Abstract
Phospholipids are subjected to remodeling through the Lands cycle to attain appropriate FA compositions. In recent years, at least two families of lysophospholipid acyltransferases have been identified. Acyl-CoA lysocardiolipin acyltransferase 1 (ALCAT1) was initially identified as a microsomal lysocardiolipin acyltransferase. However, the physiological relevance of how this enzyme is involved in cardiolipin remodeling has not been elucidated. We report in this study that ALCAT1 possesses acyltransferase activities toward lysophosphatidylinositol (LPI) and lysophosphatidylglycerol (LPG). Membrane preparations from human embryonic kidney 293 (HEK293) cells overexpressing human ALCAT1 demonstrated significant increases in LPI acyltransferase (LPIAT) and LPG acyltransferase (LPGAT) activities using a variety of fatty acyl-CoAs. The enzyme affinities toward LPI and LPG were determined through kinetic studies suggesting that the LPI binding affinity to ALCAT1 depends on fatty acyl-CoA. Reduced expression of ALCAT1 in Hela cells resulted in significant reductions of LPIAT and LPGAT activities, but not ALCAT activity. Through structural and functional studies, we have identified critical amino acids D168 and L169 within ALCAT1 that are potentially involved in lysophospholipid substrate binding. Our studies provide the molecular basis for future investigations of the physiological function of ALCAT1 and offer evidence of critical amino acids involved in substrate binding for the family of glycerolipid acyltransferases.
Keywords: lysophosphatidylinositol acyltransferase, lysophosphatidylglycerol acyltransferase, membrane bound O-acyltransferase
Phospholipids (PLs) are essential membrane components of lipid bilayers, and some PLs also serve as important signaling molecules. There are more than 1,000 PLs, differing in the phosphoryl head groups and the FAs of both the sn-1 and sn-2 positions in chain lengths and degrees of saturation (1). These PLs are unevenly distributed within plasma and subcellular membranes, and the relative quantities may fluctuate depending on physiological needs. For instance, phosphatidylcholine (PC) is the principle PL of plasma membranes, whereas phosphatidylserine (PS) and phosphatidylethanolamine (PE) are enriched within the inner leaflet of plasma membranes. Phosphatidylglycerol (PG) and cardiolipin (CL) are more enriched in mitochondria inner membranes, whereas phosphatidylinositide (PI) is primarily found within membranes of the endoplasmic reticulum (ER) and plasma membranes (1, 2). The unevenly distributed PLs may determine membrane fluidity and curvature and their respective physiological functions. The physiological significance of the existence of so many PL species is currently not fully elucidated and is becoming an area of active exploration.
The generation of different PL species may arise from the glycerolipid biosynthetic pathway and PL biosynthesis. For instance, most PLs have saturated or monounsaturated FAs at the sn-1 position, and mitochondrial glycerol-3-phosphate acyltransferase 1 (GPAT1) prefers saturated fatty acyl-CoAs as the substrates in generating lysophosphatidic acid (LPA) (1, 3–5). The enzymes in the glycerolipid biosynthetic pathway to convert LPA to phosphatidic acid (PA), acyl-CoA:acylglycerol-3-phosphate acyltransferases 1 and 2 (AGPAT1 and AGPAT2), on the other hand, do not appear to have any significant selectivity in incorporating FAs onto the sn-2 position of the glycerol backbone (6, 7). The dephosphorylation of PA generates diacylglycerol, which can be utilized as a substrate for either triacylglycerol synthesis or PL biosynthesis (PC, PE). PA can also be incorporated into the biosynthetic pathway of anionic PLs (PI, PG, and CL) (2).
An alternative pathway through PL remodeling contributes significantly as well to the generation of multiple PL species differing in chain lengths and degrees of saturation primarily at the sn-2 position. The FA at the sn-2 position in PLs is often remodeled through the Lands cycle, i.e., deacylation through a member of the phospholipase A2 family and reacylation through lysophospholipid (lysoPL) acyltransferases (lysoPLATs) (8–10). In recent years, two families of lysoPLATs have been discovered owing primarily to genomic efforts (11–19). These enzymes exhibit two distinct features. First, multiple enzyme isoforms exist for a single biochemical activity. Although the exact physiological roles for these different isoforms are not clear at the moment, the existence of multiple isoforms with different tissue distributions, differential substrate preferences, and differential regulation patterns suggests potential differential physiological roles for these isoforms. For instance, lysophosphatidylcholine acyltransferase 1 (LPCAT1) is primarily expressed in aveolar cells in lung with fatty acyl-CoA preference toward saturated fatty acyl-CoAs (13, 14). This observation is consistent with the hypothesis that the enzyme may be primarily responsible for generation of PC within surface surfactants in lung, in which PC has mostly saturated FAs at both the sn-1 and sn-2 positions. LPCAT2, on the other hand, has a high level of expression in macrophage cells and may be involved in the inflammatory response (15). LPCAT3 is mainly expressed in tissues that are metabolically active, and its regulation by proxisome proliferator-activator receptor α (PPARα) suggests its potential role in lipid homeostasis (16, 17). Second, the majority of the lysoPLATs discovered so far have relatively broad lysoPL substrate specificities. For example, LPCAT3 can use lysophosphatidylcholine (LPC), lysophosphatidylserine (LPS), and lysophosphatidylethanolamine (LPE) as substrates (16, 17), thus having the potential to remodel PC, PS, and PE. LPCAT2 was also reported to have an acetyl-CoA:lyso-platelet-activating factor acyltransferase activity (15). A recently identified lysophosphatidylethanolamine acyltransferase 2 (LPEAT2) was originally reported to have LPCAT activity (18, 20). These lysoPLATs belong to two families of proteins based on their primary structures. One is the GPAT/AGPAT family, in which the members share four conserved motifs that are intimately involved in substrate binding and catalysis (5, 21). The so-called MBOAT (membrane-bound O-acyltransferase) family of proteins contains the rest of the lysoPLATs (22). The biochemical activity of this family of proteins is to catalyze the transfer of fatty acyl-CoAs to either protein or lipid substrates.
