Abstract
Glycerophospholipids are structural and functional components of cellular membranes as well as precursors of various lipid mediators. Using acyl-CoAs as donors, glycerophospholipids are formed by the de novo pathway (Kennedy pathway) and modified in the remodeling pathway (Lands' cycle). Various acyltransferases, including two lysophosphatidic acid acyltransferases (LPAATs), have been discovered from a 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) family. Proteins of this family contain putative acyltransferase motifs, but their biochemical properties and physiological roles are not completely understood. Here, we demonstrated that mouse LPAAT3, previously known as mouse AGPAT3, possesses strong LPAAT activity and modest lysophosphatidylinositol acyltransferase activity with a clear preference for arachidonoyl-CoA as a donor. This enzyme is highly expressed in the testis, where CDP-diacylglycerol synthase 1 preferring 1-stearoyl-2-arachidonoyl-phosphatidic acid as a substrate is also highly expressed. Since 1-stearoyl-2-arachidonoyl species are the main components of phosphatidylinositol, mouse LPAAT3 may function in both the de novo and remodeling pathways and contribute to effective biogenesis of 1-stearoyl-2-arachidonoyl-phosphatidylinositol in the testis. Additionally, the expression of this enzyme in the testis increases significantly in an age-dependent manner, and β-estradiol may be an important regulator of this enzyme's induction. Our findings identify this acyltransferase as an alternative important enzyme to produce phosphatidylinositol in the testis.
Keywords: 1-acylglycerol-3-phosphate O-acyltransferase, phosphatidic acid, phosphatidylinositol, β-estradiol
Tissues maintain distinct content and composition of various glycerophospholipids, such as phosphatidic acid (PA), phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol (PI), phosphatidylserine, and cardiolipin (1–4). They are formed by two pathways using acyl-CoAs as donors. One is the de novo pathway (Kennedy pathway) in which glycerophospholipids are formed from glycerol 3-phosphate (5). The other is the remodeling pathway (Lands' cycle), where the concerted activation of phospholipase A2s and lysophospholipid acyltransferases (LPLATs) occurs (6–10). These pathways are the basis of membrane asymmetry and diversity. In general, saturated and monounsaturated fatty acids are esterified at the sn-1 position, whereas polyunsaturated fatty acids are at the sn-2 position. The combinations of fatty acids at the sn-1 and sn-2 positions vary among different classes of phospholipids. In the rat liver and brain, for example, PA possesses a low arachidonic acid content (11–13), whereas arachidonic acid is a major component of PI (4, 13, 14).
Extensive studies of acyltransferases have been conducted over the last decade, mostly using homology searches (6, 7, 15–35). Several acyltransferase families have been proposed, including the 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) family. At least seven AGPAT family members have been identified in mouse (21, 36, 37), and each of them contains a highly conserved putative catalytic motif (NHX4D) and putative substrate binding motif (EGTR) (38–40) (Fig. 1A). Some of the AGPAT family members are relatively well characterized. Lysophosphatidic acid acyltransferase (LPAAT) activity of mouse LPAAT1 (previously known as mouse AGPAT1) and mouse LPAAT2 (or mouse AGPAT2) is well documented (21, 37), and mutations in human LPAAT2 cause congenital generalized lipodystrophy (41). Recently, mouse AGPAT6 was shown to have glycerol-3-phosphate acyltransferase activity (33). Mouse AGPAT3 has been investigated in the past, but the characterization was far from being done (37). The existence of at least seven members in AGPAT family raises questions as to the specific role of each member. Therefore, investigation of their tissue distributions or biochemical properties will be important in understanding their biological roles.
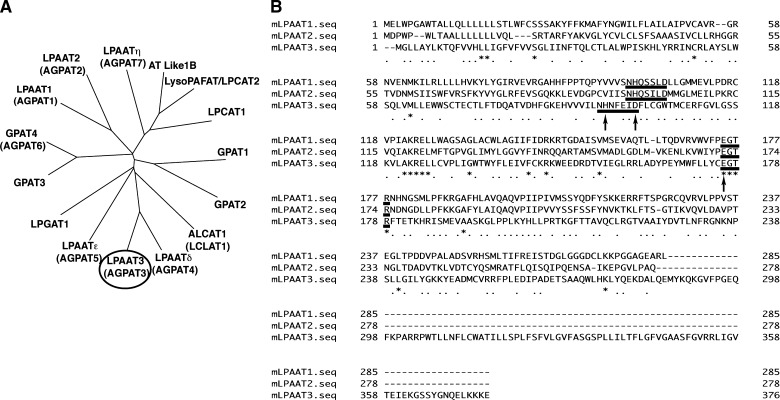
Fig. 1.
Phylogenetic tree of AGPAT family members and alignment of mLPAAT1, 2, and 3. A: A phylogenetic tree was drawn by using ClustalW, DDBJ (http://www.clustalw.ddbj.nig.ac.jp/top-j.html). Sequences of mouse acyltransferases are available in the DDBJ/EMBL/GenBank databases. mLPAAT3 is circled. B: mLPAAT1, mLPAAT2, and mLPAAT3 sequences were aligned using Genetic-Mac software. Conserved putative catalytic motif NHX4D and binding motif EGTR are underlined. Mutated amino acids are indicated by arrows (see Fig. 5). The accession numbers are shown as follows: GPAT1 (NP_032175), GPAT2 (NP_001074558), GPAT3 (NP_766303), GPAT4 (NP_061213), LPAAT1 (NP_061350), LPAAT2 (NP_080488), LPAAT3 (AB377215), LPGAT1 (NP_758470), ALCAT1 (acyl-CoA:lysocardiolipin acyltransferase 1; also called as LCLAT1) (Q3UN02), LPCAT1 (BAE94687), LysoPAFAT/LPCAT2 (BAF47695), LPAATδ (NP_080920), LPAATɛ (NP_081068), LPAATη (NP_997089), and AT Like 1B (NP_081875).
We present here, to our knowledge, the first detailed biochemical and biological characterization of mouse AGPAT3 (mAGPAT3). Surprisingly, mAGPAT3 possesses both LPAAT and lysophosphatidylinositol acyltransferase (LPIAT) activities and prefers arachidonoyl-CoA as a donor, indicating its dual roles in the de novo and remodeling pathways. Point mutations in highly conserved motifs NHX4D or EGTR completely suppressed both LPAAT and LPIAT activities. The enzyme was localized in the endoplasmic reticulum (ER) and expressed in the liver, kidney, and testis. In the testis, cytidine diphosphodiacylglycerol (CDP-diacylglycerol) synthase 1 is highly expressed and particularly converts 1-stearoyl-2-arachidonoyl-PA to CDP-diacylglycerol, a phospholipid precursor (42, 43). This might suggest that mLPAAT3 produces PI effectively. Additionally, mAGPAT3 expression in the testis increases significantly in an age-dependent manner. Since β-estradiol induced this enzyme in testicular cell line, mAGPAT3 may play an important role in the testis coupled with sex hormone. We renamed this enzyme as LPAAT3 according to a proposal for the standardization of LPLAT nomenclature by Shindou and Shimizu (6).
MATERIALS AND METHODS
Materials
DMEM, 12F-HAM, and RPMI1640 were obtained from Sigma-Aldrich (St. Louis, MO). TLC silica gel plates (type 5721) were purchased from Merck (Darmstadt, Germany). Various lysophospholipids and acyl-CoAs were from Avanti Polar Lipids (Alabaster, AL). [1-14C]Oleoyl-CoA (1.924 GBq/mmol), [1-14C]Linoleoyl-CoA (2.035 GBq/mmol), and [1-14C]Arachidonoyl-CoA (2.035 GBq/mmol) were purchased from Moravec Biochemicals (Mercury Lane, CA). [1-14C]Palmitoyl-CoA (2.22 GBq/mmol) and [3H]acetyl-CoA (185 GBq/mmol) were obtained from GE Healthcare (Buckinghamshire, UK).
