Abstract
Pregnane X receptor (PXR) is an important nuclear receptor xenosensor that regulates the expression of metabolic enzymes and transporters involved in the metabolism of xenobiotics and endobiotics. In this study, ultra-performance liquid chromatography (UPLC) coupled with electrospray time-of-flight mass spectrometry (TOFMS), revealed altered urinary metabolomes in both Pxr-null and wild-type mice treated with the mouse PXR activator pregnenolone 16α-carbonitrile (PCN). Multivariate data analysis revealed that PCN significantly attenuated the urinary vitamin E metabolite α-carboxyethyl hydroxychroman (CEHC) glucuronide together with a novel metabolite in wild-type but not Pxr-null mice. Deconjugation experiments with β-glucuronidase and β-glucosidase suggested that the novel urinary metabolite was γ-CEHC β-D-glucoside (Glc). The identity of γ-CEHC Glc was confirmed by chemical synthesis and by comparing tandem mass fragmentation of the urinary metabolite with the authentic standard. The lower urinary CEHC was likely due to PXR-mediated repression of hepatic sterol carrier protein 2 involved in peroxisomal β-oxidation of branched-chain fatty acids (BCFA). Using a combination of metabolomic analysis and a genetically modified mouse model, this study revealed that activation of PXR results in attenuated levels of the two vitamin E conjugates, and identification of a novel vitamin E metabolite, γ-CEHC Glc. Activation of PXR results in attenuated levels of the two vitamin E conjugates that may be useful as biomarkers of PXR activation.
Keywords: pregnane X receptor, pregnenolone 16α-carbonitrile, carboxyethyl hydroxychroman, metabolomics, γ-CEHC glucoside, sterol carrier protein 2, peroxisomal beta-oxidation
Pregnane X receptor (PXR, NR1I2) is a member of the nuclear receptor superfamily and highly expressed in liver and intestine of mammals. PXR was characterized as a xenosensor that regulates the expression of metabolic enzymes and transporters facilitating elimination of xenobiotics and endogenous toxic chemicals such as bile acids (1). The mechanisms of how PXR regulates numerous genes involved in the metabolism of chemicals have been investigated during the last decade. Results from those studies indicated that PXR activation is dependent on its ligands and PXR direct repeat 4 (DR-4) response elements (PXRE) usually located upstream of target genes. When a ligand binds to PXR, PXR translocates to the nucleus, heterodimerizes with retinoid X receptor (RXR, NR1B) and binds to PXREs. PXR can be activated by structurally diverse chemicals including certain pharmaceuticals, herbal medicines, dietary supplements, environmental pollutants, and endobiotics (2–4). However, PXR displays a marked species difference in response to PXR ligands; pregnenolone 16α-carbonitrile (PCN) and dexamethasone are the best known activators of rodent PXR but are weak activators of human PXR, whereas drugs such as rifampicin (RIF) and clotrimazole are strong human PXR activators but are weak activators of rodent PXR (5). This species-selective ligand specificity has led to the generation of PXR-humanized mouse models (6–8).
Subsequent genetic profiling studies have shown that PXR activation regulates the expression of a large network of genes. In genetic profiling for rats treated with the rodent-specific ligand, PCN, hundreds of genes were differentially expressed (9). In PXR-humanized mice with constitutively highly expressed human PXR, 146 genes were up- or down-regulated (10). Among the up-regulated genes by PXR activation, many were known PXR target genes. The most significantly induced are the cytochrome P450s CYP3A and CYP2B subfamilies (11, 12). Conjugating enzymes such as UDP-glucuronosyltransferase (UGT) and glutathione S-transferase and transporters, such as P-glycoprotein, multidrug resistance-related protein-3 (MRP3), and organic anion transporting polypeptide-2 (OATP2), are also induced following PXR activation (13–17). Metabolic enzymes and transporters induced by PXR activation affect the absorption, distribution, metabolism, and excretion of xenobiotics and endobiotics.
More recent studies have revealed that PXR has a physiological function in hepatic energy metabolism and plays an important role of the regulation of certain metabolic pathways. Activation of PXR represses hepatic energy metabolism by decreasing hepatic gluconeogenesis, fatty acid oxidation, and ketogenesis, and increasing triglyceride synthesis (18). This perturbation of energy homeostasis by PXR is achieved through crosstalk with various hormone responsive transcription factors such as forkhead box O1 (FoxO1), forkhead box A2 (FoxA2), and peroxisome proliferators activated receptor γ coactivator 1α (PGC1α) (19–21).
Recently, global approaches to investigate the effects on the metabolome of nuclear receptor activation have been made using the high-resolution capability of ultra-performance liquid chromatography (UPLC) coupled with the accurate mass determination of time-of-flight mass spectrometry (TOFMS). In addition, the use of genetically modified mouse models has yielded important results that help understand the physiological and toxicological consequences of nuclear receptor activation. This combination of mass spectrometry-based metabolomics and genetically modified mice has revealed that activation of peroxisome proliferators activated receptor α (PPARα) affects the metabolism of tryptophan, corticosterone, and fatty acids (22, 23), and that novel steroid metabolites in urine can be biomarkers for activation of PPARα (22).
In this study, metabolomic responses to PXR activation by PCN in Pxr-null and wild-type mice were investigated using UPLC-TOFMS coupled with multivariate data analyses. Metabolomic analysis may be useful to identify novel biomarkers for activation of PXR and to discover metabolic pathways regulated by PXR.
EXPERIMENTAL PROCEDURES
Reagents
Pregnenolone 16α-carbonitrile (PCN), RIF, β-D-glucuronidase (type H-1 from Helix pomatia), β-D-glucosidase (from almonds), ethyl levulinate, vinyl magnesium bromide, 2,3-dimethylhydroquinone, and boron trifluoride diethyl etherate were purchased from Sigma (St. Louis, MO). Tetraacetyl glucose was purchased from Wako Pure Chemical (Osaka, Japan). HPLC-grade solvents (acetonitrile, methanol, and water) were purchased from Fisher Scientific (Hampton, NH).
Animals, treatments, and urine collections
Pxr-null (kindly provided by Steven A. Kliewer and GlaxoSmithKline), wild-type, and PXR-humanized mice were maintained under a standard 12-h light/12-h dark cycle with water and a normal diet (NIH-31) provided ad libitum. The mouse background was C57BL/6. Pxr-null and PXR-humanized mice were described previously (6, 24). Groups of 8- to 12-week-old males were used for all experiments, put on synthetic purified diet (AIN-93, Bio-Serv) at day 5 before treatment and maintained with water and the same diet ad libitum. PCN was suspended in corn oil and administered at a dose of 50 mg/kg/day and administered daily by intraperitoneal (i.p.) injection to wild-type and Pxr-null mice for 4 days. Control mice were injected i.p. with vehicle (corn oil). Urine samples were collected from mice placed individually in metabolic cages for 24 h, before treatment and 24 h after the last dosing. Pxr-null and PXR-humanized mice were fed with a RIF-containing diet (approximately 10 mg/kg/day) or control diet (AIN-93) for 4 weeks. Urine samples were collected on the 28th day of treatment. All urine samples were stored at −80°C until analyzed. At the end of the study, animals were killed at 48 h after the last dose. Serum and liver tissue were collected and frozen at −80°C for further analysis. Protocols for all animal studies were approved by the National Cancer Institute Animal Care and Use Committee and were carried out in accordance with the guidelines of Institute of Laboratory Animal Resources.
UPLC-TOFMS analyses
Urine aliquots were diluted with 4 vols of 50% acetonitrile and centrifuged at 18,000 g for 20 min at 4°C to remove particles and proteins. Serum samples were diluted 20-fold with 66% acetonitrile and centrifuged. The aliquots (5 μl) were injected into a UPLC-TOFMS system using ACQUITY® UPLC and Q-TOF Premier® (Waters Corp., Milford, MA). The liquid chromatographic and mass spectrometric conditions of the UPLC-TOFMS system was the same as previously reported (22).
In vitro PCN hydroxylation
To exclude PCN-derived ions from the chromatographic mass data matrix, in vitro incubation of PCN with mouse liver microsomes was performed. The in vitro incubation mixture contained, in a total volume of 200 μl, 2 mM NADPH, 100 μg of mouse liver microsomal protein in 0.1 M sodium phosphate buffer, pH 7.4, and 50 μM PCN. Reactions were initiated by the addition of NADPH at 37°C for 30 min, and they were terminated by the addition of 200 μl of ice-cold acetonitrile. Boiled mouse liver microsomal protein was used for control reactions. MetaboLynx® (Waters) was used to generate a table of ions that derived from PCN, especially the parent compound ([M+H]+ 342.245) and hydroxylated metabolites ([M+OH]+ 358.240), and their in-source fragment ions.
Data processing and multivariate data analysis
Centroided and integrated chromatographic mass data from 50 to 800 m/z were processed by MarkerLynx® (Waters) to generate a multivariate data matrix. Ions that were obviously derived from PCN in the in vitro incubation were excluded from the data matrix. The data for each urine ion were normalized by relative creatinine concentrations on a per-sample basis. Pareto-scaled MarkerLynx matrices were analyzed by principal components analysis (PCA) and partial least-squares discriminant analysis (PLS-DA) using SIMCA-P+ 12 (Umetrics, Kinnelon, NJ). To determine which ions contribute most to the difference between control and PCN-treated wild-type mice, orthogonal partial least squares (OPLS) was used. The loadings scatter S-plots and the contribution lists were used to describe the candidate ions and urinary markers that were significant different between control and PCN-treated wild-type mice.
