Abstract
Several prostate cancer susceptibility loci have recently been identified by genome-wide association studies. These loci are candidates for susceptibility to other epithelial cancers. The aim of this study was to test these tag single nucleotide polymorphisms (SNP) for association with invasive ovarian, colorectal, and breast cancer. Twelve prostate cancer-associated tag SNPs were genotyped in ovarian (2,087 cases/3,491 controls), colorectal (2,148 cases/2,265 controls) and breast (first set, 4,339 cases/4,552controls; second set, 3,800 cases/3,995 controls) case-control studies. The primary test of association was a comparison of genotype frequencies between cases and controls, and a test for trend stratified by study where appropriate. Genotype-specific odds ratios (OR) were estimated by logistic regression. SNP rs2660753 (chromosome 3p12) showed evidence of association with ovarian cancer [per minor allele OR, 1.19; 95% confidence interval (95% CI), 1.04-1.37; Ptrend = 0.012]. This association was stronger for the serous histologic subtype (OR, 1.29; 95% CI, 1.09-1.53; P = 0.003). SNP rs7931342 (chromosome 11q13) showed some evidence of association with breast cancer (per minor allele OR, 0.95; 95% CI, 0.91-0.99; Ptrend = 0.028). This association was somewhat stronger for estrogen receptor-positive tumors (OR, 0.92; 95% CI, 0.87-0.98; P = 0.011). None of these tag SNPs were associated with risk of colorectal cancer. In conclusion, loci associated with risk of prostate cancer may also be associated with ovarian and breast cancer susceptibility. However, the effects are modest and warrant replication in larger studies.
Introduction
Breast and ovarian cancer are among the most frequent cancers in women in Western countries, and colorectal cancer is one of the most prevalent cancers that affect both men and women. The known ovarian and breast cancer susceptibility genes, such as BRCA1 and BRCA2, explain <40% of the excess familial risk of ovarian cancer and <25% of the excess familial breast cancer risk (1, 2). Similarly, the known high penetrance colorectal cancer susceptibility genes, such as APC and the mismatch repair genes, account for <5% of the overall incidence; however, it has been estimated that 35% of colorectal cancer can be due to inherited susceptibility (3, 4). It is likely that the unexplained excess familial risks for ovarian, breast, and colorectal cancer are due to a combination of multiple low/moderate penetrance genetic variants, which are associated with relatively small effects on risk in the individual but contribute substantially to the overall risk in the population.
Genome-wide association studies (GWAS) using large sets of cases and controls have proven to be an effective approach to identify the common variants that are associated with common diseases without prior knowledge of position or function. This approach has successfully identified novel loci for breast, colorectal, and prostate cancer, as well as for other complex, late-onset disorders (5-12). It is clear that some loci confer risk for more than one type of cancer. For example, the high penetrance rare deleterious variants in BRCA1 and BRCA2 increase risk of breast, ovarian, prostate, and other cancers, and deleterious mutations in the mismatch repair genes cause a spectrum of cancers including colorectal, endometrial, gastric, and ovarian. In addition, a common allele on 8q24 identified through a prostate cancer GWAS has shown also be associated colorectal and varian cancer (9, 12, 13). More than a dozen other prostate cancer susceptibility loci have now been identified from GWAS (see Table 1; refs. 8, 14-16). The aim of this study was to test 12 of these loci for evidence of association with ovarian, colorectal, or breast cancer. These loci included seven identified by Eeles and colleagues (14), two additional loci identified by Thomas and colleagues (15), and one further locus identified by Gudmundsson and colleagues (16), together with two previously identified loci on chromosome 17 (8).
Table 1.