Cloning of mLPAAT1 and mLPAAT3
The entire coding region of mLPAAT3 [DNA Data Bank of Japan (DDBJ) accession number AB377215] was identified in the National Center for Biotechnology Information (NCBI) database. A 1.1 kb cDNA clone encoding the full-length mLPAAT3 was obtained by PCR amplification using a forward primer designed to encode FLAG epitope (DYKDDDDK) in frame with the start codon of target DNA coding region (5′-CTAGCTAGCCACCATGGATTACAAGGATGACGATGACAAGGGCCTGCTTGCCTACCTGAAGACCC), and a reverse primer (CCGCTCGAGTTATTCCTTTTTCTTAAGCTCTTGGTTGCC-3′). Mouse heart cDNA was used as a template. Amplified PCR products were cloned into the pCXN2.1 vector (44) and sequenced. Similarly, mLPAAT1 (DDBJ accession number NM_018862) was identified in NCBI, and full-length mLPAAT1 was obtained by PCR amplification using a forward primer designed to encode FLAG epitope in frame (CTAGCTAGCCACCATGGATTACAAGGATGACGATGACAAGGAGCTGTGGCCCGGGGCCTGG) and a reverse primer (CCGCTCGAGTCAGAGCCGGGCTTCGCCCGCTCCCCC).
Mutagenesis of mLPAAT3
mLPAAT3 constructs with single mutations in the highly conserved motif NHX4D (His→Ala or Asp→Ala) or EGTR (Glu→Ala) were made using cloned mLPAAT3 cDNA as a template. Amplified PCR products were cloned into the pCXN2.1 vector and sequenced. Details of the method have been described previously (39).
Expression of FLAG-mLPAAT3 in Chinese hamster ovary-K1 cells
Chinese hamster ovary (CHO)-K1 cells were seeded onto 10 cm dishes, at a density of 3 × 106 cells /dish 1 day before transfection. Twelve micrograms each of pCXN2.1 vector or FLAG-mLPAAT3-pCXN2.1 was transfected using Lipofectamine 2000 (Invitrogen). At 48 h after transfection, transfected cells were scraped into 1 ml of ice-cold buffer containing 20 mM Tris-HCl (pH 7.4), 300 mM sucrose, and a proteinase inhibitor cocktail Complete (Roche Applied Science) and sonicated three times on ice for 30 s each time. Cell lysates were centrifuged at 9,000 g for 15 min. Supernatants were then centrifuged at 100,000 g for 1 h. Pellets were suspended in buffer containing 20 mM Tris-HCl (pH 7.4), 300 mM sucrose, and 1 mM EDTA. The protein concentration was measured by the method of Bradford (45) using a commercially prepared protein assay solution (Bio-Rad) and BSA (fraction V, fatty acid-free; Sigma-Aldrich) as a standard.
Production of anti-mLPAAT3 antiserum
Antiserum was generated at SCRUM (Tokyo, Japan). C-terminal peptides were used for immunization of rabbits (EKGSSYGNQELK and FPGEQFKPARRPWT). Specificity of the antiserum was examined by Western blot analysis using mircosomes from vector- and mLPAAT3-transfected cells. Microsomal fractions from mAGPAT4- or AGPAT5-transfected cells were also used as negative controls.
Western blot analysis
Cell lysates were centrifuged at 800 g for 10 min. Supernatants were centrifuged at 9,000 g for 15 min. Supernatants were then centrifuged at 100,000 g for 1 h. Initial 9,000 g pellets were homogenized again and centrifuged at 9,000 g. The pellets were used as 9,000 g pellets. Two micrograms each of 9,000 g pellets, 100,000 g pellets, and 100,000 g supernatants were resolved by 10% SDS-PAGE and transferred to a Hybond ECL nitrocellulose membrane (GE Healthcare UK). The membrane was blocked with 5% skim milk, incubated with anti-FLAG M2 mAb (IBI/Kodak) or anticytochrome c oxidase antibody (Invitrogen), and washed and incubated with horseradish-peroxidase-labeled anti-mouse IgG (GE Healthcare UK). After washing, the membrane was exposed to ECL reagents (GE Healthcare UK) and X-ray film (GE Healthcare UK) to visualize immunoreactive proteins. Expression of the FLAG-tagged target protein was confirmed. For the examination of endogenous mLPAAT3 subcellular localization, rabbit anti-mLPAAT3 antiserum and horseradish-peroxidase-labeled anti-rabbit IgG (GE Healthcare UK) were used as primary and secondary antibodies, respectively.
Confocal microscopy
CHO-K1 cells transfected with pCXN2.1 vector or FLAG-mLPAAT3-pCXN2.1 cells were fixed with 4% paraformaldehyde and permeabilized with methanol/acetone solution (1:1, v/v). Cells were incubated with 10 μg/ml primary antibody for 30 min. As a marker, M5 anti-FLAG mouse mAb or anti-FLAG rabbit antibody (Sigma-Aldrich) for FLAG epitope, anti-calnexin antibody (BD Biosciences) for ER, anticytochrome c oxidase antibody (Invitrogen) for mitochondria, and anti-GM130 (Invitrogen) for Golgi were used. Cells were blocked with 1% BSA and then incubated with 10 μg/ml Alexa Fluor 546 goat anti-rabbit IgG (Eugene, OR) and Alexa Fluor 488 goat anti-mouse IgG (Eugene, OR). Confocal microscopy was performed using an LSM510 laser scanning microscope (Carl Zeiss, Germany) equipped with a ×63 water immersion objective lens (numerical aperture = 1.2). Cells were observed by excitation at 543 nm with a He/Ne laser and emission through a 585-nm long-pass filter for the detection of red fluorescence. For the detection of green fluorescence, the excitation was at 488 nm with an argon laser, and emissions were collected using a 505 to 550 nm band-pass filter.
Assay of LPLAT activity
Acyltransferase activity was measured by the transfer of [1-14C]acyl-CoAs or [3H]acetyl-CoA to lysophospholipids to form phospholipids. Reaction mixtures contained 100 mM Tris-HCl (pH 7.4), 1 mM EDTA, and indicated concentrations of acyl-CoA, lysophospholipids, and enzyme (100,000 g pellets) in a total volume of 100 μl. The amount of total protein and concentrations of acceptors and donors are described in corresponding figure legends. After incubation at 37°C for 10 min, reactions were stopped by the addition of 300 μl of chloroform:methanol (1:2, v/v). The reaction progressed linearly at least for 10 min. Total lipids were extracted using the Bligh-Dyer method (46) and subsequently analyzed by TLC in chloroform:methanol:acetic acid:water (50:25:8:4, v/v/v/v). Bands at positions corresponding to the expected products were visualized with I2 vapor, cut off the plate, placed in Microscinti-O (Perkin-Elmer Life Sciences), and analyzed in an LS6500 liquid scintillation counter (Beckman).
Quantitative real-time PCR
Total RNA was prepared using RNeasy Mini Kit (Qiagen). First-strand cDNAs were synthesized using Superscript II (Invitrogen). PCR was conducted in microcapillary tubes, in 20 μl reaction volumes consisting of 2 μl of cDNA solution, 1× FastStart DNA Master SYBR Green I (Roche Applied Science), and 0.5 μM each of the forward and reverse primers. Sequences of primers used in PCR are shown in supplementary Table I.
Stimulation of testicular cell line TM4 cells with various sex hormones
Testicular cell line TM4 cells were cultured in F12-HAM:DMEM (1:1,v/v) containing 5% horse serum (Gibco) and 2.5% FBS. Cells (1 × 105) were incubated with either mock, 100 nM β-estradiol (Sigma-Aldrich), dihydrotestosterone (Sigma-Aldrich), or testosterone (Sigma-Aldrich) for 24 h and collected.
Animals
C57BL/6J mice were obtained from Clea Japan (Tokyo, Japan). Mice were maintained at 21°C in a light-dark cycle with light from 08:00 to 20:00. Mice were fed with a standard laboratory diet and water ad libitum. All animal studies were conducted in accordance with the guidelines for Animal Research at the University of Tokyo and were approved by the University of Tokyo Ethics Committee for Animal Experiments.
Statistics
Data are presented as mean + SD. P values < 0.05 were considered statistically significant. All statistical calculations were performed using Prism 4 (GraphPad Software).
