Abstract
Plasma lipoprotein(a) (Lp[a]) level is an independent risk factor of cardiovascular disease that is under strong genetic control. We conducted a genome-wide association study of plasma Lp(a) in 386 members of a founder population that adheres to a communal lifestyle, proscribes cigarette smoking, and prepares and eats meals communally. We identified associations with 77 single nucleotide polymorphisms (SNPs) spanning 12.5 Mb on chromosome 6q26-q27 that met criteria for genome-wide significance (P ≤ 1.3 × 10−7) and were within or flanking nine genes, including LPA. We show that variation in at least six genes in addition to LPA are significantly associated with Lp(a) levels independent of each other and of the kringle IV repeat polymorphism in the LPA gene. One novel SNP in intron 37 of the LPA gene was also associated with Lp(a) levels and carotid artery disease number in unrelated Caucasians (P = 7.3 × 10−12 and 0.024, respectively), also independent of kringle IV number. This study suggests a complex genetic architecture of Lp(a) levels that may involve multiple loci on chromosome 6q26-q27.
Keywords: genome-wide association study, LPA, carotid artery disease, kringle IV
Lipoprotein (a) [Lp(a)] is recognized as an independent risk factor for atherosclerotic cardiovascular disease (1, 2). The mechanisms underlying this pathogenesis are poorly understood, although proatherogenic, prothrombotic, and inflammatory pathways contribute. Moreover, plasma Lp(a) levels are not responsive to statins and other cholesterol-lowering drugs, except for niacin, for which the long-term efficacy and safety is not yet established (3). Lp(a) is produced in the liver (4) and circulates in the plasma as an LDL particle having as a protein moiety apolipoprotein(a) [apo(a)], encoded by the LPA gene, linked by a disulfide bond to an apolipoprotein B-100 particle, on a 1:1 molecular basis (5). While apolipoprotein B-100 remains relatively constant in size, apo(a) varies in size due to polymorphism in the number of tandemly repeated kringle IV type 2 domains encoded by sequences in exons 1 and 2 of the LPA gene (6).
The human LPA gene arose as a duplication of the PLG gene in the primate lineage and retains 80% sequence identity to PLG (7), which has only a single kringle IV structure. The number of kringle IV repeats in Lp(a) is under genetic control and inversely correlates with plasma levels of Lp(a), likely as result of the lower secretion rate in hepatocytes of apo(a) isoforms with larger numbers of kringle IV repeats (8, 9). The LPA locus accounts for 70–90% of the variability in Lp(a) levels in worldwide populations, with the kringle IV size polymorphism accounting for approximately half of this effect (10–15). Polymorphisms in LPA other than the kringle IV repeats have been associated with circulating levels of Lp(a) (16, 17), but the known variants in LPA do not account for the remaining heritability of this trait, indicating that other, as yet unidentified, variation at this locus influences Lp(a) levels and the corresponding risk for cardiovascular disease.
To more fully dissect the genetic architecture of Lp(a) cholesterol, we measured Lp(a) protein in plasma after an overnight fast from 386 Hutterites (18, 19) (Table 1), as previously described (20, 21). The heritability of plasma Lp(a) protein levels is high in the Hutterites (H2 = 0.77; SEM 0.08) and similar to estimates in other populations (22). The subjects in our studies live on communal farms in South Dakota and are related to each other through multiple lines of descent in a 3,028-person, 13-generation pedigree with 62 founders (22). The small number of founding genomes reduces genetic heterogeneity, whereas their communal lifestyle ensures that nongenetic factors are remarkably uniform between individuals. In particular, all food is prepared and eaten in a communal kitchen, smoking is prohibited, and other lifestyle factors that could influence disease risk vary little between individuals (18, 20).
TABLE 1.
Characteristics of the Hutterite sample
| Males |
Females |
|||
|---|---|---|---|---|
| 15–40 yrs | ≥40 years | 15–40 years | ≥40 years | |
| Sample size | 109 | 66 | 133 | 78 |
| Mean age (Range) | 26 (15–39) | 54 (40–89) | 26 (15–39) | 52 (40–83) |
| Mean BMI (kg/m2) | 25.47 (5.06) | 30.37 (5.33) | 23.88 (4.85) | 29.50 (5.41) |
| Mean SBP (mmHg) | 126 (10.95) | 137 (14.88) | 113 (10.27) | 127 (17.48) |
| Mean DBP (mmHg) | 78 (8.30) | 84 (9.91) | 71 (7.16) | 78 (8.53) |
| Mean Lp(a) protein (mg/dl) | 2.41 (3.32) | 2.79 (3.48) | 3.24 (3.88) | 3.18 (4.09) |
| Mean Lp(a)-cholesterol (mg/dl) | 3.14 (4.32) | 3.62 (4.53) | 4.22 (5.04) | 4.14 (5.32) |
| Mean LDL-cholesterol (mg/dl) | 128 (47.22) | 144 (28.11) | 117 (32.03) | 149 (36.07) |
| Mean true LDL (mg/dl) | 125 (47.48) | 141 (27.70) | 113 (32.05) | 145 (36.60) |
| Mean HDL-cholesterol (mg/dl) | 46 (13.11) | 39 (10.86) | 52 (13.61) | 52 (13.71) |
| Mean total cholesterol (mg/dl) | 199 (48.03) | 217 (35.59) | 189 (35.87) | 231 (39.15) |
| Mean non-HDL-cholesterol (mg/dl) | 153 (49.73) | 178 (35.84) | 136 (36.53) | 179 (39.30) |
| Mean triglycerides (mg/dl) | 130 (87.54) | 194 (126.33) | 101 (63.28) | 153 (72.68) |
| No. with type 2 diabetes (No. on medications) | 0 | 17 (12) | 0 | 11 (1) |
| No. with hypertension (No. on medication) | 2 (2) | 20 (18) | 1 (1) | 23 (21) |
Lp(a) is expressed in terms of Lp(a) protein; Lp(a) cholesterol was calculated as Lp(a) protein multiplied by 1.3. True LDL does not include Lp(a) cholesterol. Twenty-six individuals did not have LDL-cholesterol measurements; one did not have blood pressure measurements, and seven did not have BMI values. Means (±SD) shown for each trait.
According to National Cholesterol Education Program guidelines (40) the normal ranges for each phenotype are as follows: Lp(a) (0.1–6.5 mg/dl), LDL-cholesterol (60–129 mg/dl), HDL-cholesterol (40–80 mg/dl), total cholesterol (120–199 mg/dl), triglycerides (30–149 mg/dl), and BMI (<25 kg/m2).
Here, we identify variation in at least six genes on chromosome 6q26-q27, in addition to the LPA gene, that significantly influence plasma Lp(a) levels in the Hutterites independent of the kringle IV size polymorphism and show that a novel intronic single nucleotide polymorphism (SNP) in LPA is associated with carotid artery disease in unrelated Caucasian cases and controls.