Association of prostate SNPs with colorectal, ovarian, and breast cancer
| SNPs | Base change* | Gene/Chr. | MAF† | Reported GWAS prostate cancer |
Colorectal cancer (2,148 cases/2,265 controls) |
Ovarian cancer‡ (1,973 cases/3,419 controls) |
Breast cancer (4,339 cases/4,552 controls) |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)§ | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||||
| rs5945619 | T/C | ChrX | 0.36 | 1.19 (1.07-1.31) | 1.01 (0.93-1.11) | 0.76 | 1.08 (0.99-1.17) | 0.09 | 1.06 (0.99-1.12) | 0.08 |
| rs2660753 | G/A | Chr3 | 0.09 | 1.18 (1.06-1.31) | 0.92 (0.83-1.02) | 0.26 | 1.19 (1.04-1.37) | 0.012 | 1.06 (0.96-1.17) | 0.26 |
| rs10993994 | G/A | Chr10 | 0.39 | 1.25 (1.17-1.34) | 0.95 (0.89-1.01) | 0.25 | 1.05 (0.96-1.14) | 0.32 | 0.99 (0.93-1.05) | 0.81 |
| rs7931342|| | C/A | Chr11 | 0.49 | 0.84 (0.79-0.90) | 0.96 (0.91-1.02) | 0.39 | 1.01 (0.93-1.09) | 0.96 | 0.95 (0.91-0.99) | 0.028 |
| rs9364554 | C/T | SLC22A3 | 0.30 | 1.17 (1.08-1.26) | 0.98 (0.91-1.04) | 0.59 | 1.06 (0.97-1.16) | 0.17 | 1.03 (0.96-1.10) | 0.40 |
| rs2735839 | G/A | Chr19 | 0.15 | 0.83 (0.75-0.91) | 1.08 (0.99-1.17) | 0.22 | 0.99 (0.89-1.11) | 0.87 | 0.99 (0.91-1.07) | 0.74 |
| rs6465657 | A/G | LMTK2 | 0.46 | 1.12(1.05-1.20) | 1.00 (0.94-1.06) | 1.00 | 0.96 (0.89-1.04) | 0.25 | 1.00 (0.95-1.06) | 0.91 |
| rs7501939 | G/A | TCF2 | 0.40 | 0.84 (0.79-0.89) | 1.02(0.96-1.08) | 0.65 | 0.96 (0.89-1.04) | 0.29 | 1.03 (0.97-1.09) | 0.41 |
| rs1859962 | T/G | Chr17 | 0.48 | 1.20 (1.14-1.27) | 0.98 (0.92-1.04) | 0.60 | 0.98 (0.90-1.06) | 0.67 | 0.99 (0.93-1.05) | 0.70 |
| rs10486567 | G/A | JAZF1 | 0.24 | 0.78 (0.71-0.85) | 1.01 (0.94-1.09) | 0.84 | 1.01 (0.92-1.12) | 0.98 | 0.95 (0.89-1.02) | 0.19 |
| rs12769019¶ | A/G | CTBP2 | 0.28 | 1.25 (1.16-1.35) | 1.03 (0.97-1.11) | 0.48 | 1.00 (0.90-1.10) | 0.94 | 0.99 (0.92-1.06) | 0.72 |
| rs2710646 | C/A | EHBP1 | 0.20 | 1.15 (1.10-1.21) | 0.93 (0.87-1.01) | 0.22 | 1.05 (0.94-1.19) | 0.36 | 1.00 (0.93-1.07) | 0.96 |
The most common allele in controls is given first.
MAF: minor allele frequency in breast cancer controls.
White subjects of European ancestry only.
Per rare allele ORs with 95% CI are presented. ORs for the first seven SNPs in the table are from Eeles and colleagues (14), rs7501939 and rs1859962are from Gudmundsson and colleagues (8), rs10486567 and rs12769019 (in perfect LD with rs4962416) are from Thomas and colleagues (15), and rs2710646 (in perfect LD with 721048) is from Gudmundsson and colleagues (16).
rs7931342was genotyped in breast cancer validation set as well with a total of 8139 cases and 8547 controls.
Genotyping failed in the MALOVA and UKOPS ovarian cancer studies for rs12769019. Data highlighted with bold text are the statistically significant results.
