Abstract
Heat stress transcription factors (Hsfs) are the central regulators of defense response to heat stress. We identified a total of 25 rice Hsf genes by genome-wide analysis of rice (Oryza sativa L.) genome, including the subspecies of O. japonica and O. indica. Proteins encoded by OsHsfs were divided into three classes according to their structures. Digital Northern analysis showed that OsHsfs were expressed constitutively. The expressions of these OsHsfs in response to heat stress and oxidative stress differed among the members of the gene family. Promoter analysis identified a number of stress-related cis-elements in the promoter regions of these OsHsfs. No significant correlation, however, was found between the heat-shock responses of genes and their cis-elements. Overall, our results provide a foundation for future research of OsHsfs function.
Keywords: Heat shock, Transcription factors, Rice, Protein structure, Expression analysis
INTRODUCTION
Heat stress transcription factors (Hsfs) are the central regulators of defense response. They control the expression of heat shock proteins (HSPs) in plants by specific binding to the highly conserved heat shock element (HSE) characterized by palindromic motifs of nGAAn (Miller and Mittler, 2006). The basic structure and promoter recognition of Hsfs are highly conserved throughout the eukaryotic kingdom (von Koskull-Döring et al., 2007). A typical Hsf protein contains a modular structure with an N-terminal DNA-binding domain (DBD), an adjacent bipartite oligomerization domain composed of heptads repeat of hydrophobic amino acid residues (HR-A/B), a nuclear localization signal (NLS) essential for nuclear uptake of the protein, a nuclear export signal (NES), and in many cases a less conserved C-terminal activation domain (CTAD) rich in aromatic, hydrophobic and acidic amino acids (AHA) that have been reported to be crucial for activation function (Nover et al., 2001). Based on the conservative DBD and the HR-A/B regions, 21 putative Hsfs from the Arabidopsis, 23 from rice, and 18 from tomato have been identified through the genome-wide analysis (Baniwal et al., 2004). Plant Hsf gene family is divided into three classes, HsfA, HsfB, and HsfC, according to their protein structures (Nover et al., 2001). HsfA and HsfC have insertions of 21 and 7 amino acids, between the hydrophobic regions HR-A and HR-B, respectively. HsfB and HsfC are also characterized by lack of AHA motifs in their C-terminal regions (CTRs).
Evidence from several important representatives in tomato and Arabidopsis displayed that plant Hsfs diversify in their biological functions (Kotak et al., 2007b; von Koskull-Döring et al., 2007). HsfA members are capable of transcriptional activation, while HsfB members act as repressors or as co-activators (e.g., HsfB1) of HsfA members (Bharti et al., 2004; Czarnecka-Verner et al., 2004). However, AtHsfA4 activity was reported to be repressed by AtHsf5, which belongs to class A Hsfs (Baniwal et al., 2007). Overexpression of Hsf genes in transgenic plants resulted in an up-regulation of heat stress-associated genes and an enhancement of thermotolerance, whereas the down-regulation of Hsf genes leads to a reduction in the thermotolerance (Charng et al., 2007; Mishra et al., 2002; Schramm et al., 2008). In addition to control of heat stress response, Hsfs have also been reported to be involved in the defense response to pathogen attack, oxidative stress, heavy metals, dehydration and salinity, and in certain processes of development and differentiation (Larkindale et al., 2005; von Koskull-Döring et al., 2007).
Rice is the most important cereal crop, which feeds more than a half of the world’s population (Jeon et al., 2008). Molecular dissection of rice Hsf gene family would help to unravel the stress response mechanism in rice. Compared with the extensive studies done in Arabidopsis Hsf genes, only a few researches have involved monocots, such as rice and maize (Fu et al., 2006; Yamanouchi et al., 2002; Yokotani et al., 2008). Although 23 Hsfs were identified in the Oryza japonica previously, the structure and expression profile of these OsHsfs have not been elucidated. In this study, we identified and classified 25 rice Hsf genes from both O. japonica and O. indica genomes. In addition, the expression of the individual genes was investigated through both digital expression analysis and semi-quantitative reverse-transcript polymerase chain reaction (RT-PCR). Our work will facilitate the function analysis of the OsHsfs genes.
MATERIALS AND METHODS
Search for Hsf genes in rice genome and gene annotation
Consensus amino acid sequences of Arabidopsis heat shock factors, including the DNA-binding domain and HR-A/B region, were used to search the GenBank (National Center for Biotechnology Information (NCBI), Bethesda, MD, USA; http://www.ncbi.nlm.nih.gov), the International Rice Genome Sequencing Project (IRGSP; http://rgp.dna.affrc.go.jp), and Beijing Genomics Institute (BGI; http://btn.genomics.org.cn/rice), using an E-value cutoff of 1.0. The full length of cDNA sequence, 1 kb DNA sequence upstream of the start-codon, and bacterial artificial chromosomes (BACs) or phage artificial chromosomes (PACs) containing OsHsf genes were obtained from the NCBI, IRGSP, or BGI. Expressed sequence tag (EST) sequences of all OsHsf genes collected from dbEST database (http://www.ncbi.nlm.nih.gov/dbEST/) were used for the identification of the tissue specific expression patterns of the OsHsfs (Audic and Claverie, 1997). Finally, we compared all the OsHsf genes to identify redundant sequences. Promoters were analyzed by using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Sequence alignment and phylogenic analysis
Amino acid sequences of DBD and HR-A/B regions were used for multiple alignments by using ClustalX version 1.83 (Hicks et al., 1997). To produce preferable alignments, the parameters were set as followings: for pairwise parameters, gap opening cost=30, gap extension cost=0.3; for multiple parameters, gap opening cost=20, gap extension cost=0.15; the Gonnet series were applied for protein weight matrices and defaulting parameters were used for other settings. Phylogenetic tree of OsHsf gene family was constructed using the N-J method. The ScHsf1 gene from Saccharomyces cerevisiae was selected as outgroup and bootstrap analysis was performed to measure the robustness of all nodes.
