Abstract
High-throughput gene analysis would benefit from new approaches for delivering DNA or RNA into cells. Here we describe a simple system that allows any molecular biology laboratory to carry out multiple, parallel cell transfections on microscope coverslip arrays. By using magnetically defined positions and PCR product–coated paramagnetic beads, we achieved transfection in a variety of cell lines. Beads may be added to the cells at any time, allowing both spatial and temporal control of transfection. Because the beads may be coated with more than one gene construct, the method can be used to achieve cotransfection within single cells. Furthermore, PCR-generated mutants may be conveniently screened, bypassing cloning and plasmid purification steps. We illustrated the applicability of the method by screening combinatorial peptide libraries, fused to GFP, to identify previously unknown cellular localization motifs. In this way, we identified several localizing peptides, including structured localization signals based around the scaffold of a single C2H2 zinc finger.
Site-localized transfection in microarrays1,2 is a powerful technique to facilitate large-scale genetic analysis or to screen protein libraries3. The increasing application of genome-wide experiments, involving RNAi4 or the study of gene interactions5, requires efficient multicomponent gene delivery and would therefore benefit from new methods to construct transfection arrays6. By defining spatial patterns of gene expression on a culture plate, gene activities can be rapidly assayed, either individually or combinatorially. Furthermore, by expressing genes in different locations in a cell culture, one could construct elaborate interacting networks of ‘differentiated’ cells, or synthetic gene networks7,8, for artificial tissue-engineering applications.
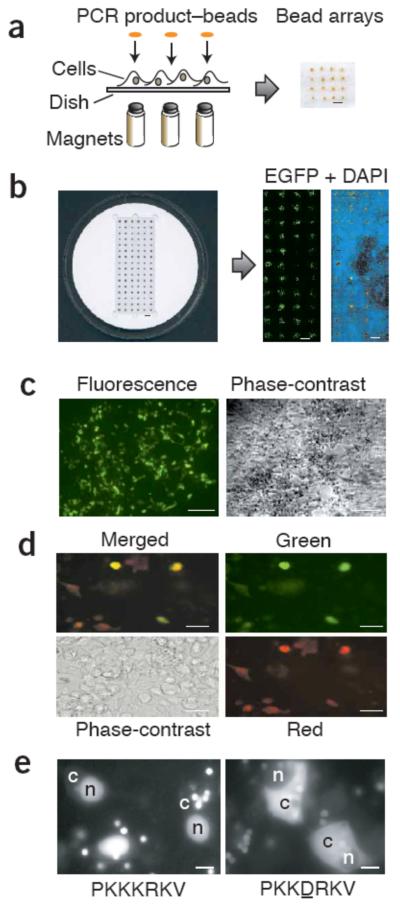
To construct transfection arrays, we have developed a PCR-based method that takes advantage of streptavidin-biotin linkage to position genes. We generated PCR products of mammalian gene expression constructs using a biotinylated forward primer, which can bind to streptavidin-coated paramagnetic beads. Thus, by arraying ‘spots’ of beads over magnets, fixed either in a plasticine matrix or in a specially constructed transfection chamber, we can define transfection patterns at will (Fig. 1a,b).
Figure 1.
Magnetically defined transfection arrays. (a) Array construction illustrating magnets under a cell-coated dish or coverslip. Magnets may be fixed in either plasticine or a solid plastic matrix. PCR product–coated beads were overlaid by injection. An example of a resulting 4 × 4 spot array is shown (viewed from above). Scale bar, 1 mm. (b) A 96-magnet transfection chamber constructed out of an acrylic disc (left). A cell-coated 24 × 60 mm coverslip fits within the central rectangular recess. The resulting EGFP transfection arrays (right) are detected with a fluorescence array scanner at 5 μm resolution. DAPI staining for nuclear DNA (blue) allows visualization of the cell distribution over the array. Scale bars, 3 mm. (c) An EGFP transfection spot, viewed by fluorescence and phase-contrast microscopy. Scale bars, 100 μm. (d) Results of cotransfection in an array of beads coated with both EGFP and HcRed PCR products are shown as a (clockwise) merged image, view through green and red filters, and a phase-contrast micrograph. Scale bars, 20 μm. (e) Mutant phenotype screen: a GFP with an SV40 nuclear localization signal (PKKKRKV; left) and GFP with a mutant NLS; a point mutation was introduced into the gene by PCR (PKKDRKV; right). Cytoplasm and nucleus in sample cells are indicated by ‘c’ and ‘n’, respectively. Note the multiple beads adjacent to cells. Whether beads can be seen depends on the imaging procedure: beads are generally best visualized in a fluorescence microscope (using a >40× objective) or in a confocal system without any background subtraction. Scale bars, 10 μm.
