Abstract
Various gene amplifications have been observed in gliomas. Prognostic-genomic correlations testing simultaneously all these amplified genes have never been conducted in anaplastic oligodendrogliomas. A set of 38 genes that have been reported to be amplified in gliomas and investigated as the main targets of amplicons were studied in a series of 52 anaplastic oligodendrogliomas using bacterial artificial chromosome–array based comparative genomic hybridization and quantitative polymerase chain reaction. Among the 38 target genes, 15 were found to be amplified in at least one tumor. Overall, 27% of anaplastic oligodendrogliomas exhibited at least one gene amplification. The most frequently amplified genes were epidermal growth factor receptor (EGFR) and cyclin-dependent kinase 4/sarcoma amplified sequence (CDK4/SAS) in 17% and 8% of anaplastic oligodendrogliomas, respectively. Gene amplification and codeletion of chromosome arms 1p/19q were perfectly exclusive (p = 0.005). In uni- and multivariate analyses, gene amplification was a negative prognostic factor for progression-free survival and overall survival in anaplastic oligodendrogliomas, providing complementary information to the classic prognostic factors identified in anaplastic oligodendrogliomas (extent of surgery, KPS, and chromosome arms 1p/19q status).
Keywords: amplification, anaplastic, gene, oligodendroglioma, prognosis
Grade III oligodendroglioma (OIII) is a well- characterized tumor in the WHO classification.1 Molecular studies have shown that this pathological group is composed of distinct biological entities that show different prognoses and responses to treatment.2–4 The most important molecular predictor of a favorable prognosis and response to chemotherapy is the codeletion of chromosome arms 1p/19q, which corresponds to a translocation.5–6
Although other genetic alterations have been reported in OIIIs, including various gene amplifications,7–8 prognostic-genomic correlations testing simultaneously all genes reported amplified in gliomas have never been conducted in anaplastic oligodendrogliomas. We searched the Web site of the U.S. National Library of Medicine (http://www.ncbi.nlm.nih.gov/sites/entrez) using the keywords “astrocytoma,” “oligodendroglioma,” “glioma,” “glioblastoma,” “amplification,” and/or “gene” and the limits “humans” and “all adults: 19+ years.” From several studies we selected validated or candidate target genes of genomic amplification that were reported to be amplified and overexpressed in gliomas.9–40 To better understand this issue, we used a 0.7 megabase resolution bacterial artificial chromosome (BAC) array–based comparative genomic hybridization (aCGH) approach, allowing the identification of gene amplification along the whole genome in a large series of OIIIs.
Materials and Methods
Selection of Patients
All patients with a primary glioma fulfilling the following inclusion criteria were selected from our brain tumor database: (1) A histological diagnosis of OIII according to the 2000 WHO classification of tumors of the CNS (i.e., a cellular diffusely infiltrating high-grade glioma featuring uniform rounded hyperchromatic nuclei, peri-nuclear halos on paraffin sections [“honeycomb appearance”], few cellular processes, and displaying micro-calcifications, mucoid/cystic degeneration, and a dense network of branching capillaries).41 Microvascular proliferation and/or areas of necrosis were acceptable, if an otherwise typical appearance of OIII was present.1 All specimens were agreed on by two experienced neuro-pathologists with specific attention given to exclusion of anaplastic oligoastrocytoma (i.e., high-grade glioma comprising both significant astrocytic and oligodendroglial components without area of necrosis nor prominent microvascular proliferation), glioblastoma with oligodendroglial component (i.e., cellular glioma composed of poorly differentiated, often pleomorphic, astrocytic tumor cells with marked nuclear atypia, brisk mitotic activity, prominent microvascular proliferation, and/or necrosis and displaying foci resembling oligodendroglioma), small cell glioblastoma (i.e., highly monomorphic glial tumor characterized by small, round to slightly elongated, densely packed cells displaying high mitotic activity, microvascular proliferation, and necrosis, including pseudopalisading).42 (2) Available high-quality tumor DNA that would allow an aCGH experiment. All patients gave written informed consent, as requested by the law, allowing molecular, genetic, and translational research studies on cancer tissue samples. The analysis was performed on anonymized data.