Materials and Methods
Cancer case-control studies
Two UK breast cancer case-control studies were used for this analysis. SEARCH (breast; 6,640 cases and 6,832 controls) started in 1996. SEARCH is an ongoing, UK population-based study of different epithelial cancers ascertained through the Eastern Cancer Registration and Information Centre (formerly East Anglian Cancer Registry). Eligible cases were those diagnosed under the age of 55 between 1991 and 1996 and those diagnosed under age 70 y since 1996 (17). Controls were randomly selected from the Norfolk, United Kingdom, component of European Prospective Investigation of Cancer study (EPIC). The ethnic background of cases and controls is similar with >98% being self-reported as White. The Genetics of Familial Breast Cancer Study (GFBCS; 1,499 cases and 1,715 controls) has been described previously in detail (18). It comprises unrelated cases (index cases) with a family history of breast cancer in close relatives but no close relative with ovarian cancer. These were ascertained through regional cancer genetics clinics in the United Kingdom. Mutations and large deletions/duplications in BRCA1 and BRCA2 had been excluded in the index cases. Controls were ascertained from the 1958 Birth Cohort Study, which is an ongoing study of all persons born in Great Britain, 3 to 9 March 1958 (currently of age 50 y). The study included a recent biomedical assessment during 2002 to 2004 at which blood samples and informed consent were obtained for the creation of a genetic resource. All cases were White, and at least 98% of the controls were of White ethnicity.
To reduce genotyping costs, the breast study samples were divided into two sets: the first set comprising 4,339 cases and 4,552 controls from SEARCH and the second set comprising a further of 2,301 cases and 2,280 controls from SEARCH and the GFBCS samples. Single nucleotide polymorphisms (SNP) that showed marginally significant association in the initial set (Ptrend < 0.05) were also genotyped in the second set.
The colorectal study, also from SEARCH, comprises 2,148 cases ages between 18 and 69 y and diagnosed since the year 1996. Cancer-free controls (2,265) were recruited from general practices from around the Eastern UK region (19). Eligible individuals were sex and frequency matched in 5-y age bands to cases. More than 98% of participants are White.
Invasive epithelial ovarian cancer cases and controls came from 4 different studies (total cases 2,085 and 3,486 controls) described in detail elsewhere (20, 21). Briefly, these were SEARCH (ovarian) from United Kingdom (1,013 cases and 1,235 controls, among them 947 cases and 1,229 controls are White) comprises cases diagnosed under age 70 y since 1998 with controls selected from EPIC as described above. The UKOPS ovarian cancer study (303 cases and 606 controls, among them 293 cases and 600 controls are White) is recruiting incident cases from gynecological oncology National Health Service centers throughout the United Kingdom (January 2006 onwards) and healthy postmenopausal women from the United Kingdom Collaborative Trial of Ovarian Cancer Screening as controls. MALOVA is a population-based study from Denmark comprises 681 incident cases diagnosed from 1994 to 1999 and 1,460 matching controls, from the municipalities of Copenhagen and Frederiksberg, and surrounding counties, 446 cases and 1,221 controls (all are White) were used in this analysis. Finally, the GEOCS study from the United States (325 cases and 429 controls, among them 287 cases, 369 controls are White subjects of European ancestry) comprises consecutively collected cases diagnosed from 1997 to 2002 in the Greater Bay Area Cancer Registry of San Francisco, and matched controls from the same study area using random-digit dial identification. We restricted the analysis to the White subjects of European ancestry for all the ovarian studies.
All studies has ethical approval and all participants provided written, informed consent.
Genotyping
All samples were genotyped using the Taqman 7900HT Sequence Detection System according to the manufacturer’s instructions. Assays were carried out in 384-well plates and included 12 duplicate samples in each plate for quality control. Genotypes were determined using Allelic Discrimination Sequence Detection Software (Applied Biosystems). Each assay was carried out using 10 ng DNA in a 2.5 μL reaction using TaqMan universal PCR master mix (Applied Biosystems), forward and reverse primers, and FAM and VIC-labeled probes designed by Applied Biosystems (ABI Assay-by-design). Details of primer and probe sequences and assay conditions used for each polymorphism analyzed are available upon request. Where discordant genotypes were observed in duplicates, the genotyping was repeated. Overall concordance between duplicate samples was over 98%. Individual samples with failed calls were not repeated. Hence, there are variations in the number of samples successfully genotyped for each polymorphism.