RESULTS
Cloning of mLPAAT3
To identify novel LPLATs, we focused on AGPAT family proteins. A phylogenetic tree was drawn by pairwise comparisons of the amino acid sequences of LPAAT family members using ClustalW, DDBJ (http://www.clustalw.ddbj.nig.ac.jp/top-j.html) (Fig. 1A) (47). The amino acid sequences of mLPAAT1, mLPAAT2, and mLPAAT3 are shown in Fig. 1B. A 1.1 kb cDNA clone encoding the full-length mLPAAT3 enzyme was obtained by PCR amplification. mLPAAT3 encodes a 376 amino acid protein of 43.3 kDa, containing four transmembrane domains, predicted by ConPred II (48), and the conserved motifs (NHX4D and EGTR) found in members of AGPAT family (Fig. 1B). The protein also possesses the C-terminal sequence motif KKXX (49), suggesting that mLPAAT3 localizes to the ER, similar to LPCAT1 and LysoPAFAT/LPCAT2 (15, 16).
Tissue distribution of mLPAAT3 and mMBOA7
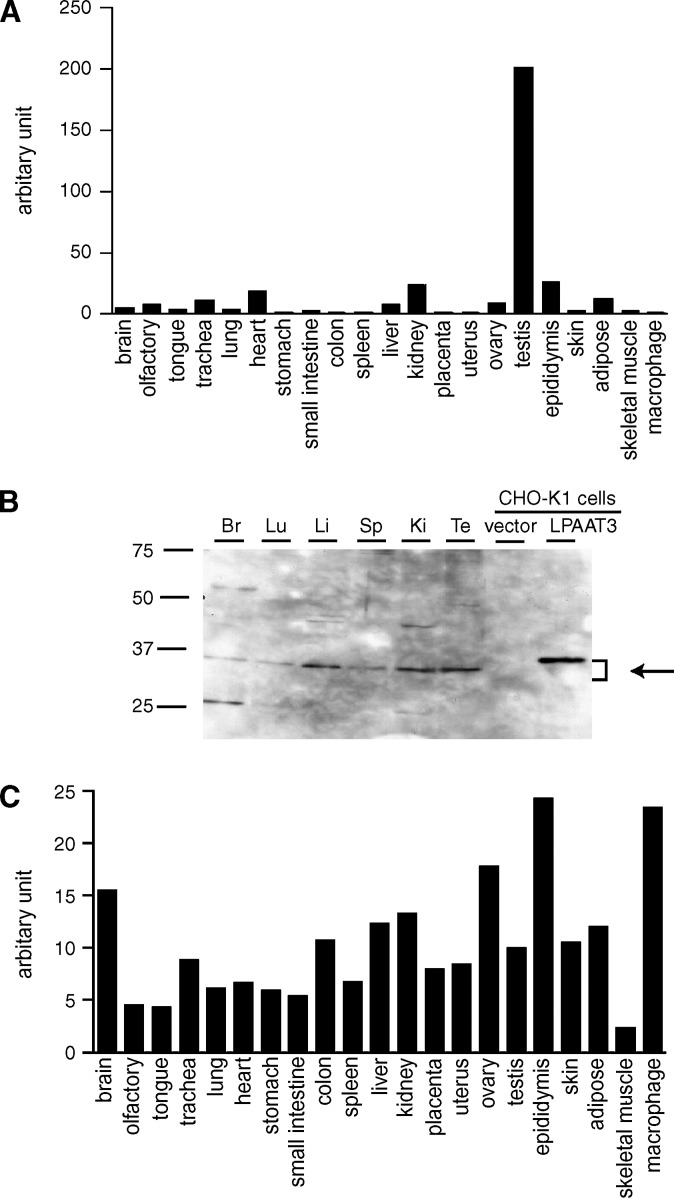
The tissue distribution of mLPAAT3 mRNA was analyzed by quantitative real-time PCR analysis. mLPAAT3 was predominantly expressed in the testis (Fig. 2A), whereas mMBOA7 was ubiquitously expressed (Fig. 2C). We examined mLPAAT3 expression profile in protein level by Western blot analysis using rabbit anti-mLPAAT3 antiserum. mLPAAT3 was highly expressed in the testis (Fig. 2B). mLPAAT3 was also expressed in the liver and kidney (Fig. 2B). Discrepancy between mRNA and protein expression level of mLPAAT3 may possibly be due to the difference in translational efficiency from mRNA into protein and/or half-life of this enzyme within tissues.
Fig. 2.
Expression profile of mLPAAT3 and mMBOA7 (LPIAT1) in mice. Expression levels of mLPAAT3 mRNA (A) and mMBOA7 (LPIAT1) mRNA (C) in 21 tissues from C57BL/6J mice were analyzed using quantitative real-time PCR. mLPAAT3 (A) was expressed predominantly in the testis, whereas mMBOA7 (LPIAT1) (C) was ubiquitously expressed. Similar results were obtained in a separate independent experiment. B: Expression of mLPAAT3 was analyzed at the protein level by Western blots using anti-mLPAAT3 antiserum. Three micrograms of 100,000 g pellets from various tissues were loaded in each lane. Br, Lu, Li, Sp, Ki, and Te stand for brain, lung, liver, spleen, kidney, and testis, respectively. mLPAAT3 was highly expressed in the testis. High expression was noted in the liver and kidney as well. The results are representative of three independent experiments.
Subcellular localization of FLAG-mLPAAT3 and endogenous mLPAAT3
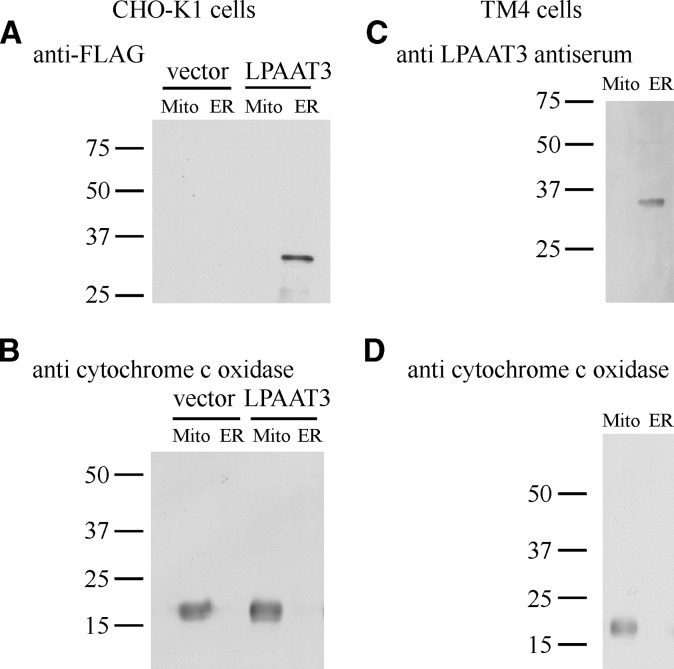
To facilitate immunocytochemical and Western blot analyses of mLPAAT3, we constructed an mLPAAT3 fusion protein that contains the FLAG epitope fused in frame to the N terminus of mLPAAT3. FLAG-mLPAAT3 was transfected into CHO-K1 cells, and the subcellular localization was examined by confocal microscopy after 48 h. Cells were stained for ER (anticalnexin N-terminal), Golgi (anti-GM130), or mitochondrial (anticytochrome c oxidase) markers. The subcellular distribution pattern of FLAG-mLPAAT3 was similar to that of calnexin N-terminal (see supplementary Fig. I). Neither Golgi nor mitochondrial marker protein distributions overlapped with mLPAAT3 (data not shown). To confirm these observations, CHO-K1 cells were transiently transfected with FLAG-mLPAAT3 and homogenized, and differential subcellular fractions were collected using an ER extraction kit (Sigma-Aldrich). When the subcellular fractions were analyzed by Western blots using anti-FLAG M2 antibody, the enzyme was found in ER fraction (Fig. 3A), consistent with the data obtained by confocal microscopy (see supplementary Fig. I). FLAG-mLPAAT3 had an apparent molecular mass of ∼37 kDa, slightly less than the predicted molecular mass of 43.3 kDa. The discrepancy in molecular mass was observed in other acyltransferases possessing multiple membrane spanning domains as previously described (17). To examine subcellular localization of endogenous mLPAAT3, subcellular fractions of testicular cell line TM4 cells were analyzed by Western blots using rabbit anti-mLPAAT3 antiserum. Consistent with the finding in the FLAG-mLPAAT3 overexpression system, endogenous mLPAAT3 was predominantly localized to the ER fraction (Fig. 3B).