RESULTS
PCR transfection arrays with fluorescent protein genes
Initially, we tested the PCR product–bead transfection method with genes expressing the fluorescent proteins EGFP9 and HcRed10 (Fig. 1). For optimal mammalian expression, templates included a 5′ human cytomegalovirus (CMV) immediate early promoter and a 3′ SV40 early mRNA polyadenylation signal11. We typically immobilized 300 ng of purified PCR product DNA (1.6 kilobase pairs) with 5 μl beads, incubated with transfection reagent (Effectene; Qiagen), diluted appropriately (typically to 50 μl) and dispensed in 1-μl spots over magnet positions in a HEK293 cell monolayer (Fig. 1b,c). We dispensed beads manually, with a pipetting technique very similar to loading an electrophoretic gel.
We judged transfection by visual inspection to detect green fluorescent cells. As expected, GFP was only detected over magnetic spots (generally, we observed 250–400 transfected cells per spot when using the standard protocol for HEK293 cells). In control transfections with non–DNA coated magnetic beads or with beads incubated with the plasmid encoding GFP (but otherwise treated identically, including washing steps), we observed no transfectants. We were also unable to detect transfection in cells between magnetic spot areas, indicating that the magnets attracted the beads sufficiently strongly to prevent transfection outside the magnet boundary. Moreover, the parallel north-south pole arrangement of magnets created mutually repulsive fields between spots, ensuring that beads remained tightly localized and did not migrate between adjacent magnets.
For estimating transfection efficiencies, we counted cells by eye in an inverted microscope, using a 40× objective, with 100–200 cells in each field of view. For three independent experiments (three spots each, three fields of view per spot), the transfection efficiencies in HEK293 with 6 ng PCR product per spot were 57 ± 5%; 58 ± 4% and 61 ± 6% (mean ± 1 s.d.).
We tested varying dosages of PCR product–coated beads to establish optimal transfection conditions. For the majority of concentrations tested (range: 0.5–12 ng PCR product per spot), the average transfection efficiency was 62 ± 7% (mean ± 1 s.d.). Notably, the efficiency varied with bead distribution: the use of fewer beads resulted in reduced spot size, but did not reduce the local efficiency in bead-rich areas. However, for higher concentrations of beads with DNA (≥15 ng per spot) the transfection efficiency dropped substantially, perhaps because of transfection reagent toxicity. Notably, transfections were observed even in the absence of transfection reagent, albeit with very low efficiency (data not shown).
Transfection efficiencies in various cell lines
Previous studies of transfection microarrays with plasmids1 or small interfering RNAs (siRNAs)2 have primarily used HEK293 cells because they transfect with high efficiency, although their low adherence can be a problem in handling arrays. Using other cell lines, 30–50% efficiency of transfection with plasmids in HeLa and A549 cells has been reported1, and another group has achieved higher efficiency with siRNAs: COS7L (70%), MCF7 or HeLa (80–90%) and HEK293 (95%)2. To compare our PCR product–bead methods to these systems, we carried out transfections with COS7, NIH3T3 and HeLa cells. These cell lines were initially harder to transfect (giving <100 transfectants per spot with the standard protocol, compared with 250–400 for HEK293). By optimizing the protocol (see Methods, alternative transfection method), we achieved 100–200 transfected cells per spot. Although this corresponds to transfection efficiencies around 30%, the number of transfectants in these cell lines would be sufficient for peptide screening applications, such as those described below. HEK293 cells were used in all of the following experiments unless otherwise stated.