METHODS
The Hutterites of South Dakota
The Hutterites in our mapping studies have been described previously (20, 23). This study focused on 386 Hutterites living in nine South Dakota (Schmiedeleut) colonies with plasma Lp(a) measured (Table 1), as previously reported (21). All subjects gave written informed consent, and the project was approved by the University of Chicago Institutional Review Board.
Genotyping and quality control methods
A total of 357 (92.5%) individuals with plasma Lp(a) measurements were genotyped with the Affymetrix GeneChip® 500k Mapping Array. Whole-genome SNP genotyping was performed in the Hutterite samples using both the early access and commercial Affymetrix GeneChip 500k Mapping Array at the University of Chicago, as described (24). A set of 421,374 autosomal SNPs were present on both sets of chips. Another 1,423 nonsynonymous SNPs were genotyped at the NHLBI Resequencing and Genotyping Service (Johns Hopkins University) using a custom 1,536 SNP oligo pool and BeadArray method, as previously described (25). In the combined set of SNPs, 131,049 were not further studied because either they were monomorphic (n = 52,732) or had minor allele frequencies <5% (n = 58,152) in the Hutterites. The remaining 310,490 SNPs were subjected to quality control checks. An additional 20,165 SNPs were excluded because either they had call rates <90% (n = 3,614), they deviated from Hardy-Weinberg expectations at P < 0.001 (correcting for the Hutterite inbreeding and population structure) (n = 5,082), or because they generated ≥5 Mendelian errors (n = 11,469), yielding a set of 290,325 markers. Lastly, we included two SNPs in the LPA gene (rs1853021 and rs1800769) that were previously genotyped in our lab using a PCR and immobilized probe assay, as described by Cheng et al. (26). The final marker set of 290,327 SNPs had a median intermaker spacing of 4.3 kb (range 17–22.97 kb).
Associated SNPs were genotyped by TaqMan (rs9384296, rs6917698, rs9364496, rs8191829, rs6919346, rs14224, and rs2022991) or Illumina (rs4252125) in the CLEAR replication cohort. Associated SNP rs7745725 in SYNE1 was not genotyped in this sample.
Association testing in the Hutterites
The natural log of plasma Lp(a) protein was used for the heritability and association studies (20). The heritability of plasma Lp(a) was estimated using a variance component maximum likelihood method (27). At each SNP, we used the general two-allele model test of association in the entire pedigree, keeping all inbreeding loops intact, as described (28). SNP-specific P values were determined based on Gaussian theory (28); genome-wide P values were determined by a Monte Carlo permutation-based test that preserves the covariance structure due to relatedness of individuals and assesses significance in the presence of multiple, dependent tests while guarding against deviations from normality in the data. We used 100 permutations to generate the empirical distribution of P values and considered a P value to be genome-wide significant if it was equal to or smaller than the 5% quantile of the permutation-based empirical distribution of the global minimum P value.
To estimate effect size of associated alleles in the Hutterites, we performed a generalized linear regression of the transformed phenotype values on the covariates, using the estimated covariance matrix [obtained as described in reference (28)] as weights in the analysis. This was performed twice for each associated allele, once under the null hypothesis, with only age and sex as covariates, and once under the alternative hypothesis, with genotype data included as an additional covariate. To estimate the percentage of variance explained by an allele, we calculated the residual sum of squares (RSS) for each regression and used the equation (RSSnull − RSSalt)/RSSnull.
Replication Studies in the CLEAR cohort
The replication sample consisted of 1,054 unrelated subjects from the CLEAR cohort (29). This sample included 306 subjects with >80% stenosis of one or both internal carotid arteries on duplex ultrasound or prior endarterectomy, 214 subjects with between 15 and 79% stenosis, and 534 age-matched controls with <15% internal carotid artery stenosis bilaterally and no known coronary or peripheral vascular disease. Lp(a) concentration was measured by a direct binding double monoclonal antibody-based enzyme-linked immunoassay developed at the Northwest Lipid Research Center (30). The cohort is described in additional detail elsewhere (29), and characteristics of this cohort are shown in Table 4. All subjects gave written informed consent, and the project was approved by the University of Washington, Virginia Mason Medical Center, and Puget Sound Veterans Affairs Heath-care System Institutional Review Boards.
TABLE 4.
Characteristics of the CLEAR case and control samples
| All | Cases | Controls | P Value | |
|---|---|---|---|---|
| n | 1,053 | 493 | 560 | |
| Mean age (range) | 67.0 (31–92) | 70.0 (46–89) | 64.3 (31–92) | 9.60 × 10−25 |
| Mean BMI | 28.4 (4.9) | 27.8 (4.8) | 29 (4.9) | 0.0004 |
| Mean total cholesterol (mg/dl) | 189.2 (37.6) | 182.6 (38.0) | 194.8 (36.3) | 3.23 × 10−08* |
| Mean LDL-cholesterol (mg/dl) | 111.3 (31.7) | 105.6 (31.3) | 116.5 (31.3) | 1.65 × 10−07* |
| Mean LDL-B100 (mg/dl) | 64.7 (18.5) | 60.9 (17.9) | 68.2 (18.3) | 1.42 × 10−09* |
| Mean HDL-cholesterol (mg/dl) | 46.3 (14.6) | 43.3 (12.9) | 48.9 (15.5) | 2.83 × 10−11* |
| Mean APOAI (mg/dl) | 136.5 (25.5) | 130.8 (24.2) | 141.3 (25.8) | 6.14 × 10−12* |
| Mean triglycerides (mg/dl) | 149.0 (108.8) | 157.3 (103.8) | 142.0 (112.6) | 0.0003* |
| Mean VLDL-cholesterol (mg/dl) | 29.5 (20.3) | 31.3 (20.1) | 27.8 (20.3) | 0.0002* |
| Mean Lp(a) (nmole/L)a | 65.2 (86.4) | 89.0 (105.5) | 45.7 (60.0) | 5.00 × 10−10* |
| Percentage on lipid meds | 431 | 312 | 119 | 1.66 × 10−45 |
| Percentage of current smokers | 192 | 139 | 53 | 2.51 × 10−15 |
| Percentage diabetic | 146 | 97 | 49 | 2.23 × 10−07 |
| Percentage on hypertensives | 633 | 406 | 227 | 1.75 × 10−45 |
Means (±SD) are show for each trait. P value for comparison between cases and controls using t-test or χ2 test; *comparison performed on log-transformed values. All are Caucasian males.
Approximately equivalent to 5.2 (6.9) mg/dl (All), 7.1 (8.4) mg/dl (Cases), and 3.6 (4.8) mg/dl (Controls).