Data analysis
Deviation of genotype frequencies from those expected under Hardy-Weinberg equilibrium (HWE) were assessed by χ2 tests (one degree of freedom) for each set of controls as part of the genotyping quality control. The primary tests of association were comparison of genotype frequencies in cases and controls using a trend test for each SNP and ovarian, colorectal, and breast cancer. This was done using unconditional logistic regression stratified by study where appropriate. Odds ratios (OR) for allele dosage and associated 95% confidence intervals (95% CI) were also estimated by unconditional logistic regression. We tested for heterogeneity between study strata by comparing logistic regression models with and without a genotype-stratum interaction term using likelihood ratio tests. A case-only analysis was used to compare genotype-specific risks by disease subgroup.
Results
We genotyped the 12 SNPs in the ovarian, colorectal, and breast cancer case-control studies. The Taqman assay for SNP rs4962416, the SNP used in Thomas and colleagues (15) failed manufacture, and so we replaced it with an assay for a perfectly correlated SNP, rs12769019 (r2 =1, in the HapMap CEU population). Genotype distributions in controls were consistent with HWE except for rs7501939 in GEOCS (ovary; P = 0.01) and rs12769019 in SEARCH (breast; P = 0.02). Inspection of the cluster plots indicated good discrimination between genotypes, suggesting that these deviations from HWE are likely to be chance observations.
Genotype-specific ORs and tests of association are presented in Table 1. The observed genotype frequencies for each of the data sets are presented in Supplementary Table S1. There was no association in controls between age and genotype frequency for any of the SNPs, and age-adjusted genotype-specific ORs were similar to unadjusted ORs (data not shown). None of SNPs tested was associated with colorectal cancer. One SNP, rs2660753 on chromosome 3p12, showed evidence of association with ovarian cancer and another SNP, rs7931342 on chromosome 11q13, was associated with breast cancer.
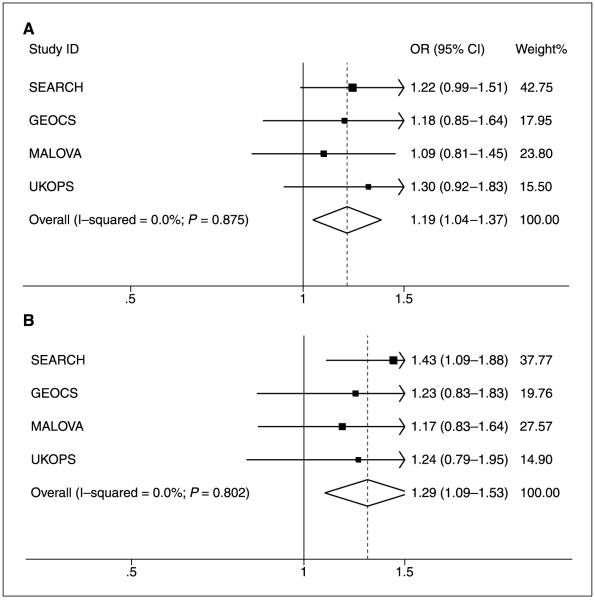
Carriers of the minor allele of rs2660753 were at increased risk of ovarian cancer (per minor allele OR, 1.19; 95% CI, 1.04-1.37; Ptrend = 0.012). There was no evidence for between-study heterogeneity (P = 0.88). The log-additive, codominant model fit the data best, but the recessive model also fitted reasonably well (OR minor allele homozygote versus common allele carrier, 2.04; 95% CI, 1.08-3.84; P = 0.028). We limited subgroup analysis to the serous subtype as numbers of the other histologic subtypes were low. The genotypic-specific risks of serous type ovarian cancer, estimated from the combined data, are presented in Table 2. These were similar to the overall ovarian cancer risks, although the risk for the minor allele of rs2660753 was somewhat stronger: per minor allele OR, 1.29; 95% CI, 1.09-1.53; (Ptrend=0.003); minor allele homozygote OR, 2.74; 95% CI, 1.35-5.55 (P = 0.005) compared with the common allele carrier. Figure 1 shows the genotype-specific ORs for each ovarian study and for the combined analysis for rs2660753.
