Abstract
Pediatric ependymomas are enigmatic tumors, and their clinical management remains one of the more difficult in pediatric oncology. The identification of biological correlates of outcome and therapeutic targets remains a significant challenge in this disease. We therefore analyzed a panel of potential biological markers to determine optimal prognostic markers. We constructed a tissue microarray from 97 intracranial tumors from 74 patients (WHO grade II–III) and analyzed the candidate markers nucleolin, telomerase catalytic subunit (hTERT; antibody clone 44F12), survivin, Ki-67, and members of the receptor tyrosine kinase I (RTK-I) family by immunohistochemistry. Telomerase activity was determined using the in vitro–based telomere repeat amplification protocol assay, and telomere length was measured using the telomere restriction fragment assay. Primary tumors with low versus high nucleolin protein expression had a 5-year event-free survival of 74% ± 13% and 31% ± 7%, respectively. Multivariate analysis identified low nucleolin expression to be independently associated with a more favorable prognosis (hazard ratio = 6.25; 95% confidence interval, 1.6–24.2; p = 0.008). Ki-67 and survivin correlated with histological grade but not with outcome. Immunohistochemical detection of the RTK-I family did not correlate with grade or outcome. Telomerase activity was evident in 19 of 22 primary tumors, with telomere lengthening and/or maintenance occurring in five of seven recurrent cases. Low nucleolin expression was the single most important biological predictor of outcome in pediatric intracranial ependymoma. Furthermore, telomerase reactivation and maintenance of telomeric repeats appear necessary for childhood ependymoma progression. These findings require corroboration in a clinical trial setting.
Keywords: ependymoma, nucleolin, pediatric, telomerase, telomere
From the clinical and biological perspective, intracranial ependymomas of childhood remain enigmatic and challenging tumors to treat. The most widely accepted prognostic factor is the degree of surgical resection, with complete resection associated with a better prognosis. However, a small number of studies have failed to demonstrate this relationship except for supratentorial disease.1,2 Other prognostic factors include age at diagnosis and location, with children older than 3 years at diagnosis and those with supratentorial tumors having a better prognosis. Whether this relates to concerns over choice of adjuvant therapy in younger children or the degree of surgical resectability or underlying biology is uncertain.
Though widely considered to be a “surgical disease,” a significant proportion of patients relapse even following complete surgical removal of the tumor. Adjuvant therapy is presently an important part of the overall treatment strategy.3–8 However, such treatments bring late effects. Use of radiotherapy creates concerns regarding neurocognitive development, growth, endocrinopathies, and second cancers.9–12 Chemotherapy induces late effects via specific toxicities such as ototoxicity, as well as more generic problems.13 It may be that some patients are curable without radiotherapy or chemotherapy following a complete surgical removal, particularly those with supratentorial tumors, but reliable clinical and biological correlates of outcome on which treatment decisions can be made are still needed.14
The relationship between outcome and histological grading according to WHO criteria15 remains a controversial prognostic factor. Some studies have found that anaplasia carries a poorer prognosis,3,5,16–18 whereas others have not.1,4,19–22 Differentiation between grade II and grade III ependymoma requires interpretation of a spectrum of pathological features; an international consensus on the interpretation of these factors and tumor grade as a prognostic factor is now required. In the meantime, we need other biological markers that will predict tumor outcome.
A number of “biological” markers have now been identified and reported to be of prognostic significance, including Ki-67, survivin, the receptor tyrosine kinase I (RTK-I) family, and hTERT, the catalytic subunit of telomerase. The proliferation marker Ki-67 has been widely reported as a predictor of outcome in ependymoma, although few studies have been conducted exclusively in pediatric cases.17,23–29 Survivin has been reported to function as a mitotic regulator,30 with two studies reporting its prognostic significance in ependymoma, with contrasting results. In one study that analyzed pediatric ependymomas and choroid plexus tumors, low levels of nuclear survivin were shown to be a marker of more aggressive disease and/or tumor grade.31 Another study that analyzed both adult and pediatric intracranial ependymomas found high survivin expression to be associated with a poor outcome.32 These findings highlight the need to further evaluate the prognostic value of survivin in ependymoma. The RTK-I family includes epidermal growth factor receptor (EGFR), ERBB2 (erythroblastic leukemia viral oncogene homolog 2; HER2), ERBB3, and ERBB4, which are involved in many cellular processes, including cell division, cell survival, and cell motility.33,34 Coexpression of ERBB2 and ERBB4, high Ki-67 labeling index (LI), and degree of surgical resection are reported to be predictive of poor prognosis in ependymoma.28
A recent study has reported that hTERT expression was the strongest predictor of outcome in pediatric ependymoma, independent of other clinical and pathological prognostic markers.35 However, more recent reports have reevaluated the target of the hTERT antibody clone 44F12, providing evidence to support specificity toward nucleolin.36 Nucleolin is abundant in tumor cells,37,38 and its prognostic role in ependymoma has yet to be ascertained.
There is a clear need to better understand the underlying biology of this disease in order to improve therapeutic options and outcomes.39,40 Many of the “biological” studies to date have looked at a single biological marker in isolation and/or have conducted the studies on mixed cohorts of adult and pediatric ependymomas. We therefore sought to correlate the expression of a comprehensive panel of putative biological markers with clinical parameters and, through multifactorial analysis, to better understand and predict the behavior of pediatric intracranial ependymomas.
Materials and Methods
Between 1961 and 2006, 114 primary and recurrent intracranial tumors were identified for entry into the study from the Children’s Cancer and Leukaemia Group (CCLG), West Midlands Children’s Tumor Registry (WMCTR), and pathology archives at Birmingham Children’s Hospital. Histologies of all cases were centrally reviewed (M.-A.B., D.E.) and graded according to the 2000 WHO classification.15 Only cases of WHO grade II and III intracranial ependymoma were included in this study. Data on date of birth, age at diagnosis, degree of surgical resection, further treatment, and current status were obtained from the CCLG data center, WMCTR, and the case notes.
A tissue microarray (TMA) was constructed from routinely processed formalin-fixed paraffin-embedded (FFPE) tumor material. Areas of viable and representative tumor following review of all blocks were marked by a pathologist (M.-A.B.) prior to inclusion into the TMA. Typically, cores 3 × 0.6 mm for each tumor were included; however, to account for the heterogeneity of tumor histology, in 15 of 97 tumors (15%), up to three sets of triplicate cores were taken from representative areas. Corresponding frozen material was included for follow-up with hematoxylin and eosin smear verified by a pathologist to confirm viable tumor prior to inclusion in the study.
Immunohistochemistry
Five-micrometer sections were obtained from the TMA. Briefly, the slides were incubated at 37°C overnight, deparaffinized in xylene, and hydrated through decreasing concentrations of ethanol. For Ki-67, survivin, ERBB2, ERBB4, 44F12, and nucleolin, antigen retrieval was performed in a pressure cooker for 1 min at full pressure in sodium citrate buffer (pH 6.0). The sections were incubated with normal goat serum, followed by an endogenous peroxidase block (Dako, Cambridgeshire, UK). Ki-67 (MIB-1) mouse monoclonal antibody (Dako) was incubated for 1 h at room temperature (1:50). Survivin (sc-10811) rabbit polyclonal (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:1,000), ERBB2 (NCL-CB11) mouse monoclonal (Novocastra, Newcastle, UK; 1:1,000), ERBB4 (c-18) rabbit polyclonal (Santa Cruz Biotechnology; 1:4,000), 44F12 mouse monoclonal (Novocastra; 1:100), and nucleolin (4E2) rabbit monoclonal (Ab13541; Abcam, Cambridgeshire, UK; 1:2,000) antibodies were all incubated overnight at 4°C. Target antigen was detected using the Dako Chemate Envision Detection Kit with diaminobenzidine chromogen for visualization, according to manufacturer’s instructions. Sections were then counterstained with Harris hematoxylin (Surgipath, Cambridgeshire, UK), dehydrated, and mounted for analysis. Detection of EGFR and validation of ERBB2, using a second detection method, were carried out using the PharmDx (Dako) and HercepTest (Dako) kits, respectively, according to manufacturer’s instructions. Additionally, immunohistochemical detection of ERBB2 was independently analyzed (R.G.) in our cohort, as previously described.28
Appropriate controls were included for each antibody: Ki-67, tonsil; survivin, tonsil; EGFR, HT-29 (2+ intensity) adenocarcinoma cell line (PharmDx) and liver tissue; ERBB2, MDA-231 breast cancer control cell line (HercepTest); ERBB4, appendix; 44F12, tonsil; nucleolin, tonsil. Expression was verified prior to data analysis. For negative controls, primary antibody was replaced with antibody diluent (Dako).