BAC-Array Experiment and Localization of Target Genes on BAC-Array CGH
DNA was extracted from frozen tumors using a QIAamp DNA Mini Kit procedure (Qiagen, Courtaboeuf, France). After digestion with DpnII (Ozyme, Saint Quentin en Yvelines, France) and column purification (Qiaquick PCR Purification Kit; Qiagen), tumor DNA was labeled by the random priming method (Bioprime DNA labeling system; Invitrogen, Cergy-Pontoise, France) with cyanine-5 (Perkin-Elmer, Wellesley, MA, USA). Using the same procedure, we labeled control DNA with cyanine-3. After ethanol-coprecipitation with 140 μg of Human Cot-1 DNA (Invitrogen), resuspension in hybridization buffer (50% formamide), denaturation at 95°C for 10 min and prehybridization at 37°C for 90 min, probes were cohybridized on the aCGH. The aCGH (Integragen, Paris, France) analyzed 4,500 sequence-validated BACs. The 4,500 BACs were spotted in quadruplicate on the array. Among those, 500 were selected because they contained genes involved in oncogenesis, particularly well-known oncogenes, and tumor-suppressor genes. The other BACs were selected randomly across the genome to provide a mean resolution of 0.7 megabase. After washing, arrays were scanned using a 4000B scanner (Axon, Union City, CA, USA). Image analysis was performed with the GenePix 6.0 software (Axon) and ratios of Cy5/Cy3 signals were measured. Data were normalized with microarray normalization (MANOR); BAC statuses (gained, lost, amplified, or normal) were determined using gain and loss analysis of DNA (GLAD); and final results were visualized with visualization and analysis of array-CGH, transcriptome, and other molecular profiles (VAMP) software. Those steps were performed within the CAPweb platform.43 The ratio of Cy5/Cy3 of the BAC including or contiguous to the target gene investigated was attributed to that gene. A ratio above at least 3.0 was necessary to conclude that a gene amplification occurred. The status of the chromosome arms 1p/19q was evaluated with the aCGH results.
qPCR Experiment and Validation of BAC-Array CGH Results
Gene amplification of tumor DNA was detected by using SYBR Green real-time quantitative polymerase chain reaction (qPCR) analysis (Absolute SYBR Green Rox Mix; Abgene, Paris, France). The gene primers (forward and reverse) amplified 70 to 217 base pair (bp) genomic fragments. The reference primers amplified two genomic fragments from 1q32 and 2q12.1, according to the status of this region based on the aCGH results (Table 1). The sequence of the reference amplicon checked using PCR with computer software was unique (http://genome.ucsc.edu/cgi-bin/hgPcr?command=start). Real-time qPCR reactions were performed according to the manufacturer’s instructions. Real-time qPCR cycles were as follows: initial denaturation at 95°C for 15 min; 95°C for 30 sec and annealing at 60°C for 1 min through 40 cycles.
Table 1.
Real-time quantitative polymerase chain reaction primers
| Genomic Region Amplified | Primer Forward | Primer Reverse | Amplicon Size (bp) |
|---|---|---|---|
| 1q32.2 | CGAGAGGAGGAGAACGCTGAGG | CGGAGGCGGGAATGACCTG | 217 |
| 2q12.1 | TTGGGCAGAAAGGGAGGAGGA | GCTCGCAGGGCAGGTTGAAG | 100 |
| CCND1 | AGGTCTGCGAGGAACAGAAG | AGGAAGCGGTCCAGGTAGTT | 70 |
| CCND3 | CTTCCCTGCCTGAGACTTTG | CACCTCCCAGTCCTGAAAAA | 101 |
| CDK4 | GCCACTAAGCAGTAACCATTCAACT | CGGCTTCAGAGTTTCCACAGAA | 102 |
| EGFR | GTGCAGATCGCAAAGGTAATCAG | GCAGACCGCATGTGAGGAT | 79 |