Acyl-CoA lysocardiolipin acyltransferase 1 (ALCAT1) was originally identified as a lysocardiolipin acyltransferase with ER localization (11). Because the final steps of CL synthesis are within mitochondria (2), it is not clear at present how the microsomal ALCAT1 remodels CL within mitochondria. On the other hand, both PG and PI are synthesized within the ER (1). Here we report novel LPI acyltransferase (LPIAT) and LPG acyltransferase (LPGAT) activities of ALCAT1 suggesting its potential role in PI and PG remodeling. In addition, we report a potential lysoPL substrate binding site for the family of proteins through structure-activity studies using ALCAT1 as a model.
MATERIALS AND METHODS
Materials
Various lysoPLs and acyl-CoAs were purchased from Avanti Polar Lipids (Alabaster, AL). 14C-labeled lysoPLs, acyl-CoAs, and PLs were ordered from American Radiolabeled Chemicals (St. Louis, MO).
Plasmid construction
The full-length human ALCAT1 coding cDNA (National Center for Biotechnology Information accession number NM_182551) was purchased from Invitrogen (Carlsbad, CA). The human LPCAT1 (TC112001), mitochondrial glycerol-3-phosphate acyltransferase (GPAM) (TC106982), AGPAT2 (TC116102), and LPGAT (TC108742) cDNAs were purchased from Origene Technologies (Rockville, MD). Human Leng4 cDNA was cloned from human prostate marathon ready cDNA library (BD Bioscience). The C-terminally FLAG-tagged LPCAT3 was generated as described by Zhao et al. (16). A FLAG tag was inserted at the carboxyl terminus of LPGAT, Leng4, and ALCAT1 through a PCR-based method and subcloned into mammalian expression vector p3xFLAG-CMV-14. Point mutants were generated by PCR using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). For this, cDNA encoding ALCAT1-FLAG was used as template. To make mutants from the template DNA by PCR, 0.2 mM of each PCR primer, 0.2 mM of deoxynucleoside triphosphate, 2.5 units of Pfu Turbo DNA polymerase, and Pfu DNA polymerase reaction buffer were used (95°C for 1 min; 95°C for 50 s, 60°C for 50 s, and 68°C for 7 min, for 18 cycles; and then 68°C for 7 min) in a total volume of 50 μl. Each reaction was incubated with 10 units of DpnI at 37°C for 1 h. Sequences of all constructs were confirmed by sequencing from both directions.
Cell culture, transfection, and membrane protein preparation
Human embryonic kidney 293 (HEK293), COS-7, and Hela cells were purchased from American Type Culture Collection (Manassas, VA) and cultured in DMEM/F-12 (3:1) medium supplemented with 5% FBS in a humidified air atmosphere containing 5% CO2. For transfection, 5 × 105 HEK293 cells, 1 × 105 COS-7 cells, or 2 × 105 Hela cells were seeded in 6-well plates (Falcon, Corning, NY). In each experiment, 1 μg of DNA was tranfected using Fugene 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. After 48 h of transfection, cells were scraped into 150 μl of ice-cold buffer containing 20 mM Tris-HCl (pH 7.5), 250 mM sucrose, 1 mM EDTA, and a protease inhibitor cocktail (Roche Diagnostics), and then sonicated three times on ice for 30 s. After centrifugation for 10 min at 1,000 g, the supernatant was centrifuged at 100,000 g for 1 h. The pellet containing the membrane fraction was resuspended in the same buffer and frozen at −80°C until use. Protein concentrations were determined using a commercially prepared protein assay kit (Pierce, Rockford, IL) with BSA as a standard. Expression of FLAG-tagged ALCAT1, Leng4, and LPGAT1 was verified by Western blotting analysis with a mouse anti-FLAG monoclonal antibody (Sigma, St. Louis, MO).