Table 2.
Genotype-specific risks (95%CI) for serous-type invasive ovarian cancers in combined data for White subjects of European ancestry
| dbSNP | Controls |
Cases |
Per rare allele OR (95% CI) | P het | P trend | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA* | Aa† | aa‡ | Total | AA* | Aa† | aa‡ | Total | ||||
| rs5945619 | 1,355 | 1,565 | 433 | 3,353 | 362 | 407 | 139 | 908 | 1.07 (0.96-1.19) | 0.18 | 0.24 |
| rs2660753 | 2,760 | 525 | 18 | 3,303 | 717 | 170 | 14 | 901 | 1.29 (1.09-1.53) | 0.004 | 0.003 |
| rs10993994 | 1,298 | 1,574 | 483 | 3,355 | 337 | 434 | 137 | 908 | 1.04 (0.93-1.15) | 0.77 | 0.50 |
| rs7931342 | 852 | 1,709 | 807 | 3,368 | 243 | 446 | 223 | 912 | 1.00 (0.90-1.11) | 0.63 | 0.96 |
| rs9364554 | 1,585 | 1,397 | 297 | 3,279 | 430 | 381 | 90 | 901 | 1.06 (0.94-1.18) | 0.54 | 0.36 |
| rs2735839 | 2,455 | 850 | 68 | 3,373 | 682 | 205 | 26 | 913 | 0.95 (0.82-1.10) | 0.09 | 0.50 |
| rs7501939 | 1,164 | 1,612 | 571 | 3,347 | 300 | 439 | 169 | 908 | 1.07 (0.96-1.19) | 0.45 | 0.21 |
| rs6465657 | 955 | 1,653 | 734 | 3,342 | 275 | 435 | 186 | 896 | 0.93 (0.84-1.04) | 0.38 | 0.20 |
| rs1859962 | 860 | 1,670 | 826 | 3,356 | 243 | 438 | 219 | 900 | 0.97 (0.87-1.07) | 0.67 | 0.53 |
| rs10486567 | 1,922 | 1,183 | 177 | 3,282 | 545 | 308 | 49 | 902 | 0.94 (0.83-1.07) | 0.50 | 0.34 |
| rs2710646 | 2,095 | 1,017 | 121 | 3,233 | 601 | 262 | 48 | 911 | 1.01 (0.89-1.15) | 0.06 | 0.85 |
| rs12769019 | 806 | 641 | 121 | 1,568 | 265 | 191 | 49 | 505 | 1.02 (0.87-1.20) | 0.23 | 0.79 |
AA, common homozygote.
Aa, heterozygote.
aa, rare homozygote. Data highlighted with bold text are the statistically significant results.
Figure 1.
Genotype-specific risks of SNP rs2660753 for ovarian cancer by study in White subjects of European ancestry. A, all ovarian cancer subtypes included; B, analysis restricted to serous type ovarian cancers.