Synthesis of (±)-γ-CEHC glucoside
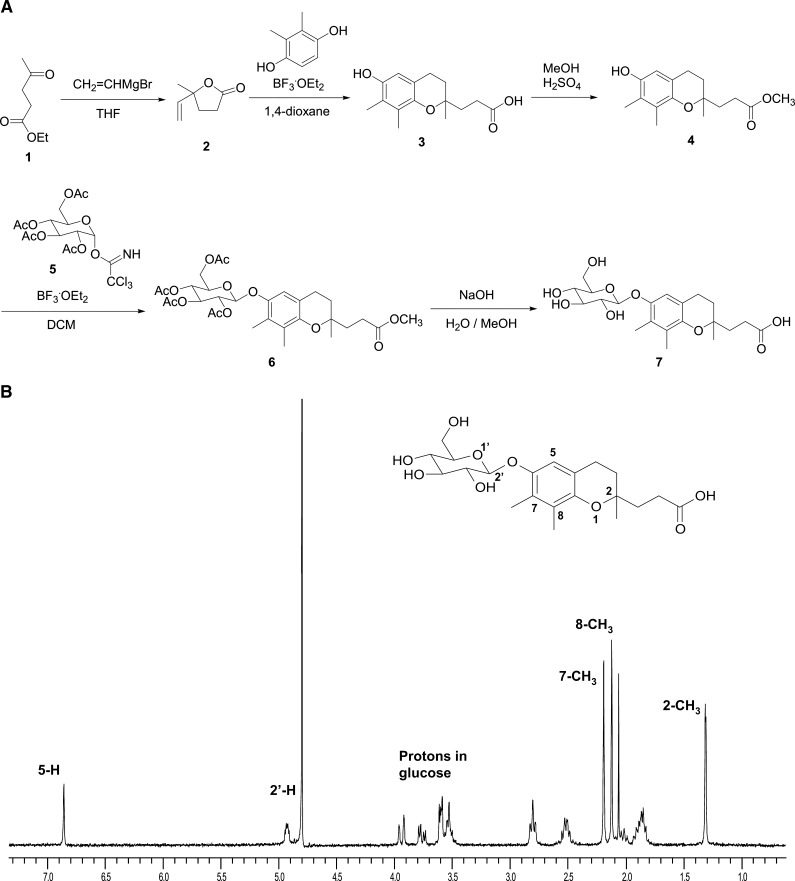
The novel endogenous vitamin E metabolite, 2,7,8-trimethyl-2-(β-carboxyethyl)-6-acetylhydroxychroman (γ-CEHC) glucoside (Glc), has not been reported. As shown in Fig. 3A, racemic (±)-γ-CEHC was synthesized using a previously described method (25) and then the glucose conjugate of (±)-γ-CEHC was synthesized. Briefly, ethyl levulinate [1] was converted to γ-methyl-γ-vinylbutyrolactone [2] using vinyl magnesium bromide, followed by reaction with 2,3-dimethylhydroquinone in the presence of boron trifluoride diethyl etherate to yield (±)-γ-CEHC [3]. The glucose conjugate of (±)-γ-CEHC was synthesized using standard glycosylation methodology (26). The carboxylic acid of (±)-γ-CEHC was protected as the methylester [4] prior to glycosylation of the phenol, through acidic esterification of (±)-γ-CEHC using methanol with catalytic sulfuric acid (27). Tetraacetyl glucosyl trichloroimidate [5] was prepared from 2,3,4,6-tetra-O-acetyl-β-D-glucopyranose and trichloroacetonitrile in dichloromethane (28). The methyl ester [4] was treated with the tetraacetyl glucosyl trichloroimidate [5] in the presence of boron trifluoride diethyl etherate at −20°C to yield the protected (±)-γ-CEHC Glc [6] in good yield. The final (±)-γ-CEHC Glc [7] was obtained by saponification of the protected (±)-γ-CEHC Glc [6] with sodium hydroxide in water and methanol. It was isolated by column chromatography in methylene chloride/methanol/acetic acid (25:5:1). In the same elution solvent, the Rf value of [6] was 0.40 on silica gel thin-layer chromatography. Although, [7] was prepared as a mixture of epimers, it was sufficient for use as a mass spectroscopy standard. In addition, (±)-α-CEHC glucuronide was synthesized by similar synthetic routes to the (±)-γ-CEHC Glc.
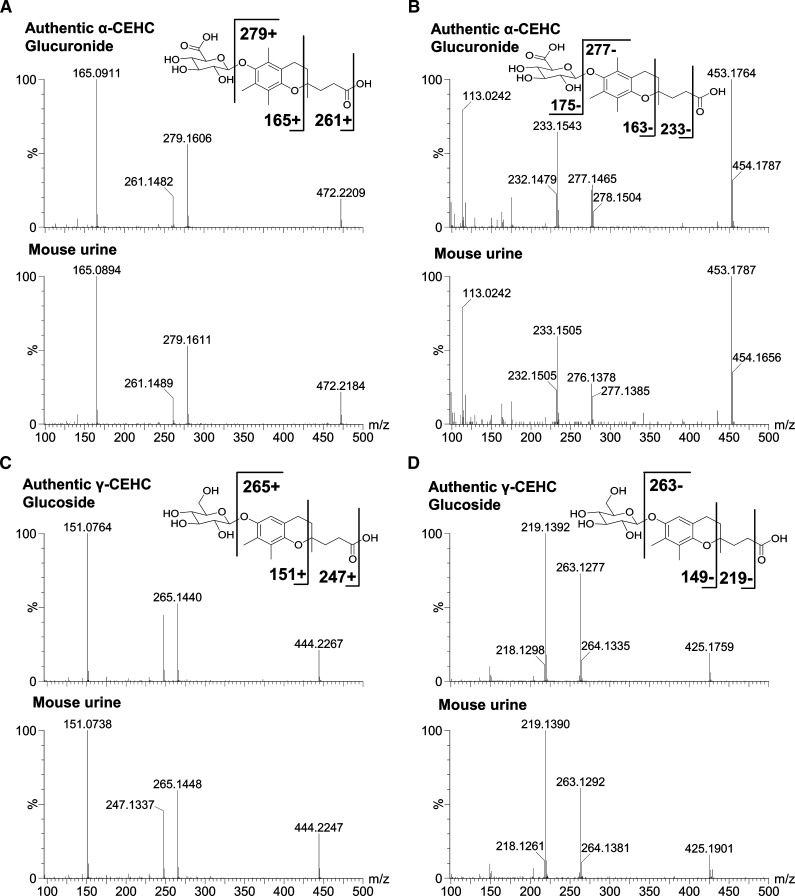
Fig. 3.
LC-MS/MS structural elucidation of α-CEHC glucuronide and γ-CEHC Glc in mouse urine. MS/MS fragmentation was conducted with collision energy ramping from 5 to 35 eV. Major daughter ions from fragmentation were interpreted in the inlaid structural diagrams. A: MS/MS spectrum of authentic α-CEHC glucuronide and α-CEHC glucuronide of mouse urine in positive ion mode. B: MS/MS spectrum of authentic α-CEHC glucuronide and α-CEHC glucuronide of mouse urine in negative ion mode. C: MS/MS spectrum of authentic γ-CEHC Glc and γ-CEHC Glc of mouse urine in positive ion mode. D: MS/MS spectrum of authentic γ-CEHC Glc and γ-CEHC Glc of mouse urine in negative ion mode.
Structure identification of CEHC metabolites
To identify the structure of high-contribution score metabolites, elemental compositions were generated with MassLynx® (Waters) based on the exact mass of metabolites with the mass tolerance of 10 ppm. In addition, the mass-based search in Madison Metabolomics Consortium Database (http://mmcd.nmrfam.wisc.edu) was used to identify potential candidate biomarkers, especially α-CEHC and γ-CEHC with the mass tolerance of 10 ppm (29). Authentic standards at 5–20 μM in 50% acetonitrile were used to confirm the identities of the markers with UPLC-MS/MS, and then MS/MS fragmentation spectra of authentic standards were compared with those of urinary metabolites.
Deconjugation of CEHC metabolites
3-Acetylumbelliferyl β-D-Glc (AUGlc) and phenolphthalein β-D-glucuronide (PPTGlu) were used as positive control substrates for β-D-glucosidase and β-D-glucuronidase, respectively. The deconjugation system for positive control substrates included 10 U of β-D-glucosidase or 40 U of β-D-glucuronidase, 10 μM AUGlc, 10 μM PPTGlu, 200 mM sodium acetate buffer (pH3.8), and total volume was 200 μl. The deconjugation system for urinary CEHC glycosides contained 40 μl of mouse urine, 10 U of β-D-glucosidase or 40 U of β-D-glucuronidase, 200 mM sodium acetate buffer (pH3.8), and total volume was 200 μl. The deconjugation mixtures were incubated for 6 h at 37°C and reactions terminated by adding an equal volume of acetonitrile, and centrifuged at 18,000 g for 20 min at 4°C. Aliquots (5 μl) were injected onto UPLC-TOFMS system. The peaks of glycons and aglycones were extracted in negative ion mode; AUGlc ([M-H]- 365.088), 3-acetylumbelliferone ([M-H]- 203.035), PPTGlu ([M-H]- 493.115), phenolphthalein ([M-H]- 317.079), α-CEHC_Glu ([M-H]- 453.174), α-CEHC ([M-H]- 277.142), γ-CEHC_Glc ([M-H]- 425.179), and γ-CEHC ([M-H]- 263.125).