Subjects from the CLEAR study were genotyped for seven of the eight SNPs associated with Lp(a) in Hutterites using either Illumina or Taqman methods. Associations between Lp(a) levels and each SNP genotype were tested using multivariate linear regression, assuming an additive model and including body mass index (BMI), censored age, and current smoking status as covariates. This sample included 306 cases with carotid artery diseases and 534 controls. Associations between the each SNP and carotid artery disease were tested using logistic regression with the same covariates described above.
Isoform size studies
In the Hutterites, the kringle IV number in each isoform was determined in fresh plasma collected in February 2008 from 63 individuals with plasma Lp(a) levels previously measured in fresh blood collected during field trips in 1996 and 1997. The determination of kringle IV number was carried out in a high-sensitivity 4% SDS-PAGE system followed by immunoblotting using a polyclonal antibody specific for apo(a) and a standard a set of kringle IV recombinants of a defined molecular size in Chicago. The standard was kindly provided by Dr. Angles Cano (INSERM, Paris, France) and used as specified (31). The isoform size designation represents a number of kringle IV repeats present in each isoform. The more intense band was considered as the dominant form. When two bands were closely migrating and of about equal intensity, their average size was determined. Ten of 63 individuals had two bands of approximately equal intensity.
Lp(a) isoform size was determined using high-resolution SDS–agarose gel electrophoresis followed by immunoblotting in the CLEAR cohort, as previously described (32, 33), in a subset of 99 CLEAR subjects (39 cases and 60 controls). When no dominant Lp(a) isoform could be determined, average isoform size was used for association testing. When two isoforms were present, the dominant isoform was defined as >80% of the total Lp(a) mass. Nineteen subjects had only a single isoform size. The mean dominant isoform size was 23 (range = 8–36) kringle IV repeats. The dominant isoform size could not be determined in eight of the subjects.
Correlations between each SNP and dominant Lp(a) isoform size (kringle IV number) were tested using the GTAM test of association (Hutterites) or Pearson's correlation coefficient (CLEAR). Associations between each SNP and Lp(a) level were examined in each subsample using the same methods described above and also in a model that included Lp(a) isoform size (kringle IV number) as a covariate.
RESULTS
Identifying associated SNPs
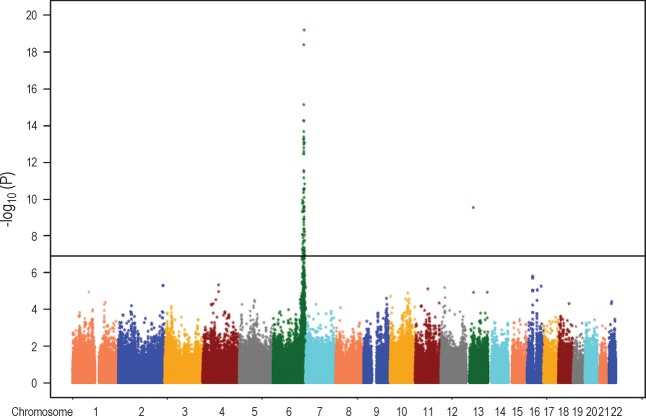
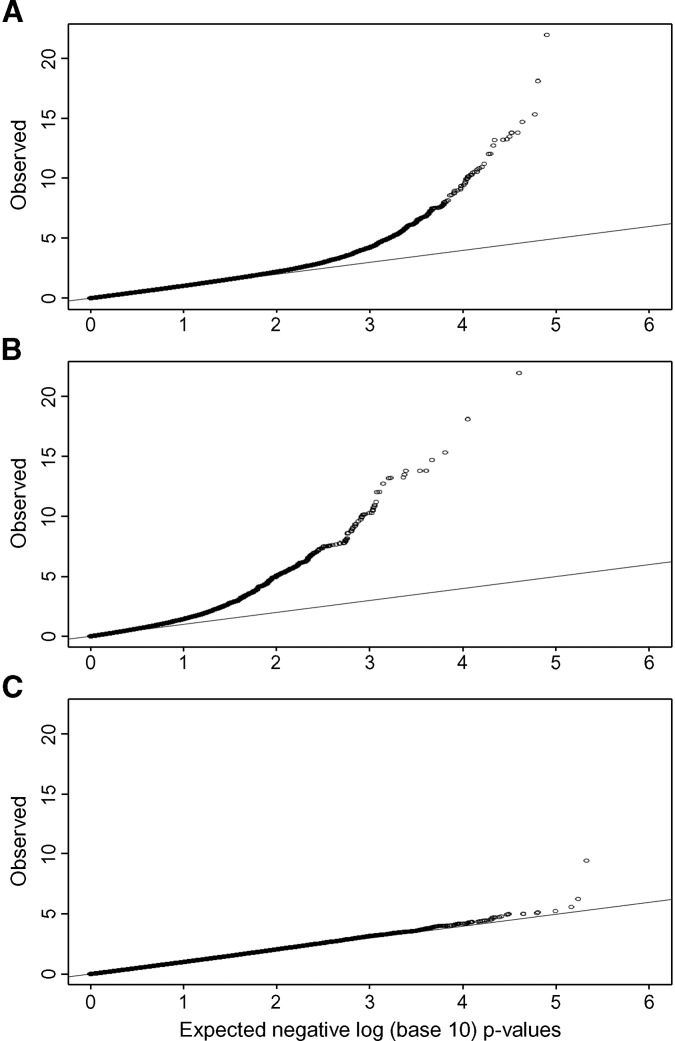
Among the 290,327 SNPs used in our genome-wide association study, 78 had single-locus association P values ≤1.3 × 10−7, which were genome-wide significant based on permutation-based empirical distributions (28) (Fig. 1). All but one of the significant SNPs were on chromosome 6q, spanning from 152,912,509 to 165,610,428 Mb (Fig. 2). The remaining significant SNP (rs17210569) was in an intergenic region on chromosome 13, ∼79 kb downstream of TRPC4 and 400 kb upstream of UFM1 (single-locus association P = 3.5 × 10−10). Despite the large number of small P values on chromosome 6, the distribution of the remaining P values across the genome was as expected (Fig. 3). Because of the large number of associated SNPs on chromosome 6q, we focused our subsequent analyses on this region. P values for all SNPs are deposited in dbGaP (Study Accession: phs000123.v1.p1).
Fig. 1.
Distribution of P values across the genome. Horizontal vertical line shows the threshold for genome-wide significance.