Digital expression analysis
Digital expression of the OsHsfs was performed using the rice dbEST database. All ESTs were sorted by the library source, and normalized libraries were delimited for expression analysis. Frequencies of the ESTs in the corresponding library were calculated to represent the gene expression level.
RT-PCR analysis
Sixty rice seedlings (Oryza sativa L. cv. Nipponbare) were grown hydroponically with a 12-h photoperiod (250 µmol photons/(m2·s)) and a day/night temperature of 28/22 °C. The 7-d-old seedlings were transferred to a growth chamber at 37 °C for heat shock treatment or immersed in 20 mmol/L H2O2 solution for H2O2 stress induction. The shoots of the seedlings were sampled at different time points (0, 1, 6, and 24 h after treatments) for isolating RNA. Total RNA was extracted with Trizol reagent (Gibco-BRL, Gaithersburg, MD, USA) according to the manufacturer’s instructions. cDNA was synthesized from 5 μg of the total RNA using moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI, USA). Rice ACTIN gene OsACT1 (accession No. X63830) was served as a housekeeping gene control. Gene specific primers for corresponding OsHsf genes were designed for semi-quantitative RT-PCR analysis (Table A1).
RESULTS
Identification of OsHsf genes in rice genome
In order to identify rice Hsf genes, we used the conserved DBD and the HR-A/B regions to conduct basic local alignment search tool (BLAST) searches against the rice genome in NCBI, IRGSP, and GPI, and identified a total of 26 homologs in the O. japonica genome, which were highly similar to HSF proteins from other plants, and 27 in the O. indica. Of these Hsf genes, two gene loci Loc_Os06g66210 (O. japonica) and OsIBCD008764 (O. indica) were manually discarded because of the truncation in DBD region and the absence of corresponding ESTs in the dbEST, respectively. Another two Hsf genes from gene loci OsIBCD028792 and OsIBCD044667, which were previously annotated as two distinct genes, were proved to be identical in their cDNA sequences (Table 1). The remaining 50 sequences were analyzed for redundancy by performing pairwise comparisons of the genomic sequences in the coding regions ranging from start to the stop codon in each cultivar. Genes that share more than 95% similarity are considered as the same gene. It was found that all Hsf genes in O. indica have corresponding orthologs in O. japonica.
Table 1.
Characteristic of the 25 OsHsf genes
| Genes | Gene loci |
BAC/PAC clone | Protein accession No. | Accession No. of cDNA | Total EST | ORF (aa) | Intron (bp) | |
| O. japonica | O. indica | |||||||
| OsHsfA1 | Os03g63750 | OsIBCD012688 | AC120506 | NP_001051938 | AK100430 | 50 | 506 | 1386 |
| AP008209 | AK106118 | |||||||
| OsHsfA2a | Os03g53340 | OsIBCD011954 | AC092558 | AAP13005 | AK069579 | 15 | 376 | 81 |
| AP008209 | AK109590 | |||||||
| OsHsfA2b | Os07g08140 | OsIBCD022526 | AP003826 | NP_001059028 | AK101824 | 10 | 372 | 1323 |
| OsHsfA2c | Os10g28340 | OsIBCD030592 | AC027658 | NP_001064617 | AK072391 | 10 | 358 | 1669 |
| AP008216 | ||||||||
| OsHsfA2d | Os03g06630 | OsIBCD008955 | AC105729 | NP_001049047 | AK066844 | 6 | 359 | 946 |
| AP008209 | ||||||||
| OsHsfA2e | Os03g58160 | OsIBCD012247 | AC092076 | NP_001051552 | AK068660 | 50 | 357 | 1199 |
| AP008209 | ||||||||
| OsHsfA3 | Os02g32590 | OsIBCD006671 | AP004777 | NP_001047003 | AK101934 | 7 | 498 | 1298 |
| OsHsfA4a | Os01g54550 | OsIBCD003303 | AP003076 | NP_001044247 | AK109856 | 21 | 440 | 582 |
| OsHsfA4d | Os05g45410 | OsIBCD018681 | AC111015 | NP_001056127 | AK100412 | 35 | 459 | 237 |
| AP008211 | ||||||||