In vitro acyltransferase activity assays
Acyltransferase activity was determined by measuring the incorporation of radiolabeled acyl-CoAs into PLs. The reaction mixture in a total volume of 100 μl contained 75 mM Tris-HCl, pH 7.5, 1 mg/ml FA-free BSA, 200 μM each lysoPL, 20 μM [1-14C]acyl-CoA, and 5 μg of membrane proteins from HEK293 cells transfected with ALCAT1, LPCAT1, LPCAT3, AGPAT2, GPAM, or empty vector. The reaction was started by the addition of the membrane proteins, incubated for 20 min at room temperature, and stopped by adding 1 ml of chloroform-methanol (2:1, v/v). The PLs were extracted by the method of Bligh and Dyer (23). The organic phase was air dried and separated by TLC using chloroform-methanol-water (65:25:4) as the solvent.
Confocal microscopy
COS-7 cells were transfected with ALCAT1-FLAG on glass-bottom culture dishes (MetTek, Ashland, MA). Forty-eight hours posttransfection, cells were incubated with 100 nM MitoTracker Orange CMTMRos (Invitrogen) for 10 min at 37°C. The cells were fixed with 4% paraformaldehyde for 20 min at 37°C and were permeabilized with 0.2% Triton X-100 in PBS. The samples were then incubated for 2 h at room temperature with a mouse anti-FLAG monoclonal antibody (Sigma) or a rabbit anti-calnexin polyclonal antibody (StressGen Biotechnologies; Victoria, BC, Canada) in PBS with 1% BSA. After a brief washing with PBS, the samples were incubated for 1 h at room temperature with Alexa Fluor 488 goat anti-mouse SFX or Alexa Fluor 555 goat anti-rabbit SFX (Invitrogen). Confocal microscopy was performed with an LSM510 laser-scanning microscope (Carl Zeiss, Germany) equipped with a 40× water immersion objective lens (numerical aperture = 1.2). ALCAT1-FLAG was monitored by excitation at 488 nm and by emission with a 505 nm–530 nm filter. ER and mitochondria markers were monitored by excitation at 543 nm and by emission with a 585 nm–615 nm filter.
Reverse transcription and real-time PCR
PolyA+ RNA was purchased from Clontech (Mountain View, CA), and cDNA was synthesized from 500 ng polyA+ RNA utilizing the Invitrogen Superscript™ III First-Strand Synthesis System for RT-PCR. The Taqman gene expression assays were ordered from Applied Biosystems (Foster City, CA): Hs00699427_A1 (human ALCAT1), Hs00383302 _A1 (human Leng4), and Hs99999905_A1 (human GAPDH). Real-Time PCR was performed on the 7900HT Fast Real-Time PCR System (Applied Biosystems) using TaqMan® Universal PCR Master Mix (Roche, Branchburg, NJ). Cycle threshold values were obtained using Applied Biosystems SDS2.3 software. GAPDH was used as a reference gene. The data were normalized by setting the average adipose expression value to 1.
siRNA transfection
Two hundred picomoles of human ALCAT1 small interfering RNAs (siRNA ID numbers 148241 and 290239; Ambion), human Leng4 siRNAs (siRNA ID numbers 112192 and 214837; Ambion), and control siRNA (silencer negative control 1; Ambion) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The siRNA transfection was performed for 2 days in Hela cells. The RNA of the transfected cells was collected for reverse transcription and real-time PCR. The membrane proteins from the transfected cells were prepared for the in vitro acyltransferase activity assay.
Statistics
The values are presented as means ± SE. Differences among groups were analyzed by one-way ANOVA with Newman-Keuls test, using Prism (GraphPad, San Diego, CA). A P value of <0.05 was considered to indicate statistical significance.
ALCAT1 mutagenic primers
ALCAT1_D168R_F
CATATTCCCAGAAGGGACTCGTCTCACAGAAAACAGCAAG
ALCAT1_D168R_R
CTTGCTGTTTTCTGTGAGACGAGTCCCTTCTGGGAATATG
ALCAT1_D168C_F
CATATTCCCAGAAGGGACTTGTCTCACAGAAAACAGCAAG
ALCAT1_D168C_R
CTTGCTGTTTTCTGTGAGACAAGTCCCTTCTGGGAATATG
ALCAT1_L169T_F
CCCAGAAGGGACTGATACCACAGAAAACAGCAAGTCTC
ALCAT1_L169T_R
GAGACTTGCTGTTTTCTGTGGTATCAGTCCCTTCTGGG
RESULTS
The primary sequence of human ALCAT1 is 89% identical to that of murine protein, and when expressed in HEK293 cells and examined with Western blotting analysis, human ALCAT1 had a molecular mass of 40 kDa (Fig. 1A). Human ALCAT1 was localized to the ER when examined by confocal microscopy (Fig. 1B), consistent with the original report using murine ALCAT1 (11). Human ALCAT1 has relatively high levels of expression in pancreas, kidney, and spleen, and moderate levels of expression in liver, lung, small intestine, and heart, as assessed by quantitative PCR (qPCR) (Fig. 1C). Although in the original report, murine ALCAT1 was regarded as a relatively specific lysocardiolipin acyltransferase using a variety of lysoPL substrates (11), further examination using lysophosphatidylinositol (LPI) and lysophosphatidylglycerol (LPG) as potential substrates revealed that human ALCAT1 possessed LPIAT and LPGAT activities. Using membrane preparations from HEK293 cells overexpressing human ALCAT1, it was demonstrated that ALCAT1 catalyzed the formation of phosphatidylinositol when the membranes expressing ALCAT1 were incubated with LPI and 14C-labeled oleoyl-CoA and examined by TLC analysis compared with vector and mitochondria GPAT1 (GPAM) as controls (Fig. 2A). Similarly, ALCAT1 also catalyzed the formation of PG when the membranes containing ALCAT1 were incubated with LPG and [14C]oleoyl-CoA (Fig. 2A). The formation of PI and PG was specific, inasmuch as omitting lysoPL substrates did not yield any PL upon incubation with membranes containing ALCAT1. Consistent with the original report, human ALCAT1 did not possess activities toward LPC and LPE, but effectively catalyzed the formation of both lysocardiolipin and CL from their respective substrates with [14C]oleoyl-CoA (Fig. 2B). Thus, it appears that ALCAT1 is capable of remodeling anionic PLs, including PI, PG, and CL.