TMA sections were blindly analyzed using a bright-field microscope (Olympus BX-41, Hertfordshire, UK) with ×10 and ×40 objectives by two independent investigators, including a pathologist (M.-A.B.). Ki-67, survivin, and nucleolin LIs were defined as the percentage of immunopositive tumor cells exhibiting nuclear staining divided by the total number evaluated. For tumors with triplicate cores (85%), the final LI was taken from the mean of the three cores. For tumors exhibiting cellular heterogeneity with more than three cores (15%), the mean LI was calculated for each set of triplicate cores with the highest LI value used for that case. 44F12 was scored as the percentage of immunopositive cells divided by the total number evaluated, including staining intensity (range, 0 [none] to 2+ [strong]). Only tumors with strong (2+) staining in >25% cells were considered positive.
Adhering to the manufacturer’s instructions (PharmDx; Food and Drug Administration [FDA] approved), EGFR protein expression was defined as complete or incomplete membranous staining of tumor cells (>1%), ranging in intensity from 1+ to 3+. ERBB2 protein expression was scored adhering to the manufacturer’s guidelines (HercepTest, FDA approved), defined as 0, no staining observed or weak membrane staining in <10% of tumor cells; 1+, faint membrane staining in >10% of the tumor cells; 2+, weak to moderate membrane staining in >10% of the tumor cells; 3+, strong complete membrane staining in >10% of the tumor cells. Only 2+ and 3+ membranous staining was considered positive. Using a separate criteria, cytoplasmic ERBB2 was also considered, which we refer to herein as cyt-ERBB2.
ERBB4 was scored as the percentage and intensity of immunopositive tumor cells exhibiting cytoplasmic staining divided by the total number evaluated.
Fluorescent In Situ Hybridization
Four-micrometer sections were cut from corresponding FFPE donor blocks for representative ERBB2 immunohistochemical expression cases included in the TMA. ERBB2 gene amplification was detected using the Path-Vysion HER-2 DNA probe kit adhering to the manufacturer’s instructions (Abbott Laboratories, Berkshire, UK). HER-2/Spectrum Orange probe hybridizes to the ERBB2 gene locus (17q11.2–q12), and chromosome enumeration probe-17 (CEP-17)/Spectrum Green hybridizes to the centromeric region of chromosome 17. Sections were counterstained with DAPI, coverslipped, and stored in the dark at 4°C prior to analysis. Fluorescence in situ hybridization (FISH) sections were examined with a Leica DMRB multifilter fluorescence microscope (Leica, Milton Keynes, UK; ×100 oil immersion objective). Sections were analyzed by two investigators, including an independent pathology service (Medical Solutions, Not-tinghamshire, UK). NIS-Elements (Melville, NY) software (version 2.30) was used for merging of the images. ERBB2 and CEP-17 signals were counted for a minimum of 40 tumor cells in each sample. The ratio score was then calculated by dividing the mean score for the ERBB2 signal by the CEP-17 signal. A signal ratio score of >2 was considered gene amplification.
Telomerase Assay
Telomerase activity was analyzed using the telomere repeat amplification protocol (TRAP) assay as described previously,41 using the TRAPeze telomerase detection kit (Millipore, Hertfordshire, UK). Briefly, 0.1 μg protein from homogenized tumor tissue that had been flash-frozen was used. Telomerase extension was achieved by heating samples at 30°C for 30 min. Amplification of the telomerase product was subsequently carried out using a thermal cycle of 94°C for 30 min, followed by 59°C for 30 min, repeated 30 times. Products were resolved on a 10% polyacrylamide gel, detected using SyBR Green 1 and visualized with a Fujifilm FLA-2000 phosphoimager (Amersham Biosciences, Buckinghamshire, UK).
Telomere Measurement
Mean telomere length was determined using the telomere restriction fragment (TRF) Southern blot technique (TeloTAGGG) according to the manufacturer’s guidelines (Roche, Burgess Hill, UK). Briefly, 3 μg genomic DNA was digested with a Hinf1/Rsa1 frequent cutter mix, separated on 0.8% agarose, and transferred to a nylon membrane by Southern blotting. Pulse-field gel electrophoresis was conducted at 60 V for 16 h using 1% agarose. Fragments were hybridized to a digoxigenin (DIG)-labeled telomere probe, incubated with a DIG-specific antibody, and visualized via chemiluminescent signal. The average TRF length was determined by comparing signals relative to a molecular weight standard, using ImageQuant version 5.1 software (GE Healthcare, Buckinghamshire, UK).
Statistics
SPSS version 14 (SPSS Inc., Chicago, IL, USA) was used to perform statistical analyses. Correlations of two factors were determined by the Pearson correlation test. Comparison of mean values was determined for significance by an independent-sample t-test with 95% confidence intervals. Fisher’s exact test was used to explore associations in two-way frequency tables. Comparisons of the reliability of two methods detecting the same variable were calculated using the kappa statistic (κ). Overall survival (OS) was calculated from the date of initial diagnosis to the date of death from any cause or date of last follow-up if still alive (censored). Event-free survival (EFS) was calculated from the date of initial diagnosis to the time of first event. Kaplan-Meier survival curves with significance levels determined by the log-rank test were constructed in a univariate analysis. Multiple prognostic factors were analyzed by the Cox proportional hazard regression model to determine OS comparisons with 95% confidence intervals. p-Values < 0.05 were deemed statistically significant.
Results
Patient Demographics
Following histological review (M.-A.B., D.E.) of the initial cohort of 114 cases, 10 had incorrect diagnosis and 7 had tissue processing errors beyond our control. A total of 97 intracranial tumors from 74 patients were included in the present study. Mean and median age at diagnosis was 5.4 and 3.8 years, respectively (range, 8 months to 14.9 years). Patient clinical features are summarized in Table 1.
Table 1.
Univariate analysis of clinical and biological factors on 74 primary intracranial ependymomas
| Patients
|
5-Year Overall Survival
|
5-Year Event-Free Survival
|
||||||
|---|---|---|---|---|---|---|---|---|
| Factor | n | % | % | SE | p-Value (log rank) | % | SE | p-Value (log rank) |
| Gender | ||||||||
| Male | 41 | 55 | 54 | 8 | 0.729 | 35 | 8 | 0.840 |
| Female | 33 | 45 | 58 | 9 | 40 | 9 | ||
| Age <3 years at diagnosis | ||||||||
| Yes | 34 | 46 | 53 | 9 | 0.597 | 30 | 8 | 0.122 |
| No | 40 | 54 | 58 | 8 | 44 | 8 | ||
| Histology | ||||||||
| II | 49 | 66 | 52 | 7 | 0.513 | 38 | 7 | 0.759 |
| III | 25 | 34 | 65 | 10 | 36 | 10 | ||
| Tumor location | ||||||||
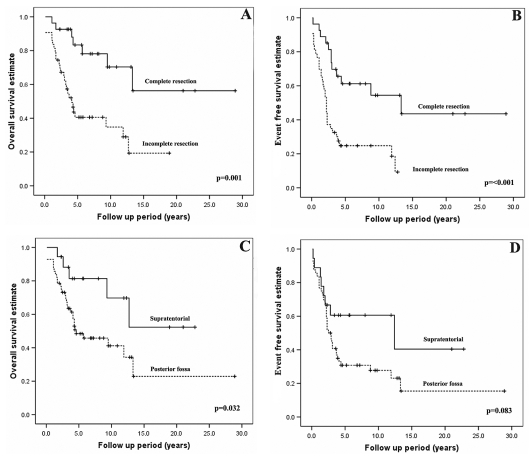
| ST | 18 | 24 | 82 | 9 | 0.032 | 61 | 12 | 0.083 |
| PF | 56 | 76 | 48 | 7 | 30 | 6 | ||
| Surgical resection | ||||||||
| C | 26 | 38 | 83 | 8 | 0.001 | 61 | 10 | <0.001 |
| IC | 43 | 62 | 41 | 8 | 25 | 7 | ||
| Radiotherapy (overall)a | ||||||||
| Yes | 54 | 75 | 61 | 7 | 0.166 | n/ab | n/ab | n/ab |
| No | 18 | 25 | 43 | 12 | n/a | n/a | ||
| Primary radiotherapy | ||||||||
| Yes | 29 | 53 | 55 | 10 | 0.47 | 38 | 10 | 0.039 |
| No | 26 | 47 | 48 | 10 | 26 | 9 | ||
| Chemotherapy | ||||||||
| Yes | 47 | 65 | 62 | 8 | 0.188 | 34 | 7 | 0.656 |
| No | 25 | 35 | 45 | 10 | 42 | 10 | ||
| Ki-67 LI | ||||||||
| Low | 35 | 52 | 66 | 8 | 38 | 9 | ||
| Intermediate | 22 | 33 | 60 | 11 | 0.132c | 43 | 11 | 0.792c |
| High | 10 | 15 | 36 | 7 | 30 | 14 | ||
| Survivin LI | ||||||||
| Low | 51 | 79 | 66 | 7 | 44 | 7 | ||
| Intermediate | 11 | 17 | 21 | 15 | 0.110c | 12 | 11 | 0.360c |
| High | 3 | 4 | 67 | 27 | 67 | 27 | ||
| EGFR | ||||||||
| Negative | 62 | 98 | n/a | n/a | n/a | n/a | ||
| Positive | 1 | 2 | n/a | n/a | ||||
| EGFR2 | ||||||||
| Negative | 64 | 97 | n/a | n/a | n/a | n/a | ||
| Positive | 2 | 3 | n/a | n/a | ||||
| ERBB4 | ||||||||
| Negative/weak | 44 | 66 | 63 | 8 | 0.380 | 39 | 8 | 0.458 |
| Moderate/strong | 23 | 34 | 53 | 11 | 37 | 10 | ||
| Nucleolin | ||||||||
| Low expression | 12 | 19 | 90 | 9 | 0.047 | 74 | 13 | 0.007 |
| High expression | 52 | 81 | 53 | 7 | 31 | 7 | ||
| 44F12 | ||||||||
| Negative | 13 | 20 | 92 | 7 | 0.015 | 77 | 12 | 0.016 |
| Positive | 52 | 80 | 53 | 8 | 30 | 7 | ||
Abbreviations: II, classic ependymoma; III, anaplastic ependymoma; ST, supratentorial; PF, posterior fossa; C, complete resection; IC, incomplete resection; LI, labeling index.