Simultaneous transfection of multiple PCR products
To examine the potential of the system to cotransfect more than one gene, we coated beads with equimolar mixtures of PCR-amplified EGFP and HcRed genes and found that the majority of transfected cells expressed both genes (Fig. 1d; 75% of fluorescent cells apparently express both genes, 15% only EGFP, 10% only HcRed). Cotransfection is required in a variety of experiments, for example in mammalian two-hybrid screens12, in which library and reporter constructs must be simultaneously introduced into cells. The PCR product–bead approach has the potential to be applied to such experiments, and unlike with preprinted transfection arrays, the beads can be added at any time point or points during the growth of the cells on the coverslip.
Screening PCR-introduced mutations directly
Because PCR product transfection may be used to construct and screen mutants directly, we amplified EGFP with primers that introduced wild-type or mutant SV40 nuclear localization signals (NLS)13. The arrays clearly resolve the different phenotypes (Fig. 1e), proving that PCR-introduced mutations can be scanned simply, avoiding subcloning steps. The method is therefore ideally suited to carry out screenings: PCR primers can be used to introduce individual peptide tags, which can be directly screened for cellular phenotypes. Alternatively, colony PCR could be used to screen individual clones from pre-existing combinatorially randomized protein libraries14, bypassing plasmid purification steps.
Screening peptide libraries by PCR product–bead transfection
To investigate the suitability of our approach for functional sequence screening, we carried out screens from libraries of random peptides fused to EGFP3. First, we built a ‘KESL’ library, coding for 256 hexapeptide combinations of the amino acids lysine, glutamic acid, serine and lysine, linked to the C terminus of EGFP via two prolines (for example, PP-KSKLEK). This library design was expected to incorporate several potential targeting signals, including a peroxisomal localization signal (SKL15), variant NLSs16 (for example, library variant PPKKKKKL) and an endoplasmic reticulum targeting signal related to KDEL17 (library variant KEEL).
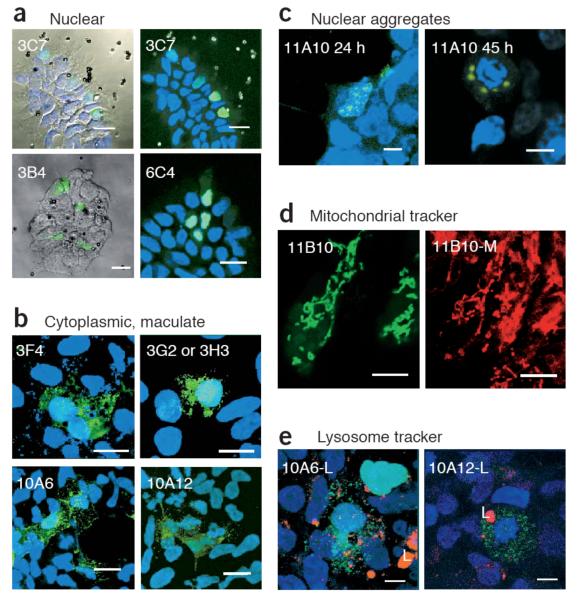
Five separate localizing species were obtained by screening 360 colony PCRs of the KESL libraries, using the 96-magnet chamber (Fig. 2 and Table 1). Two clones resulted in nuclear localization, and their products contained mainly lysine residues, which might be expected to act as NLSs (named by plate and well number: 3C7, PPKKKLKL; 3B4, PPEKKKKL). Another nuclear localizing peptide was more unusual, with two consecutive acidic residues (6C4, PPKKEEKL). Next, we found a clone with an SKL peroxisome targeting signal (3F4, PPESKLKE; Fig. 2b). This clone gave a maculate phenotype, consistent with peroxisomal distribution18. Two other clones, 3G2 and 3H3, also resulted in similar morphology and resembled the peroxisome signal sequence, encoding the peptide PPKKSLLL.
Figure 2.