SNP rs7931342was the only one associated with breast cancer risk in the first case-control set, with the minor allele of rs7931342 being associated with a decreased risk of breast cancer (per minor allele OR, 0.93; 95% CI, 0.87-0.98; Ptrend = 0.01). We therefore genotyped this SNP in the validation samples. There was no association based on these data alone (per minor allele OR, 0.99; 95% CI, 0.92-1.05; Ptrend = 0.64), but when the data were combined, the association remained significant at the 5% level (per minor allele OR, 0.95; 95% CI, 0.91-0.99; Ptrend = 0.028). There was no between-study heterogeneity (P = 0.82). Morphology (ductal or lobular) and estrogen receptor (ER) status were available for 5,822 and 3,495 breast cancer cases, respectively, from SEARCH, and we carried out analyses based on these disease subgroups. SNP rs7931342 was associated with decreased risks of ductal breast cancer (per minor allele OR, 0.94; 95% CI, 0.89-0.99; Ptrend = 0.02) but was not associated with lobular breast cancer (per minor allele OR, 1.03; 95% CI, 0.93-1.13; Ptrend = 0.57). This difference between ductal and lobular was not statistically significant (P = 0.074). SNP rs7931342 was associated with decreased risk in ER-positive breast cancer (per minor allele OR, 0.92; 95% CI, 0.87-0.98; Ptrend = 0.011) but was not associated with ER-negative breast cancer (per minor allele OR, 1.02; 95% CI, 0.91-1.14; Ptrend = 0.73). Again, the difference between risk of ER-positive and ER-negative tumors was not statistically significant (P = 0.09).
Discussion
We have evaluated 12 confirmed prostate cancer-associated loci with breast, colorectal, and ovarian cancer using tag SNPs. In all but two cases, we found no evidence of an association, and the 95% confidence limits exclude the estimated OR for prostate cancer. Thus, most of these susceptibility loci appear to be specifically associated with prostate cancer risk. Two loci, however, showed some evidence of association. We found the minor allele of rs2660753 was associated with an increased risks of invasive ovarian cancer and of serous ovarian cancer in particular. The same allele was also associated with increased prostate cancer risk, with a similar OR (Table 1). Support for a common genetic basis for prostate and ovarian cancer comes from the observation that ovarian cancer cases are more likely to report a first-degree relative with prostate cancer than controls (5.1% versus 2.4%; P = 0.00002). The minor allele of rs7931342, associated with a reduced risk of prostate cancer, was also associated with a reduced risk of breast cancer (Table 1). These results, however, need to be treated with caution. A large number of reported positive associations have not been replicated by subsequent studies (22, 23). In the literature, it has been estimated that the fraction of false-positive findings is at least 0.95 for studies of association between genetic variants and disease risks (24). Even if these loci are treated as strong candidate loci, the level of statistical significance decreases short of what would be required to establish clear evidence of association. To explore the likelihood that these results represent true associations, we have computed the false-positive report probability (FPRP) under different assumptions. FPRP depends on the prior probability that a true association exists, the observed level of significance (α), and the statistical power to detect the OR of the alternative hypothesis at the given α (25). As the genome has a very large number of common SNPs, the prior probability of association of a random SNP is very low (<1 in a million). However, the prior probability is likely to be more favorable for rs2660753 and rs7931342 because these two SNPs have already been shown to be strongly associated with a hormonally related cancer, prostate cancer (14). Among <30 common SNPs shown to be associated with common cancers, one (rs6983267) has been shown to be associated with multiple cancer types, suggesting that the prior probability for such pleiotrophy many be quite high. The FPRPs for the two associated SNPs under different prior probabilities and the power to detect the association at our observed significance level (α; assuming the true effect size is equal to that observed) are presented in Table 3. If, for example, we assume the prior to be 1 in 100, the FPRP for association of rs2660753 with serous type of ovarian cancer will be 0.33. This suggests that the association has a reasonable chance of being true and is worthy of additional follow-up (see Table 3). The evidence is weaker for rs7931342with FPRPs of 0.21 and 0.75 for priors of 1 in 10 and 1 in 100, respectively.
Table 3.
FPRP values for three disease associated SNPs
| SNP | Cancer type | OR95% CI | Statistical power* | P (α) | Prior probability |
||||
|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | |||||
| rs2660753 | Ovarian | 1.19 (1.04-1.37) | 0.66 | 0.012 | 0.05 | 0.14 | 0.64 | 0.95 | 0.995 |
| rs2660753 | Serous type ovarian | 1.29 (1.09-1.53) | 0.70 | 0.0034 | 0.014 | 0.04 | 0.33 | 0.83 | 0.98 |
| rs7931342 | Breast | 0.95 (0.91-0.99) | 0.56 | 0.28 | 0.08 | 0.21 | 0.75 | 0.97 | 1.0 |
Trend test P value (one degree of freedom). Data highlighted with bold text are FPRP < 0.5.