Fig. 1.
Expression, subcellular localization, and tissue distribution of human acyl-CoA lysocardiolipin acyltransferase 1 (ALCAT1). A: Expression of human ALCAT1. A FLAG tag was inserted at the carboxyl terminus of respective enzymes, and the expression levels of human ALCAT1-FLAG, Leng4-FLAG, and LPGAT1-FLAG were examined using Western blotting analysis with a mouse anti-FLAG monoclonal antibody 24 h after transient transfection in human embryonic kidney 293 (HEK293) cells. B: Subcellular localization study. Forty-eight hours after transfection, COS-7 cells were processed for mitochondria staining with MitoTracker Orange CMTMRos (b) or indirect immunofluorescence staining with FLAG antibody (a, d) or calnexin (e), a transmembrane protein served as an endoplasmic reticulum marker. Shown in c and f are the merged images of a and b, as well as d and e, respectively. The yellow color indicates colocalization. C: Tissue expression of human ALCAT1. The relative expression of human ALCAT1 in multiple tissues was measured using GAPDH as a reference gene and normalized to the adipose expression. Quantitative RT-PCR was performed as described in Materials and Methods.
Fig. 2.
Enzymatic analysis of the recombinant human ALCAT1 expressed in HEK293 cells. A: Lysophospholipid acyltransferase activities of recombinant ALCAT1 against LPI, LPA, LPC, LPS, LPE and LPG. B: Lysophospholipid acyltransferase activities of recombinant ALCAT1 against MLCL and DLCL. The acyltransferase assays were conducted by incubating 20 μM [14C]palmitoyl-oleoyl-coA with 200 μM each of the lysophospholipids (lysoPLs) in the presence of 5 μg of membrane proteins from HEK293 cells transfected with pCMV-XL5 (vector), GPAM mitochondria glycerol-3-phosphate acyltransferase (GPAT1), lysophosphatidylcholine acyltransferase (LPCAT3), acyl-CoA:acylglycerol-3-phosphate acyltransferase 2 (AGPAT2), LPG acyltransferase 1 (LPGAT1), or ALCAT1 expression plasmid, followed by lipid extraction and TLC analysis. Representative TLC data are shown. The migrating positions of each of the phospholipids were confirmed by the authentic standards. MLCL, monolysocardiolipin; CL, cardiolipin; PC, phosphatidylcholine; PS, phosphatidylserine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PA, phosphatidic acid; PI, phosphatidylinositol; LPI, lysophosphatidylinositol; LPA, lysophosphatidic acid;LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPG, lysophosphatidylglycerol; LPS, lysophosphatidylserine.
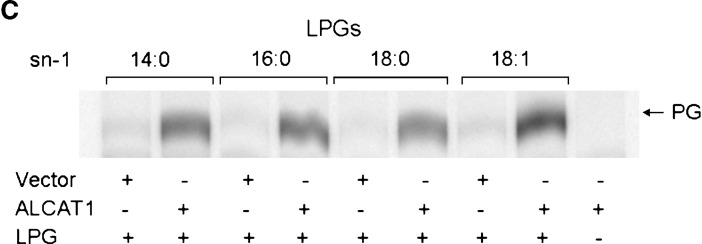
The fatty acyl-CoA specificity was then evaluated using 14C-labeled fatty acyl-CoAs with different chain lengths and degrees of saturation (Fig. 3). The LPIAT activity from ALCAT1 had relatively higher activity utilizing oleoyl-CoA as a substrate, whereas in general, the enzyme was active using a variety of fatty acyl-CoAs, including arachidonyl-CoA, as substrates (Fig. 3A). The fatty acyl-CoA specificity for LPG was examined similarly, and LPGAT activity appeared to prefer stearoyl- and oleoyl-CoA, whereas no activity was observed when shorter-chain-length fatty acyl-CoA (8:0) or arachidonyl-CoA was used as a substrate (Fig. 3B). The specificity was further examined using LPGs that differ in the FAs at the sn-1 position as substrates. The results shown in Fig. 3C indicate that all LPGs with various chain lengths at the sn-1 position were good substrates for LPGAT activity of human ALCAT1.