| OsHsfA5 | Os02g29340 | OsIBCD006509 | AP004999 | NP_001046889 | AK072210 | 16 | 475 | 1475 |
| AP008208 | AK065643 | |||||||
| OsHsfA6 | Os06g36930 | OsIBCD021096 | AP005456 | NP_001057889 | − | − | 331 | 122 |
| OsHsfA7 | Os01g39020 | OsIBCD002361 | AP003308 | NP_001043378 | AK064271 | 7 | 402 | 1406 |
| OsHsfA9 | Os03g12370 | OsIBCD009329 | AC107226 | NP_001049429 | AK072571 | 24 | 406 | 1231 |
| AP008209 | ||||||||
| OsHsfB1 | Os09g28354 | OsIBCD028792a | AP006057 | NP_001063364 | AK101182 | 36 | 302 | 5880 |
| OsIBCD044667a | AP008215 | AK061433 | ||||||
| OsHsfB2a | Os04g48030 | OsIBCD015287 | AL663003 | NP_001053591 | AY344483 | 4 | 305 | 102 |
| OsHsfB2b | Os08g43334 | OsIBCD027305 | AP004163 | NP_001062423 | AK101700 | 29 | 390 | 101 |
| OsHsfB2c | Os09g35790 | OsIBCD029172 | AP005681 | NP_001063726 | AK106545 | 19 | 454 | 121 |
| AK106525 | ||||||||
| AK105409 | ||||||||
| OsHsfB4a | Os08g36700 | OsIBCD026821 | AP004693 | BAD09860 | − | − | 380 | 93 |
| OsHSFB4b | Os07g44690 | OsIBCD024341 | AP005292 | NP_001060424 | AK063952 | 26 | 310 | 1665 |
| AP008213 | AK099354 | |||||||
| OsHsfB4c | Os09g28200 | OsIBCD028788 | AP005655 | NP_001063356 | AK241190 | 5 | 394 | 88 |
| OsHsfB4d | Os03g25120 | OsIBCD010276b | AC125784 | ABF96133 | − | 1 | 305 | 1450 |
| OsIBCD010275b | AP008209 | |||||||
| OsHsfC1a | Os01g43590 | − | AP002744 | AAQ23067 | AK069479 | 1 | 348 | 109 |
| OsHsfC1b | Os01g53220 | OsIBCD003218 | AP003309 | NP_001044160 | AK066316 | 9 | 250 | 79 |
| OsHsfC2a | Os02g13800 | OsIBCD005620 | AP004070 | NP_001046370 | AK106488 | 9 | 298 | 86 |
| OsHsfC2b | Os06g35960 | OsIBCD021036 | AP003682 | NP_001057843 | − | 1 | 278 | 1128 |
BAC: bacterial artificial chromosome; PAC: phage artificial chromosome; EST: expressed sequence tag: ORF: open reading frame.
Same genes with two cDNAs separated by intron;
Two OsHsfB4d gene loci annotated in O. indica genome
Protein structure and classification of OsHsfs
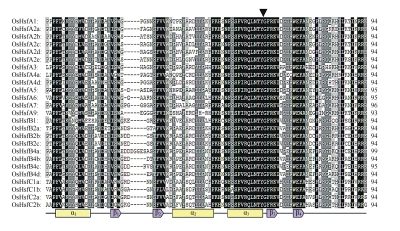
Protein sequence analysis detected a conserved DBD in the N-terminal region (Fig.1) and an adjacent HR-A/B region in each of the OsHsf proteins (Fig.2). The DBD domain contained three α-helix bundles and a small four-stranded antiparallel β-sheet as previously described in LpHsf24 (Fig.1) (Schultheiss et al., 1996). There was a conserved intron located near the 3′-end of the third helix (Fig.1) with variant length (Table 1) in all the genes.
Fig. 1.
Multiple sequence alignment of DNA binding domains
All OsHsfs have only one intron between the invariant tyrosine residue and glycine residue, which is indicated by arrow. The secondary structure elements are showed below the sequence alignment: α1 to α3, α-helix; β1 to β4, β-sheet
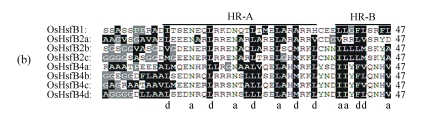
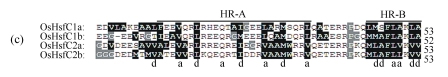
Fig. 2.
Multiple sequence alignment of HR-A/B domains
Based on the distance between HR-A and HR-B domains, OsHsfs are grouped into three classes: (a) OsHsfA, (b) OsHsfB, and (c) OsHsfC. The hydrophobic residues and unique amino acids (glycine and proline) are colored black and gray respectively. The positions a and d of heptapeptide repeats are marked below
Further analyses of the HR-A/B domain revealed that there were, in the majority of cases, three or more repeated heptads in the HR-A domain and two incompletely repeated heptads in the HR-B domain (Fig.2). HR-B in classes A and C had a very characteristic structure of overlapping sets of hydrophobic heptapeptide repeats (Fig.2). According to the linker region between the HR-A and HR-B domains, OsHsfs family (Nover et al., 2001) was divided into three classes, including 13 OsHsfAs, 8 OsHsfBs, and 4 OsHsfCs (Table 2).
Table 2.