Confocal microscopy images of selected clones from the colony-PCR bead-transfection screen of the EGFP peptide libraries. (a) Nuclear localized clones 3C7, 3B4 and 6C4 from the EGFP-KESL library, viewed by reflection, with the green EGFP signal and/or DAPI superimposed (left), or viewed by fluorescence microscopy with blue DAPI staining for nuclei (right). (b) Cytoplasmically distributed clones with maculate morphology. 3F4, 3G2 and 3H3 are from the KESL library, whereas 10A6 and 10A12 are from the Zif library. Note that 3F4 contains an SKL peroxisomal targeting signal. (c) The 11A10 clone, from the Lrm library, forms aggregates that are generally located next to the nuclear membrane (24 h) and increase size with time. When the cell is in mitosis, aggregates remain around the DNA (45 h). (d) The Lrm 11B10 clone, viewed with a green filter for EGFP (left) or viewed with red filters for MitoTracker dye (right; 11B10-M), reveals colocalization of the EGFP signal with mitochondria. (e) LysoTracker staining for lysosomes (L; red) shows that the green maculate distribution of the zinc fingers 10A6 and 10A12 is not lysosomal. Green, red and DAPI signals are superimposed. Scale bars: 20 μm (a,b); 8 μm (c,d,e).
Table 1.
List of selected clones from the transfection library screens, showing localization properties and sequence identity with proteins in the UNIPROT database as found by FASTA321 (percentage identity and expectation values, E(), are given for each sequence). Bold letters highlight matching residues. The α-helices in the zinc finger sequences are underlined. For reference, the design of the Zif library framework is shown, with X denoting randomized amino acid positions; helical positions −1, 2, 3 and 6 are shown above this sequence. Note that two clones (3B4 and 3F4) contained mixed populations of sequence in their PCR product.
| Clone | Source | Localization | Sequence | % identity, E() |
|---|---|---|---|---|
| 3C7 | KESL library | Nuclear | PPKKKLKL | |
| 3B4 | KESL library | Nuclear | PPEKKK(K/E)L | |
| 6C4 | KESL library | Nuclear | PPKKEEKL | |
| 3F4 | KESL library | Peroxisomal | PPE(S/L)KLKE | |
| 3G2 | KESL library | Cytoplasmic, maculate | PPKKSLLL | |
| 3H3 | KESL library | Cytoplasmic, maculate | PPKKSLLL | |
| 11A10 | Lrm library | Perinuclear | MARLSRRLIRLRCR YLDCSSVIRLVFGVF FSSFLLFRLIMGI |
|
| UNIPROT:Q8F2J3 | Putative Leptospira sp. protein |
— | MDLAVETISIVRVIF LVFFTELILFLISLGV |
39%, 7.2 |
| 11B10 | Lrm library | Mitochondrial | MAHRRLTKKLLNQ VHPRTLRNI—RIST RNTNNKSIRALNIA CWRSG |
|
| UNIPROT:Q7QQY8 |
Giardia lamblia genomic sequence |
— | RRLWKRTLGQVHS FNLRRKLCWRRLS CAGAIHQGIRSLCIV CY |
40%, 2.3 |
| Zif library framework | — | Helical position: −1, 2,3 & 6 AEERPYXCXXXSCDRRFSXXXXLXXHXXXHTGQK |
||
| 10A6 | Zif library | Cytoplasmic, maculate | AEERPYKCGGSSCD RRFSHLSCLLFHFYC HTGQK |
|
| UNIPROT:AAH66874 | Mouse hypothetical protien |
— | TQEKPYKC—NQCG KAFLHLSCLRVHER THTGEK |
61%, 7.3 |
| 10A12 | Zif library | Cytoplasmic, maculate | AEERPYNCGKVSC DRRFSALLVLVAH LACHTGQK |
|
| UNIPROT:Q8BSI2 | Mouse Zif-11 | — | KREKPHKC—EECG RAFSALSVLTQHRIT HTGEK |
61%, 39 |
Next, we constructed a more complex peptide library (library random; Lrm). We built the Lrm gene cassette through PCR of an 85-bp oligonucleotide containing NNS codon repeats (where S = G or C; see Methods) and blunt-cloned it to the N terminus of EGFP. Although the theoretical library size is large, and it would be impossible to clone such a library exhaustively, even extremely low-coverage sampling of a complex library can yield interesting results. In fact, screening only 48 Lrm colonies was sufficient to discover two clones with distinct phenotypes.
First, the clone 11A10 gene product showed a characteristic nuclear aggregate distribution (Fig. 2c). Small aggregates were detectable next to the nuclear membrane after 24 h, and larger bodies were observed around condensed mitotic DNA by 45 h. Notably, the protein sequence of 11A10 (Table 1) is particularly hydrophobic, which may favor protein aggregation.
