse. Our study compares the phenotypic impact and development of the crf4 mutation against wild-type morphology and development at adult, neonatal and embryonic stages in mouse. We hypothesize that the Crf4 phenotype is established in the embryo prior to the fusion of the facial prominences and affects growth during the chondrocranial period (late embryonic and fetal). This would suggest that the mutant Crf4 phenotype is a product of abnormal early developmental processes such as disrupted signaling, cell proliferation, apoptosis and/or cell aggregation in the developing face. The results of this morphometric study will guide subsequent work directed at the nature and timing of these developmental processes.
The Crf4 mice are smaller than the wild-type strain. Changes in size usually produce correlated or allometric changes in shape. A question that arises, therefore, is whether the Crf4 shape effects are a secondary effect of a reduction in cranial size. This question cannot be answered by simply examining the correlation between size and shape (or statistically removing size) because if the mutation affects both size and shape, the severity of those effects are likely to be correlated. For example, if the mutation reduces the size of the face and also reduces cranial size, individuals with smaller heads will also have smaller faces, not because of the scaling of shape (allometry) but because the mutation affects both traits. We take a different approach here and instead compare the effects of the crf4 mutation to the effects of a mutation on a similar genetic background which affects the somatic growth axis. To address the contribution of allometric scaling to the shape effects of the crf4 mutation, we therefore compare the crf4 mutation to ‘little mice’, which have a null mutation in the growth hormone releasing hormone receptor (ghrhr).
A secondary goal of this study is to determine whether the crf4mutation influences the phenotypic variance as well as the mean. This is true of many mutations for reasons that remain only partly known (Scharloo, 1964, 1991; Hallgrímsson et al. 2002, 2006).
Materials and methods
Mouse samples
Homozygous Crf4 (crf4/crf4) male and female mice were obtained from the Baylor Mouse Mutagenesis Resource (http://www.mouse-genome.bcm.tmc.edu/ENU/ENUMutantSources.asp). The short-faced Crf4 mutant was developed on the C57BL/6J (wt/wt) (Jackson Labs) background by chemical induction through N-ethyl-N-nitrosourea mutagenesis (Kile et al. 2003). Although the gene or genes involved remain undefined, the crf4mutation is a non-lethal, recessive mutation located on chromosome 13 (MGI:3026658). The crf4/crf4 mice are also heterozygous for a 24CM inversion on chromosome 11 (Kile et al. 2003). This inversion, representing about 2% of the mouse genome, is derived from 129/SvEv mice and may thus contain unknown genetic variants that exert dominant or semi-dominant effects on craniofacial morphology. However, given the significant phenotypic effect of the crf4mutation, we assume for the purpose of this study that the differences between the crf4/crf4 mice and C57BL/6J wildype mice are due to the mutation and not to 129/SvEv *C57BL6/J heterozygosity in the region of the inversion on chromosome 11. This assumption is given additional support by the fact that the other mutants identified through the Baylor ENU mutagenesis screen are also heterozygous for the inversion on chromosome 11 but these pedigrees do not exhibit the short-faced crf4 phenotype.
We opted to compare crf4 mutants with the C57BL/6J wild-type strain, from which these mutants are derived, rather than to littermates from a Crf4 *C57BL/6J cross. As the gene is unknown, genotyping for such a sample based on phenotype would have created circularity for the phenotypic comparisons, particularly if the phenotypic distributions of the genotypes overlap.
To test for the possibility that the shape differences between Crf4 and wildype C57BL/6J are a consequence of reduction in overall growth, we also obtained mice with a null mutation in the growth hormone releasing hormone receptor (ghrhr–/–) on the C57BL/6J background (Jackson Labs). These ‘little’ mice are reduced in body size and the difference in shape between them and the wild-type strain is an allometric consequence of overall reduction in growth because the mutation affects body size and not craniofacial development directly.
Mice were housed under standard animal care facility conditions and sacrificed at 90 days of age via CO2 asphyxiation. Table 1 shows the sample sizes used for this study. Neonates were obtained at p0 (day of birth) and sacrificed under isoflurane anesthesia by intra-cardiac injection of 0.25–0.5 mL Euthanyl (240 mg mL−1 sodium pentobarbital). Data were collected from Crf4 (n = 22) and C57BL/6J (n = 23) neonate skulls of indeterminate sex. Mouse embryos were collected on gestational days (GD) 10–12 based on initial detection of a vaginal plug; the litter was assumed to be aged GD 0.5 that morning. Embryos were dissected and staged by tail somite (TS) number. Due to high inter- and intra-litter variation in timing of development, staging by TS rather than by GD afforded the most accurate and fine-scaled indication of developmental stage (Young et al. 2007; Parsons et al. 2008). Embryos were immediately fixed in Bouin's solution as previously described (Wang & Diewert, 1992a). Bouin's fixation is a long-standing accepted method of embryo fixation for morphometric analysis (Trasler, 1968). This fixative enhances embryo density, thus optimizing micro-computed tomography (µ-CT) scanning of the embryo, and facilitates scanning of specimens – normally stored in solution – in a non-aqueous environment as is required. We collected µ-CT scan data from Crf4 (n = 101) and C57BL/6J (n = 97) embryos of indeterminate sex between GD 10 and GD 12.
Table 1.
Sample sizes
| Strain | Adults (90 d) female | Adult (90 d) male | Neonate (p 0) sex unknown | Embryo (GD10–12) sex unknown |
|---|---|---|---|---|
| Crf4 | 12 | 8 | 22 | 101 |
| C57BL/6J | 14 | 16 | 23 | 97 |
Computed tomography
We collected landmark data from our specimens using micro-computed-tomography (µ-CT), which facilitates three-dimensional (3D) morphometric analyses via the high-resolution data capture of minute objects combined with digital reconstructions of high accuracy. Adult and neonate crania were scanned in the Scanco Medical vivaCT 40 with a conebeam protocol at 55 kVp, 109 µA, 35 µm resolution and a 0.36 rotation step. The CT data were then imported into analyze 5.0 software (Mayo Foundation for Medical Education and Research, Rochester, MN, USA) for landmarking. Following Parsons et al. (2008), embryonic heads were scanned in the Skyscan 1072 X-Ray Micro-Tomograph with a conebeam protocol at 6.25 µm resolution, 100.0 kV, 98.4 µA, 0.90 rotation step, 2 frame averaging and a 3.8-s exposure time (corrected for flat field errors and random movement errors). Reconstructions were completed in the program ‘Cone_Rec’ with post-alignment correction and ring-artifact reduction. Reconstructed image stacks were cropped in ImageJ and loaded into analyze software for landmarking.
3D Landmarking
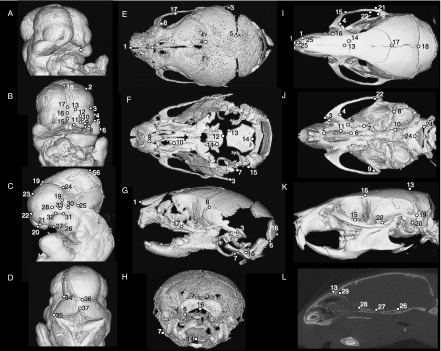
Figure 2 and Tables 2–4 detail the 3D landmarks used for this study at all three developmental stages. Type 1 and Type 2 landmarks (Bookstein, 1991) were chosen according to the guidelines presented by Zelditch et al. (2004c). Type 1 landmarks (points at discrete juxtapositions of tissues; limited on three axes) were simple to locate in adult and neonatal skulls. However, in the unossified embryonic face, a greater proportion of Type 2 landmarks (local minima and maxima of curvatures of local structures) were used. The use of Type 2 (fuzzy) landmarks has been validated (Valeri et al. 1998) for the sampling and analysis of anatomical size and shape information in 3D. To increase landmarking precision on our embryos, points on a curve were acquired from a single, consistent view. The x-axis was standardized by tilting the head so that the superior border of the frontonasal mass was perpendicular to the plane of view. The y-axis was standardized by rotating the head to centre its midline in the frontal view, and the medial nasal prominences were superimposed exactly in the lateral views.