Function motifs of rice OsHsfs
| Hsfs | NLS | NES | AHA motifs |
| HsfA1 | (231)RRR-5aa-KKRRLPK | (493)LTEQMGLL | AHA (449)DSFWEQFLVA |
| HsfA2a | (255)RNR-7aa-KKRRR | (366)LVQSIYHL | AHA (318)ESFWMQLLSL |
| HsfA2b | (239)RK-7aa-KKRRRR | (356)LSEKMGYL | AHA (328)DNFWEELLNE |
| HsfA2c | (227)KEKRK-7aa-KKRRR | (345)LAQQLGYL | AHA (315)DDFWAELLVE |
| HsfA2d | (223)KMK-7aa-KKRTR | (346)LAQKLGYL | AHA (315)DDFWEELLNE |
| HsfA2e | (224)RK-7aa-KKRRRR | (340)LLSQKMGYL | AHA (314)EDFWEDLLHE |
| HsfA3 | (247)RQQK-7aa-KRKFLK | (472)LDDGDLQL | AHA (463)KFWELDFQAL |
| HsfA4a | (200)RKKRR | (429)EKLGLL | AHA1 (375)DGFWQQFLTE |
| AHA2 (416)HLWWGKRNVE | |||
| HsfA4d | (210)KKRRVPK | (445)EQMGHL | AHA (397)DVFWERFLTE |
| HsfA5 | (214)NKKRR | (470)VEQLKL | AHA (422)DKFWEQFLTE |
| HsfA6 | (231)RKKRR | (319)LVQQIDCL | AHA1 (271)MIWYELLGEE |
| AHA2 (306)EPWEEMGEEE | |||
| HsfA7 | (282)KNGLRGAAKRQR | − | AHA1 (349)DDVWEELDAL |
| AHA2 (384)CGWVDDCPYL | |||
| HsfA9 | (246)QRR-9aa-KKRR | (371)QMGPPL | AHA (380)DYDFPQLEQD |
| HsfB1 | (263)RKRAR | − | − |
| HsfB2a | (265)REGKVRR | (275)LSDLDVLAL | − |
| HsfB2b | (318)RKRMR | − | − |
| HsfB2c | (327)LKRTR | − | − |
| HsfB4a | (346)RKRS | − | − |
| HsfB4b | (291)RKKRAHR | − | − |
| HsfB4c | (269)RKKP | (363)LVLECDDLSL | − |
| HsfB4d | (286)KKRRVQL | − | − |
| HsfC1a | (229)RKRRR | − | − |
| HsfC1b | (209)KRRR | − | − |
| HsfC2a | (213)KRPRLLL | − | − |
| HsfC2b | (219)KRARLLL | − | − |
Numbers in brackets indicate positions of the putative function motifs; NLS: nuclear localization signal; NES: nuclear export signal; AHA: aromatic, large hydrophobic and acidic amino acids. Aromatic and large hydrophobic residues of AHA are set in boldface type
Within the OsHsfs, the nuclear localization signals (NLS) composed of a cluster of arginine and lysine residues were detected adjacent to the HR-A/B domain (Table 2). There was a leucine-rich nuclear export signal (NES) at the C-terminus of all OsHsfAs except OsHsfA7, and of a small portion of OsHsfBs and OsHsfCs (Table 2). The C-terminal activation domain (CTAD) was only found in OsHsfA, suggesting that only class A Hsfs can activate autonomously.
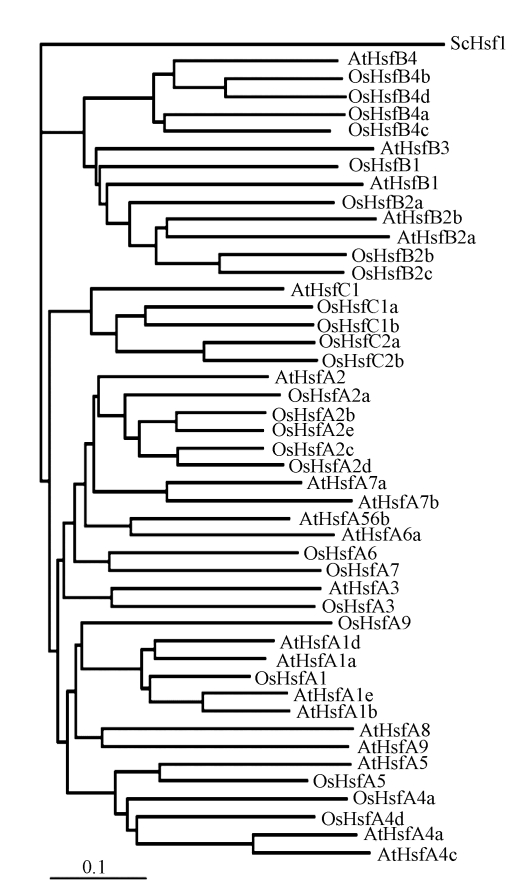
Phylogenetic analysis of OsHsfs
To determine the phylogenetic relationship among the OsHsfs, neighbor-joining phylogenetic trees were constructed using the amino acid sequences of DBD, the HR-A/B region, and the linker between them (Fig.3). Arabidopsis Hsf genes were included in the phylogenetic tree as reference to classify rice Hsfs. As expected, the classes A, B and C Hsfs formed three individual clusters. Furthermore, the class A Hsfs were divided into two sub-clusters (Fig.3). In a previous study, the N-terminal part and C-terminal part of DBD and HR-A/B regions were used separately to draw phylogenetic trees (Nover et al., 2001). Although most proteins fixed their positions in the different phylogenetic trees, a few Hsfs changed their positions (Nover et al., 2001). Similar phenomenon was also observed on the OsHsfs (data not shown). A more convinced relationship of the Hsfs was revealed by combining the DBD, HR-A/B, and the flexible linker between DBD and HR-A/B (Fig.3).
Fig. 3.