Second, the clone 11B10 gene product was mitochondrially distributed (Fig. 2d). Sequence analysis, using the sublocalization predictor TargetP19, gave a very high score for mitochondrial localization (0.891). Furthermore, the helical predictor AGADIR20 revealed that regions within the peptide (LTKKLLNQV and SIRALNIACWR) have a propensity for forming amphiphilic helices, which are associated with mitochondrial localization. Notably, a FASTA3 (ref. 21) search of the UNIPROT database for the 11B10 protein sequence found a degree of identity (40% identity; expectation value (E()) = 2.3) for a putative Giardia lamblia protein (UNIPROT Q7QQY8 (ref. 22); Table 1).
We also constructed a more structured peptide library around a scaffold of finger 1 of the zinc finger protein, Zif268 (ref. 23). By screening 60 clones with the PCR product–bead method, we found two clones with a maculate cytoplasmic distribution, very similar to the peroxisomal distribution seen for 3F4 from the Lrm library (10A6 and 10A12; Fig. 2b and Table 1). The zinc fingers had noticeably hydrophobic α-helices, perhaps involved in membrane or protein-protein interactions24, and a FASTA3 identity search21 revealed 61% identity to uncharacterized murine zinc finger proteins (Table 1; UNIPROT AAH66874, E() = 7.3, and UNIPROT Q8BSI2, E() = 39; although the latter E() value is poor, the residues in α-helical positions −1, 2, 3 and 6 of a zinc finger are functionally the most important14, and these residues matched well).
Although we did not characterize the specific cytoplasmic sublocalization of the 10A6 and 10A12 zinc fingers, staining for lysosomes or mitochondria ruled out targeting to these compartments (Fig. 2e). To our knowledge, these two clones provide the first example of non-nuclear cellular localization signals in classical C2H2 zinc fingers25. Even though C2H2 zinc fingers are the second most common protein motif in the human genome, found in over 700 proteins26, the majority of their functions remain largely unknown. It would therefore be interesting to characterize clones 10A6 and 10A12 further, along with their mouse zinc finger homologs.
DISCUSSION
We have developed a new method to allow simultaneous transfection of multiple gene constructs in a cell culture, with spatial definition. Unlike conventional transfection methods, which use plasmid DNA, this method uses PCR products, thereby greatly reducing the amount of time it takes to mutagenize or screen peptide libraries in an array format. Recently, an elegant method of creating plasmid microarrays by printing gelatin-DNA mixtures with a robotic arrayer was reported1,27. Although this printing technique produces higher-density arrays than the magnetic arrays we used, we have found PCR product–bead transfection to be a practical alternative, which can be implemented by any laboratory without sophisticated robotic equipment. Although hand-pipetting gelatin-DNA mixtures onto a glass coverslip is also possible, we have not been able to generate very efficient or reliable transfections using PCR products with this method. Our best results achieve no more than 20 transfectants per DNA spot (with 60 ng/μl DNA spotting solution), using an alternative lipid-DNA method (see Supplementary Information in ref. 1).
An alternative to coverslip microarrays is to use well-based transfection, which is established for high throughput28. We have achieved PCR product transfection in 96-well plates with slightly higher efficiencies than with bead spots, although lower than for standard plasmid transfection (PCR transfection efficiencies in wells: HEK 293: 65%, NIH3t3: 44%, Cos7: 56%, HeLa: 46%; efficiencies using GFP-plasmid with Effectene: HEK 293: 65–80%, NIH3t3: 40–50%, Cos7: 40–50%, HeLa: 50–70%).
Plates are potentially easier to handle than coverslips because assays can be automated with standard pipetting robots and sample cross-contamination is prevented by the well boundaries. However, in principle, pipetting onto magnets could be automated by a modified gel loading system and magnet-bead attraction also prevents cross-contamination between adjacent samples; all localized constructs in our peptide screens were retransfected separately to ensure beads had not been misplaced and in no case was cross-contamination detected. Bead spots are also more economical, requiring at least 17-fold less DNA per sample, and optical-quality plates cost 160-fold more than coverslips, which cost around ten cents each. Another advantage of having all the transformants on one coverslip is that it may be easier to apply automated confocal microscopy with pattern recognition software to scan and recognize features such as fluorescently labeled subcompartments29. Finally, because beads can be added to cells at any stage of the transfection process (unlike preprinted reverse-transfection arrays), it is possible to apply diverse treatments to the cells in situ before adding beads and to add beads progressively with different constructs or time delays.