Fig. 2.
Landmarks on mouse embryo head (A–D) in para-frontal (A), frontal (B), lateral (C) and occipital (D) views. Landmark positions on neonate (E–H) and adult (I–L) mouse skulls, in dorsal (E,I), ventral (F,J), lateral (G,K), occipital (H) and internal (L) views.
Table 2.
Adult skull landmarks
| Number | Abbreviation | Landmark name |
|---|---|---|
| 1 (R/L) | MSI | Midline superior incisor |
| 2 (R/L) | AIF | Anterior margin of incisive foramen |
| 3 (R/L) | AIZ | Anterior inferior zygomatic |
| 4 (R/L) | PM | Point of greatest curvature on the posterior margin of the malar process |
| 5 (R/L) | ASA | Anterior superior alveoli |
| 6 (R/L) | PIF | Posterior incisive foramen |
| 7 (R/L) | LPP | Lateral palatal-pterygoid junction |
| 8 (R/L) | AIA | Anterior inferior auditory bulla |
| 9 (R/L) | PZA | Point of greatest curvature along posterior edge of zygomatic process of temporal bone |
| 10 (R/L) | OAS | Occipital-auditory-sphenoid junction |
| 11 (R/L) | MMP | Medial maxilla-premaxilla junction |
| 12 (R/L) | LNS | Anteriormost point along lateral zygomatic-frontal suture |
| 13 (M) | NAS | Nasion |
| 14 (R/L) | LFS | Lateral point along frontal suture |
| 15 (R/L) | MS | Superior margin of suture of temporal and zygomatic processes of zygomatic arch |
| 16 (R/L) | FTP | Frontal-temporal-parietal junction |
| 17 (M) | BRG | Bregma |
| 18 (M) | LAM | Lambda |
| 19 (R/L) | MST | Point along occipomastoid suture |
| 20 (R/L) | TYM | Supero-posterior extremity of tympanic ring |
| 21 (R/L) | PTZ | Posterior temporal-zygomatic junction |
| 22 (R/L) | ATZ | Anterior temporal-zygomatic junction |
| 23 (M) | BAS | Basion |
| 24 (R/L) | OAJ | Occipital atlas junction |
| 25 (R/L) | ANM | Most anterior point along premaxilla nasal junction |
| 26 (R/L) | SOS | Spheno-occipital synchondrosis |
| 27 (M) | MSS | Midline presphenoid sphenoid synchondrosis |
| 28 (M) | SES | Spheno-ethmoidal synchondrosis |
| 29 (M) | FC | Foramen caecum |
R, right; L, left; M, midline.
Table 4.
Neonate skull landmarks
| Number | Landmark description |
|---|---|
| 1 (R/L) | Most anterior-medial point of the nasal bone |
| 2 (R/L) | Most posterior-superior medial point of the nasal bone |
| 3 (R/L) | Most posterior point of the squamosal bone |
| 4 (R/L) | Most medial intersection of the frontal and parietal bones (at the suture) |
| 5 (R/L) | Most posterior-medial point on the parietal bone, near the posterior fontanelle at the point where the parietal comes in closest contact medially with the inter-parietal bone |
| 6 (R/L) | Most inferior-lateral point on the occipital bone |
| 7 (R/L) | Most posterior-superior point of the posterior aspect of the annulus |
| 8 (R/L) | Most superior point on the squamosal temporal at the intersection of the coronal suture with the squamosal |
| 9 (R/L) | Most anterior point of the incisive foramen |
| 10 (R/L) | Most posterior point of the incisive foramen |
| 11 (R/L) | Posterior tip of medial pterygoid process |
| 12 (M) | Most posterior-inferior medial point on the basi-sphenoid |
| 13 (R/L) | Most anterior-lateral point on the corner of the basi-occipital bone, left side |
| 14 (M) | Basion |
| 15 (R/L) | Most anterior-inferior tip of the lateral inferior process of the occipital condyle |
| 16 (M) | Opisthion: posterior-medial point of foramen magnum |
| 17 (R/L) | Most superior-anterior point on the zygomatic bone |
| 18 (R/L) | Point of greatest curvature on the posterior margin of the malar process |
Statistical shape analyses
Our statistical procedures address four hypotheses. The first is that the adult morphology of the Crf4 mutants is statistically distinct from that in C57BL6/J wild-type mice.The second and third hypotheses are that the differences between the strains are detectable in neonates and embryos, respectively. Finally, we test the hypothesis that the morphological variance is altered in crf4/crf4 mice. We employed a combination of three different standard statistical shape analyses to test these four hypotheses.
Geometric morphometric methods quantify and compare size and shape using landmark-based data (Bookstein, 1991; Lele & Richtsmeier, 1991; Zelditch et al. 2004c; Slice, 2005). We quantified size and shape differences using a combination of Procrustes-based and Euclidean distance matrix analysis (EDMA) methods (Lele & Richtsmeier, 2001). Procrustes coordinates were calculated using the generalized Procrustes analysis method (GPA) (Goodall, 1991) using the software packages Morpheus (Slice, 1994–1999), Morphologika (O’Higgins & Jones, 1998) or MorphoJ (Klingenberg, 2008).
Embryo shape and size change dramatically with ontogeny and can vary significantly even with litters. Ontogenetic shape variation, therefore, must be quantified and removed for comparison of shape variation between strains. To this end, Procrustes coordinates were regressed on tail somite (TS) number for embryos using the pooled within-sample multivariate regression of TS on shape in MorphoJ or a pooled multivariate regression of TS on shape in IMP (Sheets, 2004b) as described in Zelditch et al. (2004b). The pooled within-sample regressions obtain slopes within each group and thus take into account potential differences among strains in the relationship between tail somite stage and shape. We also performed a univariate regression of the Procrustes shape distance from the mean against TS as described in Zelditch (2004b).
To compare mean shape for both the stage standardized and unstandardized samples we used a battery of tests. We used both the Goodall's F-test (Goodall, 1991) and the bootstrap F-test in Simple3D (Sheets, 2004a). In MorphoJ, we used the permutation tests for both the Mahalanobis and Procrustes distances between-group (10 000 permutations). Finally, we use the Procrustes manovain MorphoJ to compare groups. The reason for the battery of statistical tests is that the tests have different limitations. The Procrustes distance-based methods such as Goodall's F-test assume that the variation about each landmark in the dataset is isotropic. This assumption is unrealistic (Klingeber & Monteiro, 2005) but it is not fully understood to what extent violation of this assumption affects significance testing. Procrustes manova, on the other hand, requires that the number of samples is greater than the number of landmarks (Zelditch et al. 2004a). This is the case for most but not all comparisons in this study. It can be assumed that all tests reported as significant were significant with all tests unless otherwise noted. The same methods have been used to compare embryo craniofacial data in mice (Parsons et al. 2008).
For the embryo sample, the very large amount of shape variation poses a problem as this can introduce distortions when the Procrustes coordinate data are projected onto the tangent plan as partial warps (Dryden & Mardia, 1998). To mitigate this issue, we conducted comparisons of ontogenetic subsets, each of which involves much less overall shape variation and thus less distortion.
Mean shapes were also visualized using the method of Kristensen et al. (2008). In this method, individual scans are first scaled isotropically to the sample mean and then superimposed using a rigid intensity-based registration with the fit measured by Matte's mutual information metric. Once superimposed, the datasets are summed and averaged to create an average volumetric dataset that can be thresholded for visualization.
To compare shape variances, we obtained the Procrustes distances of each individual from the group mean (mean Procrustes landmark configuration). The variance of these distances is our measure of shape variance for each sample. These distributions were compared using the Delta V permutation test in Simple3D and Levene's test following previous analyses (Willmore et al. 2006a,b; Zeditch et al. 2004b).