Patients who received primary or delayed radiotherapy at any point during the course of disease.
Analysis for event-free survival was not carried out because not all patients were irradiated prior to first event (primary radiotherapy).
Low versus intermediate/high expression.
Nine of 26 patients (35%) who had complete resection relapsed with recurrent disease. In total, 35 patients (47%) relapsed (mean and median time to first event, 2.5 and 2.2 years, respectively). Twenty-three recurrent tumors from 19 patients were included in the study. Thirty-eight patients (51%) were alive, with mean and median follow-up times of 8.3 and 6.3 years, respectively (range, 2.0–28.9 years), and 36 patients (49%) had died, with mean and median survival times of 3.7 and 2.9 years, respectively (range, 1.2 months to 13.3 years). Five-year OS and EFS were 56% ± 6% and 37% ± 6%, respectively.
Incomplete resection of the primary tumor was significantly associated with a worse OS and EFS (p = 0.001 and p ⩽ 0.001, respectively; Fig. 1A,B). Posterior fossa location was significantly associated with a worse OS and showed a tendency toward a worse EFS (p = 0.032 and p = 0.083, respectively; Fig. 1C,D).
Fig. 1.
(A and B) Kaplan-Meier survival analysis of clinical factors for overall survival (OS) and event-free survival (EFS) for resection status. Incomplete resection (indicated by the dashed line) has a significantly less favorable outcome in terms of OS and EFS compared to complete resection (p = 0.0001 and p ⩽ 0.001, respectively). (C and D) Posterior fossa tumors (indicated by the dashed line) have a significantly less favorable outcome in terms of OS (C; p = 0.032) and a trend toward a poorer EFS (D; p = 0.083) compared with supratentorial tumors. The distribution of tumor location was not significantly associated with resection status (p = 0.569) by Fisher’s exact test.
Adjuvant therapy was administered to 65 of 72 patients. Eighteen patients received radiotherapy alone, 11 had chemotherapy alone, and 36 had a combination of radiotherapy and chemotherapy. Treatment information was unavailable for two patients. Adjuvant chemotherapy (n = 47) did not confer a survival advantage in terms of OS and EFS (p = 0.188 and p = 0.656, respectively). Five-year OS for irradiated patients (n = 54) was 61% ± 7%, which although not significant (p = 0.166), did show a trend toward a more favorable OS compared to nonirradiated patients (n = 18; 5-year OS, 43% ± 12%). Comprehensive radiotherapy data were available for 55 patients. Patients who were electively irradiated as part of primary treatment (n = 29) versus those who did not receive radiotherapy or had delayed radiotherapy (i.e., following the first event; n = 26) had a significantly more favorable EFS (p = 0.039) but not OS (p = 0.47). Other clinical factors, including gender, age <3 years at diagnosis, and histology, were not associated with a worse outcome.
Overall, 10% (range, 9%–15%) of cases were excluded from immunohistochemical analysis due to core dropout, nonviable tumor, or <50% intact cores following processing. Expression levels of the biological factors are summarized in Table 1.
Ki-67
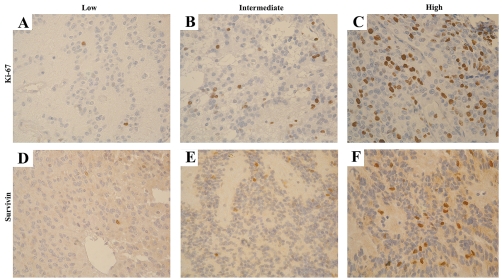
Sixty-seven primary tumors (100%) demonstrated Ki-67 staining, with mean and median LIs of 2.9% and 1.0%, respectively (range, <1% to 27%). Tumors were categorized into low (⩽1%), intermediate (2%–4%), or high (⩾5%) Ki-67 expression levels. Thirty-five (52%) tumors exhibited low Ki-67 expression, 22 (33%) intermediate, and 10 (15%) high (Fig. 2A–C). Univariate analysis did not show Ki-67 levels to be a contributor to worse OS or EFS when considering low versus high, low versus intermediate/high (OS, p = 0.132; EFS, p = 0.792), and low/intermediate versus high (Table 1). Stratification of Ki-67 by tumor location also showed no prognostic significance for posterior fossa tumors (OS, p = 0.184; EFS, p = 0.547). Data from 82 primary and recurrent tumors showed higher Ki-67 LI values correlated with grade III tumors (p = 0.02). The mean Ki-67 expression level for grade II and III tumors was 2.2% ± 0.6% and 4.3% ± 0.8%, respectively.
Fig. 2.
Positive Ki-67 (A–C) and survivin (D–F) immunostaining was categorized into three expression groups based on the labeling index, as indicated by positive immunostained nuclei: <1%, low (A, D); 2–4%, intermediate (B, E); >5%, high (C, F). Original magnification, ×10.
Survivin
Sixty-five primary tumors (100%) demonstrated survivin expression, with mean and median LIs of 1.1% and 0.5%, respectively (range, <1% to 6%). Tumors were categorized into low (⩽1%), intermediate (2%–4%), and high (⩾5%) survivin expression levels (Fig. 2D–F). Fifty-one tumors (79%) exhibited low expression, 11 intermediate (17%), and 3 high (4%). A strong correlation was observed between survivin and Ki-67 LI values (R2 = 0.753, p ⩽ 0.001).
Higher survivin LI also correlated with grade III tumors (p ⩽ 0.001) but was of no prognostic value when considering low versus high, low versus intermediate/high (OS, p = 0.110; EFS, p = 0.360), and low/intermediate versus high (Table 1). Stratification of survivin by tumor location also showed no prognostic significance for posterior fossa tumors (OS, p = 0.363; EFS, p = 0.390).
RTK-I (ERBB) Family
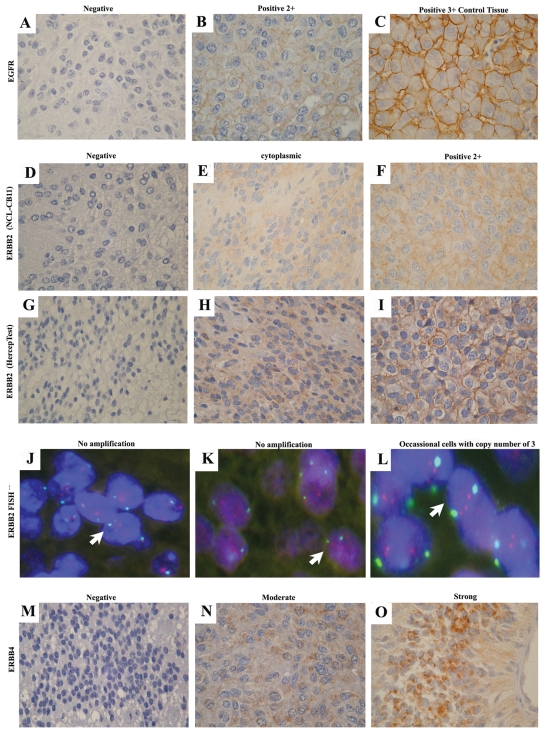
Sixty-two tumors (98%) were negative for EGFR protein expression, and one tumor (2%) was positive (2+ intensity) (Fig. 3A,B). Because EGFR was detected in only one case, further analysis was not carried out.
Fig. 3.
RTK-I family expression. (A–C) epidermal growth factor receptor–negative immunostaining was identified in 98% of tumors (A), 2+ intensity positive staining was detected in one tumor (B), and 3+ intensity was achieved in hepatocyte control tissue (C). (D–L) Tumor negative by immunostaining and with no gene amplification (D, G, and arrow in J; ratio score, 1.1), tumor exhibiting cytoplasmic staining and with no gene amplification (E, H, and arrow in K; ratio score, 1.2), and tumor demonstrating 2+ intensity by immunohistochemistry and occasional cells with ERBB2 copy number of 3, considered negative overall (F, I, and arrow in L; ratio score, 1.33). In J–L, red indicates ERBB2 probe (locus 17q11.2–q12), and green, CEP-17 probe. ERBB2:CEP-17, ratio score >2 considered amplification. (M–O) ERBB4 was absent in 66% of tumors (M), with expression detected in 34% of tumors with either moderate (N) or strong (O) expression.