Quantification of urinary metabolites and serum α-tocopherol
QuanLynx® (Waters) was used to quantify urinary creatinine, α-CEHC_Glu, and γ-CEHC_Glc metabolites and serum α-tocopherol from their peak areas. Five μM of debrisoquine ([M+H]+ 176.1180) was included as an internal standard. Calibration curves were constructed from 31.25 to 1,000 μM of creatinine ([M+H]+ 114.0670) and 0.78 to 100 μM of (±) α-CEHC_Glu ([M+NH4]+ 472.2198), and (±) γ-CEHC_Glc ([M+NH4]+ 444.2236). All determined correlation coefficients were >0.95 for each analyte. The concentration of each analyte in mouse urine was determined from the calibration curves and expressed as μM/mM creatinine. To quantify α-tocopherol in serum, a calibration curve was constructed from 1.56 to 100 μM of α-tocopherol ([M+H]+ 431.3811) and the determined correlation coefficient was >0.95. Concentration of α-tocopherol was expressed as μM.
Gene expression analysis
Quantitative real-time PCR (qPCR) was performed using cDNA generated from 1 μg total mRNA SuperScript II Reverse Transcriptase kit (Invitrogen, Carlsbad, CA). Primers were designed for qPCR using the Primer Express software (Applied Biosystems, Foster City, CA) based on GenBank sequence data: Cyp3a11: forward 5′-ttctgtcttcacaaaccggc-3′; reverse 5′-gggggacagcaaagctctat-3′; Cyp4f14: forward 5′-tccgatctatcctcaatgcc-3′, reverse 5′-gtcaccagcactcaccaaga-3′; Ugt1a1: forward 5′-tcagaaaaagcccctatccc-3′; reverse 5′- gagaccatggatcccaaaga-3′; Ugt1a10: forward 5′-tgtgatgcccaatgtgatct-3′; reverse 5′-cagaggcgttgacataggct-3′; Cpt1a: forward 5′-gcccatgttgtacagcttcc-3′; reverse 5′-agtggcctcacagactccag-3′; Hmgcs2: forward 5′-gaaacaaccagcctttcacc-3′, reverse 5′-tcattgaacatcaaccgagc-3′; Scp2: forward 5′-tcacttacgaagcactgggg-3′; reverse 5′-aggcctccactagggttgat-3′; Acox1: forward 5′-tcgcagaccctgaagaaatc-3′; reverse 5′-cctgattcagcaaggtaggg-3′; and Actb: forward 5′-ttctttgcagctccttcgtt-3′, reverse 5′-atggaggggaatacagccc-3′. qPCR reactions contained 25 ng of cDNA, 150 nM of each primer and 5 μl of SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a total volume of 10 μl. All reactions were performed in duplicate on an Applied Biosystems Prism 7900HT Sequence Detection System. Relative mRNA levels were calculated by the comparative threshold cycle method using β-actin (Actb) as the internal control.
Statistical analysis
Each group consisted of five to seven animals for the PCN treatment study. Three to eight animals were assigned for each group for the RIF treatment study. All values are expressed as the means ± SD. Statistical analysis was performed by one-way ANOVA combined with Bonferroni's multiple-comparisons test using Prism 5 (GraphPad Software Inc., San Diego, CA). A P value of less than 0.05 was considered statistically significant.
RESULTS
Phenotypes of PXR activation by PCN
Male wild-type and Pxr-null mice did not show significant differences in percent changes of body weight in PCN-treated versus corn oil-treated (control) mice during treatment and 24-h urine collection. Liver to body weight ratio on day 6 of PCN treatment was significantly increased to 140% in PCN-treated wild-type mice compared with control wild-type mice (data not shown). However, there was no significant difference in liver/body weight between PCN-treated and control groups of Pxr-null mice. These phenotypes are consistent with previously reported wild-type and Pxr-null mice after PXR activation by PCN (17).
Metabolomic analysis of mouse urine
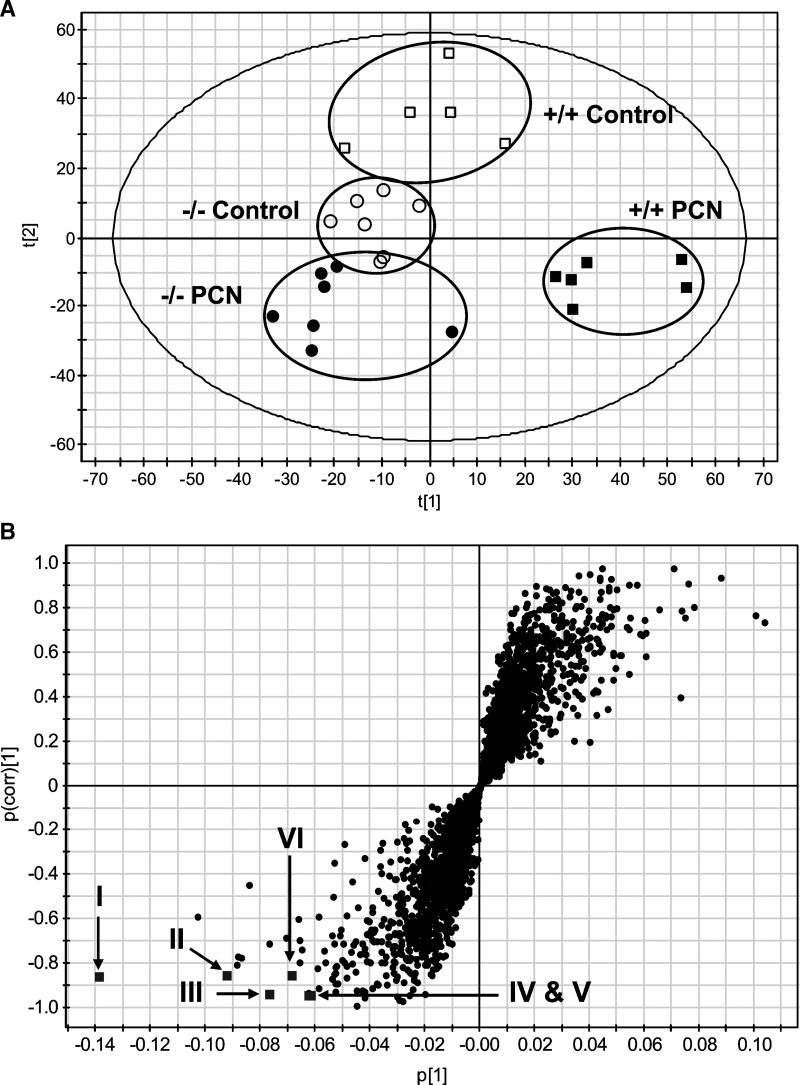
Urine samples for 0–24 h were collected twice (i.e., before and on 5 days of PCN treatment as described in Experimental Procedures). Urine collection before treatments was performed for acclimation of mice to the metabolic cages, and the urine samples on 5 days of treatment were analyzed by UPLC-TOFMS operating in positive ionization mode. A large data matrix containing approximately 3,300 positive ions was produced by Markerlynx. PCN-derived ions, especially the parent compound ([M+H]+ 342.245) and hydroxylated form ([M+OH]+ 358.240), and their in-source fragment ions were excluded from the data matrix and the data matrix subjected to both PCA and PLS-DA multivariate data analyses. Unsupervised PCA did not yield a good separation of the data sets from the control and PCN-treated groups in both wild-type and Pxr-null mice (data not shown). However, a supervised PLS-DA model with two components successfully discriminated the differences between all four groups of mice, having fitness (R2 value) of 0.98 and prediction power (Q2 value) of 0.75. Figure 1A shows a PLS-DA scores plot, representing a clear separation between control and PCN-treated groups in wild-type mice in component 1 (X-axis) but not in the Pxr-null mice. This result revealed specific metabolic phenotypes associated with the PXR activation by PCN treatment. After OPLS analysis of control and PCN-treated groups in wild-type mice, a loadings S-plot showed ions with the highest confidence and greatest contribution to separation between control and PCN-treated group in wild-type mice (Fig. 1B). The significant ions decreased after PCN treatment were in the lower-left quadrant and increased in the upper-right quadrant (Fig. 1B). The decreased positive ions of m/z = 316.2109, 449.1788, and 477.1737 ranked as the first, second, and third most important variables from the loadings plot, are labeled in Fig. 1B as I, II, and III, respectively. To identify the structure of the above ions, chemical formula calculations using elemental composition and mass-based search in Madison Metabolomics Consortium Database were performed. The 449.1788 (II; [M+Na]+) and 477.1737 (III; [M+Na]+) ions correspond to sodium adducts of γ-CEHC Glc and α-CEHC glucuronide, respectively. However, the chemical structure of 316.2109 (I) ion remained unknown. Among the other highest contributing ions were in-source fragment ions and ammonium adducts of γ-CEHC Glc and α-CEHC glucuronide, shown in Fig. 1B and listed in Table 1.
Fig. 1.
