Horizontal gene transfer and clonal expansion contribute substantially to the dissemination of resistant strains
Keywords: Salmonella enterica, drug resistance, multidrug resistance, integrons, horizontal gene transfer, research
Abstract
Salmonella enterica bacteria have become increasingly resistant to antimicrobial agents, partly as a result of genes carried on integrons. Clonal expansion and horizontal gene transfer may contribute to the spread of antimicrobial drug–resistance integrons in these organisms. We investigated this resistance and integron carriage among 90 isolates with the ACSSuT phenotype (resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline) in a global collection of S. enterica isolates. Four integrons, dfrA12/orfF/aadA2, dfrA1/aadA1, dfrA7, and arr2/blaOXA30/cmlA5/aadA2, were found in genetically unrelated isolates from 8 countries on 4 continents, which supports a role for horizontal gene transfer in the global dissemination of S. enterica multidrug resistance. Serovar Typhimurium isolates containing identical integrons with the gene cassettes blaPSE1 and aadA2 were found in 4 countries on 3 continents, which supports the role of clonal expansion. This study demonstrates that clonal expansion and horizontal gene transfer contribute to the global dissemination of antimicrobial drug resistance in S. enterica.
Salmonella enterica bacteria are a leading cause of foodborne disease worldwide (1,2). In the United States, as many as 1.4 million cases of S. enterica–associated disease occur annually (3,4). While usually self-limiting, salmonellosis may require antimicrobial drug treatment in infants, the elderly, or immunocompromised persons. However, antimicrobial drug resistance has become increasingly common in S. enterica, which can complicate therapy. The National Antimicrobial Resistance Monitoring System reported that in 2004, 15.0% of non-Typhi isolates were resistant to >2 classes of antimicrobial drugs, and 8.1% were resistant to >5 classes. The most common S. enterica multidrug-resistance pattern in 2004 was ACSSuT (resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline) (5).
Antimicrobial drug resistance can occur by point mutations in the bacterial genome or through horizontal transfer of genetic elements carrying resistance genes. Resistance may be disseminated through clonal expansion of drug-resistant strains or through horizontal transfer of genetic elements coding for resistance determinants. S. enterica populations change through the introduction of strains that expand and displace existing populations (6,7). Such population dynamics enable antimicrobial drug resistance in S. enterica to spread as a result of clonal expansion. The global dissemination of the multidrug-resistant (MDR) S. enterica serovar Typhimurim phage type DT104 clone is an example of the role of clonal expansion in the spread of antimicrobial drug resistance determinants across multiple countries and continents (8). Clonal expansion is also probably responsible for the dissemination of nalidixic acid resistance in S. enterica serovar Typhimurium isolates obtained in southern Asia and Africa (9).
Horizontal transfer of genetic material among S. enterica or from other bacterial species also plays an important role in the dissemination of drug resistance in this pathogen (10). Evidence indicates that horizontal gene transfer plays a major role in the dissemination of antimicrobial drug resistance in other bacterial species, such as Escherichia coli (11) and Stenotrophomonas maltophilia (12). The location of antimicrobial drug–resistant genes on mobile genetic elements, such as plasmids, transposons, and integrons, facilitates the mobilization of resistance from one organism to another (13).
Integrons are genetic structures capable of capturing and excising gene cassettes, which usually encode antimicrobial drug resistance determinants. Although integrons are not self-mobilizable, they are usually found in association with transposons and are often located on plasmids, facilitating their mobility (13). Integrons are thus ideally suited for the dissemination and recombination of antimicrobial drug–resistance genes. Integrons are common in S. enterica and make an important contribution to the extent of antimicrobial resistance in this species (10,13,14). Because of their plasmid and transposon association, integrons are assumed to be mobilized predominantly through horizontal gene transfer (10). However, the clonal nature of S. enterica suggests that clonal expansion may also play a role in dissemination of drug resistance. An example of clonal expansion of integron bearing S. enterica is the global distribution of the serovar Typhimurium DT104 clone, which harbors a genetic resistance island known as the Salmonella genomic island 1 (SGI1). This region contains a number of drug resistance elements including 2 integrons with the gene cassettes blaPSE1 and aadA2 and genes for tetracycline and chloramphenicol resistance, which are not integron associated (15).