Three different expression groups were observed for ERBB2: (1) negative staining, (2) cytoplasmic staining including either an epithelioid “dot-like” pattern or diffuse granular staining, and (3) membranous staining of 2+ intensity (Fig. 3D–F). Analysis of ERBB2, when adhering to the FDA scoring criteria, identified 2 positive (2+ membranous staining; 3%) and 64 (97%) negative tumors. Forty-seven corresponding primary tumors screened for ERBB2 with the Dako HercepTest gave good concordance with the results when adhering to the FDA scoring criteria, confirming the two positive cases. When considering cyt-ERBB2 staining, 22 tumors (34%) exhibited immunopositivity with the ERBB2 antibody. Comparatively, with the HercepTest, of 15 representative positive cyt-ERBB2 cases demonstrated with the ERBB2 antibody, 10 (67%) showed similar staining patterns, and 5 (33%) were negative (Fig. 3G–I). Reliability between the two techniques was good (κ = 0.63, p ⩽ 0.001). The results with the ERBB2 antibody were confirmed by independent analysis (data not shown).
Gene copy number in whole-tissue sections from seven representative cases from the ERBB2 immunohistochemistry was determined by FISH. Despite a small number of tumor cells with a gene copy number of 3 for ERBB2, the two tumors that demonstrated 2+ membranous staining were considered negative by FISH (ratio scores, 1.28 and 1.33). No gene amplification was observed in two ERBB2-negative tumors or in the three cases that exhibited cyt-ERBB2 expression (ratio score range, 1.1–1.37; Fig. 3J–L).
Univariate analysis on the FDA-scored ERBB2 expression data was not possible in this cohort due to the small number of positive cases (n = 2); however, additional consideration with cyt-ERBB2, demonstrated in 22 cases (34%), did not contribute to a worse outcome compared to negative tumors (OS, p = 0.179; EFS, p = 0.246).
Three groups of cytoplasmic staining were observed for ERBB4 expression, categorized into negative, moderate, and strong. Forty-four (66%) tumors were negative for ERBB4 expression, and 23 (34%) exhibited moderate/strong expression (Fig. 3M–O). Kaplan-Meier survival analysis showed that moderate/strong ERBB4 expression was not a predictor of outcome when compared to negative expression (OS, p = 0.380; EFS, p = 0.458; Table 1). When considering coexpression of cyt-ERBB2 with ERBB4 and/or cases with high Ki-67 levels, no prognostic significance was found.
44F12
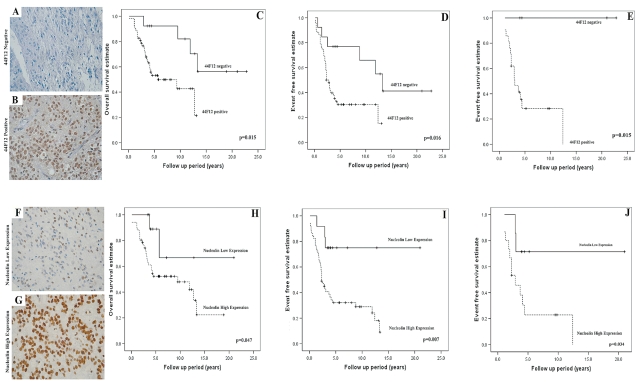
From 65 primary tumors, 52 (80%) were positive for 44F12 and 13 (20%) were negative. Initially, Fisher exact tests determined no association of 44F12 expression with clinical factors, including tumor location (p = 1.0), histology (p = 0.51), resection status (p = 0.53), and age<3 years at diagnosis (p = 0.341). Kaplan-Meier survival analysis showed that 44F12-positive tumors contributed to both a worse OS and a worse EFS (p = 0.015 and p = 0.016, respectively; Fig. 4A–D, Table 1). Five-year OS and EFS were 92% ± 7% and 77% ± 12%, respectively, for 44F12-negative tumors and 53% ± 8% and 30% ± 7% for 44F12-positive tumors. Stratification of radiotherapy status by 44F12 status was also analyzed for 51 patients. Of the 25 of 51 patients (49%) who received primary radiotherapy, the 5-year EFS for 44F12-positive tumors (n = 21) and 44F12-negative tumors (n = 4) was 28% ± 11% and 100% ± 0%, respectively (p = 0.015; Fig. 4E). All of the patients who received primary radiotherapy with 44F12-negative tumors are alive and event free, with mean and median follow-up times of 13 ± 5.1 and 12.7 years, respectively.
Fig. 4.
Immunohistochemical detection of 44F12 (A–E) and nucleolin (F–J). (A and B) Tumors were grouped as 44F12 negative or positive by immunostaining. (C and D) Kaplan-Meier survival analysis identified a poorer overall survival (OS; p = 0.015) and event-free survival (EFS; p = 0.016) for 44F12 positive tumors, as indicated by the dashed line. (E) Patients who received primary radiotherapy were stratified by 44F12 status. A poorer EFS for 44F12-positive tumors was also observed (p = 0.015), as indicated by the dashed line. The distribution of 44F12-positive and -negative tumors was not significantly associated with tumor location (p = 0.743) or resection status (p = 0.528) by Fisher’s exact test. (F and G) Tumors were grouped into low or high nucleolin expression by immunostaining. (H and I) Kaplan-Meier survival analysis identified a poorer OS (p = 0.047) and EFS (p = 0.007) for tumors with high nucleolin expression, as indicated by the dashed line. J. Patients who received primary radiotherapy were stratified by nucleolin status. A poorer EFS for tumors with high nucleolin expression was also observed (p = 0.034), as indicated by the dashed line. The distribution of high-nucleolin and low-nucleolin tumors was not significantly associated with tumor location (p = 1.000) or resection status (p = 0.745) by Fisher’s exact test.
Nucleolin
Nucleolin protein expression was evident in 64 (100%) primary tumors with a mean LI of 68% (range, 3.0%–92%). Two distinct groups were observed, with 50% LI used as the cutoff to distinguish between high and low expression. Fifty-two cases (81%) demonstrated high expression (mean LI, 75% ± 1.5%), and 12 cases (19%) demonstrated low expression (mean LI, 35% ± 3.6%). Initially, Fisher exact tests determined no association of nucleolin expression with clinical factors, including tumor location (p = 1.0), histology (p = 0.51), resection status (p = 0.53), and age at diagnosis (p = 0.34). Kaplan-Meier survival analysis showed that high nucleolin expression contributed to a worse OS and EFS (p = 0.047 and p = 0.007, respectively; Fig. 4F–I, Table 1). Five-year EFS for high and low nucleolin expression was 31% ± 7% and 74% ± 13%, respectively. Furthermore, 10-year EFS for high and low nucleolin expression was 28% ± 7% and 74% ± 13%, respectively. Of those patients that were 44F12 negative and exhibited low expression of nucleolin (n = 5), all were alive and event-free, with a mean follow-up time of 9.2 years (range, 3.6–21.0 years). We next sought to stratify resection and radiotherapy status by nucleolin expression. Paired data of nucleolin expression and resection status were available for 59 primary tumors. For completely resected primary tumors demonstrating low nucleolin expression (n = 5), the majority of patients (80%) were event-free with mean and median follow-up times of 8.9 ± 4.1 and 5.5 years, respectively (range, 3.6–21.0 years; 5-year EFS, 75% ± 22%). Ten of the 19 completely resected cases with high nucleolin expression (47%) relapsed, with a mean and median times to progression of 4.0 ± 1.3 and 2.7 years, respectively (range, 0.2–13.3 years; 5-year EFS, 56% ± 12%). For incompletely resected primary tumors demonstrating high nucleolin expression (n = 29), the majority of these cases (90%) relapsed, with mean and median times to progression of 2.3 ± 0.6 and 1.7 years, respectively (range, 0.1–12.4 years; 5-year EFS, 17% ± 7%). For incompletely resected primary tumors demonstrating low nucleolin expression (n = 6), the majority of cases (83%) were event-free, with mean and median follow-up times of 6.0 ± 1.7 and 4.4 years, respectively (range, 3.4–12.8 years; 5-year EFS, 83% ± 15%).
Patients who received primary radiotherapy and who had tumors demonstrating high or low nucleolin expression (n = 22) had a 5-year EFS of 23% ± 11% and 70% ± 18%, respectively (p = 0.034; Fig. 4J). Five of seven (71%) irradiated patients demonstrating low nucleolin expression were event-free, with mean and median follow-up times of 7.7 ± 3.3 and 4.4 years, respectively (range, 3.9–21.0 years). By contrast, 12 of 15 (80%) irradiated patients with high nucleolin expression relapsed, with mean and median times to progression of 3.2 ± 0.89 and 2.3 years, respectively (range, 1.1–12.4 years). Additionally, 16 of 22 (73%) nonirradiated patients demonstrating high nucleolin expression relapsed, with mean and median times to progression of 1.2 ± 0.27 and 0.9 years, respectively (range, 0.1–3.1 years). Analysis of nonirradiated patients with low nucleolin expression could not be carried out because the numbers were too small.