Fig. 3.
Subcellular localization of FLAG-mLPAAT3 in CHO-K1 cells and endogenous mLPAAT3 in TM4 cells. A: At 48 h after transfection, proteins from CHO-K1 cells were subjected to Western blot analysis using anti-M2 FLAG antibody. Expression of FLAG-mLPAAT3 was confirmed by Western blots. Homogenates of pCXN2.1 vector or FLAG-mLPAAT3-pCXN2.1-transfected CHO-K1 cells were separated by differential centrifugation using an ER extraction kit (Sigma-Aldrich) as described in Materials and Methods. Two micrograms of 9,000 g pellets (indicated as Mito) and 100,000 g pellets (ER) were loaded in the lanes indicated. Molecular sizes are indicated on the left in kilodaltons. Results are representative of two independent experiments. B: As a marker of mitochondria, anticytochrome c oxidase antibody was used. Molecular sizes are indicated on the left in kilodaltons. Results are representative of two independent experiments. C: Subcellular localization of endogenous mLPAAT3 in testicular cell line TM4 cells was confirmed by Western blots. One and a half micrograms of 9,000 g pellets (Mito) and 100,000 g pellets (ER) were loaded in the lanes indicated. mLPAAT3 was detected using rabbit anti-mLPAAT3 antiserum. Results are representative of two independent experiments. D: As a marker of mitochondria, anticytochrome c oxidase antibody was used. Molecular sizes are indicated on the left in kilodaltons. Results are representative of two independent experiments.
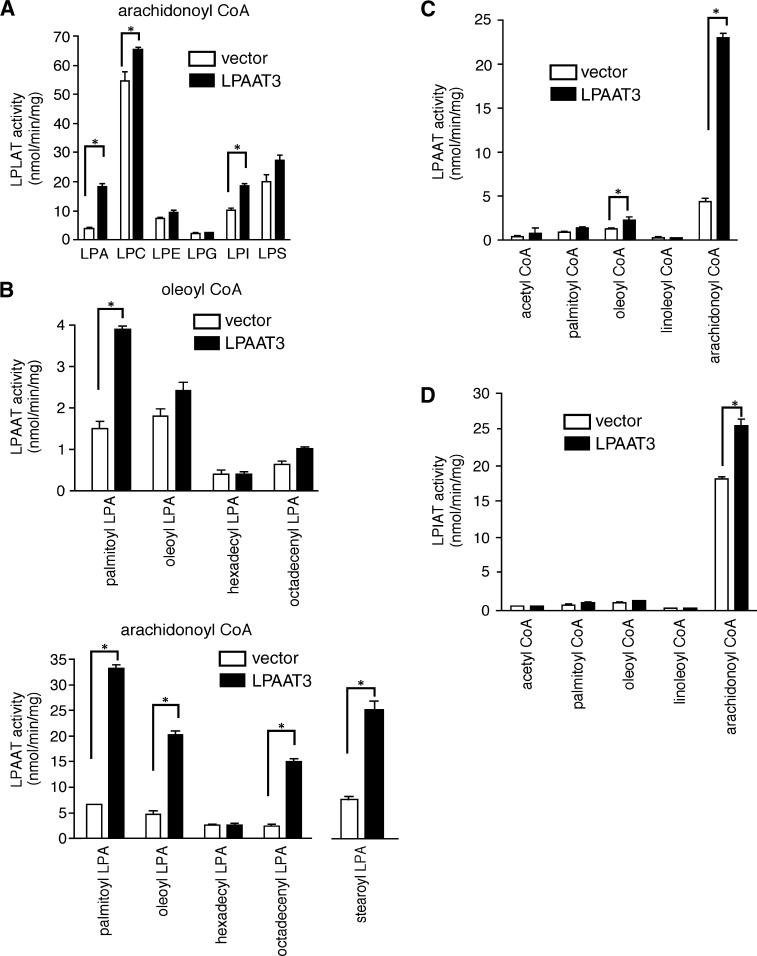
Substrate selectivity of mLPAAT3
Using [1-14C]arachidonoyl-CoA as an acyl donor, we analyzed the substrate specificity of mLPAAT3 using a variety of lysophospholipid acceptors (Fig. 4A). The microsomal fraction from CHO-K1 cells transfected with mLPAAT3 had detectable LPLAT activity for LPA, LPC, and LPI (Fig. 4A). However, its LPC acyltransferase activity was not pursued further because of its very limited activity. We also checked substrate selectivity using [1-14C]oleoyl-CoA as an acyl donor and did not observe any significant LPLAT activity of mLPAAT3 (data not shown).
Fig. 4.
Substrate selectivity of mLPAAT3. A: Lysophospholipid preferences of mLPAAT3 were determined. Acyltransferase activity was examined using 2 μg protein (100,000 g pellet), 25 μM [1-14C]arachidonoyl-CoA (33,000 dpm), and 50 μM lysophospholipids. Data are shown as mean + SD of triplicate measurements. Statistical significance was analyzed using ANOVA with Tukey post hoc pairwise comparisons. *P < 0.05. B: The preference of mLPAAT3 for various LPA acceptors was examined using oleoyl-CoA or arachidonoyl-CoA as a donor. Acyltransferase activity was examined using 2 μg protein, 25 μM [1-14C]oleoyl-CoA (33,000 dpm), and 50 μM lysophospholipids. Data are shown as mean + SD of triplicate measurements. Statistical significance was analyzed using ANOVA with Tukey post hoc pairwise comparisons. Only for stearoyl-LPA group, t-test was used for analysis. *P < 0.05. C: The acyl-CoA selectivity of mLPAAT3 was examined using palmitoyl-LPA as an acceptor. Acyltransferase activity was examined using 2 μg protein, 25 μM acyl-CoAs (33,000 dpm), and 50 μM palmitoyl-LPA, with the exception that 100 μM acetyl-CoA (111,000 dpm, 185MBq / mmol) was used. Data are shown as mean + SD of triplicate measurements. Statistical significance was analyzed using ANOVA with Tukey post hoc pairwise comparisons. *P < 0.05. D: The acyl-CoA selectivity of mLPAAT3 was examined for LPIAT activity. Acyltransferase activity was examined using 2 μg protein, 25 μM acyl-CoAs (33,000 dpm), and 50 μM LPI with exception of acetyl-CoA. The concentration of acetyl-CoA used was 100 μM (111,000 dpm, 185MBq/mmol). Data are shown as mean + SD of triplicate measurements. Statistical significance was analyzed using ANOVA with Tukey post hoc pairwise comparisons. *P < 0.05. In A–D, results are representative of two independent experiments.
Next, we examined the preference of mLPAAT3 for various LPA acceptors using [1-14C]oleoyl-CoA or [1-14C]arachidonoyl-CoA as acyl donors. As seen in Fig. 4B, mLPAAT3 exhibited strong LPAAT activity using arachidonoyl-CoA as a donor with palmitoyl-LPA, stearoyl-LPA, oleoyl-LPA, and octadecenyl-LPA as acceptors (Fig. 4B). We then examined the acyl-CoA selectivity of mLPAAT3 using palmitoyl-LPA as an acceptor. mLPAAT3 demonstrated a clear preference for arachidonoyl-CoA as a donor (Fig. 4C). We also examined the acyl-CoA selectivity of mLPAAT3 using 1-acyl-LPI as an acceptor. mLPAAT3 showed LPIAT activity using arachidonoyl-CoA as a donor (Fig. 4D). Other LPI acyltransferases, such as MBOA7, would explain the high endogenous activity with vector-transfected cells.
Kinetics of mLPAAT3 expressed in CHO-K1 cells
We examined the acyltransferase activity of mLPAAT3 using palmitoyl-LPA and arachidonoyl-CoA. The pH optimum for the reaction was between 7.4 and 10, and the reaction did not require Ca2+ (data not shown). A kinetic analysis was conducted by measuring acyltransferase activity in the microsomal fraction derived from vector- and mLPAAT3-transfected CHO-K1 cells, using increasing concentrations (1.5–100 μM) of [1-14C]arachidonoyl-CoA in the presence of 50 μM palmitoyl-LPA or using increasing concentrations (6.25–100 μM) of palmitoyl-LPA in the presence of 25 μM [1-14C]arachidonoyl-CoA (see supplementary Fig. IIA, B). The Km values of mLPAAT3 were 15.9 μM for arachidonoyl-CoA and 26.3 μM for palmitoyl-LPA. The corresponding Vmax values were 50.4 and 21.8 nmol/min/mg.
