Abstract
Glioblastoma multiforme (GBM) is the most frequent primary brain tumor in adults. Prognosis is poor. Using a series of 214 GBM patients, we observed an effect of the variant 5,10-methylenetetrahydrofolate reductase (MTHFR) c.677C>T on overall survival. This effect was strongest in patients younger than 60 years at diagnosis (overall survival, median ± SE: genotype CC, 13 ± 1 months; CT, 11 ± 2 months; TT, 7 ± 3 months; multivariate Cox regression analysis, Wald = 8.58, p = 0.007). In addition, the MTHFR genotype significantly influenced the overall survival of patients with a postoperative Karnofsky score >70 (CC, 12 ± 2 months; CT, 11 ± 1 months; TT, 10 ± 4 months; Wald = 5.89, p = 0.015). These data suggest the MTHFR c.677C>T variant is a risk factor for survival in GBM patients.
Keywords: glioblastoma, glioma, homocysteine, MTHFR, survival
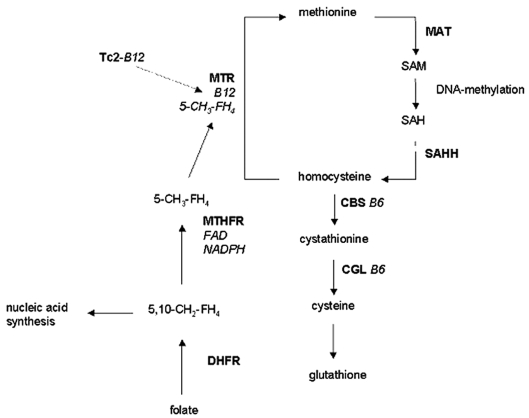
Methionine metabolism plays an essential role in DNA methylation (Fig. 1). The enzyme 5-methyltetrahydrofolate-homocysteine S-methyltransferase (MTR) catalyzes the remethylation of homocysteine to methionine. The latter is activated by ATP to S-adenosylmethionine (SAM), which provides the methyl groups used for DNA methylation. MTR depends on cobalamin (vitamin B12) and 5-methyltetrahydrofolate as cofactors. The most important transporter protein of cobalamin is transcobalamin 2 (Tc2), and 5-methyltetrahydrofolate is synthesized out of 5,10-methylenetetrahydrofolate by 5,10-methylenetetrahydrofolate reductase (MTHFR). Because 5,10-methylenetetrahydrofolate is also directed to the synthesis of purines and thymidine, MTHFR activity regulates whether a one-carbon unit of tetrahydrofolate is utilized for methionine synthesis or for nucleic acid synthesis and thus for DNA synthesis, stability, and repair.1
Fig. 1.
Human methionine metabolism. Methionine becomes activated to S-adenosylmethionine (SAM) via methionine adenosyltransferase (MAT). SAM is a ubiquitous methyl group donor, for example, for the synthesis of biogenic amines and DNA methylation. The degradation product of SAM is S-adenosylhomocysteine (SAH), which is hydrolyzed to homocysteine via homocysteine hydrolase (SAHH). Homocysteine can be transsulfurated to cystathionine and cysteine via vitamin B6–dependent cystathionine beta-synthase (CBS) and cystathionine gamma-lyase (CGL); alternatively, homocysteine can be remethylated to methionine via 5-methyltetrahydrofolate-homocysteine S-methyltransferase (MTR; also called methionine synthase), which needs a derivate of vitamin B12 (methylcobalamin) and a derivate of folate (5-methyltetrahydrofolate; 5-CH3-FH4) as cofactors. Vitamin B12 is transported by transcobalamin 2 (Tc2), and 5-CH3-FH4 is synthesized by flavin adenine dinucleotide (FAD)–dependent 5,10-methylenetetrahydrofolate reductase (MTHFR) from 5,10-methylenetetrahydrofolate (5,10-CH2-FH4), which is synthesized from folate by dihydrofolate reductase (DHFR) in two subsequent steps.