Transfection arrays are beginning to find many uses, for example in functional genomics screens using RNAi30,31. Using magnets for transfection arrays allows great control over pattern shape and size. Ultimately, one could gain precise control over single cells using single magnetic beads, placed by electromagnetic tweezers. Moreover, localized cotransfection could enable the design and delivery of multiple gene network cassettes, for systems biology applications7,8. For example, one could envision designing spatially defined signaling networks of interacting cells. We anticipate that such systems may be conveniently constructed and manipulated using PCR product–bead transfection.
METHODS
Chamber construction
We constructed transfection chambers by fixing 96 stirrer-bar magnets (8 × 1.5 mm; VWR International), from which the plastic coating had been stripped before use, into drilled holes in a solid, 80-mm-diameter acrylic disc (Fig. 1b). The disc fit inside a cell culture dish and contained an etched recess to fit a 24- × 60 mm cell-coated coverslip. The same magnetic pole always faced the cells growing on the coverslip. This parallel north-south arrangement is very important to create distinct bead spots; misinserted magnets create field lines between adjacent magnets, redistributing the beads. It is useful to check the uniformity of the magnetic field before permanently fixing the magnets with epoxy resin by spreading iron filings on a piece of paper over the array, to visualize the magnetic field. Note that magnets on the outside rows of a parallel magnet array form field lines away from the center of the array, resulting in diffuse edge spots. Therefore, to get good spot definition for an array with A rows and B columns, use (A + 2) × (B + 2) magnets. To construct a functional chamber simply and rapidly, we have also successfully used a plasticine bed to fix the magnets, sealing with epoxy resin for direct use in cell culture.
Standard transfection method (for HEK293 cells)
Because the central magnetic positions gave the best-defined magnetic spots in the array, we typically transfected in a 12- × 4-spot array, in the 96-magnet chamber. For a typical 48-spot array, we washed 37 μl Dynabeads from a KilobaseBinder kit (Dynal) and resuspended in 510 μl KilobaseBinder binding solution (Dynal), according to the manufacturer’s instructions. We pipetted 10 μl bead aliquots in a 96-well plate, adding 5 μl of water plus 5 μl of PCR product per well. After mixtures were incubated for 3 h at 22 °C, with mixing at 800 r.p.m, a magnetic particle concentrator (MPC; Dynal) was used to pellet the beads. After washing twice in KilobaseBinder washing solution and once in 10 mM Tris-Cl, pH 8.5, we resuspended beads in 30 μl EC buffer (Effectene kit; Qiagen). We added 10 μl of diluted Enhancer solution (diluted 0.6 μl per 10 μl EC buffer) per well, followed by 10 μl of diluted Effectene reagent (2 μl plus 10 μl EC buffer). We incubated the plate for 45 min at 22 °C with mixing at 800 r.p.m and used the MPC to concentrate the beads down to 10 μl, in the same solution. We dispensed 1 μl of the resulting suspension over each magnet position. Thus, each preparation was sufficient for at least eight replicate arrays. The bead pipetting technique is very similar to loading an electrophoresis gel.
We made bead ‘spots’ on top of HEK 293 cell monolayers grown on 24 × 60 mm polylysine-coated glass coverslips, consisting of 5 × 106 exponentially growing cells per coverslip loaded 24 h in advance, and incubated in 100-mm-diameter cell culture dishes with 10 ml growth medium (complete DMEM from Gibco with 10% FCS, 50 U/ml penicillin and 50 μg/ml streptomycin). For bead arraying, we first removed the coverslips, inserted in a magnetic platform (presterilized with 70% ethanol and UV light for 15 min) and covered with 3 ml fresh medium. After arraying, we incubated the entire platform 16–48 h at 37 °C. When indicated, we added Lysotracker and Mitotracker (Molecular Probes Inc.) to the cell culture medium 5 min before fixation (5 nM and 100 nM, respectively). We fixed samples by rinsing the coverslip twice in 1× PBS, once in 1× PBS with 4% formaldehyde and then for 20 min in 1× PBS with 4% formaldehyde. In certain cases, DAPI DNA staining followed fixing, and we mounted slides using Mowiol. In general, we observed fluorescence 16–48 h after transfection with a Leica TCS SP2 AOBS confocal microscope. For array composition, we used large-field biomapping, with a 10× objective. Alternatively, we used a laser fluorescence scanner (ScanArray 5000) at 5 μm resolution.