The role of the highly conserved motifs NHX4D and EGTR on enzyme activity
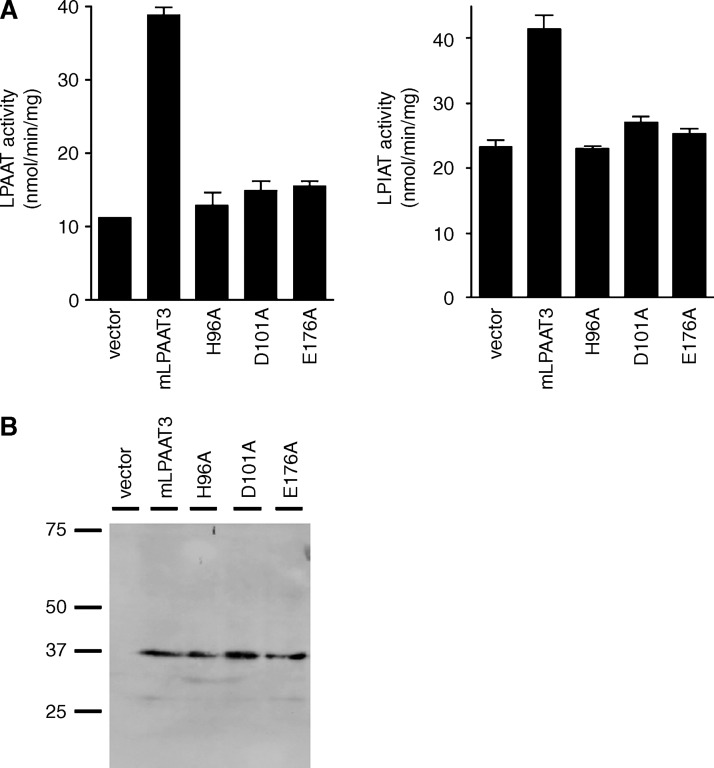
We constructed three single point mutants of mLPAAT3 (H96A, D101A, and E176A; arrows in Fig. 1B). Both LPAAT and LPIAT activities of mLPAAT3 were completely suppressed by these mutations (Fig. 5A). Expression of wild-type, H96A, D101A, and E176A mutants was confirmed by Western blot analysis (Fig. 5B). These results indicate that the motifs are critical for the enzymatic activity and that both enzymatic activities (LPAAT and LPIAT) reside on a single protein.
Fig. 5.
The role of highly conserved motifs NHX4D and EGTR in LPLAT activity of mLPAAT3. We constructed H96A, D101A, and E176A mutants of mLPAAT3 by mutating single amino acids in highly conserved motifs NHX4D or EGTR (Fig. 1B). A: The acyltransferase activity of mLPAAT3 wild type and three mutants was measured using 2 μg protein (100,000 g pellet), 50 μM palmitoyl-LPA, or LPI as an acceptor, and 25 μM arachidonoyl-CoA (33,000 dpm) as a donor. Data represent mean + SD of triplicate samples measurements. Statistical significance was analyzed using ANOVA with Tukey post hoc pairwise comparisons. *P < 0.05. The results are representative of two independent experiments. B: Expression of mLPAAT3 wild type and three mutants was confirmed by Western blot analysis. The results are representative of two independent experiments.
Age-dependent mLPAAT3 expression and LPAAT activity in the testis
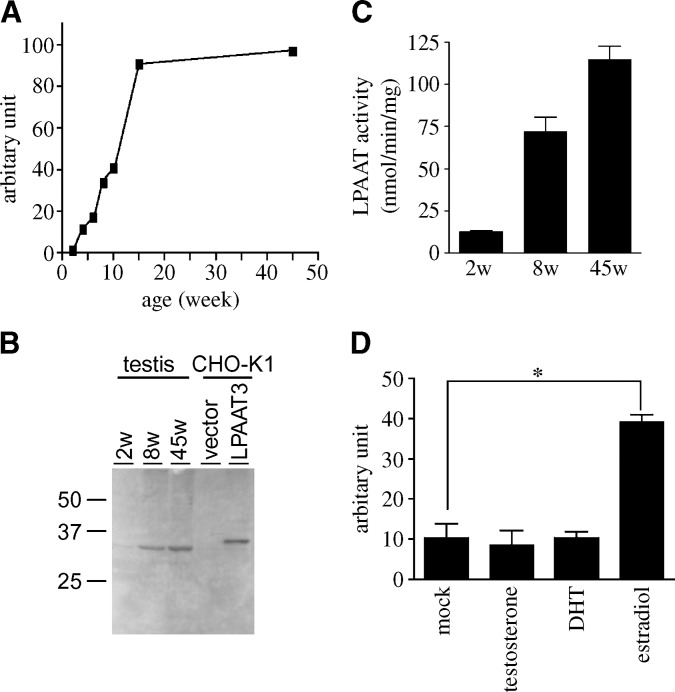
Total RNA of the testis was prepared from C57BL/6J mice at various ages. Interestingly, mLPAAT3 mRNA level was upregulated significantly until 15 weeks and then increased only slightly (Fig. 6A). In another independent experiment, after 15 weeks, mLPAAT3 expression level decreases slightly (data not shown). The trend of this age-dependent mLPAAT3 change was observed in protein levels as well by Western blot analysis (Fig. 6B). Additionally, LPAAT activity of the testis at 2, 8, and 45 weeks was checked using palmitoyl-LPA as an acceptor and arachidonoyl-CoA as a donor. We observed that LPAAT activity increased in an age-dependent fashion (Fig. 6C).
Fig. 6.
Age-dependent expression of mLPAAT3 in the testis and mLPAAT3 induction in testicular cell line. A: mLPAAT3 mRNA expression in the testis at various ages was compared using real-time quantitative PCR. mLPAAT3 mRNA expression is enhanced significantly in an age-dependent manner until 15 weeks of age. The results are representative of two independent experiments. B: mLPAAT3 protein expression in the testis at different ages was compared by Western blots using anti-mLPAAT3 antiserum. Four micrograms each of 100,000 g pellets at different ages were loaded. Results are representative of two independent experiments. C: LPAAT activity of 2, 8, and 45 weeks testis was examined using 1 μg protein (100,000 g pellet), 25 μM arachidonoyl-CoA (33,000 dpm), and 50 μM palmitoyl LPA. Data represent mean + SD of triplicate samples measurements. The results are representative of two independent experiments. D: Testicular cell line TM4 cells were treated with either mock, 100 nM β-estradiol, dihydrotestosterone (DHT), or testosterone for 24 h. mLPAAT3 mRNA level was compared using real-time quantitative PCR. β-Estradiol induced mLPAAT3 significantly. Data represent mean + SD of three independent experiments. Statistical significance was analyzed using ANOVA with Tukey post hoc pairwise comparisons. *P < 0.05.
The effect of sex hormones on mLPAAT3 expression
Since the level of sex hormones changes with age (50–56), we investigated whether age-dependent mLPAAT3 expression is derived from the induction by sex hormones. In testicular cell line TM4 cells, mLPAAT3 was upregulated significantly with β-estradiol treatment for 24 h (Fig. 6D).
The substrate selectivity of mLPAAT1
We examined the acyl-CoA selectivity of mLPAAT1 using palmitoyl-LPA as an acceptor. mLPAAT1 demonstrated LPAAT activity using palmitoyl-LPA as an acceptor and palmitoyl-CoA, oleoyl-CoA, and arachidonoyl-CoA as a donor (see supplementary Fig. III). Thus, at least mLPAAT1 seems to be different from mLPAAT3 in acyl-CoA preference.