Fig. 3.
Substrate selectivity of the recombinant human ALCAT1 expressed in HEK293 cells. A: Acyl selectivity of the recombinant human ALCAT1 toward different acyl-CoAs, including n-octanoyl-CoA (8:0), lauroyl-CoA (12:0), palmitoyl-CoA (16:0), stearoyl-CoA (18:0), oleoyl-CoA (18:1), linoleoyl-CoA (18:2), and arachidonyl-CoA (20:4), was analyzed by incubating 200 μM of sn-1-palmitoyl-LPI with 20 μM each of the [14C]acyl-CoAs in the presence of 5 μg of membrane proteins from HEK293 cells transfected with empty vector (vector) or ALCAT1 expression vector (ALCAT1), followed by lipid extraction and TLC analysis. B: Acyl selectivity of the LPGAT activity of the recombinant human ALCAT1 toward different acyl-CoAs as analyzed in A. C: Acyl selectivity of the recombinant human ALCAT1 toward various LPGs with different acyl groups at the sn-1 position, including myristoyl (14:0), palmitoyl (16:0), stearoyl (18:0), and oleoyl (18:1), in the presence of 20 μM [14C]palmitoyl-oleoyl-coA and 200 μM each of the LPGs, respectively.
To evaluate the relative substrate binding affinities, we carried out kinetic studies. Different fatty acyl-CoAs yielded different Km values of LPI, suggesting cooperative binding of both LPI and fatty acyl-CoAs. The exact Km values are given in Table 1. Of note, oleoyl-CoA and linoleoyl-CoA resulted in low-affinity binding of LPI to ALCAT1, whereas other fatty acyl-CoAs led to relatively high-affinity binding of LPI to ALCAT1 (Table 1 and Fig. 4A). Similar studies were carried out to characterize LPGAT activity. Distinct from what was observed with LPIAT activity, different fatty acyl-CoAs gave comparable LPG binding affinities to ALCAT (Table 2 and Fig. 4B).
TABLE 1.
Apparent kinetic studies of LPIAT activity of ALCAT1
| [14C]acyl-CoA | Km | Vmax | Vmax/Km |
|---|---|---|---|
| μM | nmol/min/mg protein | ||
| Arachidonoyl-CoA (20:4) | 39 | 3,230 | 82.82 |
| Oleoyl-CoA (18:1) | 289 | 7,489 | 25.91 |
| Linoleoyl-CoA (18:2) | 134 | 4,696 | 35.04 |
| Stearoyl-CoA (18:0) | 34 | 1,078 | 31.71 |
| Palmitoyl-CoA (16:0) | 53 | 11,203 | 211.38 |
LPIAT, lysophosphatidylinositol acyltransferase; ALCAT1, acyl-CoA lysocardiolipin acyltransferase 1. Kinetic data shown in Fig. 3A were used to calculate apparent Km and Vmax.
Fig. 4.
Effects of acyl composition on enzyme kinetics of the recombinant human ALCAT1 expressed in HEK293 cells. A: Lysophosphatidylinositol acyltransferase (LPIAT) assays were performed by incubating increasing concentrations of sn-1 palmitoyl-LPI with radiolabeled palmitoyl-CoA (C16:0), stearoyl-CoA (C18:0), oleoyl-CoA (C18:1), linoleoyl-CoA (C18:2), or arachidonoyl-CoA (C20:4), respectively, for 20 min under the same assay conditions. B: LPGAT assays were performed by incubating increasing concentrations of sn-1 palmitoyl-LPG with radiolabeled palmitoyl-CoA (C16:0), stearoyl-CoA (C18:0), oleoyl-CoA (C18:1), or linoleoyl-CoA (C18:2), respectively, for 20 min under the same assay conditions. Lysophospholipid acyltransferase activity was measured by TLC and quantified. All enzyme activity data were derived from two independent experiments and are shown as mean ± SE.
TABLE 2.
Apparent kinetic studies of LPGAT activity of ALCAT1
| [14C]Acyl-CoA | Km | Vmax | Vmax/Km |
|---|---|---|---|
| μM | nmol/min/mg protein | ||
| Oleoyl-CoA (18:1) | 54 | 5,514 | 102.11 |
| Linoleoyl-CoA (18:2) | 36 | 1,358 | 37.72 |
| Stearoyl-CoA (18:0) | 70 | 3,530 | 50.43 |
| Palmitoyl-CoA (16:0) | 34 | 1,673 | 49.21 |
LPGAT, lysophosphatidylglycerol acyltransferase. Kinetic data shown in Fig. 3B were used to calculate apparent Km and Vmax.