| GAS16 | AGGTGAGGTCATGTGGGAGA | GCTCGTGTGTGACCACTCTG | 149 |
| GAS41 | AACCATTGCACAGATTTGACA | AGCATTGCATTGGTGTCTGA | 92 |
| GAS64 | CGCTTTAGGGACACTTTGGA | TCTGTTGCCTTGCTCTTCCT | 113 |
| GAS89 | TGGAAATGGGGGAATGTAAA | CAGACTCCAGGTCCTTCTCG | 144 |
| MDM2 | TTGGTTTCTAGACCATCTACCTCATCT | AAAAGCTGTGTGAATGCGTCAAAT | 92 |
| MYCN | ACCAAGGCTGTCACCACATT | GGGAAGGCATCGTTTGAG | 105 |
| PDGFR-α | TGTCCTGGTTGTCATTTGGA | AGCTCCTTCTCTGTGCCAAG | 115 |
| PIKE | GGAAGTCATCGCTCATCCAC | TCTCCACACCTGCCTAATCA | 145 |
| MDM4 | GGGGGTATAGGGGAGGTGT | GACCACTTATGCCCAGGAAA | 120 |
| SAS | GGGAAATTGAGGCACTTGAA | TTTCCAGTTCATCCCGAGTC | 132 |
| TNFSF13B | CAAAGCCTCCCATTTTGTGT | GTCCCATGGCGTAGGTCTTA | 89 |
The 2−ΔΔCT method was used to determine the relative copy number. The fluorescent signals were measured after each primer-annealing step at 60°C. The calculation of the relative amounts of the studied gene DNA compared to the reference gene DNA was done by the Relative Quantification Software (Mx Pro 3000P, Strat-agene, Amsterdam, The Netherlands). Final results were expressed as a ratio of the studied gene and reference gene copies in the sample, normalized with the ratio of the reference gene copies in the calibration DNA. A ratio above at least 3.0 was regarded as being positive for gene amplification. Discordant results between aCGH and qPCR were reexamined twice.
Statistical Analysis
Association of gene amplification with pathological features of tumors was evaluated using the chi-square test. Nondiscrete age and KPS were compared using the Mann-Whitney test. Overall survival (from diagnosis to death) curves were obtained using the Kaplan-Meier method and compared with a log-rank test. A p-value of <0.05 was considered as significant. Significant prognostic factors in univariate analyses were included as candidate variables in a multivariate Cox proportional-hazards regression model analysis.
Results
Fifty-two patients with a primary OIII fulfilled the inclusion criteria: 26 men and 26 women (sex ratio = 1:1). The median age of the patients at diagnosis was 50.3 years (range: 22.9–83.5 years). Patients received standard treatment consisting of maximal possible surgery and radiotherapy. Chemotherapy (procarbazine, lomustine, and vincristine [PCV] or temozolomide) was administrated in 23 patients.
Gene Amplification and Correlation with Chromosome Arms 1p/19q Codeletion
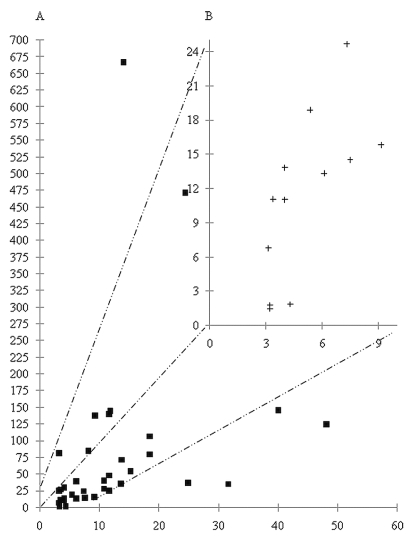
Among the 37 gene amplifications detected by aCGH in the whole series (n = 52), qPCR could confirm 34 of them, indicating a very good concordance (Fig. 1). Only genes found to be amplified using the two techniques were analyzed. Overall, 15 of the 38 tested genes were amplified in at least one OIII (Fig. 2). In contrast, 23 genes, c-MYC, MYCL-1, ELF3, PIK3C2B, GAC-1, TAX-1, TGFα, STIM-2, KIT, VEGFR2, c-fos, IL-6, CDK6, MDM4, MET, MYC, GLI, GADD153, COL4A2, AKT-1, HER2/neu, STK-15, and DCR-3, were not amplified in this series.
Fig. 1.
(A) Figure representing correlation of results obtained using quantitative polymerase chain reaction (qPCR) and bacterial artifi-cial chromosome (BAC) array–based comparative genomic hybridization (aCGH) ratios. For each gene amplification (a black square in 1A and a black cross in 1B), x-axis and y-axis indicate ratios observed using qPCR and aCGH, respectively. (B) Zoom on 1A as indicated by the broken lines.
Fig. 2.