Instability of genomic DNA and impaired DNA methylation are important for the development and progression of many tumors.2 Therefore, functional genetic variants of methionine metabolism are attractive candidate factors influencing the development and clinical course of human cancers. Indeed, the MTHFR missense variant c.677C>T (A222V) and the MTR missense variant c.2756A>G (D919G) have been correlated with the incidence of various malignancies.3 Furthermore, functional synergisms have been suggested for the variants MTR c.2756A>G and Tc2 c.776C>G (P259R).4 However, only a few studies have investigated the role of the genetic variants of methionine metabolism as a prognostic factor in patients with cancer. In the present study, we analyzed the effects of four functional polymorphisms of methionine metabolism on the clinical course of 214 patients with glioblastoma multiforme (GBM). Although our previous results did not identify the MTHFR c.677C>T variant as a risk factor for the incidence of GBM,3 the present study suggests an influence of MTHFR c.677C>T on survival.
Materials and Methods
Patients
This study was approved by the Ethics Committee of the Medical Faculty of the University of Bonn.
We analyzed 214 GBM patients (41% female; median age at diagnosis, 62 years; range, 23–80 years; age at diagnosis, 58 ± 12 years, mean ± SD) who underwent surgery in the Department of Neurosurgery of the University Hospital Bonn (Bonn, Germany) between 1994 and 2003. All histopathological diagnoses were made at the Department of Neuropathology/German Brain Tumor Reference Center at the University of Bonn using the WHO criteria.5 A gliosarcoma was diagnosed in 6% of the patients and a giant cell GBM in 3%. Surgical treatment consisted of total resection in 52% of the patients, subtotal/partial resection in 43%, and biopsy in 5%. Eighty-two percent of patients underwent standard fractionated postoperative radiotherapy, and 11% underwent additional adjuvant chemotherapy. Thus, adjuvant therapy after surgery was categorized as none (surgery only), radiotherapy, and radiation and chemotherapy. For 107 patients, treatment at tumor relapse could not be ascertained. Hence, therapy for recurrent tumors was not included in the statistical analysis. Median overall survival of all patients (± SD) was 10 ± 1 months (95% confidence interval, 9–11 months), with 7% censored.
Genotyping
The genotypes of four missense variants involved in methionine metabolism, MTHFR c.677C>T, MTHFR c.1298A>C, MTR c.2756A>G, and Tc2 c.776C>G, were determined by PCR amplification of genomic DNA prepared from peripheral leukocytes with subsequent restriction analysis as published previously.4
Statistical Analysis
The log rank test and Kaplan-Meier curves were used to analyze the effect of the four variants on overall survival. Owing to multiple testing, α was set to 0.0125. The effect of the MTHFR variant c.677C>T on overall survival was further investigated by Cox regression, with simultaneous analysis of MTHFR genotype, age, sex, postoperative Karnofsky score, extent of resection, and adjuvant therapy as covariables. In additional explorative analyses (univariate log rank tests and multivariate Cox regression analysis with the covariables listed above), the effect of the MTHFR c.677C>T variant was analyzed in subgroups of patients defined by adjuvant therapy, age (<60 years vs. >60 years, the mean and median age at diagnosis in our population and prior studies6), and postoperative Karnofsky score (<70 vs. ⩾70). Data are reported as median ± SE.
Results
Log rank testing demonstrated a significant influence of the MTHFR c.677C>T variant, but not of the other three genetic polymorphisms, on overall survival: CC, 10 ± 1 months; CT, 10 ± 1 months; TT, 7 ± 3 months (log rank = 9.21, p = 0.010; Table 1).
Table 1.