To assess respective cellular lysoPLAT activities, we managed to reduce ALCAT1 expression through siRNA knockdown in Hela cells. Two RNA interference oligos were used and demonstrated more than 80% knockdown of ALCAT1 expression compared with control oligos in Hela cells as assessed by qPCR (Fig. 5A). Surprisingly, no reductions in ALCAT activity were observed using either monolysocardiolipin or dilysocardiolipin as substrates when incubating with membrane preparations from Hela cells along with radiolabeled oleoyl-CoA (Fig. 5D). However, LPGAT activity was reduced by about 80% (Fig. 5B). Significant reduction in LPIAT activity was also observed when ALCAT1 expression was reduced by either RNAi oligo. We also used two different RNAi oligos to knock down the human ortholog of Caenorhabditis elegans LPIAT1 (also named MBOAT 7 or Leng4) (19) and these two oligos managed to reduce LPIAT1 expression by 80% and 70%, respectively. Accordingly, LPIAT activity was reduced by 60% and 40%, respectively, by these two oligos (Fig. 5C). These experiments indicated that both LPIAT1 and ALCAT1 contributed to the cellular LPIAT activity in Hela cells, whereas ALCAT1 was largely responsible for cellular LPGAT activity.
Fig. 5.
Reduced LPGAT and LPIAT activities in Hela cells after ALCAT1 knockdown. Two human ALCAT1 siRNAs and a control siRNA were transfected into Hela cells, respectively. After 48 h, mRNA level (A), LPGAT activity (B), LPIAT activity (C), and lysocardiolipin acyltransferase activity (D) were measured as described in Materials and Methods. Two human Leng4 siRNAs were transfected in Hela cells as positive controls for LPIAT activity (A, C). The quantified data in B, C, and D represent the mean ± SE of three independent measurements. * P < 0.05.
The identification of LPIAT and LPGAT activities from ALCAT1 provided the opportunity to study potential substrate binding sites. PI, PG, and CL are regarded as anionic PLs, whereas PC carries a positively charged choline group. The ALCAT1 primary sequence suggests that it belongs to the mitochondria GPAT1 family, and the proteins in this family share four conserved sequence motifs that are potentially important for either substrate binding or catalysis (11, 21). Notably, motif 2 and motif 3 were hypothesized as binding motifs to substrates, and recently, Shindou and colleagues (15) identified motif 2 as the fatty acyl-CoA binding site. Thus, we focused on the sequence alignment in motif 3 of ALCAT1, LPCAT1, LPCAT2, AGPAT7 (a novel LPCAT/LPEAT), AGPAT1, and AGPAT2 from several different species, including Drosophila melanogaster, Mus musculus, Rattus norvegicus, Xenopus laevis, Homo sapiens, C. elegans, Gallus gallus, Saccharomyces cerevisiae, and Canis lupus familiaris (Fig. 6A). We reasoned that the conserved sequences were possibly involved in binding to common components of lysoPL and fatty acyl-CoAs, whereas nonconserved residues may represent the amino acids responsible for different lysoPL substrate binding. We chose D168 and L169, the two amino acids that are the most proximal to the conserved region of motif 3, for structural activity analysis. Changes of aspartic acid at 168 to either arginine (D168R) or cysteine (D168C) completely abolished both LPIAT and LPGAT activity, suggesting the critical role of aspartic acid 168 in substrate binding or catalysis (Fig. 6B, C). A threonine-to-leucine mutant (T169L) lost its LPIAT activity. However, LPGAT activity was comparable to that of the wild-type control (Fig. 6B, C). All three mutants were expressed at levels comparable to those of the wild type in HEK293 cells (Fig. 6D). These data highlight the importance of the D168/T169 region in potential substrate binding and/or catalysis.
Fig. 6.

Identification of amino acid residues important for lysoPL binding in ALCAT1. A: Alignment of lysoPL acyltransferase sequences near a region (motif 3) that is conserved among species but divergent for distinct lysoPL substrates. B: Representative acyltransferase activities of ALCAT1 mutants using 20 μM of sn-1 palmitoyl-LPI or LPG and oleyol-CoA as substrates. HEK293 cells were transfected with plasmids expressing either wild-type or mutant proteins in triplicate, and the membranes were prepared to analyze the LPIAT and LPGAT activities by TLC as described in Materials and Methods. C: Quantitative representation of LPIAT and LPGAT activities of ALCAT1 wild-type or mutant proteins. * P < 0.05. The quantified data in C represent the mean ± SE of three independent measurements. D: Expression levels of wild-type ALCAT1 and ALCAT1 mutants transfected in HEK293 cells were compared using Western blotting analysis with an anti-FLAG monoclonal antibody. Blot of β-actin was also included as a loading control.
DISCUSSION
ALCAT1 was originally identified as a lysocardiolipin acyltransferase with fatty acyl-CoA substrate preference toward linoleoyl-CoA and oleoyl-CoA (11). Although it was proposed to have a potential role in remodeling CL based on the in vitro data, no experimental evidence has been presented thus far to explain how ALCAT1 as a microsomal enzyme accesses CL, which is primarily synthesized and localized within the inner membrane of mitochondria. Our studies identifying novel activities in LPIAT and LPGAT activities of ALCAT1 may thus provide plausible physiologically relevant functions of this enzyme. Indeed, PI and PG are both synthesized within the ER, and thus these substrates are readily available to ALCAT1 for the ensuing remodeling for specific functions. Furthermore, in Hela cells, reduction of ALCAT1 expression reduced LPIAT and LPGAT activities, but not lysocardiolipin acyltransferase activity (Fig. 5), suggesting that LPIAT and LPGAT are potentially physiologically relevant activities in cells.