To visualize differences between groups, we used both principal components analysis (PCA) and canonical variates analysis (CVA) to visualize shape variation between groups. PCA finds the central axes of covariation in the pooled samples. Usually, but not always, the first principal components capture differences between groups. CVA maximally discriminates between group means by scaling each within-group variance in proportion to, and along the axis of, the greatest variance in the pooled (averaged) within-group variances. We present the results of the CVA visualization as this method specifically captures the shape differences between groups.
To localize shape differences, we used EDMA (Lele & Richtsmeier, 1991). EDMA unambiguously describes form (size and shape) by using coordinate landmark data to calculate every possible inter-landmark (Euclidean) distance in a form-matrix. For shape analyses, mean forms computed through an algorithm (Lele, 1993) were scaled by the geometric mean of all distances (scaling factor) in each sample to define the shape matrices (Lele & Cole, 1996). A shape difference matrix (SDM) was then defined by the difference between the two shape matrices. With this method, localized shape differences and even differences in single inter-landmark distances can be statistically tested for significance. EDMA was performed in Win EDMA (Cole III, 2003). We visualize values significantly above and below +/– 3% (threshold of 0.03) in the SDM. Although a larger sample of interlandmark difference is significantly different, this arbitrary threshold is sufficient to visualize the major regional differences. EDMA analyses could not be applied to the embryo sample as it does not allow for the standardization of shape to ontogenetic stage.
To further characterize the adult and embryonic phenotypes, we also compared the key measurements listed in Table 3. Endocranial volume was calculated from image datasets subjected to median (radius = 2) and maximum filtering (radius = 1) in ImageJ to remove pseudoforamina and sutures. Virtual endocranial casts were then created and measured in analyze 3D 5.0 and edited to remove the brain stem and any non-endocranial projections. Measurement error for endocranial volume represented 4.6% of the total variance or 3.2% of the mean. For the adult sample, we compared these variables using two-way anovas by genotype and sex.
Table 3.
Adult measurements used
| Variable | Description |
|---|---|
| Cranial base angle | Midsagittal angle (ventral aspect) between chord from basion (BAS) to the mid-sphenoidal synchondrosis (MSS), and chord from the foramen caecum (FC) to the mid-sphenoidal synchondrosis (MSS). |
| Posterior basicranial length | Distance from basion (BAS) to the mid-sphenoidal synchondrosis (MSS). |
| Anterior basicranial length | Distance from the mid-sphenoidal synchondrosis (MSS) to foramen caecum (FC). |
| Posterior basicranial width | Width of the skull at the external auditory meati taken from landmarks on the mid-superior rim of the tympanic bulla (TYM). |
| Anterior basicranial width | The average of the distances between the foramina ovale (AFO) on each side and a landmark at the intersection of the frontal suture and the orbital rim on each side (IOS). |
| Endocranial volume | The three-dimensional internal volume of the cranial vault (virtual endocast). |
| Neurocranial height | Distance from the mid-sphenoidal synchondrosis (MSS) to lambda (LAM). |
| Neurocranial length | Distance between lambda (LAM) and the midpoint of a line between a landmark at the junction between the frontal, parietal and temporal bones (FTP). |
| Neurocranial width | Width of the skull at the occipitomastoid suture (MST). In mice, this point most frequently represents the widest point of the neurocranium. |
| Facial centroid size | The centroid size of the configuration of all landmarks in the face. |
| Facial length | The average of two measures of facial length: upper facial length [the distance between the alveolar rim of the maxillary incisor (MSI) and nasion (NAS)] and palatal length [(the distance between the incisive foramen (AIF) and the posterior alveolar tubercle (PSA)]. |
| Facial Height | The distance between the midpoint of a line between the posterior alveolar tubercle (PSA) on each side and foramen caecum (FC). |
Abbreviations refer to landmarks listed in Table 2.
Allometry
To determine whether the difference in shape between adult Crf4 and C57BL/6J mice is due to allometry, we compared both groups to C57BL6/J mice homozygous for the ghrhr null mutations, also known as ‘little mice’. These mice are small and also on a C57BL6J background. The difference in craniofacial shape between them and the wild-type strain is thus presumably due to size-related shape variation or allometry. We compared three groups using multivariate regression of shape on centroid size. In this analysis, if the shape effects of the crf4 mutation are due to allometry, Crf4 mice should differ from C57BL/6J mice along the size–shape relationship defined by the comparison between the ghrhr null and C57BL/6J wild-type mice. Further, removing the allometric component of shape variation by obtaining the residuals of the multivariate regression of the Procrustes coordinates on log centroid size, the samples should all superimpose if they differ only in the shape characteristics that are associated with size. We also used Form Analysis to compare the groups (Mitteroecker et al. 2004). In this method, ln centroid size is added back into the data matrix before computation of the variance/covariance matrix and subsequent extraction of principal components. If the same simple reduction in overall growth that produces the shape effects seen in the ghrhr null mice explains the shape effects of the Crf4 mutation, then the Crf4 mice should fall along a shape–size continuum represented by the ghrhr null and C57BL/6J wild-type mice.
Developmental trajectory
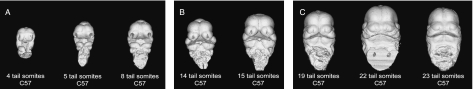
We compared the developmental trajectories of the Crf4 mutants and C57BL6/J wild-type mice to determine whether shape differences in the embryonic face might result from developmental delay rather than altered pattern of growth. We compared the relationship between centroid size and tail somite (TS) stage among the strains using analysis of covariance. To compare ontogenetic shape trajectories, we used regression of total shape variation on TS stage for the pooled sample. We also defined and compared three ontogenetic subsets (TS 4–10, TS 11–18, TS 19–31), which capture pre-fusion, fusion, post-fusion and late stages of facial formation (Trasler, 1968). These comparisons, although involving smaller samples, capture stage-specific differences between the strains during face formation and early outgrowth and have the additional advantage of avoiding the statistical issues created by the large amount of shape variation in the whole sample.