Kaplan-Meier analysis of the effect of genotype on overall survival: all patients
| Variant | Median Overall Survival (± SE, months) | Log Rank (2 degrees of freedom) | ||
|---|---|---|---|---|
| MTHFR c.677C>T | CC: 10 ± 1 | CT: 10 ± 1 | TT: 7 ± 3 | 9.21, p = 0.010 |
| MTHFR c.1298A>C | AA: 10 ± 1 | AC: 9 ± 1 | CC: 14 ± 4 | 0.60, p = 0.742 |
| MTR c.2756A>G | AA: 10 ± 1 | AG: 9 ± 1 | GG: 8 ± 5 | 0.07, p = 0.967 |
| Tc2 c.776C>G | CC: 8 ± 2 | CG: 10 ± 1 | GG: 9 ± 2 | 1.52, p = 0.469 |
Abbreviations: MTHFR, 5,10-methylenetetrahydrofolate reductase; MTR, 5-methyltetrahydrofolate-homocysteine S-methyltransferase; Tc2, transcobalamin 2. Owing to multiple testing, the threshold was defined with α = 0.0125. Censored for MTHFR c.677C>T: CC, n = 9; CT, n = 6; TT, n = 0.
Multivariate Cox regression including the MTHFR c.677C>T genotype and the clinical data indicated that important prognostic factors were younger age (linear variable; Wald = 7.10, p = 0.008), higher Karnofsky score after surgery (linear variable; Wald = 21.2, p = 0.001), and adjuvant therapy (none, 2 ± 0.35 months; radiotherapy, 10 ± 0.36 months; radiation and chemotherapy, 11 ± 0.82 months; Wald = 7.81, p = 0.005). The extent of resection had a statistically significant influence on survival in the univariate log rank test (total resection, 10 ± 0.69 months; subtotal/partial resection, 8 ± 0.85 months; biopsy, 2 ± 1.58 months; log rank = 10.3, p = 0.006) but not in the multivariate Cox regression analysis (Wald = 0.68, p = 0.447). In the multivariate analysis, the effect of the MTHFR c.677C>T genotype on survival was not statistically significant (Wald = 1.01, p = 0.316).
Univariate log rank subgroup analysis revealed a highly significant association of the MTHFR c.677C>T polymorphism with survival for patients younger than 60 years at diagnosis (37% of our study sample): CC, 13 ± 1 months; CT, 11 ± 2 months; TT, 7 ± 3 months (log rank = 9.94, p = 0.007; Table 2). Multivariate Cox regression analysis showed that these data were robust; that is, the effect of the MTHFR c.677C>T variant on survival in this patient subset was as strong as the effects of some of the clinical covariables investigated simultaneously: MTHFR c.677C>T (Wald = 8.58, p = 0.003), extent of resection (Wald = 2.10, p = 0.148), Karnofsky score after surgery (Wald = 12.3, p < 0.001), and adjuvant therapy (Wald = 3.32, p = 0.086). No significant effect was seen for the MTHFR c.677C>T polymorphism for patients >60 years of age (Wald = 0.27, p = 0.601).
Table 2.
Influence of MTHFR c.677C>T genotype on median overall survival: univariate (log rank) and multivariate analyses
| Median Overall Survival (±SE, months)
|
|||||
|---|---|---|---|---|---|
| Patient Group | CC | CT | TT | Log Rank | Cox Regression (Wald) |
| All | 10 ± 1 | 10 ± 1 | 7 ± 3 | 9.21, p = 0.010 | 1.01, p = 0.316 |
| Age <60 years | 13 ± 1 | 11 ± 2 | 7 ± 3 | 9.94, p = 0.007 | 8.58, p = 0.003 |
| Age >60 years | 9 ± 8 | 8 ± 6 | 6 ± 5 | 2.56, p = 0.278 | 0.27, p = 0.601 |
| KPI >70 | 12 ± 2 | 11 ± 1 | 10 ± 4 | 5.23, p = 0.051 | 5.89, p = 0.015 |
| KPI <70 | 5 ± 1 | 4 ± 2 | 5 ± 3 | 0.42, p = 0.896 | 0.01, p = 0.951 |
Abbreviation: MTHFR, 5,10-methylenetetrahydrofolate reductase. Covariables included age (except in the subgroups defined by age), sex, postoperative KPI (except in the subgroups defined by KPI), extent of resection, and adjuvant therapy.