Hidden population stratification is an alternative explanation for a spurious association. This occurs when allele frequencies differ between population subgroups and case and controls are drawn differentially from those subgroups. It seems unlikely that population stratification is important in this association study because we restricted our analysis to White subjects with European ancestry for the four ovarian cancer studies used. The two breast cancer studies reported here were both from United Kingdom and largely drawn from the same ethnic groups (>98% were of European ancestry). The extent of population stratification in the British population has been found to be generally modest (5).
Assuming the results represent true associations, they may either be due to a direct causative effect of the SNPs tested, or may be because these SNPs are markers in linkage disequilibrium (LD) with a functional variant. Neither SNP rs2660753 nor rs7931342 are located within known genes. rs2667053 is situated on chromosome 3p12. The nearest gene is VGLL3 (~70 kb away), which encodes colon carcinoma-related protein. The nearest alternative candidate genes are CHMP2B and POU1F1 in the same side of SNP rs2660753 and are 166 and 198 kb away, respectively. POU1F1 encodes POU domain class 1 transcription factor 1. POU1F1 is a pituitary-specific transcription factor centrally involved in regulating growth hormone (GH) synthesis. It is expressed in normal and human breast tumors and regulates GH secretion and cell proliferation (26). CHMP2B encodes chromatin-modifying protein 2B. CHMP2B belongs to the chromatin-modifying protein/charged multivesicular body protein family. It has been reported that a mutation in CHMP2B leads to aberrant mRNA splicing in tissue samples from affected individuals with familial frontotemporal dementia (27). SNP rs7931342is situated on chromosome 11q13, an area where rearrangements are frequently observed in human cancers. The nearest gene, MYEOV, is 67 kb away but in different haplotype block with SNP rs7931342. MYEOV is a putative oncogene that is frequently amplified in breast tumors and esophageal carcinomas (28). It often coamplifies with the cell cycle control gene CCND1 (~360 kb away from MYEOV). MYEOV amplification is correlated with estrogen and progesterone receptor-positive breast cancer, the lobular carcinoma subtype, and axillary nodal involvement (29). This is consistent with our finding that the association was stronger for ER-positive cases but not with the observation that the association was restricted to cases of the ductal subtype. Increased MYEOV expression is also associated with cell proliferation and invasion in colon cancer cell lines (30). The effect of this SNP in colorectal cancer was in the same direction and of a similar magnitude to that in breast cancer, but our sample size was much smaller for colorectal cancer and the association was not significant. Although it is plausible that the effects of these functional variants at these loci is to regulate one or more of the local genes, only functional tests will determine if this is the mode of action and which genes are having an active role in cancer development.
We found no evidence of association with colorectal cancer for any of the other 11 SNPs analyzed in our study. Our colorectal cancer study (2,148 cases of 2,265 controls) was able to provide at least 86% power at a type I error of 0.01 to detect a codominant allele with a frequency of 0.3 that confers a relative risk of 1.2. Thus, at the present time, SNP rs6983267 on 8q seems to be unique in being strongly associated with both prostate and colorectal cancer. However, we cannot exclude the possibility that the alleles investigated are associated with smaller risks with colorectal cancer.
In conclusion, we have genotyped 12 prostate cancer-associated SNPs in colorectal, ovarian, and breast cancer case-control studies. We found some evidence for association of SNP rs2660753 on chromosome 3p12with ovarian cancer and of SNP rs7931342on chromosome 11q13 with breast cancer risk. None of the 12 SNPs tested were associated with colorectal cancer. The observed associations with ovarian and breast cancer warrant confirmation in larger studies.
Supplementary Material
Acknowledgments
Grant support: Cancer Research-UK. D.F. Easton is a Principal Research Fellow of Cancer Research UK. P.D.P. Pharaoh is Cancer Research UK Senior Clinical Research Fellow. S.J. Ramus is supported by the Mermaid/Eve Appeal.