Metabolomic analysis of control and PCN-treated mouse urine. Wild-type and Pxr-null mice (n = 5∼7) were treated with vehicle (corn oil) or PCN (50 mg/kg/day, i.p.) for 4 days. Urinary metabolites were analyzed by ultra-performance liquid chromatography time-of-flight mass spectrometry (UPLC-TOFMS) and data processing and multivariate data analysis conducted using MarkerLynx and SIMCA-P+ software, respectively. A: Scores scatter plot of PLS-DA model of urine from the control and PCN-treated groups in wild-type (+/+) and Pxr-null (−/−) mice. A two-component PLS-DA model was constructed to characterize the relationship among four mouse groups, including wild-type (control, □; PCN treated, ▪) and Pxr-null (control, ○; PCN, •). The t(1) and t(2) values represent the scores of each sample in principal components 1 and 2, respectively. The fitness (R2 value) and prediction power (Q2 value) of this two-component model are 0.98 and 0.75, respectively. B: OPLS loading S-plot comparing control vs. PCN-treated group in wild-type mice. The x-axis is a measure of the relative abundance of the ions, and the y-axis is a measure of the correlation of each ion to the model. Positive ions located in the lower left quadrant are decreased when comparing PCN-treated group with control group in wild-type mice. Labeling of significant ions decreased (▪) is the same as in Table 1: II, III, IV, V, and VI.
TABLE 1.
Positive ions of vitamin E metabolites identified in the LC-MS-based metabolomic analysis of the urine samples from PCN-treated and control wild-type mice (n = 5∼7)
| Symbol | [M+H]+ | Retention time (min) | Ion rank | Empirical formula | Mass error (ppm) | Identity |
|---|---|---|---|---|---|---|
| II | 449.1788 | 4.79 | 2 | C21H30O9Na+ | 0.4 | γ-CEHC Glc Na+ adduct |
| III | 477.1737 | 4.99 | 3 | C22H30O10Na+ | 1.9 | α-CEHC Glucuronide Na+ adduct |
| IV | 472.2198 | 4.99 | 8 | C22H34NO10+ | 4.9 | α-CEHC Glucuronide NH4+ adduct |
| V | 165.0925 | 4.99 | 10 | C10H13O2+ | 6.1 | α -CEHC in-source fragment ion |
| VI | 444.2236 | 4.79 | 11 | C21H34NO9+ | 1.4 | γ-CEHC Glc NH4+ adduct |
Glc, glucoside. Conditions for ultra-performance liquid chromatography time-of-flight mass spectrometry (UPLC-TOFMS) analysis were described in Experimental Procedures. Ion rank from a loading S-plot of OPLS analysis showed the rank of ions with the highest confidence and greatest contribution to separation between control and PCN-treated group in wild-type mice. Symbol is the same as in Fig. 1B.
Identification of CEHC metabolites in urine
Authentic compounds of (±)-γ-CEHC Glc and (±)-α-CEHC glucuronide were required to confirm the structural identification of these metabolites by UPLC-MS/MS. The γ-CEHC Glc as a novel urinary metabolite of γ-tocopherol was synthesized following Fig. 2A. The synthetic (±)-γ-CEHC Glc was confirmed by NMR and mass fragmentography analysis. 1H-NMR resonances corresponding to the aromatic proton appears 6.86 ppm and the glucose ring protons appear 3.50–3.96 and 4.92 ppm in Fig. 2B.
Fig. 2.
Synthesis of (±)-γ-CEHC glucoside (Glc). A: Synthetic route to epimeric (±)-γ-carboxyethyl hydroxychroman (CEHC) Glc from ethyl levulinate. THF, tetrahydrofuran; BF3·OEt2, boron trifluoride etherate; MeOH, methanol; DCM, dichloromethane B: NMR spectrum of synthetic (±)-γ-CEHC Glc. Proton spectra were recorded using a Jemini-300MHz spectrometer and chemical shifts were reported as ppm.
The MS/MS structural elucidation of α-CEHC glucuronide ([M+NH4]+ = 472.2198, [M-H]- = 453.1719) in both positive and negative ion mode is shown in Fig. 3A, B, respectively. The major daughter ions 279, 261, and 165 in positive ion mode and 277, 233, and 163 in negative ion mode were interpreted in the inlaid structural diagram. In addition, the MS/MS structural elucidation of γ-CEHC Glc ([M+NH4]+ = 444.2236, [M-H]- = 425.1868) in both positive and negative ion mode is shown in Fig. 3C, D, respectively. The major daughter ions 265, 247, and 151 in positive ion mode and 263, 219, and 149 in negative ion mode were interpreted in the inlaid structural diagram. MS/MS fragmentation of sodium adducts of these metabolites was not detected, so ammonium adducts were used in positive ion mode.
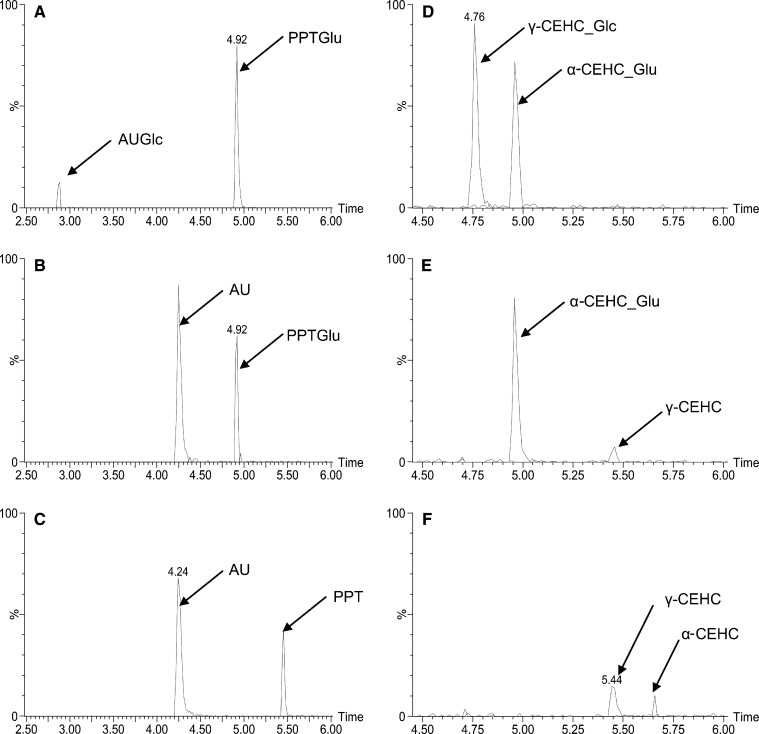
The identity of the novel γ-CEHC Glc (γ-CEHC_Glc) was further elucidated by specific enzymatic hydrolysis of the conjugates. Both β-D-glucosidase and β-D-glucuronidase were used for deconjugation analysis. AUGlc and phenolphthalein β-D-glucuronide (PPTGlu) were used as positive control substrates for β-glucosidase and β-glucuronidase, respectively. After incubation with β-glucosidase for 6 h, the AUGlc and γ-CEHC_Glc peaks completely disappeared and 3-acetylumbelliferone (AU) and γ-CEHC peaks appeared, whereas both glucuronide peaks (i.e., PPTGlu and α-CEHC_Glu) were intact (Fig. 4B, E). After incubation with β-glucuronidase for 6 h, all peaks of glucuronides and Glc disappeared and their aglycone peaks appeared (Fig. 4C, F), which indicated that β-glucuronidase was able to hydrolyze glucose-conjugated compounds in addition to glucuronides. These deconjugation data were consistent with the tandem MS data, which verified unequivocally the identity of γ-CEHC Glc.
Fig. 4.
Deconjugation of urinary α-CEHC glucuronide and γ-CEHC Glc. 3-Acetylumbelliferyl β-D-Glc (AUGlc) and phenolphthalein β-D-glucuronide (PPTGlu) were used as positive control substrates for β-D-glucosidase and β-D-glucuronidase, respectively. Details of deconjugation and peak extraction in chromatogram were described in Experimental Procedures. A: Chromatograms of AUGlc, PPTGlu, 3-acetylumbelliferone (AU), and phenolphthalein (PPT) after incubation of AUGlc and PPTGlu without enzyme. B: Chromatograms of AUGlc, PPTGlu, AU, and PPT after incubation of AUGlc and PPTGlu with β-D-glucosidase for 6 h. C: Chromatograms of AUGlc, PPTGlu, AU, and PPT after incubation of AUGlc and PPTGlu with β-D-glucuronidase for 6 h. D: Chromatograms of α-CEHC glucuronide (α-CEHC_Glu), γ-CEHC Glc (γ-CEHC_Glc) and their aglycones after incubation of wild-type mouse urine without enzyme. E: Chromatograms of α-CEHC_Glu, γ-CEHC_Glc, α-CEHC, and γ-CEHC after incubation of wild-type mouse urine with β-D-glucosidase for 6 h. F: Chromatograms of α-CEHC_Glu, γ-CEHC_Glc, α-CEHC, and γ-CEHC after incubation of wild-type mouse urine with β-D-glucuronidase for 6 h.