Genes reported to be amplified in the present series of anaplastic oligodendrogliomas according to genomic position. Each dot represents a tumor.
As expected, EGFR was the most frequently amplified gene, observed in 17% of OIII. CDK4, SAS, and GAS89 were amplified in 8% of OIIIs. Moreover, CDK4 and SAS were always coamplified. The other amplified target genes are shown in Fig. 2. Five OIIIs exhibited at least two gene amplifications.
Chromosome arms 1p/19q codeletion was found in 15 of 52 patients (29%). There was a complete exclusion between gene amplification and chromosome arms 1p/19q codeletion.
Correlations between Gene Amplification and Clinical Course
In a univariate analysis, progression-free survival (PFS) was significantly shorter in patients whose tumor had a gene amplification (Table 2). Indeed, the median PFS was 3.1, 1.2, and 0.6 years in 1p/19q codeleted OIII patients, nonamplified and non-1p19q codeleted OIII patients, and patients whose tumor had a gene amplification, respectively (p = 0.002) (Fig. 3a). Extent of surgery (p = 0.04) and chromosome arms 1p/19q codeletion (p = 0.02) also significantly influenced PFS, while age, gender, and KPS had no significant effect (Table 2). When the prognostic factors identified in univariate analyses were included in a multivariate analysis, gene amplification remained the only significant variable in OIII (Table 2). Indeed, the relative risk of progression of patients whose OIII exhibited a gene amplification was 3.3 in comparison with patients without gene amplification (p = 0.0003) (Table 2).
Table 2.
Progression-free survival (PFS): patient and tumor characteristics and prognostic factors in the entire population of grade III oligodendrogliomas
| Median PFS (years) | Univariate Analysis
|
Multivariate Analysis
|
||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | n | % | p-Value | HR (95%CI) | p-Value | RR (95% CI) | ||
| Age | ⩽50.3 years | 26 | 50 | 1.2 | ||||
| >50.3 years | 26 | 50 | 1 | NS | 0.78 (0.38–1.51) | |||
| Gender | Men | 26 | 50 | 1.3 | ||||
| Women | 26 | 50 | 0.9 | NS | 1.17 (0.60–2.37) | |||
| Extent of surgery | Resection | 35 | 67 | 1.3 | ||||
| Biopsy | 17 | 33 | 0.6 | p = 0.04 | 0.52 (0.17–0.97) | NS | ||
| KPS | ⩾90 | 26 | 53 | 1.3 | ||||
| <90 | 23 | 47 | 0.6 | NS | 0.61 (0.28–1.18) | |||
| Gene amplification | 0 | 38 | 73 | 1.4 | ||||
| ⩾1 | 14 | 27 | 0.6 | p = 0.0006 | 2.89 (2.10–15.55) | p = 0.003 | 3.3 (1.4–7.9) | |
| Chromosomes | Codeleted | 15 | 29 | 3.1 | ||||
| 1p/19q status | Not codeleted | 37 | 71 | 0.8 | p = 0.02 | 0.42 (0.21–0.89) | NS | |
Abbreviations: HR, hazard ratio; RR, relative risk; CI, confidence interval; NS, not significant.
Fig. 3.
Kaplan-Meier curves comparing progression-free survival (A) and overall survival (B) of grade III oligodendrogliomas (OIIIs) according to their gene amplification and status of chromosome arms 1p/19q (log-rank test). The x-axis indicates survival in years and the y-axis the percentage of survivors. Continuous lines, dotted lines, and broken lines indicate chromosomes arms 1p/19q codeleted, non-1p/19q codeleted and non–gene amplified, and gene-amplified OIIIs, respectively.
In a univariate analysis, overall survival (OS) was significantly shorter in patients whose tumor had a gene amplification (Table 3). Indeed, the median OS was 6.7, 2.2, and 1.6 years in 1p/19q codeleted OIII patients, non-amplified and non-1p19q codeleted OIII patients, and patients whose tumor had a gene amplification, respectively (p = 0.05) (Fig. 3b). Extent of surgery (p < 0.05), KPS (p < 0.05), and chromosome arms 1p/19q codeletion (p = 0.05) also significantly influenced OS, while age and gender did not affect OS (Table 3). When the prognostic factors identified in univariate analyses were included in a multivariate analysis, gene amplification remained the unique significant variable (Table 3). Indeed, the relative risk of death of patients whose OIII exhibited a gene amplification was 2.5 in comparison with patients without gene amplification (p = 0.03) (Table 3).