Clonal expansion of integron-bearing S. enterica would account for the occurrence of a particular genetic lineage with a specific integron in a variety of regions. Horizontal gene transfer would account for the existence of identical integrons in isolates of different genetic lineages. To explore the roles of clonal expansion and horizontal gene transfer in the dissemination of antimicrobial drug resistance caused by class 1 integrons, we investigated the integron structure and genetic lineage of 90 MDR nontyphoidal S. enterica isolates from a global collection comprising >1,900 isolates from 13 countries and 6 continents. A goal of this study was to improve our understanding of the contributions of clonal expansion and horizontal gene transfer to the dissemination of integrons carrying antimicrobial drug–resistance genes in S. enterica to enable the development of improved strategies for the control of antimicrobial drug resistance in this organism as well as other emerging pathogens of public health importance.
Materials and Methods
Bacterial Isolates
A total of 1,920 S. enterica isolates were investigated; 1,743 isolates were collected by laboratories in Argentina, Australia, Belgium, Canada, Denmark, Germany, Italy, the Philippines, South Africa, Spain, Taiwan, Uganda and the United States during September 2001–August 2002 (Table 1). These isolates were collected as part of a separate study that attempted to identify a genetically and geographically diverse group of S. enterica isolates with reduced susceptibilities to fluoroquinolones. The isolates were not selected but rather were collected consecutively, without regard to their antimicrobial drug susceptibility. In addition, 179 isolates were collected by the Allegheny County Department of Health in Pennsylvania during 2002–2003 as part of routine surveillance. Serotyping was performed by the collecting laboratories, except for isolates from Taiwan, which were serotyped by the Pennsylvania Department of Health.
Table 1. Laboratories that provided Salmonella enterica isolates for this study.
| Country | Institution | Contact person |
|---|---|---|
| United States | Centers for Disease Control and Prevention Foodborne and Diarrheal Diseases Laboratory Section, Atlanta | Timothy J. Barrett |
| Canada | Ontario Public Health Laboratory, Toronto | Frances Jamieson |
| Canada | Laboratory for Foodborne Zoonoses, Population and Public Health Branch, Guelph | Cornelius Poppe |
| Argentina | Centro de Estudios en Antimicrobianos, Buenos Aires | Jose Maria Casellas |
| Australia | Queensland Health Scientific Services, Archerfield | John Bates |
| Belgium | Antwerp University Hospital, Antwerp | Herman Goossens |
| Germany | Bundesgesundheitsministerium für gesundheitlichen Verbraucherschutz und Veterinärmedizin, Berlin | Andreas Schroeter |
| South Africa | South African Institute for Medical Research, Johannesburg | Karen Keddy |
| Spain | Institute of Health Carlos III, Enteric Bacteria Laboratory, Madrid | Miguel Usera |
| Italy | Istituto Superiore di Sanita, Rome | Alessandra Carattoli |
| Denmark | Hvidovre Hospital, Copenhagen | Dennis Hansen |
| Taiwan | National Cheng Kung University, Tainan City | Wen-Chien Ko |
The ACSSuT resistance phenotype has become increasingly prevalent in S. enterica, and that phenotype has been commonly associated with class 1 integron carriage in this species. For these reasons, we selected a subset of isolates from the collection that exhibited the ACSSuT resistance phenotype for further investigation. Isolates selected for integron investigation were confirmed to be S. enterica by PCR with primers specific for the invA region of the inv locus (16) (Table 2).