Multivariate analysis of clinical and biological covariates (histology, tumor location, resection status, primary radiotherapy, age <3 years at diagnosis, Ki-67 LI, survivin LI, and nucleolin expression) identified incomplete resection (p = 0.001) and high nucleolin expression (p = 0.008) as the only factors to independently predict a worse EFS (Table 2).
Table 2.
Cox regression multivariate analysis of clinical and biological factors
| Event-Free Survival
|
|||
|---|---|---|---|
| Factor | HR | 95% CI | p-Value |
| Histology (II vs. III) | 1.237 | 0.449–3.406 | 0.680 |
| Tumor location (ST vs. PF) | 0.392 | 0.120–1.280 | 0.222 |
| Surgical resection (complete vs. incomplete) | 4.736 | 1.819–12.330 | 0.001 |
| Age at diagnosis (<3 vs. >3 years) | 0.430 | 0.134–1.382 | 0.156 |
| Ki-67 LI (low vs. intermediate/high) | 0.792 | 0.227–2.769 | 0.715 |
| Survivin LI (low vs. intermediate/high) | 0.722 | 0.189–2.753 | 0.633 |
| Nucleolin LI (low vs. high) | 6.252 | 1.614–24.210 | 0.008 |
| Radiationa (yes vs. no) | 0.511 | 0.206–1.267 | 0.148 |
Abbreviations: HR, hazard ratio; 95% CI, 95% confidence interval; II, classic ependymoma; III, anaplastic ependymoma; ST, Supratentorial; PF, posterior fossa; LI, labeling index.
Patient received radiotherapy prior to first event (primary radiotherapy).
Recurrent Ependymoma
Paired data for 17 primary tumors and their first recurrence revealed that mean Ki-67 in primary tumors was 3.3% ± 1.5% compared to 7.0% ± 2.8% in recurrent tumors. A similar pattern was observed for survivin LI, with a mean of 1.0% ± 0.2% for the primary tumors compared to 5.1% ± 3% in recurrent tumors.
Of the 18 patients with 44F12 data on primary and subsequent relapse tumors, only two primary tumors (11%) were negative, and eventually both patients progressed to positive status, showing that ultimately 100% of recurrent tumors were 44F12 positive.
As with primary patients, all recurrent samples (100%) were positive for nucleolin. Among 12 patients with paired data on primary and relapse tumors, two (17%) had low expression in the primary and first recurrence, but in both cases the expression levels subsequently progressed to high, showing again that, ultimately, 100% of recurrent samples expressed high levels of nucleolin.
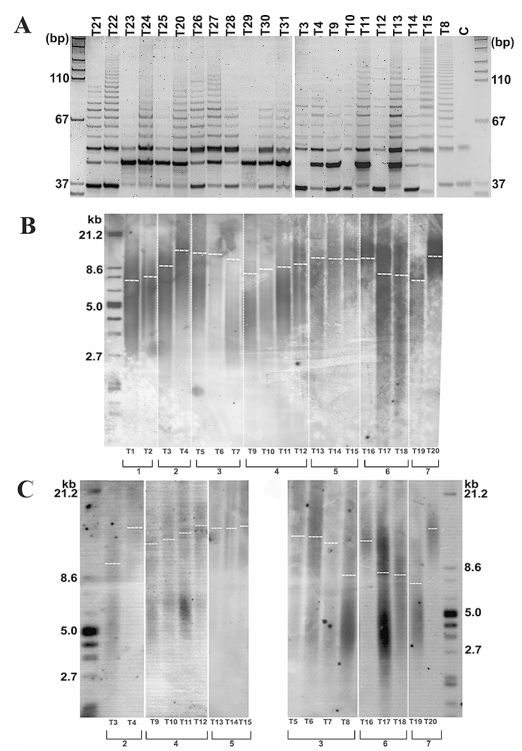
Telomerase Activity
Telomerase repeat addition processivity assay (TRAP) was performed on 22 representative tissue samples. Positive TRAP products were evident in 19 tumors (86%), indicative of a high level of telomerase repeat addition processivity (Fig. 5A). Telomerase activity was evident in all recurrent tumors analyzed by TRAP and in 11 of 14 primary tumors. There was a good agreement between telomerase activity and 44F12 detection (κ = 0.59, p = 0.02). Of six tumors negative for 44F12 detection, three (50%) were positive for telomerase activity. The reverse was never observed; of all telomerase-negative tumors, none were 44F12 positive.
Fig. 5.
Telomerase activity and telomere length in ependymoma tissue from a pediatric cohort. (A) Telomere repeat amplification protocol assay indicates variable levels of telomerase activity in pediatric ependymoma patients; 19 of 22 (86%) patients have moderate to high levels of telomerase activity, as indicated by the length and intensity of the telomere repeat amplification protocol product ladder, whereas 3 of 22 (14%) patients show an absence of any detectable telomerase activity under the conditions employed. We used 0.1 μg total protein lysate for each reaction. C, CHAPS-buffer–only control. (B) Telomere length in seven recurrent cases, encompassing 19 independent tissue samples, was determined by the TeloTAGGG assay, and mean telomere restriction fragment length was deduced by comparison to a known molecular standard. Telomere length ranged from 7.2 to 16.7 kb across all samples. Telomere lengthening is evident in four of seven cases (57%; patients 1, 2, 4, and 7), with a rapid telomere lengthening rate observed in patients 2 and 7 (see Table 3). Telomere maintenance was observed in patient 5, and telomere shortening was observed in patients 3 and 6. (C) To further validate this phenomenon of telomere lengthening, digested DNA was separated using pulsed-field gel electrophoresis (PFGE). The pattern of telomere dynamics observed using PFGE was consistent, with considerable telomere lengthening confirmed in patients 2 and 7.
Telomere Length
To determine telomere length alterations, mean telomere restriction fragment (TRF) length was calculated in seven patients with recurrent tumors (data set consisting of 6 primary tumors and 15 recurrences). Mean telomere length ranged from 7.3 to 16.7 kb, with telomere maintenance observed in five of seven patients (71%). Of these five cases, four showed telomere lengthening in relapsed tumors compared to the primary tumor (Table 3, Fig. 5C, D). A moderate increase in mean telomere length was observed in tumors T1–T2 and T9–T12 (7.3–7.6 kb and 9.2–10.3 kb, respectively). In contrast, substantial telomere lengthening was evident in tumors T3–T4 and T19–T20 (9.5–16.7 kb and 7.2–15.9 kb, respectively). Because time to recurrence was 2.9 and 2.8 years, respectively, a rapid telomere lengthening rate is inferred for these latter cases.
Table 3.
Summary of clinical factors and telomere status of recurrent tumors
| Patient | Tumor ID | Histology | Primary/Recurrence | Time to Recurrence (Years) | Nucleolin Expression | 44F12 Status | Telomerase Activity | Telomere Length (kb) | Telomere Status |
|---|---|---|---|---|---|---|---|---|---|
| 1 | T1 | II | P | 2.3 | High | + | n/a | 7.3 | L |
| T2 | III | R1 | High | + | n/a | 7.6 | |||
| 2 | T3 | II | P | 2.9 | High | + | + | 9.5 | L |
| T4 | III | R1 | n/a | + | + | 16.7 | |||
| 3 | T5 | III | P | 2.2 | High | + | n/a | 15.3 | S |
| n/a | III | R1 | 4.4 | n/a | n/a | n/a | n/a | ||
| T6 | III | R2 | 5.2 | n/a | n/a | n/a | 15.2 | ||
| T7 | III | R3 | High | + | n/a | 14.5 | |||
| n/a | III | R4 | 6.6 | n/a | n/a | n/a | n/a | ||
| T8 | III | R5 | High | + | + | 8.4 | |||
| 4 | T9 | II | P | 2.9 | Low | + | + | 9.2 | L |
| T10 | II | R1 | 4.0 | High | + | + | 9.6 | ||
| T11 | III | R2 | 4.6 | High | + | 1 | 9.8 | ||
| T12 | III | R3 | High | + | + | 10.3 | |||
| 5 | T13 | II | P | 2.3 | High | − | + | 10.2 | M |
| T14 | II | R1 | 3.1 | High | + | + | 10.2 | ||
| T15 | II | R3 | High | + | + | 10.4 | |||
| 6 | n/a | II | P | 2.0 | n/a | n/a | n/a | n/a | S |
| T16 | II | R1 | 7.9 | High | + | n/a | 14.8 | ||
| T17 | II | R2 | 8.9 | High | + | n/a | 8.2 | ||
| T18 | III | R3 | High | + | n/a | 8.1 | |||
| 7 | T19 | III | P | 2.8 | High | + | n/a | 7.2 | L |
| T20 | III | R1 | High | + | + | 15.9 |
Abbreviations: II, classic ependymoma; P, primary tumor; n/a, data not available; L, lengthening; III, anaplastic ependymoma; R, recurrent tumor; S, shortening; M, maintenance.