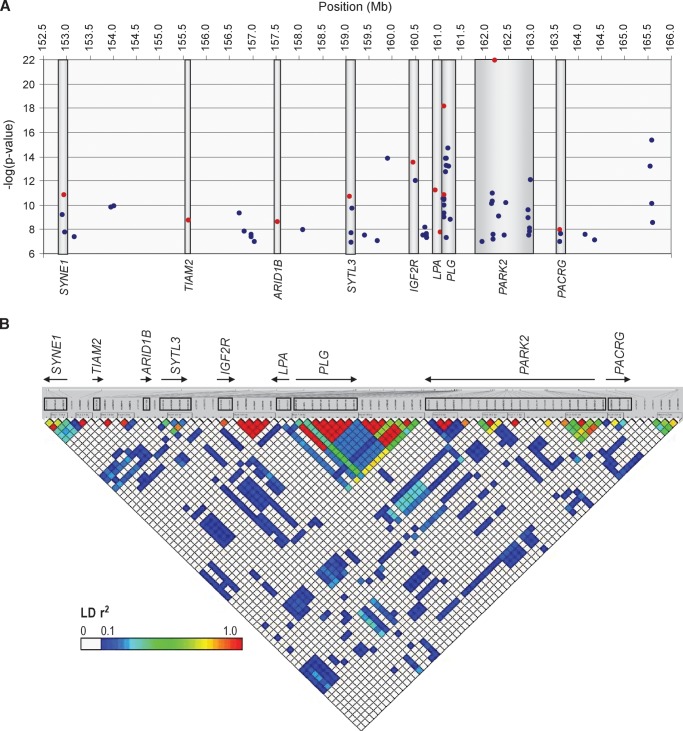
Fig. 2.
Seventy-seven SNPs on chromosome 6q that remained significant after adjusting for multiple comparisons using permutation-based empirical distributions, corresponding to association P values ≤1.14 × 10−7. The SNPs span from 152,912,509 to 165,610,428 Mb. A: SNPs reside within nine genes, shown on the bottom of the figure, and within 100 kb of an additional six genes (genes not shown). The locus (uncorrected) − log (P value) is on the y axis; SNPs selected for conditional (covariate) analyses are shown in red. B: LD (r2) between 77 associated SNPs, generated using LDselect, in 60 relatively unrelated (not first-degree relatives) Hutterites (30 males and 30 females). SNPs are equally spaced across the plot (not to physical scale). The names and direction of transcription are shown for each of the nine genes in A; SNPs within the nine genes are boxed. The comparison to LD in HapMap CEU is shown in supplementary Figure I.
Fig. 3.
Q-Q plot of P values from genome-wide association study of plasma levels of Lp(a) protein in the Hutterites. A: All autosomes; B: chromosome 6; C: all autosomes except chromosome 6.
The significant SNPs on 6q were within nine genes or in the regions between them and up to 8 Mb from the LPA locus (HapMap Release 21a) (Fig. 2A). Two associated SNPs were in LPA, rs6919346 in intron 37 (locus P = 6.3 × 10−12) and rs1853021 (+93C/T) in the 5′ untranslated region (locus P = 5.9 × 10−8). Surprisingly, eight associated SNPs were within the PLG gene, and eight were within 100 kb 3′ to PLG, and 22 associated SNPs were in the PARK2 gene. The association with the LPA +93C/T SNP and plasma Lp(a) levels has been reported in African Americans (34), but in the opposite direction as that seen in the Hutterites (23) (Table 2). SNPs in these genes each accounted for between 0.11 and 8.72% of the total variance in Lp(a) levels (Table 2).
TABLE 2.
Characteristics of SNPs in associated genes on chromosome 6q
| Allele Frequency |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Location on Chr. 6 (bp) | Gene/SNP Location | No. Genotyped/ Phenotyped | HW P Value | Call Rate | Associated Allele | Ancestral/Derived | Effect Size (%) | Association P Value | Hutt+ | CEU | YRI | CHB | JPT |
| rs7745725 | 152,944,398 | SYNE1 intron 3 | 333 | 0.679 | 1.000 | T | D | 3.01 | 1.48 × 10−11 | 0.650 | 0.808 | 0.467 | 0.778 | 0.633 |
| rs9384296 | 155,622,957 | TIAM2 intron 16 | 331 | 0.567 | 0.984 | G | A | 8.72 | 1.85 × 10−09 | 0.773 | 0.842 | 0.875 | 0.622 | 0.656 |
| rs6917698 | 157,535,466 | ARID1B intron 6 | 350 | 0.707 | 0.999 | A | A | 6.92 | 2.48 × 10−09 | 0.886 | 0.783 | 0.950 | 0.756 | 0.778 |
| rs9364496 | 159,092,493 | SYTL3 intron 5 | 331 | 0.223 | 0.989 | G | A | 0.11 | 1.91 × 10−11 | 0.647 | 0.742 | 0.825 | 0.200 | 0.159 |
| rs8191829 | 160,449,889 | IGF2R intron 21 | 349 | 0.366 | 0.994 | G | A | 2.51 | 3.27 × 10−14 | 0.611 | 0.758 | 0.695 | 0.656 | 0.636 |
| rs6919346 | 160,930,770 | LPA intron 37 | 333 | 0.476 | 1.000 | G | A | 3.84 | 6.27 × 10−12 | 0.690 | 0.825 | 0.992 | 0 | 0 |
| rs1853021 | 161,055,668 | LPA 5′UTR (+93C/T) | 364 | 0.483 | 0.915 | T | D | 0.17 | 1.60 × 10−08 | 0.111 | na | na | na | na |
| rs14224 | 161,108,190 | PLG exon 7 (C257C) | 349 | 0.352 | 0.997 | C | A | 5.83 | 7.56 × 10−19 | 0.282 | 0.433 | 0.551 | 0.523 | 0.489 |
| rs4252125 | 161,122,651 | PLG exon 11 (D472N) | 335 | 0.698 | 1.000 | G | D | 1.22 | 1.55 × 10−11 | 0.437 | 0.675 | 0.875 | 1 | 1 |
| rs2022991 | 162,218,580 | PARK2 intron 6 | 348 | 0.313 | 0.993 | G | D | 5.0 | 1.13 × 10−22 | 0.186 | 0.342 | 0.325 | 0.489 | 0.557 |
| rs11966948 | 163,610,673 | PACRG intron 5 | 349 | 0.901 | 0.999 | A | A | 2.25 | 1.06 × 10−08 | 0.438 | 0.642 | 0.450 | 0.678 | 0.578 |
Alleles associated with higher levels of plasma Lp(a) are shown (forward strand). Hutterite allele frequencies and Hardy-Weinberg test statistics were corrected for relatedness among individuals in the sample (41, 42). Allele frequencies in European (CEU), African (YRI), Chinese (CHB), and Japanese (JPT) populations are shown (HapMap). na, not typed in HapMap samples; UTR, untranslated region.