Quantification of CEHC metabolites and α-tocopherol
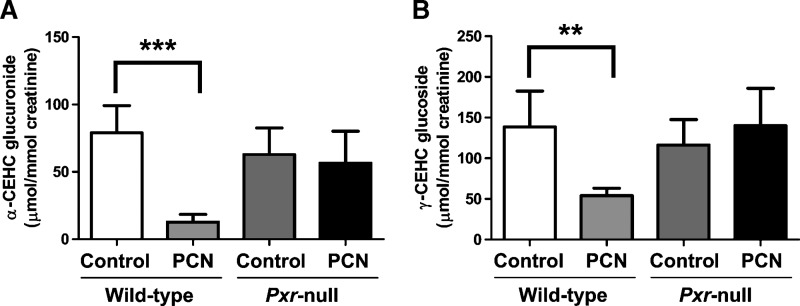
The concentrations of α-CEHC glucuronide, γ-CEHC Glc and creatinine were measured in each urine sample after construction of calibration curves using debrisoquine as an internal standard. Although synthetic authentic standard of (±)-α-CEHC glucuronide and (±)-γ-CEHC Glc were a mixture of epimers, retention times between epimers were too close to be separated under the choromatographic conditions employed, and thus these synthetic standards were used for construction of calibration curves. The urinary concentrations of α-CEHC glucuronide and γ-CEHC Glc were expressed as μmol/mmol creatinine, a variable independent of urine volume. Urinary α-CEHC glucuronide after PXR activation by PCN was significantly decreased to 16%, compared with control in wild-type mice but not in Pxr-null mice (Fig. 5A). In addition, urinary γ-CEHC Glc was significantly decreased after activation of PXR by PCN from 138.70 ± 43.86 to 54.16 ± 9.16 μmol/mmol creatinine in wild-type mice, but not changed in Pxr-null mice (Fig. 5B).
Fig. 5.
Quantification of α-CEHC glucuronide and γ-CEHC Glc in urine of wild-type and Pxr-null mice treated with vehicle (control) or PCN (50 mg/kg/day, i.p.) for 4 days. Concentrations of creatinine, α-CEHC glucuronide, and γ-CEHC Glc were determined from calibration curves of each metabolite. A: Creatinine-normalized concentration of α-CEHC glucuronide. B: Creatinine-normalized concentration of γ-CEHC Glc. Data were represented as mean value ± SD (n = 5∼7). The P-values were calculated by ANOVA with Bonferroni correction. *** P < 0.001 and ** P < 0.005 compared with control group of wild-type mice.
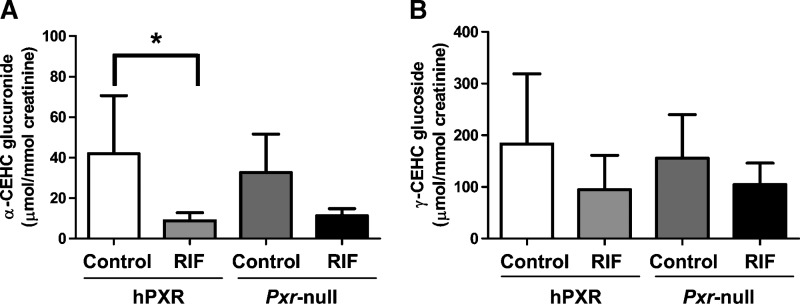
To investigate urinary excretion of these metabolites after hPXR activation in PXR-humanized mice, Pxr-null and PXR-humanized mice were fed with RIF-contained diet or control diet for 4 weeks and then 0–24 h urine samples were collected. Activation of PXR-humanized with RIF significantly decreased the α-CEHC glucuronide concentration to approximate 20% compared with control in PXR-humanized mice, whereas α-CEHC glucuronide concentration was not statistically significantly different between control and RIF-treated in Pxr-null mice (Fig. 6A). This result was consistent with the PXR activation with PCN in wild-type and Pxr-null mice. There was a trend toward a decreased level of γ-CEHC Glc after hPXR activation with RIF in PXR-humanized mice, but this was not statistically significant (Fig. 6B).
Fig. 6.
Quantification of α-CEHC glucuronide and γ-CEHC Glc in urine of PXR-humanized and Pxr-null mice fed either purified diet (control) or diet containing rifampicin (RIF) for 4 weeks. Concentrations of creatinine, α-CEHC glucuronide, and γ-CEHC Glc were determined from calibration curves of each metabolite. A: Creatinine-normalized concentration of α-CEHC glucuronide. B: Creatinine-normalized concentration of γ-CEHC Glc. Data were represented as mean value ± SD (n = 5 for control, n = 8 for RIF of hPXR mice, n = 3 for control, n = 4 for RIF of Pxr-null mice). The P-values were calculated by ANOVA with Bonferroni correction. * P < 0.01 compared with control group of PXR-humanized mice.
α-Tocopherol in serum samples of both PCN-treated and RIF-treated mice was also quantified. The level of α-tocopherol after either PXR activation with PCN or hPXR activation with RIF was decreased to 60% in wild-type mice or 83% in PXR-humanized mice, respectively, but was not statistically significant (data not shown).
Gene expression in liver
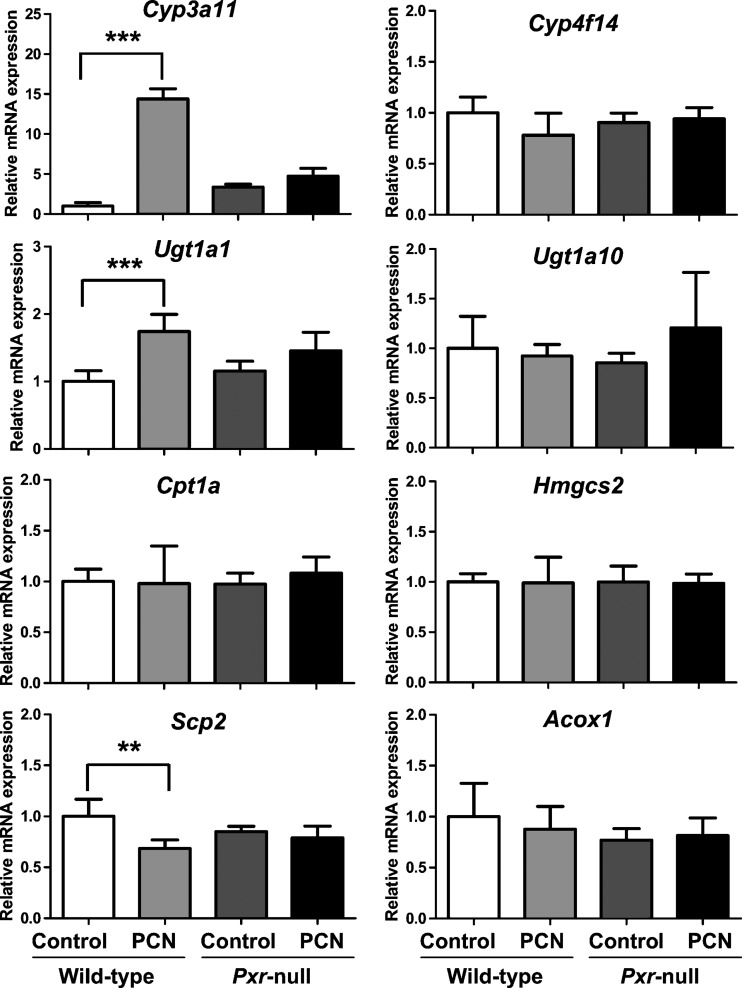
Metabolomic analysis of urine from wild-type and Pxr-null mice after administration of PXR ligand PCN suggested that PXR-specific urinary metabolic phenotype resulted from alteration of PXR target genes and metabolic activities. The expression level of genes related to metabolism of α-tocopherol and γ-tocopherol were determined in mouse liver using qPCR analysis. Relative mRNA expression of Cyp3a11, a representative PXR target gene, was elevated by approximately 15-fold in wild-type mice after PXR activation with PCN but not in Pxr-null mice (Fig. 7). Metabolism of α-tocopherol and γ-tocopherol to α-CEHC glucuronide and γ-CEHC Glc, respectively, had been reported to occur by a serial metabolic reaction (i.e., ω-oxidation, β-oxidation, and conjugation) (30). There is suggestive evidence for the involvement of CYP3A (30) and CYP4F (31) in tocopherol ω-oxidation. Thus, the mouse counterparts to these CYPs Cyp4f14 as well as Cyp3a11, may carry out ω-oxidation of tocopherols. The Ugt1a1 and Ugt1a10 genes belonging to the UGT superfamily (32) are likely involved in conjugation of CEHCs with glucuronic acid or glucose. Relative mRNA level of Ugt1a1 after PCN treatment was 1.7-fold higher than control group in wild-type (Fig. 7). Relative mRNA level of Cyp4f14 and Ugt1a10 in the PCN-treated mouse group was not different from the control mouse group in both wild-type and Pxr-null mice (Fig. 7). Cpt1a and Hmgcs2, which are major genes involved in β-oxidation in mitochondria (21), were also not differentially expressed among the various mouse groups (Fig. 7). Interestingly, Scp2 mRNA encoding an enzyme involved in peroxisomal oxidation of branched chain fatty acids, was significantly decreased in the PCN-treated groups compared with the control group in wild-type mice but not in Pxr-null mice, whereas the level of Acox1 mRNA, encoding an enzyme involved in peroxisomal oxidation of fatty acids, was not changed in any of the mouse groups (Fig. 7).
Fig. 7.
Hepatic expression of vitamin E metabolism-related genes in mouse treated with vehicle (control) or PCN (50 mg/kg/day, i.p.) for 4 days. Relative mRNA levels of ω-oxidation-related genes (Cyp3a11 and Cyp4f14), a conjugation-related gene (Ugt1a10), β-oxidation-related genes in mitochondria (Cpt1a and Hmgcs2), and β-oxidation-related genes in peroxisomes (Scp2 and Acox1) were determined by real-time PCR analysis using gene-specific primers and β-actin as the internal control as described in Experimental Procedures. Data are represented as mean value ± SD (n = 5∼7). The P-values were calculated by ANOVA with Bonferroni correction. *** P < 0.001 and ** P < 0.005 compared with control group of wild-type mice.