Neighbor-joining phylogenetic trees of OsHsf and AtHsf genes constructed using ClustalX program
The tree was generated on the basis of the amino acid sequences of the N-terminal domains of Hsfs including the DBD region, the HR-A/B region and parts of the linker between both. ScHsf1 was set as the outgroup. Bar=substitutions/site
Cis-acting elements in OsHsf promoters
One kilobase upstream regions of the OsHsf genes were analyzed for cis-acting elements using PlantCARE software (Higo et al., 1999). A total of 9 different cis-acting elements commonly existed in the promoters of OsHsfs (Table 3). Seven of these cis-acting elements were related to stress responses. Two cis-acting elements with the highest frequency were G-box/Sp1 and abscisic acid (ABA) response element (ABRE). These two elements are involved in the light responsiveness and the ABA response, respectively (Table 3). CCGTCC/CAT-box, LTR, and ARE/GC-motif are cis-acting elements related to meristem expression, low temperature response, and anaerobic induction. Compared with classes B and C OsHsf genes, class A genes contained more these two cis-elements, suggesting that class A genes may play different roles from classes B and C genes (Table 3). In addition to these two elements, other cis-elements occurred at a similar frequency among the promoters of these three classes.
Table 3.
The high frequency cis-elements of OsHsf genes
| Cis-elements |
HsfA |
T1 |
HsfB |
HsfC |
T2 | Descriptions | ||||||||||||||||||||||
| 1a | 2a | 2b | 2c | 2d | 2e | 3 | 4a | 4d | 5 | 6 | 7 | 9 | 1 | 2a | 2b | 2c | 4a | 4 | 4c | 4d | 1a | 1b | 2a | 2b | ||||
| Skn-1 and GCN4-motif | 2 | 2 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 15 | 1 | 1 | 3 | 0 | 0 | 3 | 0 | 2 | 1 | 4 | 2 | 0 | 17 | Required for endosperm expression |
| CCGTCC and CAT-box | 2 | 1 | 2 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 3 | 0 | 14 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 1 | 6 | Related to meristem expression |
| G-box and Sp1 | 5 | 8 | 17 | 12 | 3 | 6 | 14 | 7 | 3 | 3 | 17 | 6 | 5 | 106 | 10 | 7 | 2 | 10 | 7 | 5 | 7 | 7 | 13 | 8 | 7 | 19 | 102 | Involved in light response |
| HSE | 0 | 3 | 1 | 2 | 2 | 2 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 14 | 3 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 1 | 1 | 13 | Involved in heat stress response |
| LTR | 0 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | Involved in low-temperature response |
| ARE and GC-motif | 2 | 5 | 4 | 3 | 2 | 1 | 4 | 1 | 1 | 2 | 5 | 3 | 0 | 33 | 4 | 1 | 1 | 2 | 1 | 0 | 3 | 0 | 1 | 2 | 0 | 3 | 18 | Involved in the anaerobic induction |
| MBS and CCAAT-box | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 2 | 1 | 1 | 3 | 2 | 1 | 16 | 1 | 1 | 1 | 2 | 3 | 2 | 1 | 3 | 1 | 1 | 0 | 1 | 17 | MYB binding site, involved in drought-inducibility |
| ABRE | 0 | 1 | 3 | 4 | 2 | 3 | 8 | 0 | 1 | 0 | 14 | 0 | 1 | 37 | 0 | 2 | 0 | 5 | 11 | 3 | 0 | 5 | 5 | 2 | 1 | 3 | 37 | Involved in the abscisic acid response |
| CGTCA-motif | 0 | 2 | 0 | 0 | 1 | 2 | 4 | 4 | 0 | 0 | 1 | 0 | 4 | 18 | 1 | 3 | 0 | 0 | 0 | 2 | 1 | 0 | 2 | 2 | 2 | 0 | 13 | Involved in the MeJA-response |
T 1: total number of each cis-element in HsfA; T 2: total number of each cis-element in HsfB and HsfC
Expression patterns of OsHsfs
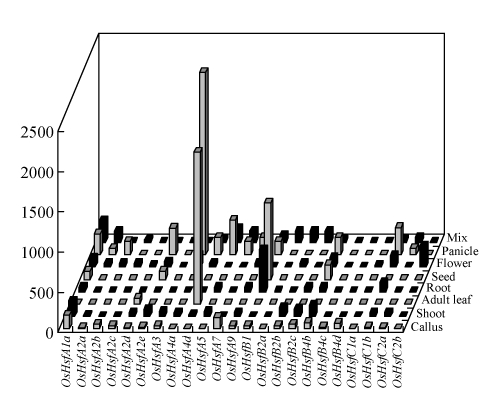
The number of ESTs for a specific gene in a cDNA library is considered to be proportional to the transcript abundance of the mRNA. We used the EST sequences downloaded from the dbEST to analyze the expression pattern of OsHsfs in various plant tissues and different developmental stages. A total of 391 OsHsf ESTs corresponding to 23 of the 25 genes were identified except the OsHsfA6 and OsHsfB4a genes (Table 1). The majority of OsHsf genes had a low to moderate level of transcripts in most tissues, while some displayed a tissue-specific manner (Fig.4). OsHsfA4a and OsHsfA4d were dominantly expressed in panicle and adult leaves, respectively. Overall, OsHsfs were expressed at a higher level in panicle and flower than in other tissues.
Fig. 4.