Linkage disequilibrium patterns and conditional analysis of 6q SNPs
The linkage disequilibrium (LD) pattern in this 12.7 Mb region was similar in the Hutterites and the HapMap CEU samples (see supplementary Fig. I), and the lack of LD (r2 < 0.30) between SNPs in different genes suggested that there are multiple independent associations on 6q (Fig. 2B). To further assess the number of independent associations on 6q, we selected the most significant SNP in each of nine associated genes and the two most significant SNPs each in LPA and PLG (Table 2) and included each SNP separately as a covariate in an association analysis. SNPs in eight of the nine genes remained significant in these conditional analyses (P < 0.001), indicating independent effects on Lp(a) levels (see supplementary Table I). The P value for three associated SNPs in PACRG became greatly reduced (conditional P = 0.003 to 0.1) when genotype at SNP rs2022991 in PARK2 was included as a covariate. As a result, SNPs in this gene were not considered further. Moreover, the P value for LPA SNP rs1853021 (+93C/T) became nonsignificant when PLG SNP rs14224 was included as a covariate (conditional P = 0.1), as did 14 other associated SNPs in the PLG gene or its downstream flanking region (conditional P = 0.06 to 0.7). Thus, the LD and covariate analyses suggested that SNPs in eight genes on 6q are independently associated with plasma Lp(a) protein levels in the Hutterites.
Studies of kringle IV number and associated SNPs
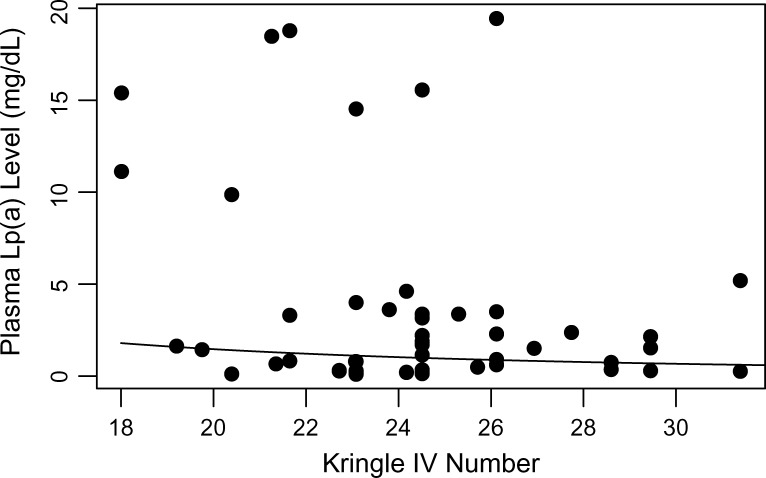
Even though there is no LD between associated SNPs in the eight genes, it remained possible that some or all of the associations were due to LD with the number of kringle IV repeats in the LPA gene. To examine this possibility, we determined kringle IV number in 63 Hutterites with measured Lp(a) levels. The mean Lp(a) levels in this subsample were 3.27 mg/dl (SD = 5.82) in 32 males and 3.55 mg/dl (SD = 5.46) in 31 females, slightly higher than mean levels in the larger sample (Table 1). Number of kringle IV was inversely correlated with plasma Lp(a) levels (P = 0.085) as expected (Fig. 4). In particular, Hutterites with high Lp(a) values (≥6.5 mg/dl) had ≤26 kringle IV (range 18–26). However, individuals with Lp(a) levels in the normal range (<6.5 ng/dl) had kringle IV numbers ranging from 19 to nearly 32.
Fig. 4.
Lp(a) isoform size (kringle IV number) and plasma Lp(a) levels in 63 Hutterites. Correlation between Lp(a) level and kringle IV number, P = 0.085 (corrected for relatedness). The curve illustrating the trend was estimated by a linear regression model of log Lp(a) on log kringle IV number.
We next examined the relationship between number of kringle IV domains and genotype at the eight SNPs that were independently associated with Lp(a) levels. In this analysis, we considered the number of kringle IV as a quantitative trait and tested for association with each SNP (Table 3). Only one SNP (rs2022991 in PARK2) was significantly associated with kringle IV number (P = 0.00054). At this SNP, the G allele was associated with small kringle IV number and with high Lp(a) levels in the larger sample (Table 2). This result suggests that some or all of the association between this SNP and Lp(a) could be due to kringle IV number in the LPA gene. The remaining seven SNPs were not associated with kringle IV number (P > 0.40).
TABLE 3.
SNP associations with kringle IV number and Lp(a) in the subsample
| Association with Lp(a) |
|||||||
|---|---|---|---|---|---|---|---|
| SNP | Gene | n | Location on Chromosome 6 (bp) | MAF | Association with Kringle IV Number P Value | P Value | P Value with Covariate |
| rs7745725 | SYNE1 | 55 | 152,893,977 | 0.49 | 0.976 | 0.00093 | 0.00064 |
| rs9384296 | TIAM2 | 55 | 155,572,536 | 0.27 | 0.990 | 0.78950 | 0.56890 |
| rs6917698 | ARID1B | 58 | 157,485,045 | 0.15 | 0.986 | 0.00065 | 0.00057 |
| rs9364496 | SYTL3 | 54 | 159,042,072 | 0.45 | 0.740 | 0.00142 | 0.00322 |
| rs8191829 | IGF2R | 58 | 160,399,468 | 0.49 | 0.505 | 0.00098 | 0.00164 |
| rs6919346 | LPA | 55 | 160,880,349 | 0.24 | 0.784 | 0.00123 | 0.00030 |
| rs14224 | PLG | 57 | 161,057,769 | 0.23 | 0.402 | 0.01255 | 0.00944 |
| rs2022991 | PARK2 | 58 | 162,168,159 | 0.19 | 0.00054 | 0.00003 | 0.00017 |
P values for associations with Lp(a) are shown without kringle IV number as a covariate and with kringle IV number as a covariate. Samples sizes for the kringle IV studies (n) are shown. Allele frequencies are adjusted for relatedness. MAF, minor allele frequency.
Lastly, we directly examined whether kringle IV number was contributing to the observed associations between Lp(a) levels and SNPs in eight genes. We first performed association studies between these eight SNPs and Lp(a) levels in the smaller sample of Hutterites. Seven of the eight SNPs that were associated with Lp(a) levels in the larger sample (Table 2) were also significantly associated with Lp(a) in this sample of <70 Hutterites (Table 3). One SNP (rs9384296 in TIAM2) was not associated with Lp(a) levels in this sample (P = 0.569). We next repeated this analysis including kringle IV number as a covariate, which would remove any effects of kringle IV number on Lp(a) levels and allow us to assess the effects of each SNP independent of isoform size. All seven SNPs, including rs6919346 in the LPA gene, remained significantly associated with Lp(a) levels in the conditional analysis, with overall little change in the P values. Even SNP rs2022991 in PARK2 that was correlated with kringle IV number remained significant, although the magnitude of the association was reduced (P = 3 × 10−5 without covariate; P = 1.7 × 10−4 with covariate).
Taken together, these data indicate that associations between SNPs in seven genes spanning an ∼7 Mb region on chromosome 6, and including the LPA gene, are associated with plasma Lp(a) levels independent of kringle IV number.