DISCUSSION
In this study, a combination of metabolomic analysis and genetically modified mouse models was used to demonstrate that activation of PXR results in attenuated vitamin E metabolites in urine possibly through down-regulation of the Scp2 gene. PXR regulates numerous genes involved in metabolism of xenobiotics and endobiotics, including oxidation and conjugation enzymes, and transporters. In addition, recent studies have shown that activation of PXR alters lipid metabolism, glucose homeostasis, and inflammation (21, 33). To our knowledge, the attenuation of vitamin E metabolism by activation of PXR is previously uncharacterized, although induction of ω-oxidation of vitamin E by RIF has been reported (30).
Metabolomic phenotypes for PXR activation were investigated in both Pxr-null and wild-type mice. Multivariate analysis by PLS-DA revealed that, the urinary metabolomes changed after PCN treatment, were distinct phenotypes associated with the PXR activation by PCN, because both control and PCN-treated Pxr-null mice were clustered together, whereas control and PCN-treated wild-type mice were separated clearly in the scores plot. Urinary metabolomes that changed consistently with activation of PXR by PCN in wild-type mice were analyzed using OPLS. The most attenuated ions after PXR activation were the vitamin E metabolites α-CEHC glucuronide and γ-CEHC Glc. α-CEHC glucuronide and γ-CEHC Glc in urine were dependent upon the chemically defined AIN 93 diet, which contained vitamin E at a concentration of 79 IU/kg diet (53 mg/kg natural vitamin E or 79 mg/kg synthetic vitamin E).
A novel endogenous metabolite of γ-tocopherol, γ-CEHC Glc was uncovered through metabolomic study of PXR activation. The major conjugated form of γ-tocopherol in mice was γ-CEHC Glc, whereas the major conjugate form of α-tocopherol was α-CEHC glucuronide. γ-CEHC glucuronide was also found in urine at a level of approximately 10% of the γ-CEHC Glc (data not shown). It should be noted that others reported that sulfated CEHCs appeared in human urine (27). However, no ions corresponding to sulfated CEHC metabolites (α-CEHC sulfate, [M-H]− 357.1008; γ-CEHC sulfate, [M-H]− 343.0851) were detected in mouse urine, at least under the UPLC-TOFMS conditions employed in this study. Thus, we cannot rule out the possibility that sulfated conjugates of CEHCs are produced in mice. To identify the chemical identities of α-CEHC glucuronide and γ-CEHC Glc, these two conjugate compounds were synthesized. Most interestingly, the γ-tocopherol metabolite, γ-CEHC Glc, was not previously been reported in biology, nor has its synthesis been published. The structural identities of the α-CEHC glucuronide and γ-CEHC Glc were verified unequivocally in both electrospray positive and negative ion modes. In addition, the identity of γ-CEHC Glc was confirmed by deconjugation analysis using β-glucosidase and β-glucuronidase. Of note, unexpectedly β-glucuronidase was found to hydrolyze glucose conjugates such as γ-CEHC Glc as well as glucuronides, whereas β-glucosidase can hydrolyze only glucose conjugates. Although the γ-CEHC Glc was one of the most abundant metabolites in urine after treatment with vitamin E, it had not been observed in previous reports on vitamin E metabolites (34). Because the urine samples in previous reports had been quantified after incubation with β-glucuronidase, which can hydrolyze Glc, most of the γ-CEHC Glc has surely been misrepresented as γ-CEHC glucuronide, which was detected in mouse urine but at much lower levels than γ-CEHC Glc (34). It is not obvious which isoforms of UGT enzymes are involved in the conjugation of CEHCs with glucuronic acid and/or glucose. Although UGT1A1 mRNA expression was induced 1.7-fold after treatment of wild-type mice with PCN compared with control mice, the marginally induced UGT1A activity by PXR activation may not significantly affect the decrease of both α-CEHC glucuronide and γ-CEHC Glc in urine.
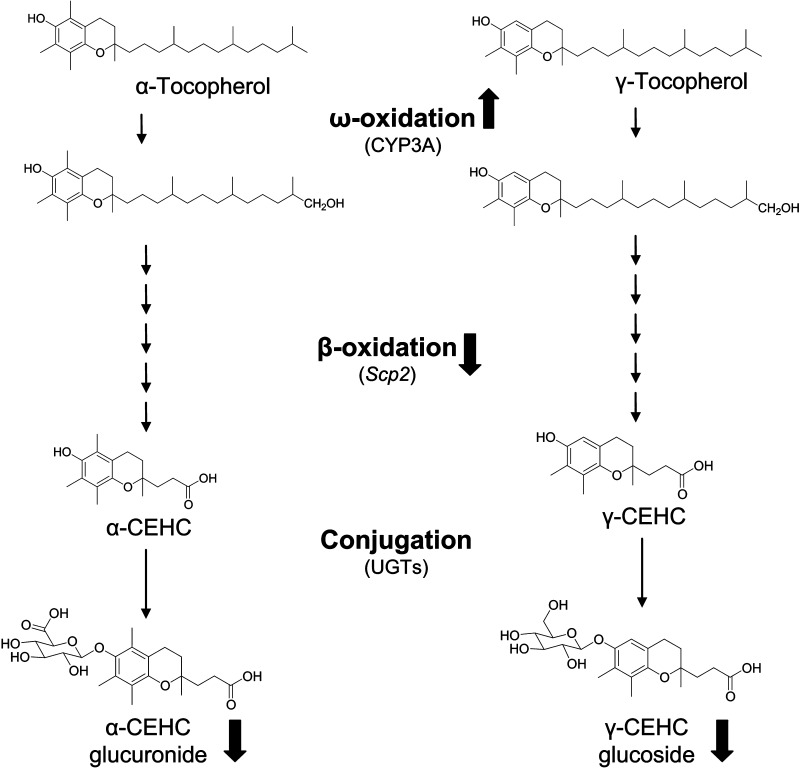
In this study, activation of PXR was found to result in the attenuation of vitamin E metabolites. The urinary excretion of α-CEHC glucuronide was significantly decreased to 16% and that of γ-CEHC Glc was attenuated to 40% in PCN-treated compared with control wild-type mice but the urinary excretion of both metabolites in Pxr-null mice were unaffected (Fig. 5). Further studies remain to validate whether attenuation of these two vitamin E conjugates in urine can serve as indicators of in vivo PXR activation. To understand the decrease of urinary vitamin E metabolites by PXR activation, hepatic expression of metabolic enzymes contributing to vitamin E metabolism and that are controlled by this nuclear receptor were examined. The scheme in Fig. 8 shows the metabolism of α-tocopherol and γ-tocopherol through the initial ω-oxidation of the phytyl side chain, five cycles of β-oxidation, and conjugation in liver, to α-CEHC glucuronide and γ-CEHC Glc. Earlier studies found that tocopherols and tocotrienols were metabolized by CYP3A4, as suggested by the detection of the intermediate precursors of CEHCs from α-, γ-, and δ-tocopherol in cultured cells (30). Additional evidence for the involvement of CYP3A4 included the finding that RIF, a CYP3A inducer, stimulated α-CEHC release by HepG2 cells and that ketoconazole, a CYP3A inhibitor, reduced vitamin E metabolism (30). In addition, others showed that CYP4F was involved in tocopherol-ω-hydroxylation (31). In this study, Cyp3a11 was induced 11-fold after treatment of wild-type mice with PCN, although Cyp4f14 expression was not changed. Induction of Cyp3a11 by PXR activation stimulates the increased ω-oxidation of α-tocopherol and results in the decreased plasma level of α-tocopherol in wild-type mice treated with PCN by approximately 40% compared with control group, although it was not statistically significant (data not shown). Consistent with these data, a recent report revealed that treatment of rats with PCN or dexamethasone, a CYP3A inducer, reduced α-tocopherol concentrations in plasma and liver but markedly reduced urinary α-CEHC excretion (35). In addition, treatment with the CYP3A inhibitor ketoconazole decreased α-CEHC levels in urine as well as increased α-tocopherol levels in liver, suggesting that CYP3A is not a major contributor to metabolism from α-tocopherol to α-CEHC although this enzyme might be a major contributor to ω-oxidation of α-tocopherol (35).
Fig. 8.
Proposed major metabolic pathways of vitamin E in wild-type mice with PXR activation. Activation of PXR increases hepatic ω-oxidation by increasing expression of ω-oxidation-related genes such as Cyp3a11 but represses peroxisomal β-oxidation by decreasing Scp2 expression, resulting in attenuated concentration of α-CEHC glucuronide and γ-CEHC Glc in urine.
After ω-oxidation of the terminal methyl group, peroxisomal oxidation of branched-chain fatty acids (BCFAs) follows to generate CEHCs (30). The peroxisomal oxidation of BCFAs is performed by a series of enzymes, including branched chain fatty acid acyl-CoA oxidase, D-bifunctional protein, and sterol carrier protein-x (SCP-x). Importantly, mRNA of Scp2 encoding SCP-x was significantly decreased by PXR activation, which might significantly contribute to attenuation of these metabolites in urine after activation of PXR. SCP-x serves as a peroxisomal 3-ketoacyl-CoA thiolase in oxidation of branched-chain lipids and facilitates in part the removal of toxic branched-chain metabolites (36). In a very recent report, Scpx-null mice treated with phytol, a source of BCFA, exhibited a decreased ability to metabolize branched-chain lipids (37). Transcription of Scp2 is positively regulated by the Forkhead transcription factor (Fox)O3a (38), but not regulated through PPARs (39, 40). Tocopherol metabolism was not enhanced by clofibrate, an inducer of peroxisomal β-oxidation, in HepG2 cells (30) and urinary CEHC metabolites were not increased in wild-type mice treated with the rodent PPARα agonist, 4-chloro-6-(2,3-xylidino)-pyrimidynylthioacetic acid (Wy-14,643) (data not shown). Thus, activation of PXR induces hepatic ω-oxidation by increasing the expression of Cyp3a11 and represses peroxisomal β-oxidation by decreasing Scp2 expression, resulting in attenuated levels of α-CEHC glucuronide and γ-CEHC Glc in urine, as shown the proposed metabolic pathways of vitamin E in Fig. 8.