The physiological role of ALCAT1 is currently not completely elucidated. PG is distributed in both the ER and mitochondria and serves as the precursor for CL biosynthesis (2). Refined PG species from PG remodeling may impact CL formation and function. PG is also a major PL component in lung, and relatively high expression of ALCAT1 in lung may thus contribute to PG diversity of molecular species for specific functions. PI derivatives are important signaling molecules, and currently it is not known whether specific PI species are required for efficient signal transduction processes. It is conceivable, however, that generation of more-refined species of PI and its derivatives may somehow impact the relative abundance of certain specific molecular species and the associated cellular functions. For example, C. elegans LPIAT1 deficiency resulted the reduction of the incorporation of PUFAs into PI and associated larval arrest and egg-laying defects (19). Evolutionarily, ALCAT1 is not expressed in species without atria (11). In zebrafish, deficiency of the ALCAT1 ortholog resulted in significant reductions of both hematopoietic and endothelial cell lineages (24). Although it was hypothesized that an acyltransferase activity toward a protein may be responsible for such observations (24), our studies point out that this enzyme may very well use lysoPLs as substrates instead of proteins. How the reduction of ALCAT1 expression led to the deficiency of hematopoietic and endothelial cell differentiation is not apparent at the moment, but may be related to the change of specific PG and/or PI species resulting from the changes of such an enzyme activity. It is also conceivable that deficiency of such a protein in mammals may result in defects of the circulatory system. Indeed, Wang et al. (25) described in their work in embryoid bodies that mouse ALCAT1 plays a key role in specification of hematopoietic and endothelial cells. Future work will focus on the elucidation of the exact physiological roles of the enzyme and its potential involvement in signal transduction and cellular functions.
It is becoming apparent that most of the lysoPLATs identified so far have relatively broad lysoPL substrate specificities. In the case of LPCAT3, it utilizes the substrates of LPC, LPE, and LPS (16, 17). Other LPCATs also appear to possess a substrate binding pocket that can adopt both LPC and LPE (17, 18, 20). ALCAT1, on the other hand, appears to have a substrate binding pocket that recognizes anionic lysoPLs. We hypothesized that certain amino acids within the substrate binding pocket are potentially responsible for such substrate specificities. Based on the previous analysis of the GPAT family of proteins and the hypothesis that motifs 2 and 3 may be involved in substrate binding (21), we chose several amino acids within motif 3 that are different from LPCATs in order to study their potential function in lysoPL substrate binding. Indeed, mutants of D168R and D168C resulted in total loss of the activity, whereas the mutant proteins were expressed at levels comparable to those of the wild-type protein. A specific mutation of L169T resulted in the loss of LPIAT activity, but retained LPGAT activity. This observation strongly indicates that the loss of LPIAT activity was not due to the loss of catalytic activity; rather, this was probably the result of the loss of LPI binding. The retained LPGAT activity suggested a very subtle conformation change, inasmuch as the mutant might have lost LPI binding but retained LPG binding. Our studies thus identify motif 3 of the GPAT family of proteins as a potential lysoPL substrate binding pocket for lysoPLATs. Our data are consistent with and complementary to the recent report that motif 2 of the GPAT family of proteins is largely responsible for fatty acyl-CoA binding (15). Our studies, along with those of Harayama et al. (26), thus provide evidence of critical amino acids involved in binding of both lysoPL and fatty acyl-CoA substrates for the family of proteins.
Acknowledgments
The authors would like to thank Dr. Shaoyou Chu for assistance with confocal microscopic study.
Abbreviations
AGPAT, acyl-CoA:acylglycerol-3-phosphate acyltransferase
ALCAT1, acyl-CoA lysocardiolipin acyltransferase 1
CL, cardiolipin
ER, endoplasmic reticulum
GPAT, glycerol-3-phosphate acyltransferase
LPCAT, lysophosphatidylcholine acyltransferase
LPG, lysophosphatidylglycerol
LPGAT, LPG acyltransferase
LPI, lysophosphatidylinositol
LPIAT, LPI acyltransferase
lysoPL, lysophospholipid
lysoPLAT, lysophospholipid acyltransferase
MBOAT, membrane-bound O-acyltransferase
PA, phosphatidic acid
PC, phosphatidylcholine
PE, phosphatidylethanolamine
PG, phosphatidylglycerol
PI, phosphatidylinositide
PL, phospholipid
PPARα, proxisome proliferator-activator receptor α
PS, phosphatidylserine
This work was supported by Lilly Research Laboratories.
Published, JLR Papers in Press, December 15, 2008.
References
- 1.van Meer G., D. R. Voelker, and G. W. Feigenson. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vance, D. E. 2002. Phospholipid Biosynthesis in Eukaryotes. 4th edition. Elsevier, St. Louis, MO.