Expression level of OsHsfs in different tissues
ESTs of each Hsf are collected from dbEST and the transcript rations of all Hsfs in each library are calculated. Transcript levels of all OsHsfs in per million transcripts are counted according to the ration. Normalized libraries and libraries with less than 5000 ESTs are excluded. Detail data are shown in Table A2
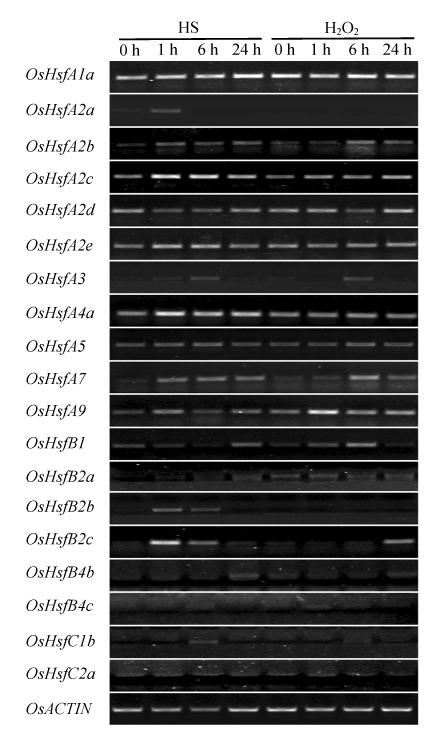
Under heat shock (HS) and H2O2 treatment, the expression of class A Hsfs was higher than those of class B and class C Hsfs (Fig.5). These results are consistent with those derived from the analyses of EST data. The lack of the detectable transcripts of several OsHsfs under our condition might have resulted from the tissue specific expression pattern (OsHsfA4d) or low expression levels of the genes (e.g., OsHsfB4a, OsHsfB4d, and so on) as suggested by the digital expression pattern. The transcription of OsHsfA1a, OsHsfA2b, OsHsfA3, OsHsfA7, OsHsfA9, OsHsfB2c, and OsHsfC1b was up-regulated by both HS and H2O2 treatments in a similar manner, while the up-regulation of OsHsfA2a, OsHsfA2c, OsHsfA2e, OsHsfA4a, OsHsfB2b, and OsSHFC1b was detected only under HS treatment, implying that there was a H2O2 independent HS responsive pathway in rice.
Fig. 5.
Expression levels of OsHsfs under heat shock (HS) and oxidative stress (H2O2) analyzed by RT-PCR
mRNA was extracted from shoots of 7-d-old O. japonica seedlings treated with heat (37 °C) and 20 mmol/L H2O2 for different time points. The transcript level of OsACTIN1 was used as the housekeep gene control. Class A Hsfs and ACTIN genes were amplified for 28 cycles, while classes B and C Hsfs were amplified for 30 cycles
DISCUSSION
Unlike yeast and Drosophila that only contain one Hsf gene, plant has a more complex family of Hsf genes (Baniwal et al., 2004; Miller and Mittler, 2006). In this study, we identified 25 OsHsfs based on the rice genome. Majority of the predicted OsHsfs were supported by the FL-cDNA and the EST sequences (Table 1). These 25 OsHsfs were divided into 3 classes (A, B and C) based on the protein structure and phylogenetic relationship (Figs.2~3). The sequence analysis identified the conserved DBD, the HR-A/B domain, and the NLS that were the common motifs of OsHSFs. The NES and the AHA motifs were also the common motifs of class A genes, but they were only detected in a small portion of classes B and C genes (Table 2).
Recently, great progress has been made in elucidating the functions of Hsfs (von Koskull-Döring et al., 2007). AtHsfA9 is involved in seed development and controlled by seed-specific transcription factor abscisic acid-insensitive 3 (ABI3) (Kotak et al., 2007a). The expression of AtHsfA3 is induced by drought and heat, and is dependent on the dehydration response element biding protein 2A (DREB2A) (Sakuma et al., 2006; Schramm et al., 2008). Promoter analysis showed that an array of OsHsf genes contains ABREs (Table 3). This indicates that an ABA pathway may be involved in Hsf induction and responsible for control of downstream processes, such as seed development or drought resistance (Kotak et al., 2007b). The transcripts of AtHsfA2 increased under several stress conditions, especially in response to high light plus heat stress (Nishizawa et al., 2006). During repeated cycles of heat stress and recovery, HsfA2 becomes a dominant Hsf in tomato and Arabidopsis (Mishra et al., 2002; Schramm et al., 2006). In contrast to tomato and Arabidopsis containing only one HsfA2, rice has five HsfA2 genes (von Koskull-Döring et al., 2007). Our results show that OsHsfA2a, OsHsfA2b, OsHsfA2c, and OsHsfA2e were induced by heat stress, while OsHsfA2b and OsHsfA2c were induced by H2O2 also (Fig.5). From the result of expression pattern, the OsHSfA2 subfamily was closely related with rice stress response. Overexpression of OsHsfA2e in Arabidopsis led to enhanced thermo and salt tolerance in transgenic plants (Yokotani et al., 2008). Further studies to elucidate the functions of the other members in the subfamily may also have potential for the development of transgenic plants with improved stress tolerance.