Results
Adult Crf4 cranial shape is distinct from and more variable than that of adult wildtype crania
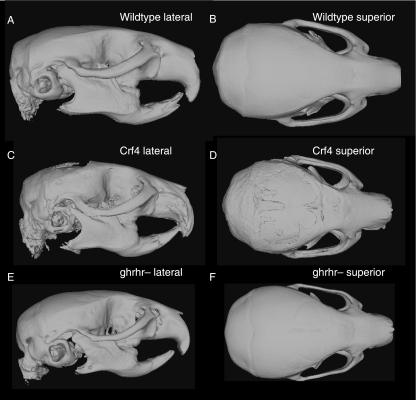
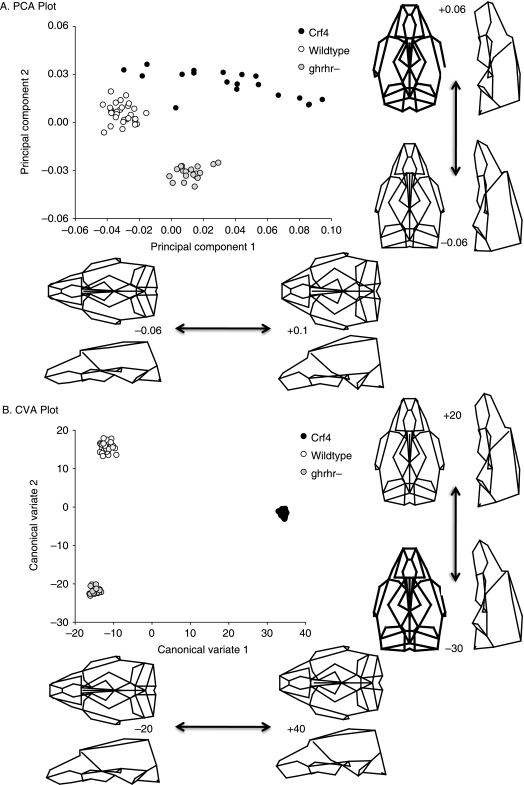
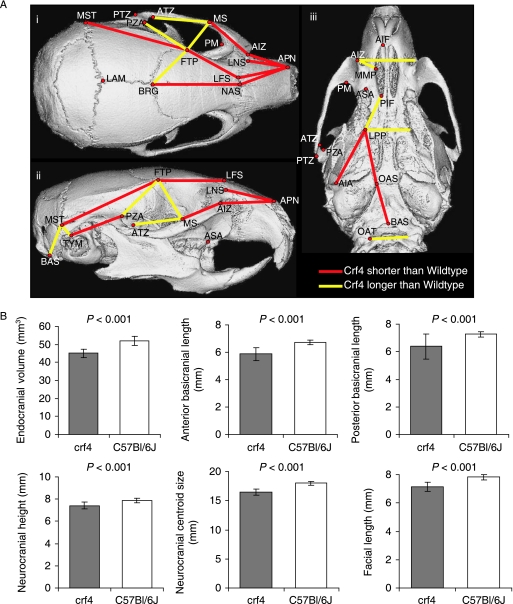
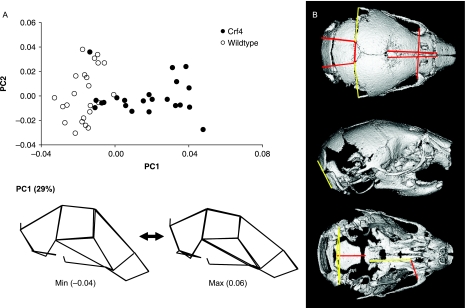
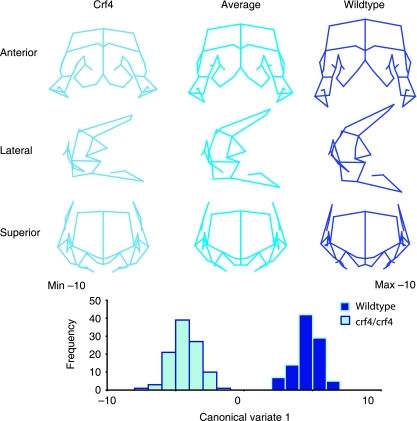
Comparisons of adult mean shape reveal large and significant differences in mean cranial shape between all three strains (Figs 4 and 5; Table 5). Principal components analysis reveals that the Crf4 and C57Bl/6J wild-type mice separate along PC1, which accounts for 40% of the variance in the sample. The C57BL/6J adult mice cluster more tightly along PC1 than do Crf4 adults (Fig. 5). The Crf4 group varies to a much greater extent along PC1 and grades into the C57BL/6J range. Along PC1, the face and basicranium are shorter, the neurocranium more globular, and the head relatively wider in Crf4 mice compared to the wild mice, although Crf4 individuals vary dramatically along this axis. In contrast, PC2 describes the difference between C57BL/6J and the Crf4 mice on the one hand and the ghrhr null mice on the other. The results of the PCA are supported by CVA, where the three strains are very well discriminated from one another and significantly different (P < 0.001) (Fig. 5b). The axis of shape variation described by CV1 agrees with that of PC1, where the adult Crf4 face and basicranium are shorter, the neurocranium taller, and entire head wider than in adult wild-type mice. These results are echoed by Euclidean matrix distance analysis: in the adult Crf4 mice, EDMA reveals a relatively shorter superior facial region and cranial base, a taller neurocranium, and wider head (Fig. 6a). All inter-landmark differences are significantly greater than 3%, with confidence intervals based on 1000 resamples (alpha level = 0.01).
Fig. 4.
Mean shapes of adult C57BL/6J wildtype (A,B), Crf4 mutant (C,D) and ghrhr-mutant (E,F) skulls in lateral and superior views.
Fig. 5.
(A) Scatterplot of principal component scores (PC1 and PC2) for adult Crf4, C57Bl/6J and ghrhr null mice along with wireframe deformations showing the shape variation along PC1 and PC2. (B) Scatterplot of CVA scores. In both graphs, the wireframe deformations are to scale (not magnified), showing the shape that corresponds to the extreme values of the graph.
Table 5.
Embryo cranial landmarks
| Number | Landmark description |
|---|---|
| 1 (R/L) | Primary choanae (or future site of primary choanae) |
| 2 (R/L) | Maximum of curvature of cerebral hemispheres as viewed from frontal aspect |
| 3 (R/L) | Maximum of superior-lateral curvature of laternal nasal prominence in frontal aspect |
| 4 (R/L) | Most lateral point (maximum of curvature) on lateral nasal prominence in frontal aspect |
| 5 (R/L) | Maximum of inferior-lateral curvature of lateral nasal prominence in frontal aspect (future nasolacrimal groove/duct) |
| 6 (R/L) | Most lateral point (maximum of curvature) on maxillary prominence in frontal aspect |
| 7 (R/L) | Most inferior-medial point of lateral nasal prominence |
| 8 (R/L) | Most inferior-lateral point of medial nasal prominence |
| 9 (R/L) | Most inferior point of nasal placode/pit (on medial nasal prominences if face is pre-fusion) |
| 10 (R/L) | Maximum of curvature of lateral nasal prominence along lateral edge of nasal pit |
| 11 (R/L) | Minimum of curvature of medial nasal prominence along medial edge of nasal pit |
| 12 (R/L) | Most superior point of nasal placode/pit (junction of medial and lateral nasal prominence) |
| 13 (R/L) | Maximum of superior-medial curvature of medial nasal prominence in frontal aspect |
| 14 (R/L) | Maximum of inferior-medial curvature of medial nasal prominence in frontal aspect |
| 15 (M) | Most inferior point of junction of the medial nasal prominences |
| 16 (M) | Most superior point of junction of the medial nasal prominences |
| 17 (M) | Midline point marking the eventual point of separation between medial nasal prominences and the cerebral hemisphere (future nasion, snout origin) |
| 18 (M) | Junction of the two cerebral hemispheres and the midbrain |
| 19 (M) | Maximum of curvature of the midbrain extending above the cerebral hemispheres in lateral aspect |
| 20 (R/L) | Maximum of curvature of medial nasal prominence in lateral aspect |
| 21 (R/L) | Maximum of curvature of lateral nasal prominence in lateral aspect |
| 22 (R/L) | Junction between medial and lateral nasal prominences and the cerebral hemispheres in lateral aspect |
| 23 (R/L) | Maximum of curvature of the cerebral hemispheres in lateral aspect |
| 24 (R/L) | Maximum of curvature and most posterior point of the cerebral hemispheres (or the most superior-anterior point of the midbrain flexure |
| 25 (R/L) | Most anterior point and point of greatest curvature of the hindbrain (most anterior point of the junction between the metencephalon and myelencephalon |
| 26 (R/L) | Most posterior point of the junction between the maxillary and mandibular prominences |
| 27 (R/L) | Maximum of the anterior-inferior curvature of the maxillary prominence (slightly anterior to the vibrissae in older embryos) |
| 28 (R/L) | Junction between anterior-superior point of primordial eye and the cerebral hemisphere and lateral nasal prominence |
| 29 (R/L) | Junction between posterior-superior point of primordial eye and the cerebral hemisphere |
| 30 (R/L) | Posterior-inferior maximum of eye |
| 31 (R/L) | Most posterior point of groove between primordial eye and maxillary prominence |
| 32 (R/L) | Junction between anterior-inferior point of primordial eye and the maxillary and lateral nasal prominence (future nasolacrimal groove/duct) |
| 33 (R/L) | Center point of primordial lens |
| 34 (R/L) | Most lateral points of the junction between the midbrain and hindbrain in dorsal aspect |
| 35 (R/L) | Most lateral points of the fourth ventricle |
| 36 (M) | Midline point of the junction between the midbrain and hindbrain |
| 37 (M) | Most superior point of the fourth ventricle |
Fig. 6.