Additionally, the MTHFR c.677C>T variant was found to significantly correlate with survival in the multivariate analysis among patients with a postoperative Karnofsky score >70 regardless of age (CC, 12 ± 2 months; CT, 11 ± 1 months; TT, 10 ± 4 months; Wald = 5.89, p = 0.015). No such effect was seen for patients with a postoperative Karnofsky score <70 (CC, 5 ± 1 months; CT, 4 ± 2 months; TT, 5 ± 3 months; Wald = 0.01, p = 0.951). The MTHFR c.677C>T polymorphism showed no significant correlation with survival in any of the subgroups defined by adjuvant therapy.
Discussion
Overall prognosis for GBM patients is grim, with a median survival of only 12–16 months even in selected series. However, certain subsets of patients (young age at diagnosis, high extent of resection, high Karnofsky score after surgery) can expect to survive for more than 2 years.7 Small therapeutic advances will more likely translate into a clinically relevant survival benefit in patients with a relatively good prognosis. Hence, the identification of prognostic survival factors may allow more selective treatment, that is, spare those with an adverse prognosis a toxic therapy, and allow for aggressive treatment in cases for which a meaningful prolongation of survival can be achieved.
Several tumor mutations have been correlated with the clinical course of GBM patients.8 In addition, a few studies have indicated that germline genetic variants may have prognostic relevance, as well, for example, the ERCC1 c.8092C>A polymorphism, the deletion of glutathione S-transferase T1 (GSTT1),9 and a polymorphism in the 5′-untranslated region of the epidermal growth factor gene (g.61G>A).10
The major finding of the present study is an association of the c.677TT genotype of the germline variant MTHFR c.677C>T (A222V) with an adverse prognosis for GBM patients. Significant effects of the MTHFR c.677C>T polymorphism were in particular seen in patient subgroups with a comparatively good prognosis. In the subgroup of patients younger than 60 years, the 6-month difference in median survival for patients homozygous for the MTHFR c.677CC genotype versus patients with the c.677TT genotype suggests that the effect size contributed by the c.677C>T genotype is clinically relevant. Accordingly, the MTHFR c.677TT genotype was also associated with a poorer prognosis within the patient subgroup with a postoperative Karnofsky score ⩾70 but not among patients with a worse performance status. The association of MTHFR c.677C>T with overall survival did not show any therapy-specific effect. Given the limitations of any association study, our findings have to be reproduced in an independent dataset.
The T-allele of MTHFR c.677C>T, in particular when in the homozygous state, leads to clearly reduced enzyme activity.11 Our findings may be explained by an effect on nucleic acid synthesis and thus on chromosomal stability.1 In addition, a lack of MTHFR activity can reduce the available SAM in several organs, including the CNS, and SAM is the key methyl group donor for DNA methylation (Fig. 1).12 The T-allele of MTHFR c.677C>T has already been reported to be associated with global hypomethylation of genomic DNA.13 Hypomethylation of the O6-methylguanine DNA methyltransferase (MGMT) promoter in GBM tumor cells has been associated with shorter survival in GBM patients receiving radiotherapy and treated with temozolomide,14 and a correlation between the T-allele of the MTHFR c.677C>T variant and a decreased MGMT promoter hypermethylation has already been observed in a series of uterine cervical cancers.15 Thus, it is tempting to speculate that differential (tumor) DNA methylation due to MTHFR c.677C>T might constitute the molecular basis of the association between the MTHFR c.677C>T polymorphism and survival of GBM patients. Accordingly, Cadieux et al.16 recently demonstrated that the MTHFR c.677C>T variant or a deletion encompassing the MTHFR gene locus on chromosome 1p36.3 was associated with global DNA hypomethylation in GBM tissue. Tumors with DNA hypomethylation or a MTHFR gene deletion exhibited an increased proliferation rate.16 Our study provides the clinical data predicted by these observations: reduced MTHFR activity due to the presence of the T-allele of the c.677C>T variant is associated with shorter overall survival. One limitation of our study was that we did not know the folate intake or plasma folate levels of the GBM patients, which might have modified the effect of the MTHFR genotype on overall survival, as the effect of the c.677C>T variant on MTHFR activity is modified by the availability of folate.11,17
Our findings may have some therapeutic implications. Because the adverse biological effects of the T-allele of the MTHFR c.677C>T variant, for example, on DNA methylation, may well become apparent only in the context of a low folate status,17 folate supplementation or dietary strategies influencing methionine and further metabolites of methionine metabolism might be interesting candidate supportive therapies for GBM patients.