Several recent studies suggested that PXR or constitutive androstane receptor (CAR) activation inhibits lipid catabolism by decreasing β-oxidation-related gene expression. For example, treatment with PCN down-regulates the mRNA level of Cpt1 in wild-type but not in Pxr-null mice in fasting mouse liver (21), although the expression of Cpt1 in our data shows no change by PCN in fed mouse liver. The transcriptional regulation of Cpt1 is tightly controlled by several transcription factors and cofactors such as the hepatocyte nuclear factor 3 β (FoxA2), PPARα, thyroid hormone receptor, and PGC1α (33). At least two independent pathways could be involved in PXR- and CAR-mediated Cpt1 gene repression by interfering with positive regulators of Cpt1 gene transcription, one involving FoxA2 and the other involving HNF4α (33). In addition to lipid metabolism, activation of PXR and CAR inhibits hepatic glucose metabolism by repressing hepatic gluconeogenic enzymes such as phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase). PXR activation could provoke PEPCK down-regulation either by interfering with HNF4α or by preventing FoxO1 from binding to its responsive element IRS in the PEPCK gene promoter (20). Accordingly, PXR activation could repress the peroxisomal β-oxidation by interfering with transcription factors such as FoxO3a involved in transcriptional regulation of the Scp2, although the mechanism remains unclear and further studies are needed.
In vitro and in vivo studies on the fate of hepatic vitamin E have shown that the various vitamin E forms such as RRR-α-tocopherol, all-rac-α-tocopherol, and γ-tocopherol are metabolized differentially. The major regulatory mechanism of the differential metabolism is α-tocopherol transfer protein (α-TTP), which functions to facilitate secretion of α-tocopherol from the liver into the plasma (41). The α-TTP has differential affinities for various forms of vitamin E with RRR-α-tocopherol (100%), SRR-α-tocopherol (11%), and γ-tocopherol (9%) (42). In this study, α-TTP mRNA was not changed after treatment of wild-type mice with PCN compared with the other groups (data not shown). Therefore, α-TTP or other forms of vitamin E may not contribute to the attenuation of vitamin E metabolites by activation of PXR. The other major consideration of vitamin E metabolism is the up-regulation of CYPs, which could be regarded as a protection mechanism against excess accumulation of vitamin E. Hepatic Cyp3a11 was increased 2.5-fold in mice fed a diet containing 20 or 200 mg/kg diet of α-tocopherol for 3 months but not by γ-tocotrienol (43). In this study, the vitamin E-containing diet (AIN 93) was fed to mice for 10 days, which might affect induction of CYPs. However, the level of Cyp3a11 mRNA was not increased in mice fed the AIN 93 diet compared with mice fed a normal diet (NIH-31) (data not shown), suggesting that the vitamin E included in the AIN 93 diet may not influence PXR activation.
Attenuated vitamin E metabolites after PXR activation were shown in PXR-humanized mice treated with RIF, a ligand for hPXR, similar to wild-type mice treated with PCN, suggesting that PXR activation by either rodent or human PXR ligand binding could regulate a common metabolic pathway for vitamin E. Because the PXR-humanized mouse model can reflect PXR transactivation in humans, the possibility exists that coadministration of vitamin E with hPXR activators such as RIF can repress the metabolism of vitamin E in humans. Indeed, it is known that vitamin E does not accumulate in the liver to toxic levels (41). However, the current data suggests that vitamin E intermediate metabolites may accumulate in human liver, because reduced activity of SCP-x prevents the phytyl chain oxidation of vitamin E. Recent meta-analyses of clinical trials on vitamin E supplementation in humans could not corroborate any benefit. In addition, an increased mortality from an intake of more than 400 IU/d vitamin E was found in a meta-analysis (44, 45). High dose vitamin E trials were commonly performed in at-risk patients that most probably were under drug therapy (46). The negative outcome in these patients could have been caused by a lower metabolism of vitamin E due to drug-induced PXR activation. However, further studies for accumulation of vitamin E metabolites in liver with PXR activation should be performed through various metabolomic approaches.
In summary, using the combination of metabolomic analysis and genetically modified mouse models, a novel vitamin E metabolite, γ-CEHC Glc, was identified activated PXR was found to significantly lower the concentration of the α-CEHC glucuronide and γ-CEHC Glc in wild-type but not in Pxr-null mice. Activation of PXR in wild-type mice was further found to inhibit expression of the hepatic Scp2 gene. PXR activation results in inhibition of peroxisomal β-oxidation of BCFA including tocopherols possibly by decreasing the expression of Scp2 through the crosstalk with other transcription factors resulting in coordinating regulating both vitamin E and lipid metabolism.
Abbreviations
AUGlc, 3-Acetylumbelliferyl β-D-glucoside
BCFA, branched-chain fatty acids
CAR, constitutive androstane receptor
CEHC, carboxyethyl hydroxychroman
Fox, forkhead transcription factor
Glc, glucoside
Glu, glucuronide
hPXR, PXR-humanized
MS/MS, tandem mass spectrometry
OPLS, orthogonal partial least squares
PCA, principal components analysis
PCN, pregnenolone 16α-carbonitrile
PLS-DA, partial least-squares discriminant analysis
PPARα, peroxisome proliferators activated receptor α
PPTGlu, phenolphthalein β-D-glucuronide
PXR, pregnane X receptor
qPCR, quantitative real-time polymerase chain reaction
RIF, rifampicin
SCP-x, sterol carrier protein-x
TOFMS, time-of-flight mass spectrometry
UGT, UDP-glucuronosyltransferase
UPLC, ultra-performance liquid chromatography
Published, JLR Papers in Press, January 20, 2009.
References
- 1.Kliewer S. A., and T. M. Willson. 2002. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J. Lipid Res. 43 359–364. [PubMed] [Google Scholar]
- 2.Poso A., and P. Honkakoski. 2006. Ligand recognition by drug-activated nuclear receptors PXR and CAR: structural, site-directed mutagenesis and molecular modeling studies. Mini Rev. Med. Chem. 6 937–947. [DOI] [PubMed] [Google Scholar]
- 3.Honkakoski P., T. Sueyoshi, and M. Negishi. 2003. Drug-activated nuclear receptors CAR and PXR. Ann. Med. 35 172–182. [DOI] [PubMed] [Google Scholar]
- 4.Carnahan V. E., and M. R. Redinbo. 2005. Structure and function of the human nuclear xenobiotic receptor PXR. Curr. Drug Metab. 6 357–367. [DOI] [PubMed] [Google Scholar]
- 5.Ma X., J. R. Idle, and F. J. Gonzalez. 2008. The pregnane X receptor: from bench to bedside. Expert Opin. Drug Metab. Toxicol. 4 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X., Y. Shah, C. Cheung, G. L. Guo, L. Feigenbaum, K. W. Krausz, J. R. Idle, and F. J. Gonzalez. 2007. The PREgnane X receptor gene-humanized mouse: a model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab. Dispos. 35 194–200. [DOI] [PubMed] [Google Scholar]
- 7.Xie W., J. L. Barwick, M. Downes, B. Blumberg, C. M. Simon, M. C. Nelson, B. A. Neuschwander-Tetri, E. M. Brunt, P. S. Guzelian, and R. M. Evans. 2000. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 406 435–439. [DOI] [PubMed] [Google Scholar]
- 8.Gong H., S. V. Singh, S. P. Singh, Y. Mu, J. H. Lee, S. P. Saini, D. Toma, S. Ren, V. E. Kagan, B. W. Day, et al. 2006. Orphan nuclear receptor pregnane X receptor sensitizes oxidative stress responses in transgenic mice and cancerous cells. Mol. Endocrinol. 20 279–290. [DOI] [PubMed] [Google Scholar]
- 9.Guzelian J., J. L. Barwick, L. Hunter, T. L. Phang, L. C. Quattrochi, and P. S. Guzelian. 2006. Identification of genes controlled by the pregnane X receptor by microarray analysis of mRNAs from pregnenolone 16alpha-carbonitrile-treated rats. Toxicol. Sci. 94 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenfeld J. M., R. Vargas, Jr., W. Xie, and R. M. Evans. 2003. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane X receptor. Mol. Endocrinol. 17 1268–1282. [DOI] [PubMed] [Google Scholar]