(A) EDMA analysis results showing inter-landmark distances that differed by more than 3% between Crf4 and C57BL/6J mice. (B) Comparisons of key measurements showing those that differ most dramatically between the two strains.
Comparison of among-individual variance reveals a greater within-strain variance (V = 0.0032, ΔV = 0.0023) in the Crf4 mice than in C57BL/6J or ghrhr null mice (Table 6). This is consistent with the larger scatter of points for the Crf4 mice evident in the plot.
Table 6.
Within-group variance between strains: adults, neonates and embryos
| Crf4 variance | C57 variance | ΔV(difference between within-group variance) | P-value | |
|---|---|---|---|---|
| Standardized (on CS) | ||||
| Adult | 0.0021 | 0.0009 | 0.0013 | < 0.001 |
| Neonate | 0.0015 | 0.0016 | 0.0002 | ns |
| Standardized (on TS) | ||||
| Embryo – pooled | 0.0123 | 0.0088 | 0.0034 | < 0.001 |
Comparisons of cranial centroid size (two-way anovaby Genotype and Sex) reveal that cranial centroid size in the adult Crf4 mice is reduced by 10% compared to the adult wild-type mice (F = 144, n = 47, P < 0.001). Again, the extent to which cranial size is reduced varies among individuals, with the Crf4 size range overlapping the wild type range. Figure 6b shows comparisons for the key cranial measurements listed in Table 3. These results show that in the Crf4 mice, endocranial volume and basicranial length are most dramatically reduced (12–13%), and facial length (8%) and face size (7%) are also significantly reduced. Neurocranial height and neurocranial size are also significantly reduced in this mutant strain. For all of these measures, variances are higher in the Crf4 mice than in the wild-type.
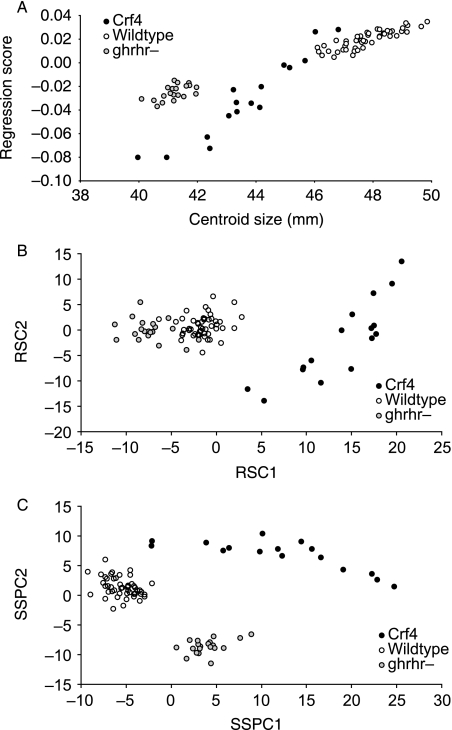
Shape differences between Crf4 and wild-type adults are not due to overall reduction in growth
To determine whether the shape difference between the crf4/crf4 mutants and C57BL/6J wild-type mice is due to the smaller heads of the mutants, we compared both these strains to the ghrhr null mice as described above. Regression of shape variation on centroid size shows that the ghrhr null and the Crf4 mice clearly differ in shape relative to size (Fig. 7a). Further, after removing the allometric component of shape variation by multivariate regression of shape on size, the Crf4 mice are clearly different in shape from the C57BL/6J and ghrhr null mice (Fig. 7b). This means that whereas the ghrhr null mice differ from C57BL/6J by allometric scaling, the Crf4 mice differ in ways differ from the expectation of allometric scaling. Finally, the form analysis (PCA including log size) also shows that all three groups differ significantly (Fig. 7c). If the shape differences between Crf4 and C57BL/6J wild-type mice are due simply to the effect of the mutation on cranial growth, then Crf4 cranial shape should be similar to ghrhr cranial shape at a comparable size. Clearly that is not the case as the Crf4 mice occupy a distinct region both of shape and of form space from the ghrhr null and wild-type mice. So we conclude that the shape effects of the crf4 mutation differ from those produced by a perturbation of the somatic-growth regulation axis.
Fig. 7.
(A) Scatterplot of PC1 and cranial centroid size (scale) for Crf4, C57Bl/6J and ghrhr null mice. (B) PCA of the residuals of multivariate regression of the Procrustes coordinates on log centroid size. Following Mitteroecker et al. (2004), we use the notation RSC (residual shape components). (C) Form analysis (PCA of Procrustes Coordinate data as well as centroid size) for all three groups. showing principal component scores for the first three components. Following Mitteroecker et al. (2004), we use the notation SSPC (Size–Shape principal components).
Wild-type and Crf4 neonates differ in mean shape
Neonatal mean shapes are significantly different between Crf4 and C57BL/6J mouse strains (F-score = 13.059, Procrustes distance = 0.042, P < 0.01). All statistics are significant. Whereas mean shape is genotype-specific, the amount of shape variance in Crf4 and C57BL/6J neonate skulls is indistinguishable. This contrasts with the greater degree of shape variance identified in Crf4 adult skulls. After a PCA, PC1 discriminates between strains, where about 29% of the total variance is described across PC1. Further, PC1 is weakly correlated with centroid size and describes skull flexure in terms of the shortening of the anterior and posterior neurocranium (Fig. 8a). CVA analysis shows the same pattern as PCA. EDMA corroborates results from the other methods: the shapes of Crf4 and C57BL/6J neonates are largely similar but can still be differentiated (Fig. 8b). At the shape difference magnitude of ±0.03, Crf4 neonates have greater neurocranium width and height at the caudal end, and shorter interparietal and occipital bone lengths (confidence intervals based on 1000 resampling iterations; alpha level of 0.05). Significant differences above and below ±0.02 show shortening of the anterior cranial vault and basicranium in the mutant mice.
Fig. 8.
(A) Plot of PC1 against PC2 for size-standardized Procrustes superimposed landmark data for the pooled neonatal sample. Below are wireframes showing shape change across PC1 within this sample. (B) EDMA results showing all inter-landmark distances that differ by more than 2%. Yellow lines mark distances in Crf4 neonates that are longer than C57 neonates. Red lines mark distances in Crf4 neonates that are shorter than C57 neonates.
Wild-type and Crf4 embryos differ in mean and variance for craniofacial shape and size during formation of the face
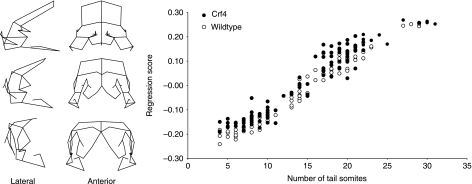
Principal components analysis of the unstandardized Procrustes coordinate data reveals, not surprisingly, that the vast majority of shape variation within both strains is ontogenetic. Figure 9 shows the regression of embryonic craniofacial shape on tail somite stage and centroid size for both groups as well as a wireframe deformation showing the shape variation along this regression. Note that the range of shape variation in this sample exceeds somewhat the range that can be analysed without distortion when the Procrustes coordinates are converted to partial warps. This issue is avoided in the ontogenetic subset comparisons below. For comparison of mean craniofacial shape, we standardized the embryo samples to TS16 by performing a pooled within-group regression for the Procrustes coordinate data on tail somite stage using genotype as the subgroup. Embryo size (CS) and developmental stage (TS) are strongly correlated (R2 = 0.959). The residuals of the TS stage regression were standardized to the TS16. Permutation tests for the Procrustes and Mahalanobis distances between groups reveal a significant difference between the two strains (10 000 permutations, P < 0.001) (Table 7). Similarly, Goodall's F-test reveal significant differences between the samples after standardization (Procrustes distance = 0.09, F = 46, df = 198, P < 0.001). Comparison of centroid size using ancova with TS as the covariate show that the crf4/crf4 embryos have slightly (2.5%) larger heads relative to TS stage (ancova, F = 4.7, df = 152, P < 0.05).