Table 3.
Overall survival (OS): Patient and tumor characteristics and prognostic factors in the entire population of grade III oligodendrogliomas
| Median OS (years) | Univariate Analysis
|
Multivariate Analysis
|
||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | n | % | p-Value | HR (95%CI) | p-Value | RR (95% CI) | ||
| Age | ⩽50.3 years | 26 | 50 | 2.5 | ||||
| >50.3 years | 26 | 50 | 1.9 | NS | 0.72 (0.31–1.55) | |||
| Gender | Men | 26 | 50 | 2.2 | ||||
| Women | 26 | 50 | 2.2 | NS | 0.83 (0.37–1.79) | |||
| Extent of surgery | Resection | 35 | 67 | 3.5 | ||||
| Biopsy | 17 | 33 | 1.7 | p < 0.05 | 0.47 (0.13–0.99) | NS | ||
| KPS | ⩾90 | 26 | 53 | 3.5 | ||||
| < 90 | 23 | 47 | 2 | p < 0.05 | 0.47 (0.19–1.00) | NS | ||
| Gene amplification | 0 | 38 | 73 | 2.8 | ||||
| ⩾1 | 14 | 27 | 1.6 | p = 0.05 | 2.07 (0.99–6.48) | p = 0.03 | 2.5 (1.1–5.7) | |
| Chromosomes 1p/19q status | Codeleted | 15 | 29 | 6.7 | ||||
| Not codeleted | 37 | 71 | 1.9 | p < 0.05 | 0.32 (0.17–0.99) | NS | ||
Abbreviations: HR, hazard ratio; CI, confidence interval; RR, relative risk; NS, not significant.
Discussion
Several confirmed or candidate target genes of genomic amplifications have been reported in gliomas. To our knowledge, the present study is the first focused on the frequency and clinical significance of such a large set of amplified genes in a series of OIIIs using a whole genome aCGH.
We found that aCGH was a useful tool to detect amplification with a good agreement with qPCR, as previously shown by several independent studies.18,20,44 Only three gene amplifications, which were detected using aCGH, were not confirmed using genomic qPCR. A technically aCGH false-positive amplicon could not be ruled out formally. In addition, presences of large copy number variations (CNVs) in the tumor or the control samples may explain some aCGH false-positive results.45
Overall, gene amplification was observed in 27% of cases of OIII. EGFR was the most frequently amplified gene in OIII (17%). Some authors consider that the presence of EGFR amplification in an OIII is characteristic of the so-called small cell glioblastoma.3,46 However, excluding the diagnosis of OIII simply because of the presence of amplified EGFR is not recognized in the recently updated WHO classification (2007). The other amplified genes that we found in OIII are mainly involved in the cell cycle and cell growth, including CCND3, CCND1, CDK4, MDM2, and PDGFRA, putatively explaining the aggressiveness of OIII harboring a gene amplification.
In this series, the frequency of 1p/19q codeletion (29%) was in agreement with the 25% figure recently found in a large prospective European study of anaplastic oligodendroglial tumors47 and somewhat lower than previous studies that reported figures ranging from 37% to 66%.6–7,48–49 In addition to possible differences among pathologists in Europe and the United States,47,49 this variation is probably influenced by heterogeneous definitions of 1p loss, 19q loss, and 1p/19q codeletion in the literature. In the present series, we selected several criteria including losses of at least 75% of the BACs on 1p and 19q and presence of centromeric breakpoints on chromosomes 1 and 19 (within the 3 juxtacentromeric BACs), which is highly suggestive of the recently described translocation (1;19) in oligodendroglial tumors with good prognosis.5–6 Indeed, OS was quite good in our 1p/19q-deleted patients (OS = 6.7 years).
We recently showed that EGFR amplification was mutually exclusive of chromosome arms 1p/19q codeletion in gliomas.50 Based on the present results, this conclusion can be extended to all gene amplifications in OIII. This suggests that chromosome arms 1p/19q codeletion does not silence genes involved in the prevention of genomic amplification, such as mismatch repair genes.51
In the present study, gene amplification was an independent negative prognostic factor for OIII, regardless of the gene involved. This finding illustrates the potential of aCGH, which has the ability to screen the whole genome and therefore detect various amplified genes, without exclusive focus on a single target gene, such as EGFR. The observation that 1p/19q codeletion was not an independent prognostic factor for PFS and OS in the present series should be taken with caution. It is probably explained by the relatively small size of the population and by the low number of events in the 1p/19q codeleted group, which require further follow up.