Alternative transfection method (recommended for COS7, NIH3T3, HeLa cells)
For cell lines other than HEK293, we found that JetSI-ENDO Cationic polymer transfection reagent (Polyplus Transfection SAS) gave higher transfection efficiencies. We modified the manufacturer’s protocol for transfecting 0.35 μg siRNA in a 24-well plate: 400 ng PCR product was bound to 5 μl Dynabeads, scaling the KilobaseBinder protocol (described above) and finally resuspending in 50 μl serum-free medium (DMEM). Meanwhile, we diluted 1 μl of JetSI-ENDO solution by vortexing briefly in 50 μl DMEM and incubating at room temperature for 10 min. We added this mixture to the resuspended beads, vortexed for 10 s and incubated at room temperature for 30 min on a rolling platform at 60 r.p.m. An MPC was used to reduce the volume to 40 μl, allowing 40 arrayed spots per sample. The growth history of the cells is critical for efficient transfection: cells should not be allowed to reach confluence in the previous two passages.
Transfection in 96-well format
We used optical-quality 96-well glass-bottom plates (Whatman), adding 1.25 × 104 cells per well, 24 h before transfection. We adapted the Effectene protocol: we used 100 ng PCR product per sample, in 30 μl EC buffer with 0.8 μl Enhancer. We vortexed Effectene (2.5 μl, diluted in 20 μl EC buffer) for 10 s with this mixture, incubated at room temperature for 10 min, diluted in 50 μl DMEM and added to cells (already in 100 μl fresh medium). We inspected cells directly after 24 h at 37 °C.
Gene library construction
We synthesized the KESL libraries from four DNA oligonucleotide cassettes coding for eight-amino-acid peptides of the form PPRRRRRT, PPRRRTRT, PPRTRTRR or PPRRTTTT, where R was the codon RAG (R = purine), coding for the amino acids lysine or glutamic acid, and T was the codon TYG (Y = pyrimidine), coding for serine or leucine. Primer extensions of template oligonucleotides were cloned via BamHI and EcoRI sites in the vector pEGFP-C1 (BD Biosciences Clontech), resulting C-terminal in-frame EGFP-fusion peptides (for example, primer: GACGTGTGGACTGACTGTGA; template: ACGTACGTGAATTCTCCUCCCRAGRAGRAGRAGRAGTYGTAAGGATCCTC ACAGTCAGTCCACACGTC; BamHI and EcoRI sites are underlined, randomised positions are in bold) We screened transformant bacterial colonies by PCR, using primers to include a 5′ CMV promoter and a 3′ SV40 early mRNA polyadenylation signal from the pEGFP-C1 vector (biotinylated forward primer: CTGTGGATAACCGTATTACCG; reverse primer: AACCACAACTAGAATGCAGTG). We used crude PCR products directly in bead transfection, as described above.
We cloned the Lrm unscaffolded peptide library from a randomized cassette, by PCR of an 85-bp oligonucleotide containing NNS repeats (where S = G or C). We blunt-cloned this cassette, in-frame and N-terminally to EGFP, using an SfrI site in a modified pEGFP-N1 vector3. Random priming PCR contributed to greater library diversity with variable insert length and base composition. Library complexity can only be estimated (at least 28 randomized amino acid positions: >1036 combinations). Therefore, we constructed only a small sample for this library: PCR colony screening was carried out on 48 transformants, as described above.