Fig. 9.
Multivariate pooled within-group multivariate regression of shape on tail somite stage for the embryonic sample. The wireframes show the variation that corresponds to the regression scores on the y-axis.
Table 7.
Mahalanobis and Procrustes distances among groups
| Mahalanobis distances among groups: | ||
| C57BL/6J WT | crf4/crf4 | |
| crf4/crf4 | 112.99 | |
| ghrhr–/– | 38.07 | 103.67 |
| Procrustes distances among groups: | ||
| Wild-type | crf4/crf4 | |
| crf4/crf4 | 0.073 | |
| ghrhr–/– | 0.055 | 0.062 |
All comparisons are significant using permutation test (P < 0.001).
Canonical variate analyses reveal that crf4/crf4 embryos differ from C57BL6/J embryos of the same TS stage in the following manner (Fig. 10). Overall, the growth of the brain is either reduced or delayed, whereas the development of the face is relatively advanced. The nasal prominences are relatively wider and formation of the primary palate is advanced relative to somite stage. Interestingly, the median nasal prominences show a greater degree of fusion at the midline in crf4/crf4 embryos relative to their size (Fig. 9). The maxillary prominences are not reduced in relative size.
Fig. 10.
Comparison of Crf4 and C57BL/6J embryos by canonical variate analysis. The samples are standardized to tail somite stage 16 by pooled within-group multivariate regression. The distribution of the canonical variate scores is shown below. The wireframes depict the variation along the canonical variate and are to scale with the histogram below.
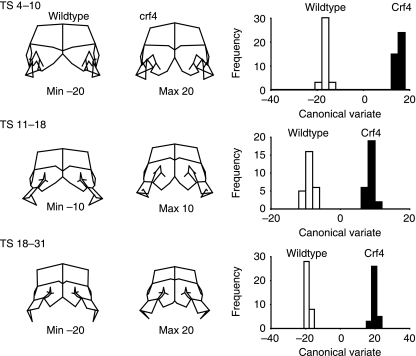
To determine more precisely the ontogenetic patterning of the effects of the crf4 mutation, we compared the two strains at the three ontogenetic stages described above (Fig. 3). Figure 11 shows the shape differences between the strains as determined by CVA of the ontogenetic subsamples. For all of these analyses, the within-stage sample was standardized to the mean TS stage using pooled within-group regression of the Procrustes coordinate data on TS stage. Significant differences in shape are evident at all three stages as determined by a permutation test for the Procrustes distance in MorphoJ (10 000 permutations, P < 0.001). At all three stages, the premature midline fusion of the medial nasal prominences is evident, as is their larger size relative to the other regions of the head.
Fig. 3.
Craniofacial development of C57Bl/6J embryos showing examples of ontogenetic subsets: (A) TS 4–10, (B) TS 11–18, (C) TS 19–25. Frontal view.
Fig. 11.
Canonical variates comparisons of Crf4 and C57BL/6J embryos for the three ontogenetic subgroups (Fig. 3). Within each ontogenetic group, shape is standardized to the median tail somite stage. The wireframes depict variation along the canonical variates and are to scale with the histograms to the right.
After standardization for TS stage using the pooled multivariate regression within subgroups, the among-individual variance in shape is significantly different between the strains (Table 6). For Crf4, the variance of the Procrustes distance from the mean was 0.0098 compared to 0.0073 for the C57BL/6J wild-type sample (Delta variance permutation test in Simple3D, 1600 permutations, P < 0.001).
Discussion
This study aims to quantitatively characterize the craniofacial skeletal morphology and morphogenesis of the Crf4 mouse mutant for which an abnormal craniofacial phenotype was suspected. The short-faced phenotype reported by the Baylor mouse mutagenesis screen suggested that this mutant might be a good model for studying the developmental determinants for variation in facial length in both evolutionary and biomedical contexts. Work using other model organisms such as birds has shown that misexpression of bone morphogenetic protein (bmp) 4 can dramatically change beak length and width in Darwin's finches (Abzhanov et al. 2004), and affect beak versus bill patterning in chicks and ducks, respectively (Wu et al. 2004). Experimental study of cichlid fish craniofacial phenotype, which varies considerably – particularly jaw length – also linked bmp4 with variation in facial morphology (Terai et al. 2002), suggesting that this protein and its pathways are important to vertebrate facial patterning in general. However, to date very little experimental embryology has been done to study facial length using a mammalian model. Our morphometric study using mouse aimed to precisely characterize the phenotype and test the hypothesis that between-strain (Crf4 versus C57BL/6J wild-type) morphological variation in adults is also manifest in embryos and/or neonates. Our results show clearly that during the formation of the face, as neonates, and as adults, the Crf4 strain is significantly different to the C57BL/6J wild-type strain from which it is derived.
During embryonic face formation, the Crf4 mutant mice differ from C57BL/6J wild-type mice in that the forebrain is relatively smaller, head size is slightly larger, and narrowing of nasal pits and the medial nasal region is advanced relative to TS stage. The facial prominences appear larger and primary palate formation is more advanced relative to stage in the Crf4 embryos. In neonates, the anterior neurocranium, basicranium and upper face are shorter in absolute terms as determined by EDMA, with the result that the neurocranium is higher and more globular in the shape analysis of the Procrustes superimposed coordinate data. In adults, the basicranium and face are clearly shorter and the neurocranium is smaller in all dimensions. Most dramatically, though, the brain is reduced in size.
Form analysis and regression of size against total shape variation show that strain-specific differences between adult Crf4 and ghrhr–/– mice and C57BL/6J wild-type mice are different (Fig. 7). The shape differences produced by the Crf4 mutation are thus different from those that are produced by a mutation that affects the rate and amount of overall somatic growth by perturbing the growth hormone axis, The shape differences seen in Crf4 mice, however, are still clearly correlated with size. Without the addition of the ‘little mouse’ sample, the shape differences seen in Fig. 7 could easily be interpreted as allometric. However, as the nature of the shape changes is different from those produced by a reduction in somatic growth, we infer that the correlation between size and shape evident in the shape comparison of Crf4 and C57Bl/6J wild-type mice is due to the correlated effects of the mutation on size and shape and not to a more generalized effect on cranial growth. In other words, as the mutation affects the size of the brain and the face, the reduction in size of those cranial components will produce a reduction in cranial size which would superficially present as allometric or size-related shape variation. Additional support for this view comes from the fact that whereas we observed a significant difference in head size between strains, body weight was statistically indistinct between Crf4 mutant and wild-type.
The Crf4 mutant thus shows the puzzling combination of a larger and more advanced face at the embryonic stage and a smaller face at later ontogenetic stages. Several possible explanations for this anatomical pattern are feasible. One is that the effects at the embryonic and later ontogenetic stages are due to separate genetic effects and thus are not related. It is possible, for instance, that the 24CM inversion on chromosome 11 which the Crf4 mice share with other strains in the Baylor Mutant Mouse project (Kile et al. 2003) carries a genetic factor or factors that influence the rate of growth of the embryonic face independently of the crf4 mutation. A search for mutations within this region with craniofacial phenotypes yields 27 hits, of which 13 involve known genes (http://www.informatics.jax.org) and there are other genes in this region that may also have functions related to craniofacial development. This possibility will be tested by work in progress that involves comparing embryos from the other Crf mutant strains to C57BL/6J wild-type embryos as done here.
A second possibility is that the mutation causes different and opposing effects on facial growth at different points in development. Work in progress on this strain will determine whether the Crf4 mice differ from the wild-type in cell proliferation, cell density and the timing of neural crest migration. This work will reveal whether the Crf4 embryonic faces are more advanced due to a direct influence on the timing and rate of facial prominence outgrowth. There are certainly many examples of mutations that affect stages of development in different ways, although direct opposite effects on facial growth at different ontogenetic stages would certainly be highly unusual.