It is interesting to note that a substantial number of OIIIs (44%) do not carry the 1p/19q codeletion or gene amplifications. Additional studies are now warranted to identify new biomarkers in this important subgroup.
In summary, our results suggest that whole genome evaluation of known target genes of genomic amplifications provides complementary prognostic information to classical prognostic factors in OIIIs. Moreover, these results encourage identification of target genes of genomic amplification whose driver gene remains unknown.
Acknowledgments
This work was supported by grants from the Institut National du Cancer and the Ligue Nationale Contre le Cancer. The BAC-array was provided by the Carte d’Identité des Tumeurs program of the Ligue Nationale Contre le Cancer.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol (Berl) 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 3.Miller CR, Dunham CP, Scheithauer BW, Perry A. Significance of necrosis in grading of oligodendroglial neoplasms: a clinicopathologic and genetic study of newly diagnosed high-grade gliomas. J Clin Oncol. 2006;24:5419–5426. doi: 10.1200/JCO.2006.08.1497. [DOI] [PubMed] [Google Scholar]
- 4.Ino Y, Betensky RA, Zlatescu MC, et al. Molecular subtypes of anaplastic oligodendroglioma: implications for patient management at diagnosis. Clin Cancer Res. 2001;7:839–845. [PubMed] [Google Scholar]
- 5.Griffin CA, Burger P, Morsberger L, et al. Identification of der(1;19) (q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol. 2006;65:988–994. doi: 10.1097/01.jnen.0000235122.98052.8f. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 7.Kitange G, Misra A, Law M, et al. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer. 2005;42:68–77. doi: 10.1002/gcc.20108. [DOI] [PubMed] [Google Scholar]
- 8.Rossi MR, Gaile D, Laduca J, et al. Identification of consistent novel submegabase deletions in low-grade oligodendrogliomas using array-based comparative genomic hybridization. Genes Chromosomes Cancer. 2005;44:85–96. doi: 10.1002/gcc.20218. [DOI] [PubMed] [Google Scholar]
- 9.Smith JS, Wang XY, Qian J, et al. Amplification of the platelet-derived growth factor receptor-A (PDGFRA) gene occurs in oligodendrogliomas with grade IV anaplastic features. J Neuropathol Exp Neurol. 2000;59:495–503. doi: 10.1093/jnen/59.6.495. [DOI] [PubMed] [Google Scholar]
- 10.Trent J, Meltzer P, Rosenblum M, et al. Evidence for rearrangement, amplification, and expression of c-myc in a human glioblastoma. Proc Natl Acad Sci U S A. 1986;83:470–473. doi: 10.1073/pnas.83.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins VP. Gene amplification in human gliomas. Glia. 1995;15:289–296. doi: 10.1002/glia.440150309. [DOI] [PubMed] [Google Scholar]
- 12.Alonso ME, Bello MJ, Arjona D, et al. Real-time quantitative PCR analysis of gene dosages reveals gene amplification in low-grade oligodendrogliomas. Am J Clin Pathol. 2005;123:900–906. doi: 10.1309/448Y-RWNC-43TE-32KQ. [DOI] [PubMed] [Google Scholar]
- 13.Arjona D, Bello MJ, Alonso ME, et al. Real-time quantitative PCR analysis of regions involved in gene amplification reveals gene overdose in low-grade astrocytic gliomas. Diagn Mol Pathol. 2005;14:224–229. doi: 10.1097/01.pas.0000177799.58336.1a. [DOI] [PubMed] [Google Scholar]
- 14.Knobbe CB, Reifenberger G. Genetic alterations and aberrant expression of genes related to the phosphatidyl-inositol-3′-kinase/protein kinase B (Akt) signal transduction pathway in glioblastomas. Brain Pathol. 2003;13:507–518. doi: 10.1111/j.1750-3639.2003.tb00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida A, Zhu XX, Vogt N, et al. GAC1, a new member of the leucine-rich repeat superfamily on chromosome band 1q32. 1, is amplified and overexpressed in malignant gliomas. Oncogene. 1998;16:2997–3002. doi: 10.1038/sj.onc.1201828. [DOI] [PubMed] [Google Scholar]