DISCUSSION
Here, we have presented the first detailed characterization of mLPAAT3. mLPAAT3, previously known as mAGPAT3, is a 43.3 kDa protein with four putative transmembrane domains and is localized in ER. Interestingly, mLPAAT3 has a clear preference for arachidonoyl-CoA as a donor in the synthesis of PA through the de novo pathway. Additionally, the enzyme exhibited LPIAT activity using arachidonoyl-CoA as a donor, suggesting that mLPAAT3 can function in the remodeling pathway as well. Prior to this report, mouse acyl-CoA:lysocardiolipin acyltransferase (ALCAT1 or LCLAT1) and mLPCAT1 were the known enzymes that could synthesize phospholipids both by the de novo and remodeling pathways (25, 57). LPT1 (otherwise known as ALE1 or SLC4) in yeast (58–61) and human MBOAT2 (62) are acyltransferases, which can synthesize phospholipids by both pathways in other species. As enzymes with LPIAT activity, LPT1 (ALE1 or SLC4) (58–61) and MBOA7 (MBOAT7 or LPIAT1) (35, 62) had been described. The highly conserved motifs NHX4D and EGTR are important for LPAAT and LPIAT activities of mLPAAT3, as point mutations in either of these motifs completely suppressed both activities.
At least seven mouse AGPAT family members exist, but their biological functions remain to be determined. At this point, even their biochemical characteristics are not well defined. Among them, LPAAT activities of mLPAAT1 and mLPAAT2 have been relatively well documented. mLPAAT1 exhibits LPAAT activity using oleoyl-LPA as an acceptor and palmitoyl-CoA, oleoyl-CoA, and arachidonoyl-CoA as donors (21). We also demonstrated that mLPAAT1 exhibits LPAAT activity using palmitoyl-LPA as an acceptor and prefers palmitoyl-CoA, oleoyl-CoA, and arachidonoyl-CoA as donors (see supplementary Fig. III). mLPAAT2 has LPAAT activity using oleoyl-LPA as an acceptor and oleoyl-CoA as a donor (37). The human homolog hLPAAT1 shows the highest activity with linoleoyl-LPA, palmitoyl-LPA, and myristoyl-LPA as acceptors and linoleoyl-CoA and palmitoyl-CoA as donors (40). hLPAAT2 exhibits LPAAT activity with palmitoyl LPA as an acceptor and myristoyl-CoA, palmitoyl-CoA, stearoyl-CoA, and arachidonyl-CoA as donors (34). Furthermore, mutations in hLPAAT2 cause congenital generalized lipodystrophy (41). To elucidate the biological roles of AGPAT family members, it will be particularly important to investigate their tissue distributions and biochemical characteristics.
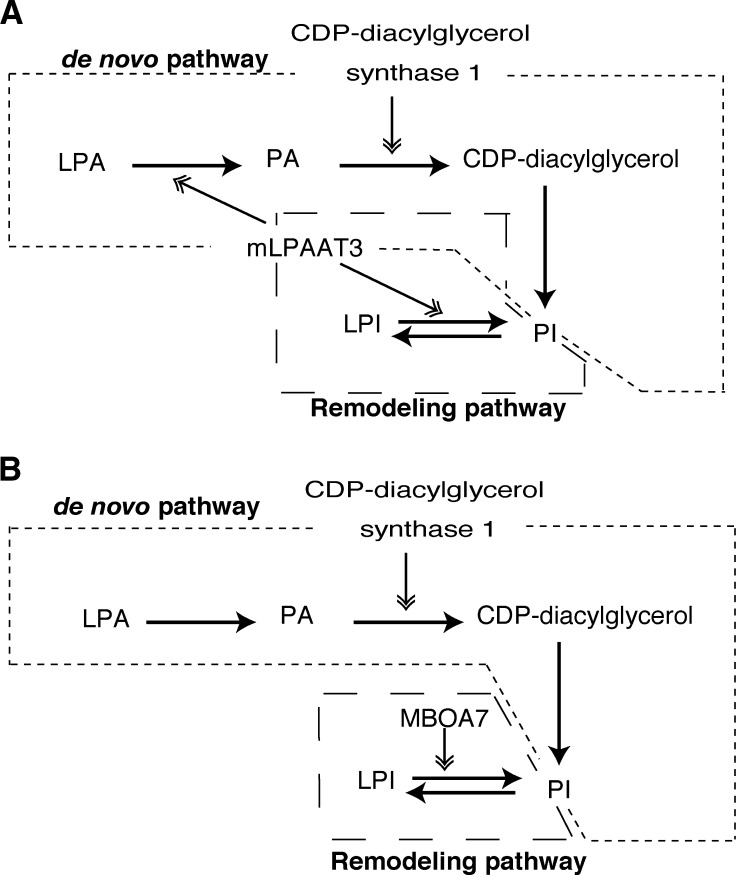
We found that mLPAAT3 possessed a strong LPAAT activity using arachidonoyl-CoA and is highly expressed in the testis (Fig. 2A, B). This is particularly interesting because: i) PA has a low arachidonic acid content at the sn-2 position in most tissues (11–13), and ii) CDP-diacylglycerol synthase 1 is highly localized to the testis and converts preferably 1-stearoyl-2-arachidonoyl-PA as a substrate to CDP-diacylglycerol, a precursor of glycerophospholipids including PI (42, 43). Since mLPAAT3 exhibits LPAAT activity using 1-stearoyl LPA and arachidonoyl-CoA, the enzyme may be functionally coupled with CDP-diacylglycerol synthase 1 to generate PI through the de novo pathway (Fig. 7A). Considering that mLPAAT3 also exhibited LPIAT activity with arachidonoyl-CoA, it can possibly help to produce PI effectively through both the de novo and remodeling pathways in the testis (Fig. 7A). Previously identified LPIAT (mouse MBOA7) is expressed ubiquitously, including in the testis (Fig. 2C). Therefore, MBOA7 and CDP-diacylglycerol synthase 1 may be functionally coupled to produce PI effectively as well (Fig. 7B). There might still exist other enzymes to assist in producing PI in the testis. Limitation of our study is that the biochemical characterization of mLPAAT3 was done using an overexpression system because the purification of this enzyme is extremely difficult due to its multiple transmembrane domains.
Fig. 7.
Proposed model for PI production in the testis. mLPAAT3 and MBOA7 (LPIAT1) may be important enzymes for producing PI in the testis. A: Both mLPAAT3 (Fig. 2A) and CDP-diacylglycerol synthase 1 (42, 43) are highly expressed in the testis. CDP-diacylglycerol synthase 1 prefers 1-stearoyl-2-arachidonoyl PA as a substrate, and mLPAAT3 shows LPAAT activity using stearoyl-LPA as an acceptor and arachidonoyl-CoA as a donor. Since 1-stearoyl-2-arachidonoyl species are major components of PI, we hypothesized that in the testis, mLPAAT3 helps to produce PI effectively through both the de novo and remodeling pathways. B: Also in the testis, CDP-diacylglycerol synthase 1 (42, 43) and MBOA7 (LPIAT1) (Fig. 2C) may generate PI effectively through the de novo and remodeling pathways, respectively. In A and B, the de novo and remodeling pathways are enclosed in dotted lines.
Additionally, mLPAAT3 expression in the testis is enhanced significantly in an age-dependent manner while relatively young and then keeps a steady level (Fig. 6A). β-Estradiol induces mLPAAT3 in TM4 cells (Fig. 6C). In males, β-estradiol is produced by aromatization of testosterone. In human, β-estradiol increases initially throughout sex maturation (50) and then either slightly increases (51), remains steady (52, 53), or even slightly decreases (54–56) with age depending on studies. The reason for these differences of β-estradiol levels in various studies is unclear but may be due to a wide range of β-estradiol level in the elderly. Considering that this induction of β-estradiol in human is similar to that of mLPAAT3, β-estradiol may be a main regulator of mLPAAT3 expression.
In conclusion, we have isolated an enzyme, mLPAAT3, which catalyzes PA and PI production in the de novo and remodeling pathways, respectively. mLPAAT3 is highly expressed in the testis and exhibits a clear preference for arachidonoyl-CoA. In conjunction with CDP-diacylglycerol synthase 1, this enzyme may play an important role in PI production in the testis (Fig. 7A). Additionally, mLPAAT3 expression increases age dependently while relatively young, and β-estradiol seems to be its important regulator. Further studies are needed to elucidate the roles of mLPAAT3 in vivo, in particular in the relationship with β-estradiol.