References
- 1.Duthie SJ, Hawdon A. DNA instability (strand breakage, uracil misin-corporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. FASEB J. 1998;12:1491–1497. [PubMed] [Google Scholar]
- 2.Beckman RA, Loeb LA. Genetic instability in cancer: theory and experiment. Semin Cancer Biol. 2005;15:423–435. doi: 10.1016/j.semcancer.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Semmler A, Simon M, Moskau S, Linnebank M. The methionine synthase polymorphism c.2756A>G alters susceptibility to glioblastoma multiforme. Cancer Epidemiol Biomarkers Prev. 2006;15:2314–2316. doi: 10.1158/1055-9965.EPI-05-0979. [DOI] [PubMed] [Google Scholar]
- 4.Linnebank M, Pels H, Kleczar N, et al. MTX-induced white matter changes are associated with polymorphisms of methionine metabolism. Neurology. 2005;64:912–913. doi: 10.1212/01.WNL.0000152840.26156.74. [DOI] [PubMed] [Google Scholar]
- 5.Kleihues P, Louis DN, Scheithauer BW, et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. [DOI] [PubMed] [Google Scholar]
- 6.Westphal M, Hilt DC, Bortey E, et al. A phase 3 trial of local chemotherapy with biodegradable carmustine (BCNU) wafers (Gliadel wafers) in patients with primary malignant glioma. Neuro-Oncology. 2003;5:79–88. doi: 10.1215/S1522-8517-02-00023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLendon RE, Halperin EC. Is the long-term survival of patients with intracranial glioblastoma multiforme overstated? Cancer. 2003;98:1745–1748. doi: 10.1002/cncr.11666. [DOI] [PubMed] [Google Scholar]
- 8.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 9.Wrensch M, Wiencke JK, Wiemels J, et al. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006;66:4531–4541. doi: 10.1158/0008-5472.CAN-05-4032. [DOI] [PubMed] [Google Scholar]
- 10.Bhowmick DA, Zhuang Z, Wait SD, Weil RJ. A functional polymorphism in the EGF gene is found with increased frequency in glioblastoma multiforme patients and is associated with more aggressive disease. Cancer Res. 2004;64:1220–1223. doi: 10.1158/0008-5472.can-03-3137. [DOI] [PubMed] [Google Scholar]
- 11.Frosst P, Blom HJ, Milos R, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 12.Surtees R, Leonard J, Austin S. Association of demyelination with deficiency of cerebrospinal-fluid S-adenosylmethionine in inborn errors of methyl-transfer pathway. Lancet. 1991;338:1550–1554. doi: 10.1016/0140-6736(91)92373-a. [DOI] [PubMed] [Google Scholar]
- 13.Shelnutt KP, Kauwell GP, Gregory JF, III, et al. Methylenetetrahydrofolate reductase 677C→T polymorphism affects DNA methylation in response to controlled folate intake in young women. J Nutr Biochem. 2004;15:554–560. doi: 10.1016/j.jnutbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 15.Kang S, Kim JW, Kang GH, et al. Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecol Oncol. 2005;96:173–180. doi: 10.1016/j.ygyno.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Cadieux B, Ching TT, Vandenberg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 17.Friso S, Choi SW, Girelli D, et al. A common mutation in the 5,10- methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]