- 11.Kliewer S. A., J. T. Moore, L. Wade, J. L. Staudinger, M. A. Watson, S. A. Jones, D. D. McKee, B. B. Oliver, T. M. Willson, R. H. Zetterstrom, et al. 1998. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 92 73–82. [DOI] [PubMed] [Google Scholar]
- 12.Xie W., J. L. Barwick, C. M. Simon, A. M. Pierce, S. Safe, B. Blumberg, P. S. Guzelian, and R. M. Evans. 2000. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 14 3014–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng S., V. Jekerle, and M. Piquette-Miller. 2003. Induction of ABCC3 (MRP3) by pregnane X receptor activators. Drug Metab. Dispos. 31 1296–1299. [DOI] [PubMed] [Google Scholar]
- 14.Mackenzie P. I., P. A. Gregory, D. A. Gardner-Stephen, R. H. Lewinsky, B. R. Jorgensen, T. Nishiyama, W. Xie, and A. Radominska-Pandya. 2003. Regulation of UDP glucuronosyltransferase genes. Curr. Drug Metab. 4 249–257. [DOI] [PubMed] [Google Scholar]
- 15.Falkner K. C., J. A. Pinaire, G. H. Xiao, T. E. Geoghegan, and R. A. Prough. 2001. Regulation of the rat glutathione S-transferase A2 gene by glucocorticoids: involvement of both the glucocorticoid and pregnane X receptors. Mol. Pharmacol. 60 611–619. [PubMed] [Google Scholar]
- 16.Geick A., M. Eichelbaum, and O. Burk. 2001. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J. Biol. Chem. 276 14581–14587. [DOI] [PubMed] [Google Scholar]
- 17.Guo G. L., J. Staudinger, K. Ogura, and C. D. Klaassen. 2002. Induction of rat organic anion transporting polypeptide 2 by pregnenolone-16alpha-carbonitrile is via interaction with pregnane X receptor. Mol. Pharmacol. 61 832–839. [DOI] [PubMed] [Google Scholar]
- 18.Konno Y., M. Negishi, and S. Kodama. 2008. The roles of nuclear receptors CAR and PXR in hepatic energy metabolism. Drug Metab. Pharmacokinet. 23 8–13. [DOI] [PubMed] [Google Scholar]
- 19.Kodama S., C. Koike, M. Negishi, and Y. Yamamoto. 2004. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol. Cell. Biol. 24 7931–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao J., S. Fang, Y. Bae, and J. K. Kemper. 2006. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J. Biol. Chem. 281 14537–14546. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura K., R. Moore, M. Negishi, and T. Sueyoshi. 2007. Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver. J. Biol. Chem. 282 9768–9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhen Y., K. W. Krausz, C. Chen, J. R. Idle, and F. J. Gonzalez. 2007. Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Mol. Endocrinol. 21 2136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idle J. R., and F. J. Gonzalez. 2007. Metabolomics. Cell Metab. 6 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staudinger J. L., B. Goodwin, S. A. Jones, D. Hawkins-Brown, K. I. MacKenzie, A. LaTour, Y. Liu, C. D. Klaassen, K. K. Brown, J. Reinhard, et al. 2001. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. USA. 98 3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wechter W. J., D. Kantoci, E. D. Murray, Jr., D. C. D'Amico, M. E. Jung, and W. H. Wang. 1996. A new endogenous natriuretic factor: LLU-alpha. Proc. Natl. Acad. Sci. USA. 93 6002–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., J. Li, D. Tuttle, J. Y. Takemoto, and C. W. Chang. 2002. The Synthesis of L-aminosugar and the studies of L-pyranoses on the ring III of pyranmycins. Org. Lett. 4 3997–4000. [DOI] [PubMed] [Google Scholar]
- 27.Pope S. A., G. E. Burtin, P. T. Clayton, D. J. Madge, and D. P. Muller. 2002. Synthesis and analysis of conjugates of the major vitamin E metabolite, alpha-CEHC. Free Radic. Biol. Med. 33 807–817. [DOI] [PubMed] [Google Scholar]
- 28.Lee H. Y., J. T. Kwon, M. Koh, M. H. Cho, and S. B. Park. 2007. Enhanced efficacy of 7-hydroxy-3-methoxycadalene via glycosylation in in vivo xenograft study. Bioorg. Med. Chem. Lett. 17 6335–6339. [DOI] [PubMed] [Google Scholar]
- 29.Cui Q., I. A. Lewis, A. D. Hegeman, M. E. Anderson, J. Li, C. F. Schulte, W. M. Westler, H. R. Eghbalnia, M. R. Sussman, and J. L. Markley. 2008. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 26 162–164. [DOI] [PubMed] [Google Scholar]
- 30.Birringer M., D. Drogan, and R. Brigelius-Flohe. 2001. Tocopherols are metabolized in HepG2 cells by side chain omega-oxidation and consecutive beta-oxidation. Free Radic. Biol. Med. 31 226–232. [DOI] [PubMed] [Google Scholar]
- 31.Sontag T. J., and R. S. Parker. 2002. Cytochrome P450 omega-hydroxylase pathway of tocopherol catabolism. Novel mechanism of regulation of vitamin E status. J. Biol. Chem. 277 25290–25296. [DOI] [PubMed] [Google Scholar]
- 32.Mackenzie P. I., K. W. Bock, B. Burchell, C. Guillemette, S. Ikushiro, T. Iyanagi, J. O. Miners, I. S. Owens, and D. W. Nebert. 2005. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genomics. 15 677–685. [DOI] [PubMed] [Google Scholar]
- 33.Moreau A., M. J. Vilarem, P. Maurel, and J. M. Pascussi. 2008. Xenoreceptors CAR and PXR activation and consequences on lipid metabolism, glucose homeostasis, and inflammatory response. Mol. Pharm. 5 35–41. [DOI] [PubMed] [Google Scholar]
- 34.Kiyose C., H. Saito, K. Kaneko, K. Hamamura, M. Tomioka, T. Ueda, and O. Igarashi. 2001. Alpha-tocopherol affects the urinary and biliary excretion of 2,7,8-trimethyl-2 (2′-carboxyethyl)-6-hydroxychroman, gamma-tocopherol metabolite, in rats. Lipids. 36 467–472. [DOI] [PubMed] [Google Scholar]
- 35.Li Y. J., and H. M. Shaw. 2007. Pregnenolone and dexamethasone, modulators of cytochrome P450–3A, not increase but reduce urinary alpha-CEHC excretion in rats. Biofactors. 31 67–76. [DOI] [PubMed] [Google Scholar]
- 36.Gallegos A. M., B. P. Atshaves, S. M. Storey, O. Starodub, A. D. Petrescu, H. Huang, A. L. McIntosh, G. G. Martin, H. Chao, A. B. Kier, et al. 2001. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog. Lipid Res. 40 498–563. [DOI] [PubMed] [Google Scholar]
- 37.Atshaves B. P., A. L. McIntosh, D. Landrock, H. R. Payne, J. T. Mackie, N. Maeda, J. Ball, F. Schroeder, and A. B. Kier. 2007. Effect of SCP-x gene ablation on branched-chain fatty acid metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 292 G939–G951. [DOI] [PubMed] [Google Scholar]
- 38.Dansen T. B., G. J. Kops, S. Denis, N. Jelluma, R. J. Wanders, J. L. Bos, B. M. Burgering, and K. W. Wirtz. 2004. Regulation of sterol carrier protein gene expression by the forkhead transcription factor FOXO3a. J. Lipid Res. 45 81–88. [DOI] [PubMed] [Google Scholar]
- 39.Fujiki Y., M. Tsuneoka, and Y. Tashiro. 1989. Biosynthesis of nonspecific lipid transfer protein (sterol carrier protein 2) on free polyribosomes as a larger precursor in rat liver. J. Biochem. 106 1126–1131. [DOI] [PubMed] [Google Scholar]
- 40.Gossett R. E., F. Schroeder, J. M. Gunn, and A. B. Kier. 1997. Expression of fatty acyl-CoA binding proteins in colon cells: response to butyrate and transformation. Lipids. 32 577–585. [DOI] [PubMed] [Google Scholar]
- 41.Traber M. G. 2004. Vitamin E, nuclear receptors and xenobiotic metabolism. Arch. Biochem. Biophys. 423 6–11. [DOI] [PubMed] [Google Scholar]
- 42.Hosomi A., M. Arita, Y. Sato, C. Kiyose, T. Ueda, O. Igarashi, H. Arai, and K. Inoue. 1997. Affinity for alpha-tocopherol transfer protein as a determinant of the biological activities of vitamin E analogs. FEBS Lett. 409 105–108. [DOI] [PubMed] [Google Scholar]
- 43.Kluth D., N. Landes, P. Pfluger, K. Muller-Schmehl, K. Weiss, C. Bumke-Vogt, M. Ristow, and R. Brigelius-Flohe. 2005. Modulation of Cyp3a11 mRNA expression by alpha-tocopherol but not gamma-tocotrienol in mice. Free Radic. Biol. Med. 38 507–514. [DOI] [PubMed] [Google Scholar]
- 44.Vivekananthan D. P., M. S. Penn, S. K. Sapp, A. Hsu, and E. J. Topol. 2003. Use of antioxidant vitamins for the prevention of cardiovascular disease: meta-analysis of randomised trials. Lancet. 361 2017–2023. [DOI] [PubMed] [Google Scholar]
- 45.Miller 3rd E. R., R. Pastor-Barriuso, D. Dalal, R. A. Riemersma, L. J. Appel, and E. Guallar. 2005. Meta-analysis: high-dosage vitamin E supplementation may increase all-cause mortality. Ann. Intern. Med. 142 37–46. [DOI] [PubMed] [Google Scholar]
- 46.Brigelius-Flohe R. 2005. Induction of drug metabolizing enzymes by vitamin E. J. Plant Physiol. 162 797–802. [DOI] [PubMed] [Google Scholar]