Table 2. Primers used for PCR amplification of Salmonella enterica integrons.
| Primer | Sequence (5′ → 3′) | Target | Reference |
|---|---|---|---|
| 5’CS | GGCATCCAAGCAGCAAGC | 5′ conserved segment | (17) |
| 3’CS | AAGCAGACTTGACCTGAT | 3′ conserved segment | (17) |
| int_F | CGATGCGTGGAGACCGAAACCTT | intI1 | (18) |
| int_R | GTAACGCGCTTGCTGCTTGGATGC | intI1 | (18) |
| invA_F | ACACAGCTCGTTTACGACCTGAAT | invA | (16) |
| invA_R | AGACGACTGGTACTGATCGATATT | invA | (16) |
| sul1_F | GCGCGGCGTGGGCTACC | sul1 | This study |
| sul1_R | CCGCAAGGCTCGCTGGAC | sul1 | This study |
| aadA1_R | CGATGACGCCAACTACCTCTGATA | aadA1internal primer | This study |
| arr2_F | ATTGTTGGCGTTGTTGAAGACTGG | arr2 internal primer | This study |
| cmlA5_F | GAATGGGAATGGGATGCCTGATAG | cmlA5 internal primer | This study |
| oxa10_R | TTTACAAAGCACGAAGACACCATT | blaOXA10 internal primer | This study |
| cmlA_F | GCAGGTCGCGAGGAAAGTAATG | cmlA 5′ forward primer | This study |
| cmlA_R | ACACCGCCCAAGCAGAAGTAGA | cmlA 3′ reverse primer | This study |
| blaOXA30_F | TCGCAAGAAATAACCCAAAAA | blaOXA30 internal primer | This study |
| aacA4_F | AAGCGGGGTTTGAGAGG | aacA4 forward primer | This study |
| aacA4_R | CGCGTACTCCTGGATCGGTTTCTT | aacA4 reverse primer | This study |
| dfr_1_F | TTTAGGCCAGTTTTTACCCAAGAC | dfrA1 internal primer | This study |
| ere_est_R | GCGCCAGCAGAATTATCCTTACAT | ereA2 internal primer | This study |
| aac(6’)IIC_F | CCGCGGGATTGACCAGT | aac(6′)IIC internal primer | This study |
| dfrA12_F | GCTGCGCATTTTGGTTCC | dfrA12 internal primer | This study |
| aadA2_R | TGTCATTGCGCTGCCATTCTCC | aadA2 internal primer | This study |
| qacH_F | GCGTCGCCGTTCTAAATCTGCTAT | qacH internal primer | This study |
| aac_R | GGGCGCCGGGTGTCTGGAG | aacA4 internal primer | This study |
| IS_F | GTCACGCCCCGACCATCACCTTCC | IS1247 internal primer | This study |
| TNP_F | CCGCGCTGGCCGACCTGAAC | Transposase A internal primer | This study |
| ere_F | CCTAACCGGGCGATTCAA | Erythromycin esterase internal primer | This study |
| cmlA_R_internal | ATCACACGCCCCATAAAACGAG | cmlA internal primer | This study |
| arr_R2 | GCGGGATCCAGAACCAGGCGACAT | arr-2 internal primer | This study |
| arr_accA_R | AGAGCGGCTTTGGTTCC | Internal primer arr-2–accA junction | This study |
| ere_F2 | CGCTGATTTCGCTGTCCTGA | ereA internal primer | This study |
| dfrA17_F | AAAAAGGCTAACAAGTCGT | dfrA17 internal primer | This study |
| cml_R2 | GCTGAATTGTGCTCGCTGTCGTA | cml internal primer | This study |
| aadA_con_F | CGACATCATYCCGTGGCGTTAT | aadA forward consensus primer | This study |
| aadA_con_R | CGGCAGCCACATCCTTC | aadA reverse consensus primer | This study |
| aacA4_F | ATGACCTTGCGATGCTCT | aacA4 internal primer | This study |
| aacA4_R | CTCGATGGAAGGGTTAGG | aacA4 internal primer | This study |
| blaOXA30_F | ACACAATACATATCAACTTCGC | blaOXA30-aadA internal primer | This study |
| aadA1_R_S | GGATAACGCCACGGAATGATGTC | aadA1 internal primer | This study |
| albany_PSE1a_F | CCTTTGGGGCCACCTACAG | blaPSE1 primer | This study |
| albany_PSE1b_F | ATCAAAATTATGGGGTTACTTACA | blaPSE1 primer | This study |
| albany_dfr1_F | ATGGTAGCTATATCGAAGAATGGA | dfr primer | This study |
| albany_dfr2_F | AAGTACTGGCTATTGCCTTAGGAG | dfr primer | This study |
| U7-L12 | ACACCTTGAGCAGGGCAAAG | SGI1 left junction | (15) |