However, telomere shortening occurred in two of seven recurrent cases (29%); in tumors T5–T8, telomeres shortened from 15.3 to 8.4 kb, and in tumors T16–T19, telomeres shortened from 14.8 to 8.1 kb (Table 3, Fig. 5B, C). Of note, T16–T19 represent tumor tissue from the oldest patient in this cohort, with final recurrence to date occurring at age 22.
We observe variability with respect to the status of the telomere and present evidence for telomere maintenance in the majority of pediatric ependymoma recurrent cases (71%), with telomere shortening evident in a minority.
Discussion
We have identified nucleolin as the single most important biological predictor of outcome in childhood intracranial ependymoma. In our cohort, univariate and multivariate analysis identified tumors with low nucleolin expression as being associated with a more favorable EFS (p = 0.008) compared to tumors with high nucleolin expression, with a 5-year event-free survival (EFS) of 74% ± 13% versus 31% ± 7%. Furthermore, five of six (83%) incompletely resected primary tumors demonstrating low nucleolin expression had a long event-free interval (mean follow-up time, 6.0 ± 1.7 years). By contrast, 26 of 29 (90%) incompletely resected tumors demonstrating high nucleolin expression had brief EFS times (mean, 2.3 ± 0.6 years). Univariate analysis of primary radiotherapy administered to patients was significantly associated with a more favorable EFS (p = 0.039), although this was not a significant factor by multivariate analysis. When stratifying patients who received primary radiotherapy by nucleolin status, we observed a significantly more favorable outcome (p = 0.034) for patients with tumors demonstrating low nucleolin expression compared to those with high nucleolin expression (most of whom relapsed), further highlighting the biological significance of nucleolin as a prognostic marker.
Nucleolin is a major retinoblastoma susceptibility (Rb) gene–related nucleolar phosphoprotein reported to interact with telomerase through protein–protein and protein–RNA interactions and to function as an hTERT chaperone between nucleolus and nucleoplasm.42–44 Others have reported the binding of nucleolin to human telomeric DNA sequence.45,46 Nucleolin expression has additionally been reported as a proliferation marker in human tumor cells47 and predicts poor prognosis in many cancers.48 Furthermore, inhibition of nucleolin has recently shown to result in cell growth arrest and a concomitant increase in apoptosis,49 providing a theoretical rationale for the involvement of nucleolin in ependymoma. Our findings are also consistent with a recent study that found hTERT expression to be the strongest predictor of outcome in pediatric intracranial ependymoma.35 The mouse monoclonal antibody (hTERT clone 44F12) that had been used to determine hTERT protein through immunohistochemistry has more recently been reevaluated and shown to detect nucleolin or a nucleolin-like protein,36 highlighting the current difficulty in hTERT immunodetection. Because nucleolin is closely associated with hTERT, it is not surprising that results using the 44F12 antibody were not questioned. Here we investigated both hTERT antibody clone 44F12 and an independent antibody raised against nucleolin.
The heterogeneous clinical behavior of pediatric ependymoma stems from variation in the proliferative potential of each tumor at the cellular level. One marker of this variable proliferation may be telomere length. We therefore investigated this possibility with respect to tumors that had recurred and explored the mechanistic basis. Dysregulated, unlimited proliferation and bypass of senescence are acquired capabilities of cancerous cells, which in part require the establishment of a telomere maintenance mechanism. Telomerase-mediated telomere maintenance is evident in virtually all types of malignant cells, and ~90% of tumors show evidence of up-regulated telomerase. 50,51 Indeed, several studies have demonstrated telomere maintenance as a valuable prognostic marker in a variety of CNS tumors.35,52–57 However, presently, no data identify telomere maintenance mechanisms in pediatric ependymoma.
In this study we measured telomere length for 21 primary and recurrent tumors from seven patients, allowing observation of telomere alterations during tumor progression. Mean telomere length ranged from 7.3 to 16.7 kb, with telomere maintenance observed in five of seven patients, four of which showed telomere lengthening in relapsed tumors compared to the primary tumor. Our results implicate telomerase-mediated telomere maintenance as a key mechanism facilitating tumor progression in pediatric ependymoma. This would be expected because all recurrent tumors analyzed were telomerase positive. Surprisingly, most cases showed telomere lengthening, and in two patients this occurred rapidly. Only two cases showed telomere shortening compared to the primary tumor. It is unclear whether elongation of telomeres directly affects tumor progression in this setting, or whether longer average telomere length in recurrent ependymoma is an epiphenomenon related to sustained telomerase activity from early development. It is conceivable that in pediatric tumors, hTERT is not transcriptionally repressed early in human development (in utero), in contrast to activation of hTERT in adult tumors, which occurs through a process of derepression.58,59 An intriguing hypothesis has emerged recently, which suggests that rather than telomerase reactivation, enzymatic activity may increase in later stages of carcinogenesis due to increased expression of hTERT or efficiency of telomerase components.60 Telomere lengthening may conceivably be the end result of this process. Our findings reflect one aspect of the genetic and molecular variability underlying unpredictable clinical behavior of childhood ependymoma.
Evaluations of a number of other biological factors, previously reported to be of prognostic value in other cohorts of ependymoma, were also analyzed in this study. Our study, albeit retrospective, has shown these to be of no prognostic value in this defined cohort.
Ki-67 protein is expressed in proliferating cells but is absent in G0 nondividing cells61 and has previously been reported as a prognostic factor in pediatric ependymoma.28,29 We have shown that higher Ki-67 LI correlates with WHO grade III, in concordance with a previous study in a cohort of mixed adult and pediatric ependymoma, with grade predicting outcome.62 This supports evidence that higher proliferative activity is one of many features associated with anaplasia, a feature of grade III ependymoma.15 However, neither grade nor Ki-67 LI predicted outcome, highlighting that the prognostic significance of grade in pediatric ependymoma remains controversial.15
Survivin is up-regulated in proliferating tumor cells, with subcellular localization to the nucleus and cytoplasm.63 Nuclear survivin acts as a mitotic regulator, whereas cytoplasmic survivin confers a cytoprotective role.30,64 One study reported low levels of nuclear survivin to be a marker of more aggressive disease and/or tumor grade in pediatric ependymomas and choroid plexus tumors,31 whereas another study found that high expression correlates to tumor grade and Ki-67 LI when analyzing both adult and pediatric intracranial ependymomas.32 Conflicting results may be reflective of low sample size and the inclusion of myxopapillary ependymomas (WHO grade I) and choroid plexus tumors in other cohorts. We found that high survivin expression correlates to WHO grade III and Ki-67 LI; however, in contrast, it did not predict outcome, consistent with tumor grade in the present cohort.
A number of studies have looked at the RTK-I family in ependymoma. Coexpression of ERBB2/ERBB4 in association with Ki-67 LI and degree of surgical resection was found to be associated with more aggressive disease behavior.28 In another study, increased EGFR protein expression in intracranial ependymomas correlated with a poor prognosis in a univariate analysis, whereas multivariate analysis revealed that EGFR over-expression was the only significant factor in grade II tumors.65 These findings, however, were from a mixed child and adult population.
In our cohort, using the FDA scoring criteria, EGFR and ERBB2 expression was detected in three tumors, whereas ERBB4 was detected in 23 (34%). Using a broader scoring criteria, including cytoplasmic staining, ERBB2 immunopositivity was observed in 22 tumors (34%). Even using “more relaxed” criteria for ERBB2 expression and the same methodology as Gilbertson,66 irrespective of coexpression with ERBB4, we were not able to correlate the RTK-I family with prognosis or with histological grade. Recognizing the limitations of immunohistochemistry in retrospective FFPE studies, immunohistochemical evaluation of ERBB2 in ependymoma is unlikely to provide robust prognostic methodology. Alternative methods to detect ERBB2 protein expression in fresh material14 may need to be considered. If we are able to inhibit tumor growth successfully by utilizing RTK-I inhibitors within pediatric CNS tumors, it is likely that we need a reevaluation and consensus for assessing patients that are eligible for treatment. Whether ERBB2 antagonists have therapeutic potential in ependymoma is presently unclear. The ongoing study conducted by the Paediatric Brain Tumor Consortium (PBTC-016) of pediatric CNS tumors using lapatinib (GW572016), a dual inhibitor of EGFR and ERBB2 receptor signaling, may resolve this question.
Our findings contribute to current understanding of ependymoma progression in children and introduce the novel concept of nucleolin detection as valuable prognostic methodology. Confirmation of these observations in a prospective study involving larger cohorts is now required and is currently ongoing in our laboratory.