Supplementary Material
Acknowledgments
The authors thank Drs. M.Nakamura, S.Ishi, Y.Kita, and all the members in the laboratory for valuable comments. The authors also thank Dr. J-I. Miyazaki for supplying pCXN2.
Abbreviations
AGPAT, 1-acylglycerol-3-phosphate O-acyltransferase
CDP-diacylglycerol, cytidine diphosphodiacylglycerol
CHO, Chinese hamster ovary
DDBJ, DNA Data Bank of Japan
ER, endoplasmic reticulum
LPAAT, lysophosphatidic acid acyltransferase
LPIAT, lysophosphatidylinositol acyltransferase
LPLAT, lysophospholipid acyltransferases
NCBI, National Center for Biotechnology Information
PA, phosphatidic acid
PI, phosphatidylinositol
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.S.). T.S. and H.S. were supported by the Center for NanoBio Integration at the University of Tokyo. H.S. was supported by Health and Labour Sciences Research Grants (Research on Allergic Disease and Immunology) supported by the Ministry of Health, Labour, and Welfare of Japan, Mitsubishi Pharma Research Foundation, and Ono Medical Research Foundation.
Published, JLR Papers in Press, December 29, 2008.
Footnotes
Nucleotide sequence data are available in the DDBJ/EMBL/GenBank databases under the accession numbers AB377215 (mouse).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table and three figures.
References
- 1.MacDonald J. I., and H. Sprecher. 1991. Phospholipid fatty acids remodeling in mammalian cells. Biochim. Biophys. Acta. 1084 105–121. [DOI] [PubMed] [Google Scholar]
- 2.Wood R., and R. D. Harlow. 1969. Structural analyses of rat liver phosphoglycerides. Arch. Biochem. Biophys. 135 272–281. [DOI] [PubMed] [Google Scholar]
- 3.Schlame M., S. Brody, and K. Y. Hosetler. 1993. Mitochondrial cardiolipin in diverse eukaryotes. Comparison of biosynthetic reactions and molecular acyl species. Eur. J. Biochem. 212 727–735. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita A., T. Sugiura, and K. Waku. 1997. Acyltransferases and transacylases involved in fatty acids remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells. J. Biochem. 122 1–16. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy E. P. 1961. Biosynthesis of complex lipids. Fed. Proc. 20 934–940. [PubMed] [Google Scholar]
- 6.Shindou, H., and T. Shimizu. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. In press. [DOI] [PubMed]
- 7.Shindou, H., D. Hishikawa, T. Harayama, K. Yuki, and T. Shimizu. Recent progress on Acyl-CoA: lysophospholipid acyltransferase research. J. Lipid Res. In press. [DOI] [PMC free article] [PubMed]
- 8.Lands W. E. 1960. Metabolism of glycerolipids. II. The enzymatic acylation of lysolecithin. J. Biol. Chem. 235 2233–2237. [PubMed] [Google Scholar]
- 9.Lands W. E., and I. Merkl. 1963. Metabolism of glycerolipids. III. Reactivity of various acyl esters of coenzyme A with alpha-acylglycerophosphorylcholine, and positional specificities in lecithin synthesis. J. Biol. Chem. 238 898–904. [PubMed] [Google Scholar]
- 10.Merkl I., and W. E. Lands. 1963. Metabolism of glycerolipids. IV. Synthesis of phosphatidylethanolamine. J. Biol. Chem. 238 905–906. [PubMed] [Google Scholar]
- 11.Possmayer F., G. L. Scherphof, T. M. Dubbelman, L. M. van Golde, and L. L. van Deenen. 1969. Positional specificity of saturated and unsaturated fatty acids in phosphatidic acid from rat liver. Biochim. Biophys. Acta. 176 95–110. [DOI] [PubMed] [Google Scholar]
- 12.Akesson B., J. Elovson, and G. Arvidson. 1970. Initial incorporation into rat liver glycerolipids of intraportally injected (3H) glycerol. Biochim. Biophys. Acta. 210 15–27. [DOI] [PubMed] [Google Scholar]
- 13.Baker R. R., and W. Thompson. 1972. Positional distribution and turnover of fatty acids in phosphatidic acid, phosphinositides, phosphatidylcholine and phosphatidylethanolamine in rat brain in vivo. Biochim. Biophys. Acta. 270 489–503. [DOI] [PubMed] [Google Scholar]
- 14.Holub B. J., and A. Kulsis. 1971. Structural and metabolic interrelationships among glycerophosphatides of rat liver in vivo. Can. J. Biochem. 49 1347–1356. [DOI] [PubMed] [Google Scholar]
- 15.Shindou H., D. Hishikawa, H. Nakanishi, T. Harayama, S. Ishii, R. Taguchi, and T. Shimizu. 2007. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:Lyso-PAF acetyltransferase. J. Biol. Chem. 282 6532–6539. [DOI] [PubMed] [Google Scholar]
- 16.Nakanishi H., H. Shindou, D. Hishikawa, T. Harayama, R. Ogasawara, A. Suwabe, R. Taguchi, and T. Shimizu. 2006. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem. 281 20140–20147. [DOI] [PubMed] [Google Scholar]
- 17.Hishikawa D., H. Shindou, S. Kobayashi, H. Nakanishi, R. Taguchi, and T. Shimizu. 2008. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA. 105 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cases S., S. J. Stone, P. Zhou, E. Yen, B. Tow, K. D. Lardizabal, T. Voelker, and R. V. Farese, Jr. 2001. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 276 38870–38876. [DOI] [PubMed] [Google Scholar]
- 19.Yen C. L., S. J. Stone, S. Cases, P. Zhou, and R. V. Farese, Jr. 2002. Identification of a gene encoding MGAT1, a monoacylglycerol acyltransferase. Proc. Natl. Acad. Sci. USA. 99 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin D. H., J. D. Paulaauskis, N. Moustaid, and H. S. Sul. 1991. Transcriptional regulation of p90 with sequence homology to Escherichia coli glycerol-3-phosphate acyltransferase. J. Biol. Chem. 266 23834–23839. [PubMed] [Google Scholar]
- 21.Kume K., and T. Shimizu. 1997. cDNA cloning and expression of murine 1-acyl-sn-glycerol-3-phosphate acyltransferase. Biochem. Biophys. Res. Commun. 237 663–666. [DOI] [PubMed] [Google Scholar]
- 22.West J., C. K. Tompkins, N. Balantac, E. Nudelman, B. Meengs, T. White, S. Bursten, J. Coleman, A. Kumar, J. W. Singer, et al. 1997. Cloning and expression of two human lysophosphatidic acid acyltransferase cDNAs that enhance cytokine-induced signaling responses in cells. DNA Cell Biol. 16 691–701. [DOI] [PubMed] [Google Scholar]
- 23.Stamps A. C., M. A. Elmore, M. E. Hill, K. Kelly, A. A. Makda, and M. J. Finnen. 1997. A human cDNA sequence with homology to non-mammalian lysophosphatidic acid acyltransferases. Biochem. J. 326 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., J. Cao, and Y. Shi. 2004. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J. Biol. Chem. 279 55866–55874. [DOI] [PubMed] [Google Scholar]
- 25.Cao J., Y. Liu, J. Lockwood, P. Burn, and Y. Shi. 2004. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 279 31727–31734. [DOI] [PubMed] [Google Scholar]
- 26.Cao J., D. Shan, T. Revett, D. Li, L. Wu, J. F. Tobin, and R. E. Gimeno. 2008. Molecular identification of a novel mammalian brain isoform of acyl-CoA:lysophospholipid acyltransferase with prominent ethanolamine lysophospholipid acylating activity, LPEAT2. J. Biol. Chem. 283 19049–19057. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y., Y. Q. Chen, T. M. Bonacci, D. S. Bredt, S. Li, W. R. Bensch, D. E. Moller, M. Kowala, R. J. Konrad, and G. Cao. 2008. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J. Biol. Chem. 283 8258–8265. [DOI] [PubMed] [Google Scholar]
- 28.Harada N., S. Hara, M. Yoshida, T. Zenitani, K. Mawatari, M. Nakano, A. Takahashi, T. Hosaka, K. Yoshimoto, and Y. Nakaya. 2007. Molecular cloning of a murine glycerol-3-phosphate acyltransferase-like protein 1 (xGPAT1). Mol. Cell. Biochem. 297 41–51. [DOI] [PubMed] [Google Scholar]