H2O2 is reported to be an essential component in the heat stress signaling pathway (Volkov et al., 2006), and the Hsf genes were considered as H2O2 sensors in plant (Miller and Mittler, 2006). Human and Drosophila Hsf1 was shown to sense hydrogen peroxide directly and assemble into a homotrimer in a reversible and redox-regulated manner (Storozhenko et al., 1998; Zhong et al., 1998). In Arabidopsis, the heat stress-induced H2O2 is required for effective expression of heat shock genes (Volkov et al., 2006), and the AtHsf4a may be an important sensors of H2O2 (Davletova et al., 2005). The intimate relationship between the HS and oxidative stress response suggests that some Hsfs might be responsive to both stresses. In this study, eight OsHsfs (OsHsfA1a, OsHsfA2b, OsHsfC1b, OsHsfA3, OsHsfA7, OsHsfA9, OsHsfB2c, and OsHsfC1b) were found to be induced by both heat shock and H2O2 (Fig.5). Heat stress led to the accumulation of H2O2 in tobacco and Arabidopsis culture cells (Volkov et al., 2006) and mustard seedlings (Dat et al., 1998). Hence, it is likely that Hsfs sense the H2O2 level in cells and transfer the stress signaling. Mutation of OsHsfA4d led to a lesion mimic phenotype in mature leaves (Yamanouchi et al., 2002). The phenotype may be caused by H2O2 accumulation that transferred a cell death signaling (Takahashi et al., 1999). It is likely that OsHsfA4d works in mature leaves to sense H2O2 levels as its homologue AtHsfA4a in Arabidopsis (Davletova et al., 2005). It is also worth noting that OsHsfA4a, the other member of OsHsfA4, has extremely high transcript abundance in panicle (Fig.4). Further research will help to elucidate the special functions of these genes.
APPENDIX: SUPPORTING ONLINE MATERIAL FOR IDENTIFICATION AND EXPRESSION ANALYSIS OF OsHsfsIN RICE
Table A1.
Gene specific primers used in this research
| Hsfs | Forward (5′ to 3′) | Reverse (5′ to 3′) |
| OsHsfA1a | TATGCCAAATGGTCAAGGTC | CAGGCAAAGAGAATACACCC |
| OsHsfA2a | GGCTTCCTCCAGATGCTCGTC | CGTCGTCATCCTCCTCGTCGTT |
| OsHsfA2b | AGCAGAGGCAACAGCAGATG | CAGACCTCCAGCATCCATAG |
| OsHsfA2c | GCAGAAACAGGTCCAGATG | TCTACTTTACCCTTCCCCAG |
| OsHsfA2d | GAGGTTGGTCAGTTCGGATT | GGTTGAGAAATGGCACTATGT |
| OsHsfA2e | ATGGCATTCTTGTCACGAGT | GGTTCCTGGTATCCTCATCG |
| OsHsfA3 | CAACACACTGAGAAGGGAGA | TGCTCTCTCCAGTGTGTTGT |
| OsHsfA4a | TGGCAGCAGTTTCTTACCGAG | AGGCACCAATGTCAGCGTTC |
| OsHsfA4d | CCCATCTCCATTTATCCACT | CATTTGCTCGGTGATCTGAT |
| OsHsfA5 | GTAAGCCTATCCACAGCCAC | ATCTTGGTCTGTCGCTGCTC |
| OsHsfA7 | TCCGAAAGGTCACTCCAGAT | TGGTCTGCTGTTGCCTTCTC |
| OsHsfA9 | GGCACTACCAGCAAACATC | CCACTGGATTTACTTGACCT |
| OsHsfB1 | GGACAACCAAACGCTGACGA | CGACGATGGAACGCTGACC |
| OsHsfB2a | GGCATACCGACGGCGATACC | CCTTCCTCCTCCTCCTCCTCCT |
| OsHsfB2b | GGCTGCTCTGCGAGATACACC | CTGCTGGGTGGAGGCGTACTTG |
| OsHsfB2c | GCAACAGAGCAACTTGTGA | CACCAACACACACACAAAGC |
| OsHsfB4b | GACGACGACGACGACGAT | TCATCACTCTTCACCAGCAT |
| OsHsfC1b | TACTTCAAGCACCGCAACT | CCGCTGCACCGCCTCGATG |
| OsHsfC2a | TGTTTGGACGAAGTGGCTG | ACAAGGCACACTCAACATTC |
| OsACTIN | TCAGCAACTGGGATGATATGGAG | GCCGTTGTGGTGAATGAGTAAC |
Table A2.