We thank all the patients and controls who took part in this study, the following for funding support: Cancer Research-UK, The Roswell Park Alliance, The Danish Cancer Society and the National Cancer Institute (CA71766 and Core Grant CA16056 and RO1 CA61107), and the Foundation Dr. Henri Dubois-Ferriere Dinu Lipatti. Some of this work was undertaken at University College London Hospitals/University of College London who received a proportion of funding from the Department of Health’s Norwegian Institute of Human Rights Biomedical Research Centres funding scheme. We thank Hannah Munday, Barbara Perkins, Mitul Shah, Clare Jordan, Judy West, Anabel Simpson, Sue Irvine, the search team: the local general practices, and nurses and the Eastern Cancer Registry for recruitment of the UK cases, and the EPIC-Norfolk investigators for recruitment of the UK controls; Kristy Driver for helping analyzing breast cancer results, Claus K. Høgdall and Jan Blaakaer for their additional contribution to the MALOVA ovarian cancer collection; and Aleksandra Gentry-Maharaj, Usha Menon, and the UKOPS team of research nurses for their contribution to the UKOPS ovarian cancer collection (funded by the OAK foundation).
Footnotes
Publisher's Disclaimer: The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Easton D. Familial risks of cancer. Eur J Cancer. 1999;35:1043–5. doi: 10.1016/s0959-8049(99)00118-5. [DOI] [PubMed] [Google Scholar]
- 2.Pharoah PD, Ponder BA. The genetics of ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2002;16:449–68. doi: 10.1053/beog.2002.0296. [DOI] [PubMed] [Google Scholar]
- 3.Bonaiti-Pellie C. Genetic risk factors in colorectal cancer. Eur J Cancer Prev. 1999;8(Suppl 1):S27–32. [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Welcome Trust Case control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 7.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 10.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 11.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 12.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 13.Ghoussaini M, Song H, Koessler T, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 15.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 17.Song H, Ramus SJ, Kjaer SK, et al. Tagging single nucleotide polymorphisms in the BRIP1 gene and susceptibility to breast and ovarian cancer. PLoS ONE. 2007;2:e268. doi: 10.1371/journal.pone.0000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–41. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 19.Koessler T, Oestergaard MZ, Song H, et al. Common variants in mismatch repair genes and risk of colorectal cancer. Gut. 2008;57:1097–101. doi: 10.1136/gut.2007.137265. [DOI] [PubMed] [Google Scholar]
- 20.Gayther SA, Song H, Ramus SJ, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67:3027–35. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 21.Ramus SJ, Vierkant RA, Johnatty SE, et al. Consortium analysis of 7 candidate SNPs for ovarian cancer. Int J Cancer. 2008;123:380–8. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 23.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 24.Colhoun HM, McKeigue PM, Davey SG. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–72. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 25.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gil-Puig C, Seoane S, Blanco M, et al. Pit-1 is expressed in normal and tumorous human breast and regulates GH secretion and cell proliferation. Eur J Endocrinol. 2005;153:335–44. doi: 10.1530/eje.1.01962. [DOI] [PubMed] [Google Scholar]
- 27.Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–8. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 28.Brecht M, Steenvoorden AC, Luf S, Bartram CR, Janssen JW. Rearrangement and expression of myeov and hst in NIH/3T3 transfectants: a caveat for the interpretation of DNA transfection analyses. Oncol Rep. 2007;17:1127–31. [PubMed] [Google Scholar]
- 29.Janssen JW, Cuny M, Orsetti B, et al. MYEOV: a candidate gene for DNA amplification events occurring centromeric to CCND1 in breast cancer. Int J Cancer. 2002;102:608–14. doi: 10.1002/ijc.10765. [DOI] [PubMed] [Google Scholar]
- 30.Moss AC, Lawlor G, Murray D, et al. ETV4 and Myeov knockdown impairs colon cancer cell line proliferation and invasion. Biochem Biophys Res Commun. 2006;345:216–21. doi: 10.1016/j.bbrc.2006.04.094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.