Comprehensive characterization of nucleolin variants in the context of ependymoma and other tumors will be required in future studies to gain better understanding of the specific role(s) nucleolin plays in tumor progression. We cannot exclude the possibility that hTERT confers prognostic value in this setting; indeed, our study, albeit retrospective, encourages the validation of nucleolin and hTERT in future prospective trials and encourages further basic research into the interactions of nucleolin and hTERT.
Acknowledgments
This work was supported by grants from the Connie and Albert Taylor Trust, Joseph Foote Foundation, and the Birmingham Children’s Hospital Special Trustees. We thank Christina Coldicott, Sue Cavanagh, Sarah Davis, Darren Redfern, and John Lane of the Histopathology Department, Birmingham Children’s Hospital, for support and technical assistance. We also thank Gavin Whyman and Sarah Leigh Nicholson of the Children’s Cancer and Leukaemia Group for assistance with clinical and biological material. We especially thank the other U.K. centers for assistance with biological material, local pathological diagnosis, and clinical details: Cambridge (Dr. Dean and Dr. Nicholson), Cardiff (Dr. Traunecker), Liverpool (Dr. Pizer), and Newcastle (Dr. Bailey).
References
- 1.Goldwein JW, Leahy JM, Packer RJ, Sutton LN, Curran WJ, Rorke LB, et al. Intracranial ependymomas in children. Int J Radiat Oncol Biol Phys. 1990;19:1497–1502. doi: 10.1016/0360-3016(90)90362-n. [DOI] [PubMed] [Google Scholar]
- 2.Akyuz C, Emir S, Akalan N, Soylemezoglu F, Kutluk T, Buyukpamukcu M. Intracranial ependymomas in childhood—a retrospective review of sixty-two children. Acta Oncol (Stockholm) 2000;39:97–100. doi: 10.1080/028418600431049. [DOI] [PubMed] [Google Scholar]
- 3.Grill J, Le Deley MC, Gambarelli D, Raquin MA, Couanet D, Pierre-Kahn A, et al. Postoperative chemotherapy without irradiation for ependymoma in children under 5 years of age: a multicenter trial of the French Society of Pediatric Oncology. J Clin Oncol. 2001;19:1288–1296. doi: 10.1200/JCO.2001.19.5.1288. [DOI] [PubMed] [Google Scholar]
- 4.Duffner PK, Horowitz ME, Krischer JP, Friedman HS, Burger PC, Cohen ME, et al. Postoperative chemotherapy and delayed radiation in children less than three years of age with malignant brain tumors. NEJM. 1993;328:1725–1731. doi: 10.1056/NEJM199306173282401. [DOI] [PubMed] [Google Scholar]
- 5.Merchant TE, Mulhern RK, Krasin MJ, Kun LE, Williams T, Li C, et al. Preliminary results from a phase II trial of conformal radiation therapy and evaluation of radiation-related CNS effects for pediatric patients with localized ependymoma. J Clin Oncol. 2004;22:3156–3162. doi: 10.1200/JCO.2004.11.142. [DOI] [PubMed] [Google Scholar]
- 6.Geyer JR, Sposto R, Jennings M, Boyett JM, Axtell RA, Breiger D, et al. Multiagent chemotherapy and deferred radiotherapy in infants with malignant brain tumors: a report from the Children’s Cancer Group. J Clin Oncol. 2005;23:7621–7631. doi: 10.1200/JCO.2005.09.095. [DOI] [PubMed] [Google Scholar]
- 7.Geyer JR, Zeltzer PM, Boyett JM, Rorke LB, Stanley P, Albright AL, et al. Survival of infants with primitive neuroectodermal tumors or malignant ependymomas of the CNS treated with eight drugs in 1 day: a report from the Children’s Cancer Group. J Clin Oncol. 1994;12:1607–1615. doi: 10.1200/JCO.1994.12.8.1607. [DOI] [PubMed] [Google Scholar]
- 8.Rogers L, Pueschel J, Spetzler R, Shapiro W, Coons S, Thomas T, et al. Is gross-total resection sufficient treatment for posterior fossa ependymomas? J Neurosurg. 2005;102:629–636. doi: 10.3171/jns.2005.102.4.0629. [DOI] [PubMed] [Google Scholar]
- 9.Copeland DR, deMoor C, Moore BD, 3rd, Ater JL. Neurocognitive development of children after a cerebellar tumor in infancy: a longitudinal study. J Clin Oncol. 1999;17:3476–3486. doi: 10.1200/JCO.1999.17.11.3476. [DOI] [PubMed] [Google Scholar]
- 10.Mulhern RK, Merchant TE, Gajjar A, Reddick WE, Kun LE. Late neurocognitive sequelae in survivors of brain tumours in childhood. Lancet Oncol. 2004;5:399–408. doi: 10.1016/S1470-2045(04)01507-4. [DOI] [PubMed] [Google Scholar]
- 11.Riva D, Giorgi C. The neurodevelopmental price of survival in children with malignant brain tumours. Childs Nerv Syst. 2000;16:751–754. doi: 10.1007/s003810000332. [DOI] [PubMed] [Google Scholar]
- 12.Spiegler BJ, Bouffet E, Greenberg ML, Rutka JT, Mabbott DJ. Change in neurocognitive functioning after treatment with cranial radiation in childhood. J Clin Oncol. 2004;22:706–713. doi: 10.1200/JCO.2004.05.186. [DOI] [PubMed] [Google Scholar]
- 13.Duffner PK. Diagnosis of brain tumors in children. Expert Rev Neu -rother. 2007;7:875–885. doi: 10.1586/14737175.7.7.875. [DOI] [PubMed] [Google Scholar]
- 14.Gajjar A, Hernan R, Kocak M, Fuller C, Lee Y, McKinnon PJ, et al. Clinical, histopathologic, and molecular markers of prognosis: toward a new disease risk stratification system for medulloblastoma. J Clin Oncol. 2004;22:984–993. doi: 10.1200/JCO.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 15.Wiestler O, Schiffer D, Coons S, Prayson R, Rosenblum MK. Ependymoma. In: Kleihues WCP, Cavenee WK, editors. Pathology and Genetics of Tumours of the Nervous System. Lyon, France: International Agency for Research on Cancer; 2000. pp. 71–81. [Google Scholar]
- 16.Korshunov A, Golanov A, Sycheva R, Timirgaz V. The histologic grade is a main prognostic factor for patients with intracranial ependymomas treated in the microneurosurgical era: an analysis of 258 patients. Cancer. 2004;100:1230–1237. doi: 10.1002/cncr.20075. [DOI] [PubMed] [Google Scholar]
- 17.Figarella-Branger D, Civatte M, Bouvier-Labit C, Gouvernet J, Gambarelli D, Gentet JC, et al. Prognostic factors in intracranial ependymomas in children. J Neurosurg. 2000;93:605–613. doi: 10.3171/jns.2000.93.4.0605. [DOI] [PubMed] [Google Scholar]
- 18.Horn B, Heideman R, Geyer R, Pollack I, Packer R, Goldwein J, et al. A multi-institutional retrospective study of intracranial ependymoma in children: identification of risk factors. J Pediatr Hematol Oncol. 1999;21:203–211. doi: 10.1097/00043426-199905000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Perilongo G, Massimino M, Sotti G, Belfontali T, Masiero L, Rigobello L, et al. Analyses of prognostic factors in a retrospective review of 92 children with ependymoma: Italian Pediatric Neuro-oncology Group. Med Pediatr Oncol. 1997;29:79–85. doi: 10.1002/(sici)1096-911x(199708)29:2<79::aid-mpo3>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 20.Pollack IF, Gerszten PC, Martinez AJ, Lo KH, Shultz B, Albright AL, et al. Intracranial ependymomas of childhood: long-term outcome and prognostic factors. Neurosurgery. 1995;37:655–667. doi: 10.1227/00006123-199510000-00008. [DOI] [PubMed] [Google Scholar]
- 21.Robertson PL, Zeltzer PM, Boyett JM, Rorke LB, Allen JC, Geyer JR, et al. Survival and prognostic factors following radiation therapy and chemotherapy for ependymomas in children: a report of the Children’s Cancer Group. J Neurosurg. 1998;88:695–703. doi: 10.3171/jns.1998.88.4.0695. [DOI] [PubMed] [Google Scholar]
- 22.Sutton LN, Goldwein J, Perilongo G, Lang B, Schut L, Rorke L, et al. Prognostic factors in childhood ependymomas. Pediatr Neurosurg. 1990;16:57–65. doi: 10.1159/000120509. [DOI] [PubMed] [Google Scholar]
- 23.Wolfsberger S, Fischer I, Hoftberger R, Birner P, Slavc I, Dieckmann K, et al. Ki-67 immunolabeling index is an accurate predictor of outcome in patients with intracranial ependymoma. Am J Surg Pathol. 2004;28:914–920. doi: 10.1097/00000478-200407000-00011. [DOI] [PubMed] [Google Scholar]
- 24.Verstegen MJ, Leenstra DT, Ijlst-Keizers H, Bosch DA. Proliferation- and apoptosis-related proteins in intracranial ependymomas: an immunohistochemical analysis. J Neurooncol. 2002;56:21–28. doi: 10.1023/a:1014471714058. [DOI] [PubMed] [Google Scholar]