Expression level of OsHsfs in different tissues
| Gene | Transcripts per million |
|||||||
| Callus | Shoot | Adult leaf | Root | Seed | Flower | Panicle | Mix | |
| OsHsfA1a | 168 | 190 | 0 | 58 | 102 | 108 | 260 | 269 |
| OsHsfA2a | 10 | 10 | 0 | 0 | 0 | 43 | 80 | 136 |
| OsHsfA2b | 56 | 0 | 0 | 0 | 0 | 43 | 166 | 0 |
| OsHsfA2c | 37 | 0 | 0 | 0 | 0 | 43 | 0 | 30 |
| OsHsfA2d | 10 | 41 | 76 | 0 | 0 | 22 | 0 | 0 |
| OsHsfA2e | 19 | 82 | 0 | 0 | 102 | 65 | 332 | 0.018 |
| OsHsfA3 | 37 | 61 | 0 | 0 | 0 | 0 | 0 | 0 |
| OsHsfA4a | 0 | 45 | 0 | 0 | 0 | 141 | 2268 | 0 |
| OsHsfA4d | 0 | 21 | 1892 | 0 | 0 | 43 | 215 | 30 |
| OsHsfA5 | 0 | 61 | 0 | 58 | 0 | 0 | 432 | 170 |
| OsHsfA7 | 143 | 0 | 0 | 0 | 0 | 0 | 166 | 142 |
| OsHsfA9 | 37 | 46 | 0 | 58 | 0 | 43 | 216 | 280 |
| OsHsfB1 | 37 | 0 | 0 | 523 | 953 | 0 | 166 | 85 |
| OsHsfB2a | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 136 |
| OsHsfB2b | 37 | 108 | 0 | 0 | 0 | 43 | 0 | 136 |
| OsHsfB2c | 56 | 93 | 0 | 58 | 0 | 0 | 0 | 136 |
| OsHsfB4b | 75 | 139 | 0 | 58 | 183 | 117 | 216 | 30 |
| OsHsfB4c | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OsHsfB4d | 67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OsHsfC1a | 0 | 0 | 0 | 0 | 0 | 22 | 0 | 0 |
| OsHsfC1b | 19 | 41 | 0 | 116 | 0 | 141 | 338 | 42 |
| OsHsfC2a | 10 | 46 | 0 | 0 | 0 | 0 | 80 | 61 |
| OsHsfC2b | 0 | 0 | 0 | 0 | 0 | 264 | 0 | 0 |
Footnotes
Project (No. 30471118) supported by the National Natural Science Foundation of China
References
- 1.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 2.Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci. 2004;29(4):471–487. doi: 10.1007/BF02712120. [DOI] [PubMed] [Google Scholar]
- 3.Baniwal SK, Chan KY, Scharf KD, Nover L. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J Biol Chem. 2007;282(6):3605–3613. doi: 10.1074/jbc.M609545200. [DOI] [PubMed] [Google Scholar]
- 4.Bharti K, von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Korbitzer A, Treuter E, Nover L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell. 2004;16(6):1521–1535. doi: 10.1105/tpc.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis . Plant Physiol. 2007;143(1):251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czarnecka-Verner E, Pan S, Salem T, Gurley WB. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol. 2004;56(1):57–75. doi: 10.1007/s11103-004-2307-3. [DOI] [PubMed] [Google Scholar]
- 7.Dat JF, Lopez-Delgado H, Foyer CH, Scott IM. Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol. 1998;116(4):1351–1357. doi: 10.1104/pp.116.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis . Plant Cell. 2005;17(1):268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu S, Rogowsky P, Nover L, Scanlon MJ. The maize heat shock factor-binding protein paralogs EMP2 and HSBP2 interact non-redundantly with specific heat shock factors. Planta. 2006;224(1):42–52. doi: 10.1007/s00425-005-0191-y. [DOI] [PubMed] [Google Scholar]
- 10.Hicks MR, Holberton DV, Kowalczyk C, Woolfson DN. Coiled-coil assembly by peptides with non-heptad sequence motifs. Folding and Design. 1997;2(3):149–158. doi: 10.1016/S1359-0278(97)00021-7. [DOI] [PubMed] [Google Scholar]
- 11.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeon J, Choi J, Park J, Lee YH. Functional genomics in the rice blast fungus to unravel the fungal pathogenicity. J Zhejiang Univ Sci B. 2008;9(10):747–752. doi: 10.1631/jzus.B0860014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Curr Opin Plant Biol. 2007;10(3):310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Kotak S, Vierling E, Baumlein H, von Koskull-Döring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis . Plant Cell. 2007;19(1):182–195. doi: 10.1105/tpc.106.048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkindale J, Hall JD, Knight MR, Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005;138(2):882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot (Lond) 2006;98(2):279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002;16(12):1555–1567. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006;48(4):535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- 19.Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6(3):177–189. doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA. 2006;103(49):18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis . Plant Mol Biol. 2006;60(5):759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- 22.Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis . Plant J. 2008;53(2):264–274. doi: 10.1111/j.1365-313X.2007.03334.x. [DOI] [PubMed] [Google Scholar]
- 23.Schultheiss J, Kunert O, Gase U, Scharf KD, Nover L, Ruterjans H. Solution structure of the DNA-binding domain of the tomato heat-stress transcription factor HSF24. Eur J Biochem. 1996;236(3):911–921. doi: 10.1111/j.1432-1033.1996.00911.x. [DOI] [PubMed] [Google Scholar]
- 24.Storozhenko S, de Pauw P, van Montagu M, Inze D, Kushnir S. The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 1998;118(3):1005–1014. doi: 10.1104/pp.118.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi A, Kawasaki T, Henmi K, Shi IK, Kodama O, Satoh H, Shimamoto K. Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 1999;17(5):535–545. doi: 10.1046/j.1365-313X.1999.00405.x. [DOI] [PubMed] [Google Scholar]
- 26.Volkov RA, Panchuk II, Mullineaux PM, Schoffl F. Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis . Plant Mol Biol. 2006;61(4-5):733–746. doi: 10.1007/s11103-006-0045-4. [DOI] [PubMed] [Google Scholar]
- 27.von Koskull-Döring P, Scharf KD, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12(10):452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci USA. 2002;99(11):7530–7535. doi: 10.1073/pnas.112209199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis . Planta. 2008;227(5):957–967. doi: 10.1007/s00425-007-0670-4. [DOI] [PubMed] [Google Scholar]
- 30.Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2(1):101–108. doi: 10.1016/S1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]