A third explanation is that these seemingly contradictory effects on facial growth are secondary or pleiotropic consequences of a consistent and common underlying effect on something else. Given the reduced forebrain size in the embryos, reduced neurocranial dimensions in neonates and the clear reduction in brain size in adults, the common factor likely involves brain development. Interactions between the growth of the brain and the face during development are known at both the structural and molecular levels (Wang & Diewert, 1992; Schneider et al. 2001; Marcucio et al. 2005). Neural crest, which forms the mesenchyme of the facial prominences, is now known to contribute to forebrain development as well (Le Douarin et al. 2007). Shh signaling in the forebrain appears to regulate or at least influence Shh expression in the developing face (Marcucio et al. 2005), indicating that direct molecular interactions between the development of the brain and face exist. These connections, however, would predict a positive correlation between the growth of the brain and face. That is the opposite of the pattern observed in Crf4 mutant.
In structural terms, the forebrain is the foundation on which the facial prominences grow forward. A smaller and, especially, narrower forebrain would mean that the facial prominences would have a smaller gap to close to form the primary palate. In other words, if the rate of growth of the prominences remains the same, a smaller brain should produce a more advanced face, which is the embryological phenotype seen in the Crf4 mutant. Increased forebrain width in relation to facial prominence outgrowth is implicated in clefting of the primary palate in A strain mice (Wang & Diewert, 1992) and correlations between brain morphology and CL/P are observed in humans as well (Nopoulos et al. 2000). If our hypothesis is correct, the more advanced face associated with a smaller brain in Crf4 mice is additional evidence for the relationship between brain development and face formation.
It is less clear why a smaller brain at later stages should produce a shorter face. The postnatal growth of the face is thought to be independent of brain size (De Beer, 1937;Moore & Lavelle, 1974). In adult mice, neurocranial size is correlated with facial width but not with length (Hallgrímsson et al. 2007). In humans, neurocranial breadth and facial breadth are clearly correlated but there is only a weak relationship between neurocranial width and facial length (Lieberman et al. 2000). There is conceivably some connection between the earlier formation of the primary palate and the smaller face at later stages. A more likely explanation, however, is that the post-embryonic reduction of the face and the reduction in brain size are pleiotropic effects of the crf4 mutation, whereas the more advanced and prognathic face size at the embryonic stage is an epigenetic (mechanical tissue interaction) effect of the reduction in brain size at that stage. Further work on developmental parameters is necessary to sort out these competing explanations for the Crf4 craniofacial phenotype.
It is clear from the morphometric analyses of three distinct developmental stages of the Crf4 mutant that the phenotype is much more complex, and possibly substantially different, than that which was reported in the initial phenotype screen. Described initially as a ‘short snout’ phenotype, the Crf4 mutant turns out to be more aptly characterized in terms of reduced brain size and basicranial dimensions. This finding raises a cautionary note about our systematic reliance on phenotype descriptions in mouse informatics resources and begs the question of whether more systematic high-throughput morphometric methods should be employed at the phenotypic screening level (Kristensen et al. in Press;Parsons et al. 2008).
A secondary finding of this study is that the Crf4 mice consistently showed higher phenotypic variances at each developmental stage. This finding is not unexpected as mutants are commonly more variable than the wild-type (Scharloo, 1964, 1991;de Visser et al. 2003;Hallgrímsson et al. 2006). This result suggests that the Crf4 mutation is incompletely penetrant and/or that it results in reduced canalization of craniofacial form.
Conclusion
This study demonstrates that the Crf4 phenotype is expressed at the embryonic stage and that it is complex, involving reduction in brain size and basicranial dimensions. Face size is reduced postnatally, but face formation and outgrowth is actually advanced during embryonic face formation. We suggest that the reduction in brain size is a primary phenotypic effect of the Crf4 mutation. If that hypothesis is correct, then the Crf4 mouse will be a valuable model for studying the interactions between brain growth and facial morphology. Comparisons of the timing of telencephalon growth and development, neural crest migration, differentiation, and proliferation via in situ hybridization, microarray and immunohistochemical analyses of cellular dynamics as well as experimental manipulations are likely to yield insights into the mechanistic basis for the interactions between the brain and the face. This is important as these interactions are a largely unexplored source of variation in craniofacial shape, relevant to both dysmorphologies and the developmental basis for the evolution of facial form. Given the variation in both brain size and facial length in primates – including modern humans – this model may of particular relevance to human evolution and development.
Acknowledgments
We acknowledge the Baylor Mouse Mutant Resource for generation of the Crf4 mouse. We thank Wei Liu for scanning and landmarking of adult mice. We are grateful to funding from National Science and Engineering grant 238992-06, Canadian Foundation for Innovation grant #3923, Alberta Innovation and Science grant #URSI-01-103-RI, Canadian Institutes of Health Research grant #131625, Genome Canada and Genome Alberta grant to B.H.
References
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Bookstein FL. Morphometric Tools for Landmark Data. Cambridge: Cambridge University Press; 1991. [Google Scholar]
- Cole TM., III WinEDMA (Windows Based Software for Euclidean Matrix Analysis) 2003. Version 1.01 beta. Copyright (c) 2003. Release Date: May 5, 2003.
- Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: insights from avian studies. J Anat. 2005;207:447–459. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer G. The Development of the Vertebrate Skull. Oxford: Clarendon Press; 1937. [Google Scholar]
- Diewert VM. Growth movements during prenatal development of human facial morphology. Prog Clin Biol Res. 1985;187:57–66. [PubMed] [Google Scholar]
- Diewert VM, Lozanoff S. A morphometric analysis of human embryonic craniofacial growth in the median plane during primary palate formation. J Craniofac Genet Dev Biol. 1993a;13:147–161. [PubMed] [Google Scholar]
- Diewert VM, Lozanoff S. Growth and morphogenesis of the human embryonic midface during primary palate formation analyzed in frontal sections. J Craniofac Genet Dev Biol. 1993b;13:162–183. [PubMed] [Google Scholar]
- Diewert VM, Lozanoff S. Animal models of facial clefting: experimental, congenital, and transgenic. In: Mooney MM, Siegel MI, editors. Understanding Craniofacial Anomalies: Etiopathogenesis of Craniosynostoses and Facial Clefting. New York: Wiley-Liss; 2002. [Google Scholar]
- Diewert VM, Shiota K. Morphological observations in normal primary palate and cleft lip embryos in the Kyoto collection. Teratology. 1990;41:663–677. doi: 10.1002/tera.1420410603. [DOI] [PubMed] [Google Scholar]
- Diewert VM, Wang KY. Recent advances in primary palate and midface morphogenesis research. Crit Rev Oral Biol Med. 1992;4:111–130. doi: 10.1177/10454411920040010201. [DOI] [PubMed] [Google Scholar]
- Dryden IL, Mardia KV. Statistical Shape Analysis. Chichester: John Wiley & Sons; 1998. [Google Scholar]
- Enlow DH. Handbook of Facial Growth. 2nd edn. Philadelphia: WB Saunders and Company; 1982. [Google Scholar]
- Enlow DH. Facial Growth. Philadelphia: WB Saunders and Company; 1990. [Google Scholar]