- 29.Wang S., D. P. Lee, N. Gond, N. M. Schwebrock, D. G. Mashek, M. R. Gonzalez-Baro, C. Stapleton, L. O. Li, T. M. Lewin, and R. A. Coleman. 2007. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2). Arch. Biochem. Biophys. 465 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao J., J. L. Li, D. Li, J. F. Tobin, and R. E. Gimeno. 2006. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 103 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yet S. F., S. Lee, Y. T. Hahm, and H. S. Sul. 1993. Expression and Identification of P90 as the Murine Mitochondrial Glycerol-3-Phosphate Acyltransferase. Biochemistry. 32 9486–9491. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y. Q., M. S. Kuo, S. Li, H. H. Bui, D. A. Peake, P. E. Sanders, S. J. Thibodeaux, S. Chu, Y. W. Qian, Y. Zhao, et al. 2008. Agpat6 is a novel microsomal glycerol-3-phosphate acyltransferase. J. Biol. Chem. 283 10048–10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagle C. A., L. Vergnes, H. Dejong, S. Wang, T. M. Lewin, K. Reue, and R. A. Coleman. 2008. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J. Lipid Res. 49 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eberhardt C., P. W. Gray, and L. W. Tjoelker. 1997. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J. Biol. Chem. 272 20299–20305. [DOI] [PubMed] [Google Scholar]
- 35.Lee H. C., T. Inoue, R. Imae, N. Kono, S. Shirae, S. Matsuda, K. Gengyo-Ando, S. Mitani, and H. Arai. 2008. Caenorhabditis elegans mboa-7, a member of the MBOAT family is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 19 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dircks L., and H. S. Sul. 1999. Acyltransferases of de novo glycerophospholipid biosynthesis. Prog. Lipid Res. 38 461–479. [DOI] [PubMed] [Google Scholar]
- 37.Lu B., Y. J. Jiang, Y. Zhou, F. Y. Xu, G. M. Hatch, and P. C. Choy. 2005. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem. J. 385 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewin T. M., P. Wang, and R. A. Coleman. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 38 5764–5771. [DOI] [PubMed] [Google Scholar]
- 39.Harayama T., H. Shindou, R. Ogasawara, A. Suwabe, and T. Shimizu. 2008. Identification of a novel noninflammatory biosynthetic pathway of platelet-activating factor. J. Biol. Chem. 283 11097–11106. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita A., H. Nakanishi, H. Suzuki, R. Kamata, K. Tanaka, K. Waku, and T. Sugiura. 2007. Topology of acyltransferase motifs and substrate specificity and accessibility in 1-acyl-sn-glycero-3-phosphate acyltransferase 1. Biochim. Biophys. Acta. 1771 1202–1215. [DOI] [PubMed] [Google Scholar]
- 41.Agarwal A. K., E. Arioglu, S. De Almeida, N. Akkoc, S. I. Taylor, A. M. Bowcock, R. I. Barnes, and A. Garg. 2002. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 31 21–23. [DOI] [PubMed] [Google Scholar]
- 42.Saito S., K. Goto, A. Tonosaki, and H. Kondo. 1997. Gene cloning and characterization of CDP-diaylglycerol synthase from rat brain. J. Biol. Chem. 272 9503–9509. [DOI] [PubMed] [Google Scholar]
- 43.Inglis-Broadgate S. L., L. Ocaka, R. Banerjee, M. Gaasenbeek, J. P. Chapple, M. E. Cheetham, B. J. Clark, D. M. Hunt, and S. Halford. 2005. Isolation and characterization of murine Cds (CDP-diacylglycerol synthase) 1 and 2. Gene. 356 19–31. [DOI] [PubMed] [Google Scholar]
- 44.Niwa H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108 193–199. [DOI] [PubMed] [Google Scholar]
- 45.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- 46.Bligh E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37 911–917. [DOI] [PubMed] [Google Scholar]
- 47.Thompson J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arai M., H. Mitsuke, M. Ikeda, J-X. Xia, T. Kikuchi, M. Satake, and T. Shimizu. 2004. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32 W390–W393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shikano S., and M. Li. 2003. Membrane receptor trafficking: evidence of proximal and distal zones conferred by two independent endoplasmic reticulum localized signals. Proc. Natl. Acad. Sci. USA. 100 5783–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson A. M., A. Juul, J. H. Petersen, J. Mueller, N. P. Groome, and N. E. Skakkebaek. 1997. Serum inhibin B in healthy pubertal and adolescent boys: relation to age, stage of puberty, and follicle-stimulating hormone, luteinizing hormone, testosterone, and estradiol levels. J. Clin. Endocrinol. Metab. 82 3976–3981. [DOI] [PubMed] [Google Scholar]
- 51.Bjørnerem A., B. Straume, M. Midtby, V. Fønnebø, J. Sundsfjord, J. Svartberg, G. Acharya, P. Oian, and G. K. Bertsen. 2004. Endogenous sex hormones in relation to age, sex, lifestyle factors, and chronic diseases in a population: the Tromsø Study. J. Clin. Endocrinol. Metab. 89 6039–6047. [DOI] [PubMed] [Google Scholar]
- 52.Muller M., I. den Tonkelaar, J. H. Thijssen, D. E. Grobbee, and Y. T. van der Schouw. 2003. Endogenous sex hormones in men aged 40–80 years. Eur. J. Endocrinol. 149 583–589. [DOI] [PubMed] [Google Scholar]
- 53.Bélanger A., B. Candas, A. Dupont, L. Cusan, P. Diamond, J. L. Gomez, and F. Labrie. 1994. Changes in serum concentrations of conjugated and unconjugated steroids in 40–80 year old men. J. Clin. Endocrinol. Metab. 79 1086–1090. [DOI] [PubMed] [Google Scholar]
- 54.Ferrini R. L., and E. Barret-Conner. 1998. Sex hormones and age: a cross-sectional study of testosterone and estradiol and their bioavailable fractions in community-dwelling men. Am. J. Epidemiol. 147 750–754. [DOI] [PubMed] [Google Scholar]
- 55.Orwoll E., L. C. Lambert, L. M. Marshall, K. Phipps, J. Blank, E. Barret-Conner, J. Cauley, K. Ensrud, and S. Cummings. 2006. Testosterone and estradiol among older men. J. Clin. Endocrinol. Metab. 91 1336–1344. [DOI] [PubMed] [Google Scholar]
- 56.van den Beld A. W., F. H. de Jong, D. E. Grobbee, H. A. Pols, and S. W. Lamberts. 2000. Measures of bioavailable serum testosterone and estradiol and their relationships with muscle strength, bone density, and body composition in elderly men. J. Clin. Endocrinol. Metab. 85 3276–3286. [DOI] [PubMed] [Google Scholar]
- 57.Chen X., B. A. Hyatt, M. L. Mucenski, R. J. Mason, and J. M. Shannon. 2006. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc. Natl. Acad. Sci. USA. 103 11724–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain S., N. Stanford, N. Bhagwat, N. Seiler, M. Costanzo, C. Boone, and P. Oelkers. 2007. Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282 30562–30569. [DOI] [PubMed] [Google Scholar]
- 59.Benghezal M., C. Roubaty, V. Veepuri, J. Knudsen, and A. Conzelmann. 2007. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J. Biol. Chem. 282 30845–30855. [DOI] [PubMed] [Google Scholar]
- 60.Tamaki H., A. Shimada, Y. Ito, M. Ohya, J. Takase, M. Miyashita, H. Miyagawa, H. Nozaki, R. Nakayama, and H. Kumagai. 2007. LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophopholipids in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 282 34288–34298. [DOI] [PubMed] [Google Scholar]
- 61.Riekhof W. R., J. Wu, M. A. Gijón, S. Zarini, R. C. Murphy, and D. R. Voelker. 2007. Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 282 36853–36861. [DOI] [PubMed] [Google Scholar]
- 62.Gijón M. A., W. R. Riekhof, S. Zarini, R. C. Murphy, and D. R. Voelker. 2008. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J. Biol. Chem. 283 30235–30245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.