Replication studies in the CLEAR cohort
We genotyped SNPs in seven genes in 1,054 unrelated Caucasian males from the CLEAR cohort, with fasting Lp(a) measurements and including 840 individuals with severe (case) or no (control) carotid artery disease (29) (Table 4). We were unable to genotype SNP rs7745725 in SYNE1 in these samples. Two SNPs were associated with Lp(a) levels: rs6919346 in the LPA gene (P = 7.3 × 10−12) and rs14224 in the PLG gene (P = 0.0047). SNP rs6919346 in LPA was also associated with carotid artery disease (P = 0.0240), as were SNPs in IGF2R and PARK2 (P = 0.0372 and 0.0139, respectively) (Table 5A). To assess the potential confounding of kringle IV number on these associations, 99 individuals were selected for isoform size studies. Among the eight SNPs genotyped for this study, only rs14224 in PLG was significantly correlated with isoform size (P = 0.0075) in this sample (Table 5B), reflecting LD between variation in PLG and kringle IV number, as previously reported (35). When the association with Lp(a) was reexamined in these 99 individuals, the P value for rs14224 was reduced from 0.0140 to 0.300 when kringle IV number was included in the model. Associations between all other PLG SNPs and plasma Lp(a) levels also became nonsignificant (P > 0.05) when isoform size was included as a covariate (data not shown). In contrast, the association between rs6919346 and Lp(a) remained significant (P = 0.0007 without isoform size and 0.0001 with isoform size included in the model). We interpret these results to indicate that the associations with PLG SNPs in the CLEAR cohort were due to long-range LD with the kringle IV size polymorphism in LPA, whereas rs6919346 in LPA is independently associated with Lp(a) levels and risk for carotid artery disease.
TABLE 5.
Association studies with plasma Lp(a) levels in the CLEAR cohort
| A |
Genotype versus Lp(a) Levels |
Genotype versus Carotid Artery Disease |
|||||
|---|---|---|---|---|---|---|---|
| Gene | rs Number | MAF | HW P | B | P Value | B | P Value |
| TIAM2 | rs9384296 | 0.15 | 0.80 | 0.164 | 0.122 | 0.8597 | 0.299 |
| ARID1B | rs6917698 | 0.22 | 0.22 | 0.063 | 0.506 | 0.9225 | 0.531 |
| SYTL3 | rs9364496 | 0.25 | 0.80 | 0.103 | 0.237 | 1.0110 | 0.926 |
| IGF2R | rs8191829 | 0.23 | 0.72 | −0.086 | 0.337 | 1.2780 | 0.0438 |
| LPA | rs6919346 | 0.16 | 0.40 | −0.663 | 3.83 × 10−11 | 0.6619 | 0.0036 |
| PLG | rs14224 | 0.41 | 0.21 | 0.219 | 0.0036 | 0.9697 | 0.767 |
|
PARK2 |
rs2022991 |
0.33 |
0.83 |
−0.026 |
0.748 |
0.7488 |
0.009 |
| B |
Genotype versus Isoform Size |
Genotype versus Lp(a) |
Genotype versus Lp(a) with Covariate (Isoform Size) |
||||
| Gene |
rs Number |
B |
P Value |
B |
P Value |
B |
P Value |
| TIAM2 | rs9384296 | 1.329 | 0.213 | −0.082 | 0.797 | 0.164 | 0.509 |
| ARID1B | rs6917698 | 0.248 | 0.815 | −0.238 | 0.470 | −0.180 | 0.482 |
| SYTL3 | rs9364496 | −0.183 | 0.872 | 0.296 | 0.399 | 0.197 | 0.273 |
| IGF2R | rs8191829 | 1.514 | 0.143 | −0.335 | 0.306 | −0.151 | 0.553 |
| LPA | rs6919346 | 0.959 | 0.376 | −0.981 | 0.002 | −0.877 | 0.0004 |
| PLG | rs14224 | −2.405 | 0.0045 | 0.676 | 0.0097 | 0.270 | 0.205 |
| PARK2 | rs2022991 | 1.003 | 0.276 | −0.136 | 0.635 | 0.102 | 0.644 |
A: Analyses in 1,054 subjects with Lp(a) measurements, including 306 cases (with carotid artery disease) and 534 controls (Table 4). B: Analyses in 99 individuals selected for isoform size studies. Last two columns are analyses adjusted for isoform size. MAF, minor allele frequency; HW P, Hardy-Weinberg P value; B, regression slope coefficient. Significant P values are shown in bold italic.
Studies of LPA enhancer elements
Two enhancers of gene expression residing between the LPA and PLG genes have been described (36, 37). To rule out LD between the Lp(a)-associated SNPs with polymorphisms in these elements, we sequenced the enhancer elements DHII and DHIII (36, 37) in DNA from 30 Hutterites who were not first-degree relatives and who represented diverse genotypes at LPA and PLG SNPs. No SNPs were identified in DHII, but two SNPs were identified in DHIII (rs9347440 and rs7758766). There was very little LD (r2 = 0–0.20) between each of these two SNPs and the 11 SNPs described in Table 2. The lack of LD between the SNPs in enhancer DHIII and Lp(a)-associated SNPs suggests that the SNPs identified in this study are not due to LD with SNPs in these enhancers of LPA expression.
DISCUSSION
Plasma Lp(a) level is one of the most heritable quantitative traits in humans, and the high heritability of this trait has been attributed to variation at the LPA locus (10–12, 15, 38). Our studies reveal a more complex genetic architecture of Lp(a) levels, with multiple contributing loci on 6q26-q27. We identified a novel SNP in the LPA gene that is associated with high Lp(a) protein levels in the Hutterites and in the CLEAR cohort, and with carotid artery disease in the latter patient population, both independent of the kringle IV number and variation in known transcriptional enhancer elements of LPA. The mechanism through which this intronic SNP acts is unknown, but the fact that it is not in LD with any other SNPs in the LPA gene (39) and that it resides within a CREB site suggest that rs6919346 could influence gene expression.
SNPs in two other genes (IGF2RA and PARK2) were associated with plasma Lp(a) levels in the Hutterites and carotid artery disease in the CLEAR cohort, and SNPs in four genes (SYNE1, ARIDB1, SYTL3, and PLG) showed associations with Lp(a) levels in the Hutterites only, all independent of kringle IV number. The fact that the latter genes were not associated with Lp(a) levels or carotid artery disease in the CLEAR cohort could be due to differences in allele frequencies, population characteristics, or ascertainment schemes in the two samples. For example, the Hutterite sample is relatively young (mean age = 35.5 years), includes approximately equal numbers of males and females, and is unselected with respect to cardiovascular phenotypes. The CLEAR cohort is older (mean age = 63.5 years), all male, and selected for the presence or absence of carotid artery disease (Table 4). Thus, studies in other populations with different ethnic, demographic, and clinical characteristics are warranted to further clarify the role of these SNPs and their corresponding genes on Lp(a) biology and carotid artery disease.