| LJ-R1 | AGTTCTAAAGGTTCGTAGTCG | SGI1 left junction | (15) |
| 104-RJ | TGACGAGCTGAAGCGAATTG | SGI1 right junction | (15) |
| 104-D | ACCAGGGCAAAACTACACAG | SGI1 right junction | (15) |
Antimicrobial Drug Resistance Testing
Antimicrobial drug resistance was determined by using the disc diffusion method on Mueller–Hinton agar (Becton, Dickinson and Co., Sparks, MD, USA), according to the manufacturer’s directions. Susceptibility to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline was determined according to the manufacturer’s breakpoints.
Integron Detection and Characterization
Genomic DNA from S. enterica isolates was prepared using the DNeasy Tissue Kit (QIAGEN, Valencia, CA, USA), according to the manufacturer’s directions. Class 1 integron carriage was determined by PCR using primers specific to the intI region of the integrase gene. Isolates positive for integrase were further characterized by PCR using primers specific for the 5′ and 3′ conserved segments (CS) of the integron structure. PCR was performed in a 50-µL volume consisting of 1× PCR buffer II (Applied Biosystems, Foster City, CA, USA), 1.5 mmol/L MgCl2, 0.1 mmol/L dNTPs, 0.33 µM forward and reverse primers, and 1.66 units Amplitaq Gold polymerase (Applied Biosystems). PCR conditions were an initial denaturation at 95°C for 5 min; 35 cycles at 95°C for 1 min, 58°C for 1 min, and 72°C for 5 min plus 5 s each cycle (5 s longer in each subsequent cycle than in the previous cycle); and a final extension at 72°C for 7 min.
Long-range PCR was performed to detect integrons >2.0 kb by using the Gene Amp HiFidelity Kit (Applied Biosystems), according to the manufacturer’s directions. Long-range PCR conditions were an initial denaturation at 94°C for 2 min; 10 cycles at 94°C for 15 s, 58°C for 30 s, and 68°C for 4 min, followed by 20 cycles at 94°C for 15 s, 58°C for 30 s, and 68°C for 4 min plus 5 s each cycle (5 s longer in each subsequent cycle than in the previous cycle); and a final extension at 72°C for 7 min. PCR products were subjected to electrophoresis on 1% agarose gels, stained with ethidium bromide, and visualized using UV illumination on a Gel Doc 2000 documentation system (Bio-Rad, Hercules, CA, USA). For isolates that amplified multiple integrons, PCR products were separated by gel electrophoresis and purified by using either the Qiaquick Gel Extraction Kit (QIAGEN) or the Quantum Prep Freeze ’N Squeeze DNA Gel Extraction Spin Column Kit (Bio-Rad). Some PCR products were cloned before sequencing by using the Topo TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s directions.
Isolates in this study were investigated for the presence of SGI1 and variant SGI1s by PCR using published primers specific for the right (104-RJ, 104-D) and left (U7-L12, LJ-R1) junctions of the chromosomal insertion site (19). Isolates were considered positive for the left or right junctions of the SGI1 if they generated PCR product of the appropriate size with primers specific for that junction. By this method, isolates with the SGI1 would be positive for the left junction but not for the right junction because of the presence of a retronphage between the 104-RJ and 104-D primer sites. Isolates with variant SGI1s would be positive for the left and right junctions.