- Goodall C. Procrustes methods in the statistical analysis of shape. J R Stat Soc B. 1991;53:285–339. [Google Scholar]
- Hallgrímsson B, Willmore K, Hall BK. Canalization, developmental stability, and morphological integration in primate limbs. Yearb Phys Anthropol. 2002;45:131–158. doi: 10.1002/ajpa.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrímsson B, Dorval CJ, Zelditch ML, German RZ. Craniofacial variability and morphological integration in mice susceptible to cleft lip and palate. J Anat. 2004;205:501–517. doi: 10.1111/j.0021-8782.2004.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrímsson B, Brown JJY, Ford-Hutchinson AF, Sheets HD, Zelditch ML, Jirik FR. The brachymorph mouse and the developmental-genetic basis for canalization and morphological integration. Evol Dev. 2006;8:61–73. doi: 10.1111/j.1525-142X.2006.05075.x. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Lieberman DE, Liu W, Ford-Hutchinson AF, Jirik FR. Epigenetic interactions and the structure of phenotypic variation in the cranium. Evol Dev. 2007;9:76–91. doi: 10.1111/j.1525-142X.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- Kile BT, Hentges KE, Clark AT, et al. Functional genetic analysis of mouse chromosome 11. Nature. 2003;425:81–86. doi: 10.1038/nature01865. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. UK: University of Manchester; 2008. MorphoJ. Faculty of Life Sciences. http://www.flywings.org.uk/MorphoJ_page.htm. [Google Scholar]
- Klingenberg CP, Monteiro LR. Distances and directions in multidimensional shape spaces: implications for morphometric applications. System Biol. 2005;54:678–688. doi: 10.1080/10635150590947258. [DOI] [PubMed] [Google Scholar]
- Kristensen E, Parsons TE, Gire J, Hallgrímsson B, Boyd S. A novel high-throughput morphological method for phenotypic analysis. IEEE Computer Graphics and Applications. doi: 10.1109/TBME.2008.923106. In Press. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Brito JM, Creuzet S. Role of the neural crest in face and brain development. Brain Res Rev. 2007;55:237–247. doi: 10.1016/j.brainresrev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Lele S. Euclidean Distance Matrix Analysis of landmark data: Estimation of mean form and mean form difference. Math Geol. 1993;25:573–602. [Google Scholar]
- Lele S, Cole TM., III A new test for shape differences when variance-covariance matrices are unequal. J Hum Evol. 1996;31:193–212. [Google Scholar]
- Lele S, Richtsmeier JT. Euclidean distance matrix analysis: a coordinate-free approach for comparing biological shapes using landmark data. Am J Phys Anthrop. 1991;86:415–427. doi: 10.1002/ajpa.1330860307. [DOI] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT. An Invariant Approach to the Statistical Analysis of Shapes. Boca Raton: Chapman & Hall; 2001. [Google Scholar]
- Lieberman DE, Pearson OM, Mowbray KM. Basicranial influence on overall cranial shape. J Hum Evol. 2000;38:291–315. doi: 10.1006/jhev.1999.0335. [DOI] [PubMed] [Google Scholar]
- Lieberman DE, McBratney BM, Krovitz G. The evolution and development of cranial form in Homo Sapiens. Proc Natl Acad Sci U S A. 2002;99:1134–1139. doi: 10.1073/pnas.022440799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Mitteroecker P, Gunz P, Bernhard M, Schaefer K, Bookstein FL. Comparison of cranial ontogenetic trajectories among great apes and humans. J Hum Evol. 2004;46:679–697. doi: 10.1016/j.jhevol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Moore WJ, Lavelle CLB. Growth of the Facial Skeleton in the Hominoidea. London: Academic Press; 1974. [Google Scholar]
- Nopoulos P, Berg S, Canady J, Richman L, Van Demark D, Andreasen NC. Abnormal brain morphology in patients with isolated cleft lip, cleft palate, or both: a preliminary analysis. Cleft Palate Craniofac J. 2000;37:441–446. doi: 10.1597/1545-1569_2000_037_0441_abmipw_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- O’Higgins P, Jones N. University College London: 1998. Morphologika. [Google Scholar]
- Parsons TE, Kristensen E, Hornung L, et al. Phenotypic variability and craniofacial dysmorphology: increased shape variance in a mouse model for cleft lip. Anat. 2008;212(2):135–143. doi: 10.1111/j.1469-7580.2007.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralphs JR. Chondrogenesis and myogenesis in micromass cultures of mesenchyme from mouse facial primordia. In Vitro Cell Dev Biol. 1992;28A:369–372. doi: 10.1007/BF02877061. [DOI] [PubMed] [Google Scholar]
- Ramirez-Yanez GO, Smid JR, Young WG, Waters MJ. Influence of growth hormone on the craniofacial complex of transgenic mice. Eur J Orthod. 2005;27:494–500. doi: 10.1093/ejo/cji028. [DOI] [PubMed] [Google Scholar]
- Scharloo W. Mutant expression and canalization. Nature. 1964;203:1095–1096. doi: 10.1038/2031095b0. [DOI] [PubMed] [Google Scholar]
- Scharloo W. Canalization: Genetic and developmental aspects. Annu Rev Ecol Syst. 1991;22:65–93. [Google Scholar]
- Schneider RA, Hu D, Rubenstein JL, Maden M, Helms JA. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development. 2001;128:2755–2767. doi: 10.1242/dev.128.14.2755. [DOI] [PubMed] [Google Scholar]
- Sears KE, Goswami A, Flynn JJ, Niswander LA. The correlated evolution of Runx2 tandem repeats, transcriptional activity, and facial length in carnivora. Evol Dev. 2007;9:555–565. doi: 10.1111/j.1525-142X.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- Sheets HD. 2004a. IMP Simple3D.
- Sheets HD. 2004b. IMP ThreeDStand6.
- Slice DE. NY: Stony Brook; 2005. Morpheus. [Google Scholar]
- Slice DE. Modern morphometrics. In: Slice DE, editor. Modern Morphometrics in Physical Anthropology. New York: Kluwer; 2005. pp. 1–45. [Google Scholar]
- Terai Y, Morikawa N, Okada N. The evolution of the pro-domain of bone morphogenetic protein 4 (Bmp4) in an explosively speciated lineage of East African cichlid fishes. Mol Biol Evol. 2002;19:1628–1632. doi: 10.1093/oxfordjournals.molbev.a004225. [DOI] [PubMed] [Google Scholar]
- Trasler DG. Pathogenesis of cleft lip and its relation to embryonic face shape in A-J and C57BL mice. Teratology. 1968;1:33–49. doi: 10.1002/tera.1420010106. [DOI] [PubMed] [Google Scholar]
- Valeri CJ, Cole TM, 3rd, Lele S, Richtsmeier JT. Capturing data from three-dimensional surfaces using fuzzy landmarks. Am J Phys Anthropol. 1998;107:113–124. doi: 10.1002/(SICI)1096-8644(199809)107:1<113::AID-AJPA9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- de Visser JA, Hermisson J, Wagner GP, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Wang K-Y, Diewert VM. A morphometric analysis of craniofacial growth in cleft lip and noncleft mice. J Craniofac Genet Dev Biol. 1992;12:141–154. [PubMed] [Google Scholar]
- Willmore KE, Leamy L, Hallgrimsson B. Effects of developmental and functional interactions on mouse cranial variability through late ontogeny. Evol Dev. 2006a;8:550–567. doi: 10.1111/j.1525-142X.2006.00127.x. [DOI] [PubMed] [Google Scholar]
- Willmore KE, Zelditch ML, Young N, Ah-Seng A, Lozanoff S, Hallgrimsson B. Canalization and developmental stability in the brachyrrhine mouse. J Anat. 2006b;208:361–372. doi: 10.1111/j.1469-7580.2006.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Jiang TX, Suksaweang S, Widelitz RB, Chuong CM. Molecular shaping of the beak. Science. 2004;305:1465–1466. doi: 10.1126/science.1098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Wat S, Diewert VM, Browder LW, Hallgrímsson B. Comparative morphometrics of embryonic facial morphogenesis: implications for cleft-lip etiology. Anat Rec (Hoboken) 2007;290:123–139. doi: 10.1002/ar.20415. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Swiderski HD, Sheets D, Fink WL. Geometric Morphometrics for Biologists: A Primer. New York: Academic Press; 2004a. [Google Scholar]
- Zelditch ML, Lundrigan BL, Garland T. Developmental regulation of skull morphology. Evol Dev. 2004b;6:194–206. doi: 10.1111/j.1525-142X.2004.04025.x. I. Ontogenetic dynamics of variance. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Swiderski HD, Sheets D, Fink WL. Geometric Morphometrics for Biologists: A Primer. New York: Academic Press; 2004c. [Google Scholar]