Our study revealed novel observations about the genetic architecture of Lp(a). Studies of kringle IV copy number in the Hutterites and the CLEAR cohort indicate that there is little LD between kringle IV size and SNPs in genes on chromosome 6q, including SNPs in the LPA gene itself. Consistent with our observation, a recent resequencing study of the LPA and PLG genes in 23 European Americans and 24 African Americans showed that kringle IV copy number was not in LD with any biallelic markers in LPA or PLG (39). Moreover, while small kringle IV number was associated with high plasma Lp(a) levels in both the Hutterites (Fig. 4) and the CLEAR cohort (data not shown), as expected, many individuals in both samples had a small number of kringle IV (≤26) and Lp(a) values in the normal range. This is consistent with numerous earlier studies indicating that the kringle IV copy number variation accounts for only ∼50% of the heritability of Lp(a) levels (10–15). However, our study further suggests that kringle IV repeats may account for nearly all of the high Lp(a) values but that additional loci maintain low circulating levels of Lp(a) in some individuals despite having small isoform sizes. Whether the other loci identified in this study are modifiers of the effects of kringle IV number remains to be proven, but any of the newly identified genes could potentially act through transcriptional, posttranscriptional, or posttranslational mechanisms to regulate plasma Lp(a) levels.
Lastly, the identification of additional loci on chromosome 6q26-q27 influencing Lp(a) levels suggests that the large and consistent linkage signal in human families at the LPA “locus” is due to the contribution of multiple linked susceptibility loci in the region. While a role for trans-acting factors on other chromosomes cannot be ruled out, for example, on chromosome 13, nearly all of the variation in Lp(a) levels in the Hutterites is accounted for by loci on chromosome 6q.
Supplementary Material
Acknowledgments
The authors thank the Hutterites for their continuing participation in our studies, Matthew Stephens and Nancy Cox for helpful discussions and comments on the manuscript, the University of Chicago General Clinical Research Center for their support, the NHLBI-funded Resequencing and Genotyping Service (Johns Hopkins University), and the SeattleSNP Program (University of Washington). The authors declare that they have no competing financial interests.
Abbreviations
apo(a), apolipoprotein(a)
BMI, body mass index
LD, linkage disequilibrium
Lp(a), lipoprotein (a)
SNP, single nucleotide polymorphism
RSS, residual sum of squares
This work was supported by grants from the National Institutes of Health (R01 HL66533, R01 HL63209, P50 HL56399, and M01 RR00055) and Hoffman-LaRoche to the University of Chicago, and National Institutes of Health grants U01 HL66682, R01 HL074366, R01HL67406, P01 HL30086, and P01 HL072262 to the University of Washington.
Published, JLR Papers in Press, January 5, 2009.
Footnotes
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and one table.
References
- 1.Danesh J., R. Collins, and R. Peto. 2000. Lipoprotein(a) and coronary heart disease. Meta-analysis of prospective studies. Circulation. 102 1082–1085. [DOI] [PubMed] [Google Scholar]
- 2.Berglund L., and R. Ramakrishnan. 2004. Lipoprotein(a): an elusive cardiovascular risk factor. Arterioscler. Thromb. Vasc. Biol. 24 2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scanu A. M., and R. Bamba. 2008. Niacin and lipoprotein(a): facts, uncertainties, and clinical considerations. Am. J. Cardiol. 101 44B–47B. [DOI] [PubMed] [Google Scholar]
- 4.Kraft H. G., H. J. Menzel, F. Hoppichler, W. Vogel, and G. Utermann. 1989. Changes of genetic apolipoprotein phenotypes caused by liver transplantation. Implications for apolipoprotein synthesis. J. Clin. Invest. 83 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koschinsky M. L., G. P. Cote, B. Gabel, and Y. Y. van der Hoek. 1993. Identification of the cysteine residue in apolipoprotein(a) that mediates extracellular coupling with apolipoprotein B-100. J. Biol. Chem. 268 19819–19825. [PubMed] [Google Scholar]
- 6.Utermann G., H. J. Menzel, H. G. Kraft, H. C. Duba, H. G. Kemmler, and C. Seitz. 1987. Lp(a) glycoprotein phenotypes. Inheritance and relation to Lp(a)-lipoprotein concentrations in plasma. J. Clin. Invest. 80 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean J. W., J. E. Tomlinson, W. J. Kuang, D. L. Eaton, E. Y. Chen, G. M. Fless, A. M. Scanu, and R. M. Lawn. 1987. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 330 132–137. [DOI] [PubMed] [Google Scholar]
- 8.Brunner C., E. M. Lobentanz, A. Petho-Schramm, A. Ernst, C. Kang, H. Diepliner, H. J. Muller, and G. Utermann. 1996. The number of identical kringle IV repeats in apolipoprotein(a) affects its processing and secretion by HepG2 cells. J. Biol. Chem. 271 32403–32410. [DOI] [PubMed] [Google Scholar]
- 9.White A. L., B. Guerra, and R. E. Lanford. 1997. Influence of allelic variation on apolipoprotein(a) folding in the endoplasmic reticulum. J. Biol. Chem. 272 5048–5055. [DOI] [PubMed] [Google Scholar]
- 10.Boerwinkle E., C. C. Leffert, J. Lin, C. Lackner, G. Chiesa, and H. H. Hobbs. 1992. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J. Clin. Invest. 90 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraft H. G., S. Kochl, H. J. Menzel, C. Sandholzer, and G. Utermann. 1992. The apolipoprotein (a) gene: a transcribed hypervariable locus controlling plasma lipoprotein (a) concentration. Hum. Genet. 90 220–230. [DOI] [PubMed] [Google Scholar]
- 12.Mooser V., D. Scheer, S. M. Marcovina, J. Wang, R. Guerra, J. Cohen, and H. H. Hobbs. 1997. The Apo(a) gene is the major determinant of variation in plasma Lp(a) levels in African Americans. Am. J. Hum. Genet. 61 402–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandholzer C., D. M. Hallman, N. Saha, G. Sigurdsson, C. Lackner, A. Csaszar, E. Boerwinkle, and G. Utermann. 1991. Effects of the apolipoprotein(a) size polymorphism on the lipoprotein(a) concentration in 7 ethnic groups. Hum. Genet. 86 607–614. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt K., H. G. Kraft, W. Parson, and G. Utermann. 2006. Genetics of the Lp(a)/apo(a) system in an autochthonous Black African population from the Gabon. Eur. J. Hum. Genet. 14 190–201. [DOI] [PubMed] [Google Scholar]
- 15.Scholz M., H. G. Kraft, A. Lingenhel, R. Delport, E. H. Vorster, H. Bickeboller, and G. Utermann. 1999. Genetic control of lipoprotein(a) concentrations is different in Africans and Caucasians. Eur. J. Hum. Genet. 7 169–178. [DOI] [PubMed] [Google Scholar]
- 16.Ogorelkova M., H. G. Kraft, C. Ehnholm, and G. Utermann. 2001. Single nucleotide polymorphisms in exons of the apo(a) kringles IV types 6 to 10 domain affect Lp(a) plasma concentrations and have different patterns in Africans and Caucasians. Hum. Mol. Genet. 10 815–824. [DOI] [PubMed] [Google Scholar]
- 17.Rubin J., H. J. Kim, T. A. Pearson, S. Holleran, R. Ramakrishnan, and L. Berglund. 2006. Apo[a] size and PNR explain African American-Caucasian differences in allele-specific apo[a] levels for small but not large apo[a]. J. Lipid Res. 47 982–989. [DOI] [PubMed] [Google Scholar]
- 18.Hostetler, J. A. 1974. Hutterite Society. Johns Hopkins University Press, Baltimore, MD.