- 16.Rickman DS, Tyagi R, Zhu XX, et al. The gene for the axonal cell adhesion molecule TAX-1 is amplified and aberrantly expressed in malignant gliomas. Cancer Res. 2001;61:2162–2168. [PubMed] [Google Scholar]
- 17.Garson JA, McIntyre PG, Kemshead JT. N-myc amplification in malignant astrocytoma. Lancet. 1985;2:718–719. doi: 10.1016/s0140-6736(85)92950-2. [DOI] [PubMed] [Google Scholar]
- 18.Hui AB, Lo KW, Yin XL, Poon WS, Ng HK. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81:717–723. doi: 10.1038/labinvest.3780280. [DOI] [PubMed] [Google Scholar]
- 19.Yung WK, Zhang X, Steck PA, Hung MC. Differential amplification of the TGF-alpha gene in human gliomas. Cancer Commun. 1990;2:201–205. doi: 10.3727/095535490820874416. [DOI] [PubMed] [Google Scholar]
- 20.Ruano Y, Mollejo M, Ribalta T, et al. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puputti M, Tynninen O, Sihto H, et al. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol Cancer Res. 2006;4:927–934. doi: 10.1158/1541-7786.MCR-06-0085. [DOI] [PubMed] [Google Scholar]
- 22.Buschges R, Weber RG, Actor B, Lichter P, Collins VP, Reifenberger G. Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol. 1999;9:435–442. doi: 10.1111/j.1750-3639.1999.tb00532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen Y, Sun ZL, Liao WM, Zhou RX. Amplification, rearrangement and over-expression of c-fos gene in human primary brain tumors. Hua Xi Yi Ke Da Xue Xue Bao. 1989;20:243–245. [PubMed] [Google Scholar]
- 24.Tchirkov A, Rolhion C, Bertrand S, Dore JF, Dubost JJ, Verrelle P. IL-6 gene amplification and expression in human glioblastomas. Br J Cancer. 2001;85:518–522. doi: 10.1054/bjoc.2001.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson JJ, Barnett D, Yang J, Assietti R, Cotsonis G, James CD. Gene amplification as a prognostic factor in primary brain tumors. Clin Cancer Res. 1998;4:215–222. [PubMed] [Google Scholar]
- 26.Costello JF, Plass C, Arap W, et al. Cyclin-dependent kinase 6 (CDK6) amplification in human gliomas identified using two-dimensional separation of genomic DNA. Cancer Res. 1997;57:1250–1254. [PubMed] [Google Scholar]
- 27.Mueller HW, Michel A, Heckel D, et al. Identification of an amplified gene cluster in glioma including two novel amplified genes isolated by exon trapping. Hum Genet. 1997;101:190–197. doi: 10.1007/s004390050612. [DOI] [PubMed] [Google Scholar]
- 28.Fischer U, Muller HW, Sattler HP, Feiden K, Zang KD, Meese E. Amplification of the MET gene in glioma. Genes Chromosomes Cancer. 1995;12:63–65. doi: 10.1002/gcc.2870120111. [DOI] [PubMed] [Google Scholar]
- 29.Knobbe CB, Trampe-Kieslich A, Reifenberger G. Genetic alteration and expression of the phosphoinositol-3–kinase/Akt pathway genes PIK3CA and PIKE in human glioblastomas. Neuropathol Appl Neurobiol. 2005;31:486–490. doi: 10.1111/j.1365-2990.2005.00660.x. [DOI] [PubMed] [Google Scholar]
- 30.Reifenberger G, Ichimura K, Reifenberger J, Elkahloun AG, Meltzer PS, Collins VP. Refined mapping of 12q13–q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets. Cancer Res. 1996;56:5141–5145. [PubMed] [Google Scholar]
- 31.Galanis E, Buckner J, Kimmel D, et al. Gene amplification as a prognostic factor in primary and secondary high-grade malignant gliomas. Int J Oncol. 1998;13:717–724. [PubMed] [Google Scholar]
- 32.Fischer U, Meltzer P, Meese E. Twelve amplified and expressed genes localized in a single domain in glioma. Hum Genet. 1996;98:625–628. doi: 10.1007/s004390050271. [DOI] [PubMed] [Google Scholar]