We constructed the randomized Zif library by annealing and extension of two primers, ZlibFWD (ACGTACGTGAATTCTGCAGAGGAACGCCCGTATNNNTGCNNNNNNNNNTC CTGCGATCGCCGCTTTTC) and ZlibREV (GACTGTGAGGATCCTTACTTCTGGCCTGTGTGNNNNNNNNNATGNNNNNN AAGNNNNNNNNNNNNAGAAAAGCGGCGATCGCAGGA). We cloned this cassette via BamHI and EcoRI sites in the vector pEGFP-C1. The library complexity is again very high (20 randomized amino acids in 13 positions: >1016 combinations). We therefore ligated a test sample, using 100 ng of vector and a 3:1 insert-to-vector ratio. We screened 60 resulting transformants, as described above.
ACKNOWLEDGMENTS
M.I. was supported by an International Research Fellowship from the Wellcome Trust, UK. M.I.S was funded by a Fundacion Carolina Postdoctoral Fellowship. We would like to thank G. DeCarcer Diez and K. Michalodimitrakis for help in the design of the transfection chamber and D. Megias Vazquez for assistance with confocal imaging.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Published online at http://www.nature.com/naturemethods/
References
- 1.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 2.Erfle H, Simpson JC, Bastiaens PI, Pepperkok R. siRNA cell arrays for high-content screening microscopy. Biotechniques. 2004;37:454–458. 460–462. doi: 10.2144/04373RT01. [DOI] [PubMed] [Google Scholar]
- 3.Bejarano LA, Gonzalez C. Motif trap: a rapid method to clone motifs that can target proteins to defined subcellular localisations. J. Cell Sci. 1999;112:4207–4211. doi: 10.1242/jcs.112.23.4207. [DOI] [PubMed] [Google Scholar]
- 4.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 5.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 6.Wu RZ, Bailey SN, Sabatini DM. Cell-biological applications of transfected-cell microarrays. Trends Cell Biol. 2002;12:485–488. doi: 10.1016/s0962-8924(02)02354-1. [DOI] [PubMed] [Google Scholar]
- 7.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 8.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 9.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 10.Gurskaya NG, et al. GFP-like chromoproteins as a source of far-red fluorescent proteins. FEBS Lett. 2001;507:16–20. doi: 10.1016/s0014-5793(01)02930-1. [DOI] [PubMed] [Google Scholar]
- 11.Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y, Batalao A, Zhou H, Zhu L. Mammalian two-hybrid system: a complementary approach to the yeast two-hybrid system. Biotechniques. 1997;22:350–352. doi: 10.2144/97222pf02. [DOI] [PubMed] [Google Scholar]
- 13.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 14.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould SG, Keller GA, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J. Cell Biol. 1987;105:2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair R, Carter P, Rost B. NLSdb: database of nuclear localization signals. Nucleic Acids Res. 2003;31:397–399. doi: 10.1093/nar/gkg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 18.Recalcati S, Menotti E, Kuhn LC. Peroxisomal targeting of mammalian hydroxyacid oxidase 1 requires the C-terminal tripeptide SKI. J. Cell Sci. 2001;114:1625–1629. doi: 10.1242/jcs.114.9.1625. [DOI] [PubMed] [Google Scholar]
- 19.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 20.Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 21.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palenik B, et al. The genome of a motile marine Synechococcus. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 23.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 24.Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem. Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 25.Isalan M. Zinc fingers. In: Lennars WJ, Lane MD, editors. Encyclopedia of Biological Chemistry. Academic Press; Elsevier, St. Louis: 2004. [Google Scholar]
- 26.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 27.Bailey SN, Wu RZ, Sabatini DM. Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discov. Today. 2002;7:S113–S118. doi: 10.1016/s1359-6446(02)02386-3. [DOI] [PubMed] [Google Scholar]
- 28.Grimm S, Kachel V. Robotic high-throughput assay for isolating apoptosis-inducing genes. Biotechniques. 2002;32:670–2. 674–7. doi: 10.2144/02323dd04. [DOI] [PubMed] [Google Scholar]
- 29.Liebel U, et al. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 2003;554:394–398. doi: 10.1016/s0014-5793(03)01197-9. [DOI] [PubMed] [Google Scholar]
- 30.Mousses S, et al. RNAi microarray analysis in cultured mammalian cells. Genome Res. 2003;13:2341–2347. doi: 10.1101/gr.1478703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva JM, Mizuno H, Brady A, Lucito R, Hannon GJ. RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:6548–6552. doi: 10.1073/pnas.0400165101. [DOI] [PMC free article] [PubMed] [Google Scholar]