- 3.Shin D. H., J. D. Paulauskis, N. Moustaid, and H. S. Sul. 1991. Transcriptional regulation of p90 with sequence homology to Escherichia coli glycerol-3-phosphate acyltransferase. J. Biol. Chem. 266 23834–23839. [PubMed] [Google Scholar]
- 4.Yet S. F., S. Lee, Y. T. Hahm, and H. S. Sul. 1993. Expression and identification of p90 as the murine mitochondrial glycerol-3-phosphate acyltransferase. Biochemistry. 32 9486–9491. [DOI] [PubMed] [Google Scholar]
- 5.Coleman R. A., and D. P. Lee. 2004. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43 134–176. [DOI] [PubMed] [Google Scholar]
- 6.Aguado B., and R. D. Campbell. 1998. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J. Biol. Chem. 273 4096–4105. [DOI] [PubMed] [Google Scholar]
- 7.Eberhardt C., P. W. Gray, and L. W. Tjoelker. 1997. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J. Biol. Chem. 272 20299–20305. [DOI] [PubMed] [Google Scholar]
- 8.Lands W. E. 1960. Metabolism of glycerolipids. II. The enzymatic acylation of lysolecithin. J. Biol. Chem. 235 2233–2237. [PubMed] [Google Scholar]
- 9.Lands W. E., and I. Merkl. 1963. Metabolism of glycerolipids. III. Reactivity of various acyl esters of coenzyme A with alpha'-acylglycerophosphorylcholine, and positional specificities in lecithin synthesis. J. Biol. Chem. 238 898–904. [PubMed] [Google Scholar]
- 10.Merkl I., and W. E. Lands. 1963. Metabolism of glycerolipids. IV. Synthesis of phosphatidylethanolamine. J. Biol. Chem. 238 905–906. [PubMed] [Google Scholar]
- 11.Cao J., Y. Liu, J. Lockwood, P. Burn, and Y. Shi. 2004. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 279 31727–31734. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., J. Cao, and Y. Shi. 2004. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J. Biol. Chem. 279 55866–55874. [DOI] [PubMed] [Google Scholar]
- 13.Chen X., B. A. Hyatt, M. L. Mucenski, R. J. Mason, and J. M. Shannon. 2006. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc. Natl. Acad. Sci. USA. 103 11724–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi H., H. Shindou, D. Hishikawa, T. Harayama, R. Ogasawara, A. Suwabe, R. Taguchi, and T. Shimizu. 2006. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem. 281 20140–20147. [DOI] [PubMed] [Google Scholar]
- 15.Shindou H., D. Hishikawa, H. Nakanishi, T. Harayama, S. Ishii, R. Taguchi, and T. Shimizu. 2007. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:LYSO-PAF acetyltransferase. J. Biol. Chem. 282 6532–6539. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y., Y. Q. Chen, T. M. Bonacci, D. S. Bredt, S. Li, W. R. Bensch, D. E. Moller, M. Kowala, R. J. Konrad, and G. Cao. 2008. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J. Biol. Chem. 283 8258–8265. [DOI] [PubMed] [Google Scholar]
- 17.Hishikawa D., H. Shindou, S. Kobayashi, H. Nakanishi, R. Taguchi, and T. Shimizu. 2008. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA. 105 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soupene E., H. Fyrst, and F. A. Kuypers. 2008. Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Natl. Acad. Sci. USA. 105 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H. C., T. Inoue, R. Imae, N. Kono, S. Shirae, S. Matsuda, K. Gengyo-Ando, S. Mitani, and H. Arai. 2008. Caenorhabditis elegans Mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 19 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao J., D. Shan, T. Revett, D. Li, L. Wu, W. Liu, J. F. Tobin, and R. E. Gimeno. 2008. Molecular identification of a novel mammalian brain isoform of acyl-CoA:lysophospholipid acyltransferase with prominent ethanolamine lysophospholipid acylating activity, LPEAT2. J. Biol. Chem. 283 19049–19057. [DOI] [PubMed] [Google Scholar]
- 21.Lewin T. M., N. M. Schwerbrock, D. P. Lee, and R. A. Coleman. 2004. Identification of a new glycerol-3-phosphate acyltransferase isoenzyme, mtGPAT2, in mitochondria. J. Biol. Chem. 279 13488–13495. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann K. 2000. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 25 111–112. [DOI] [PubMed] [Google Scholar]
- 23.Bligh E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37 911–917. [DOI] [PubMed] [Google Scholar]
- 24.Xiong J. W., Q. Yu, J. Zhang, and J. D. Mably. 2008. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ. Res. 102 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., P. W. Faloon, Z. Tan, Y. Lv, P. Zhang, Y. Ge, H. Deng, and J. W. Xiong. 2007. Mouse lysocardiolipin acyltransferase controls the development of hematopoietic and endothelial lineages during in vitro embryonic stem-cell differentiation. Blood. 110 3601–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harayama T., H. Shindou, R. Ogasawara, A. Suwabe, and T. Shimizu. 2008. Identification of a novel noninflammatory biosynthetic pathway of platelet-activating factor. J. Biol. Chem. 283 11097–11106. [DOI] [PubMed] [Google Scholar]