- 33.Maas RM, Reus K, Diesel B, et al. Amplification and expression of splice variants of the gene encoding the P450 cytochrome 25–hydroxyvitamin D(3) 1,alpha-hydroxylase (CYP 27B1) in human malignant glioma. Clin Cancer Res. 2001;7:868–875. [PubMed] [Google Scholar]
- 34.Reifenberger G, Liu L, Ichimura K, Schmidt EE, Collins VP. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 1993;53:2736–2739. [PubMed] [Google Scholar]
- 35.Fischer U, Heckel D, Michel A, Janka M, Hulsebos T, Meese E. Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum Mol Genet. 1997;6:1817–1822. doi: 10.1093/hmg/6.11.1817. [DOI] [PubMed] [Google Scholar]
- 36.Fischer U, Schutz N, Hemmer D, Meese E. GAS64, the first amplified and putative non-translated gene. Int J Oncol. 2002;20:173–176. [PubMed] [Google Scholar]
- 37.Fischer U, Hemmer D, Heckel D, et al. KUB3 amplification and overexpression in human gliomas. Glia. 2001;36:1–10. doi: 10.1002/glia.1090. [DOI] [PubMed] [Google Scholar]
- 38.Klein A, Reichardt W, Jung V, Zang KD, Meese E, Urbschat S. Over-expression and amplification of STK15 in human gliomas. Int J Oncol. 2004;25:1789–1794. [PubMed] [Google Scholar]
- 39.Arakawa Y, Tachibana O, Hasegawa M, Miyamori T, Yamashita J, Hayashi Y. Frequent gene amplification and overexpression of decoy receptor 3 in glioblastoma. Acta Neuropathol (Berl) 2005;109:294–298. doi: 10.1007/s00401-004-0956-6. [DOI] [PubMed] [Google Scholar]
- 40.Riemenschneider MJ, Knobbe CB, Reifenberger G. Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int J Cancer. 2003;104:752–757. doi: 10.1002/ijc.11023. [DOI] [PubMed] [Google Scholar]
- 41.Kleihues P, Louis DN, Scheithauer BW, et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. [DOI] [PubMed] [Google Scholar]
- 42.Ironside JW, Moss TH, Louis DN, Lowe JS, Weller RO, editors. Diagnostic Pathology of Nervous System Tumours. Edinburgh: Churchill Living-stone; 2002. [Google Scholar]
- 43.Liva S, Hupe P, Neuvial P, et al. CAPweb: a bioinformatics CGH array Analysis Platform. Nucleic Acids Res. 2006;34(Web Server issue):W477–W481. doi: 10.1093/nar/gkl215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasaki T, Arai H, Beppu T, Ogasawara K. Detection of gene amplification and deletion in high-grade gliomas using a genome DNA micro-array (GenoSensor Array 300) Brain Tumor Pathol. 2003;20:59–63. doi: 10.1007/BF02483448. [DOI] [PubMed] [Google Scholar]
- 45.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 46.Perry A, Aldape KD, George DH, Burger PC. Small cell astrocytoma: an aggressive variant that is clinicopathologically and genetically distinct from anaplastic oligodendroglioma. Cancer. 2004;101:2318–2326. doi: 10.1002/cncr.20625. [DOI] [PubMed] [Google Scholar]
- 47.van den Bent MJ, Carpentier AF, Brandes AA, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol. 2006;24:2715–2722. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 48.Cowell JK, Barnett GH, Nowak NJ. Characterization of the 1p/19q chromosomal loss in oligodendrogliomas using comparative genomic hybridization arrays (CGHa) J Neuropathol Exp Neurol. 2004;63:151–158. doi: 10.1093/jnen/63.2.151. [DOI] [PubMed] [Google Scholar]
- 49.Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006;24:2707–2714. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- 50.Idbaih A, Marie Y, Lucchesi C, et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer. 2008;122:1778–1786. doi: 10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 51.Myllykangas S, Knuutila S. Manifestation, mechanisms and mysteries of gene amplifications. Cancer Lett. 2006;232:79–89. doi: 10.1016/j.canlet.2005.07.045. [DOI] [PubMed] [Google Scholar]