- 19.Steinberg, A. G., H. K. Bleibtreu, T. W. Kurczynski, A. O. Martin, and E. M. Kurczynski. 1967. Genetic studies in an inbred human isolate. In Proceedings of the Third International Congress of Human Genetics. J. F. Crow and J. V. Neel, editors. Johns Hopkins University Press, Baltimore, MD. 267–290.
- 20.Ober C., M. Abney, and M. S. McPeek. 2001. The genetic dissection of complex traits in a founder population. Am. J. Hum. Genet. 69 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss L. A., M. Abney, R. Parry, A. M. Scanu, E. H. Cook Jr., and C. Ober. 2005. Variation in ITGB3 has sex-specific associations with plasma lipoprotein(a) and whole blood serotonin levels in a population-based sample. Hum. Genet. 117 81–87. [DOI] [PubMed] [Google Scholar]
- 22.Abney M., M. S. McPeek, and C. Ober. 2001. Heritabilities of quantitative traits in a founder population. Am. J. Hum. Genet. 68 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman D. L., et al. 2004. Are common disease susceptibility alleles the same in outbred and founder populations? Eur. J. Hum. Genet. 12 584–590. [DOI] [PubMed] [Google Scholar]
- 24.Ober C., et al. 2008. Effect of variation in CHI3L1 on serum YKL-40 level, asthma risk, and lung function. N. Engl. J. Med. 358 1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J. B., et al. 2003. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 68 69–78. [DOI] [PubMed] [Google Scholar]
- 26.Cheng S., et al. 1999. A multilocus genotyping assay for candidate markers of cardiovascular disease risk. Genome Res. 9 936–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abney M., M. S. McPeek, and C. Ober. 2000. Estimation of variance components of quantitative traits in inbred populations. Am. J. Hum. Genet. 66 629–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abney M., C. Ober, and M. S. McPeek. 2002. Quantitative trait homozygosity and association mapping and empirical genome-wide significance in large complex pedigrees: fasting serum insulin level in the Hutterites. Am. J. Hum. Genet. 70 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlson C. S., et al. 2007. TagSNP evaluation for the association of 42 inflammation loci and vascular disease: evidence of IL6, FGB, ALOX5, NFKBIA, and IL4R loci effects. Hum. Genet. 121 65–75. [DOI] [PubMed] [Google Scholar]
- 30.Marcovina S. M., J. J. Albers, B. Gabel, M. L. Koschinsky, and V. P. Gaur. 1995. Effect of the number of apolipoprotein(a) kringle 4 domains on immunochemical measurements of lipoprotein(a). Clin. Chem. 41 246–255. [PubMed] [Google Scholar]
- 31.Angles-Cano E., S. Loyau, G. Cardoso-Saldana, R. Couderc, and P. Gillery. 1999. A novel kringle-4 number-based recombinant apo[a] standard for human apo[a] phenotyping. J. Lipid Res. 40 354–359. [PubMed] [Google Scholar]
- 32.Marcovina S. M., H. H. Hobbs, and J. J. Albers. 1996. Relation between number of apolipoprotein(a) kringle 4 repeats and mobility of isoforms in agarose gel: basis for a standardized isoform nomenclature. Clin. Chem. 42 436–439. [PubMed] [Google Scholar]
- 33.Marcovina S. M., Z. H. Zhang, V. P. Gaur, and J. J. Albers. 1993. Identification of 34 apolipoprotein(a) isoforms: differential expression of apolipoprotein(a) alleles between American blacks and whites. Biochem. Biophys. Res. Commun. 191 1192–1196. [DOI] [PubMed] [Google Scholar]
- 34.Kraft H. G., M. Windegger, H. J. Menzel, and G. Utermann. 1998. Significant impact of the +93 C/T polymorphism in the apolipoprotein(a) gene on Lp(a) concentrations in Africans but not in Caucasians: confounding effect of linkage disequilibrium. Hum. Mol. Genet. 7 257–264. [DOI] [PubMed] [Google Scholar]
- 35.Drayna D. T., R. A. Hegele, P. E. Hass, M. Emi, L. L. Wu, D. L. Eaton, R. M. Lawn, R. R. Williams, R. L. White, and J. M. Lalouel. 1988. Genetic linkage between lipoprotein(a) phenotype and a DNA polymorphism in the plasminogen gene. Genomics. 3 230–236. [DOI] [PubMed] [Google Scholar]
- 36.Puckey L. H., and B. L. Knight. 2003. Sequence and functional changes in a putative enhancer region upstream of the apolipoprotein(a) gene. Atherosclerosis. 166 119–127. [DOI] [PubMed] [Google Scholar]
- 37.Wade D. P., L. H. Puckey, B. L. Knight, F. Acquati, A. Mihalich, and R. Taramelli. 1997. Characterization of multiple enhancer regions upstream of the apolipoprotein(a) gene. J. Biol. Chem. 272 30387–30399. [DOI] [PubMed] [Google Scholar]
- 38.DeMeester C. A., X. Bu, R. J. Gray, A. J. Lusis, and J. I. Rotter. 1995. Genetic variation in lipoprotein (a) levels in families enriched for coronary artery disease is determined almost entirely by the apolipoprotein (a) gene locus. Am. J. Hum. Genet. 56 287–293. [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford D. C., et al. 2008. LPA and PLG sequence variation and kringle IV-2 copy number in two populations. Hum. Hered. 66 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.National Cholesterol Education Program. 2001. Third Report of the National Cholesterol Education Program Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults. Department of Health and Human Services, National Heart Lung and Blood Institute, Bethesda, MD.
- 41.McPeek M. S., X. Wu, and C. Ober. 2004. Best linear unbiased allele-frequency estimation in complex pedigrees. Biometrics. 60 359–367. [DOI] [PubMed] [Google Scholar]
- 42.Bourgain C., M. Abney, D. Schneider, C. Ober, and M. S. McPeek. 2004. Testing for hardy-weinberg equilibrium in samples with related individuals. Genetics. 168 2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.