- 25.Prayson RA. Clinicopathologic study of 61 patients with ependymoma including MIB-1 immunohistochemistry. Ann Diagn Pathol. 1999;3:11–18. doi: 10.1016/s1092-9134(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 26.Ritter AM, Hess KR, McLendon RE, Langford LA. Ependymomas: MIB-1 proliferation index and survival. J Neurooncol. 1998;40:51–57. doi: 10.1023/a:1006082622699. [DOI] [PubMed] [Google Scholar]
- 27.Zamecnik J, Snuderl M, Eckschlager T, Chanova M, Hladikova M, Tichy M, et al. Pediatric intracranial ependymomas: prognostic relevance of histological, immunohistochemical, and flow cytometric factors. Mod Pathol. 2003;16:980–991. doi: 10.1097/01.MP.0000087420.34166.B6. [DOI] [PubMed] [Google Scholar]
- 28.Gilbertson RJ, Bentley L, Hernan R, Junttila TT, Frank AJ, Haapasalo H, et al. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin Cancer Res. 2002;8:3054–3064. [PubMed] [Google Scholar]
- 29.Bennetto L, Foreman N, Harding B, Hayward R, Ironside J, Love S, et al. Ki-67 immunolabelling index is a prognostic indicator in childhood posterior fossa ependymomas. Neuropathol Appl Neurobiol. 1998;24:434–440. doi: 10.1046/j.1365-2990.1998.00143.x. [DOI] [PubMed] [Google Scholar]
- 30.Lens SM, Vader G, Medema RH. The case for survivin as mitotic regulator. Curr Opin Cell Biol. 2006;18:616–622. doi: 10.1016/j.ceb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Altura RA, Olshefski RS, Jiang Y, Boue DR. Nuclear expression of survivin in paediatric ependymomas and choroid plexus tumours correlates with morphologic tumour grade. Br J Cancer. 2003;89:1743–1749. doi: 10.1038/sj.bjc.6601334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preusser M, Wolfsberger S, Czech T, Slavc I, Budka H, Hainfellner JA. Survivin expression in intracranial ependymomas and its correlation with tumor cell proliferation and patient outcome. Am J Clin Pathol. 2005;124:543–549. doi: 10.1309/PP2G5GAAFKV82DTG. [DOI] [PubMed] [Google Scholar]
- 33.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature Rev. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 34.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 35.Tabori U, Ma J, Carter M, Zielenska M, Rutka J, Bouffet E, et al. Human telomere reverse transcriptase expression predicts progression and survival in pediatric intracranial ependymoma. J Clin Oncol. 2006;24:1522–1528. doi: 10.1200/JCO.2005.04.2127. [DOI] [PubMed] [Google Scholar]
- 36.Wu YL, Dudognon C, Nguyen E, Hillion J, Pendino F, Tarkanyi I, et al. Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J Cell Sci. 2006;119:2797–2806. doi: 10.1242/jcs.03001. [DOI] [PubMed] [Google Scholar]
- 37.Grinstein E, Wernet P. Cellular signaling in normal and cancerous stem cells. Cell Signal. 2007;19:2428–2433. doi: 10.1016/j.cellsig.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava M, Pollard HB. Molecular dissection of nucleolin’s role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- 39.Grundy RG, Wilne SA, Weston CL, Robinson K, Lashford LS, Ironside J, et al. Primary postoperative chemotherapy without radiotherapy for intracranial ependymoma in children: the UKCCSG/SIOP prospective study. Lancet Oncol. 2007;8:696–705. doi: 10.1016/S1470-2045(07)70208-5. [DOI] [PubMed] [Google Scholar]
- 40.Bouffet E, Tabori U, Bartels U. Paediatric ependymomas: should we avoid radiotherapy? Lancet Oncol. 2007;8:665–666. doi: 10.1016/S1470-2045(07)70213-9. [DOI] [PubMed] [Google Scholar]
- 41.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 42.Grinstein E, Shan Y, Karawajew L, Snijders PJ, Meijer CJ, Royer HD, et al. Cell cycle-controlled interaction of nucleolin with the retinoblastoma protein and cancerous cell transformation. J Biol Chem. 2006;281:22223–22235. doi: 10.1074/jbc.M513335200. [DOI] [PubMed] [Google Scholar]
- 43.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112(pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 44.Khurts S, Masutomi K, Delgermaa L, Arai K, Oishi N, Mizuno H, et al. Nucleolin interacts with telomerase. J Biol Chem. 2004;279:51508–51515. doi: 10.1074/jbc.M407643200. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollice A, Zibella MP, Bilaud T, Laroche T, Pulitzer JF, Gilson E. In vitro binding of nucleolin to double-stranded telomeric DNA. Biochem Biophys Res Commun. 2000;268:909–915. doi: 10.1006/bbrc.2000.2237. [DOI] [PubMed] [Google Scholar]
- 47.Derenzini M, Sirri V, Trere D, Ochs RL. The quantity of nucleolar proteins nucleolin and protein B23 is related to cell doubling time in human cancer cells. Lab Invest. 1995;73:497–502. [PubMed] [Google Scholar]
- 48.Derenzini M. The AgNORs. Micron. 2000;31:117–120. doi: 10.1016/s0968-4328(99)00067-0. [DOI] [PubMed] [Google Scholar]
- 49.Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, et al. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol. 2007;8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 51.Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- 52.Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, et al. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 53.Poremba C, Scheel C, Hero B, Christiansen H, Schaefer KL, Nakayama J, et al. Telomerase activity and telomerase subunits gene expression patterns in neuroblastoma: a molecular and immunohistochemical study establishing prognostic tools for fresh-frozen and paraffin-embedded tissues. J Clin Oncol. 2000;18:2582–2592. doi: 10.1200/JCO.2000.18.13.2582. [DOI] [PubMed] [Google Scholar]
- 54.Sanders RP, Drissi R, Billups CA, Daw NC, Valentine MB, Dome JS. Telomerase expression predicts unfavorable outcome in osteosarcoma. J Clin Oncol. 2004;22:3790–3797. doi: 10.1200/JCO.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 55.Didiano D, Shalaby T, Lang D, Grotzer MA. Telomere maintenance in childhood primitive neuroectodermal brain tumors. Neuro-Oncology. 2004;6:1–8. doi: 10.1215/S1152851703000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tabori U, Vukovic B, Zielenska M, Hawkins C, Braude I, Rutka J, et al. The role of telomere maintenance in the spontaneous growth arrest of pediatric low-grade gliomas. Neoplasia. 2006;8:136–142. doi: 10.1593/neo.05715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morii K, Tanaka R, Onda K, Tsumanuma I, Yoshimura J. Expression of telomerase RNA, telomerase activity, and telomere length in human gliomas. Biochem Biophys Res Commun. 1997;239:830–834. doi: 10.1006/bbrc.1997.7562. [DOI] [PubMed] [Google Scholar]
- 58.Cong YS, Bacchetti S. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J Biol Chem. 2000;275:35665–35668. doi: 10.1074/jbc.C000637200. [DOI] [PubMed] [Google Scholar]
- 59.Dessain SK, Yu H, Reddel RR, Beijersbergen RL, Weinberg RA. Methylation of the human telomerase gene CpG island. Cancer Res. 2000;60:537–541. [PubMed] [Google Scholar]
- 60.Armanios M, Greider CW. Telomerase and cancer stem cells. Cold Spring Harbor Symp Quant Biol. 2005;70:205–208. doi: 10.1101/sqb.2005.70.030. [DOI] [PubMed] [Google Scholar]
- 61.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 62.Suzuki S, Oka H, Kawano N, Tanaka S, Utsuki S, Fujii K. Prognostic value of Ki-67 (MIB-1) and p53 in ependymomas. Brain Tumor Pathol. 2001;18:151–154. doi: 10.1007/BF02479429. [DOI] [PubMed] [Google Scholar]
- 63.Duffy MJ, O’Donovan N, Brennan DJ, Gallagher WM, Ryan BM. Survivin: a promising tumor biomarker. Cancer Lett. 2007;249:49–60. doi: 10.1016/j.canlet.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 64.Stauber RH, Mann W, Knauer SK. Nuclear and cytoplasmic survivin: molecular mechanism, prognostic, and therapeutic potential. Cancer Res. 2007;67:5999–6002. doi: 10.1158/0008-5472.CAN-07-0494. [DOI] [PubMed] [Google Scholar]
- 65.Mendrzyk F, Korshunov A, Benner A, Toedt G, Pfister S, Radlwimmer B, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12:2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 66.Gilbertson RJ. ERBB2 in pediatric cancer: innocent until proven guilty. Oncologist. 2005;10:508–517. doi: 10.1634/theoncologist.10-7-508. [DOI] [PubMed] [Google Scholar]