Gene Cassette Identification
Class 1 integron gene cassettes were identified by PCR and sequence analysis by using the 5′ and 3′ CS primers. Sequencing was performed with the BigDye terminator 3.1 kit (Applied Biosystems) according to the manufacturer’s instructions. Capillary sequence analysis was performed on a 3730 DNA sequence analyzer (Applied Biosystems). Sequences were analyzed and additional primers were designed by using the Lasergene 7.0.0 software package (DNAStar, Madison, WI, USA). Gene cassette homology searches were performed by using BLAST analysis (www.ncbi.nlm.nih.gov/BLAST) (20).
Ninety-one of the 121 integrons found in this study were sequenced in their entirety. In some cases, when integrons were identified that were of the same size as those previously sequenced, their gene cassettes were identified by PCR using primer pairs designed in this study (Table 2).
Multilocus Sequence Typing
Genetic relatedness of isolates was assessed using multilocus sequence typing (MLST). MLST uses sequences ≈500 bp in length from 7 housekeeping genes to define a sequence type (ST). Isolates with the same alleles at all 7 loci are considered to be genetically indistinguishable by MLST and therefore define an ST. Isolates with the same alleles at 6 loci are considered to be closely related genetically.
MLST was performed using the 7-locus scheme described on the Salmonella MLST database (http://web.mpiib-berlin.mpg.de/mlst) (21–24). Visualization of PCR products and sequencing of gene fragments was accomplished as described above for integron gene cassettes. Sequences were analyzed using Bionumerics software V 5.10 (Applied Maths, Austin, TX, USA). Alleles and STs were assigned by the Salmonella MLST database. All isolates in this study and their associated sequence types have been deposited in the Salmonella MLST database.
Definitions of Horizontal Gene Transfer and Clonal Expansion
Horizontal gene transfer was defined as S. enterica isolates belonging to different STs (STs that differ at >1 locus) but bearing the same integron. Clonal expansion was defined as S. enterica isolates bearing the same integron and belonging to the same ST or STs differing at only 1 locus but occurring in >1 location. Because isolates in this study were collected consecutively over a limited time in each location, and because source was the only epidemiologic information available, we could not determine whether genetically related isolates bearing the same integron in a given location were part of an outbreak or whether the isolates reflected clonal expansion beyond an outbreak. Therefore, S. enterica isolates collected from 1 location that belonged to the same ST and harbored identical integron structures were considered to be 1 isolate for classification as either horizontal gene transfer or clonal expansion.
Results
ACSSuT Resistance
Of the 1,920 isolates initially screened by antimicrobial drug susceptibility testing, 104 (4.9%) exhibited the ACSSuT resistance phenotype. The proportion of ACSSuT-resistant isolates ranged from 0% in Australia, Argentina, Belgium, and Canada to 19% in Taiwan and South Africa (Table 3).
Table 3. Source and ACSSuT resistance in a global collection of Salmonella enterica isolates*.
| Source | Total no. isolates | No. (%) ACSSuT-resistant isolates |
|---|---|---|
| Argentina | 148 | 0 |
| Australia | 146 | 0 |
| Belgium | 66 | 0 |
| Canada | 144 | 0 |
| Denmark | 153 | 8 (5.2) |
| Germany | 150 | 1 (0.7) |
| Italy | 156 | 3 (1.9) |
| Philippines | 67 | 6 (8.9) |
| Spain | 151 | 8 (5.3) |
| South Africa | 160 | 30 (18.8) |
| Taiwan | 150 | 29 (19.3) |
| United States/ACHD | 179 | 8 (4.5) |
| United States/CDC | 150 | 1 (0.7) |
| Uganda |
100 |
10 (10.0) |
| Total | 1,920 | 104 (5.4) |
*ACSSuT, ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline; ACHD, Allegheny County Health Department; CDC, Centers for Disease Control and Prevention.
Integron Detection and Characterization
Of the 104 nontyphoidal S. enterica isolates with the ACSSuT resistance phenotype, 90 (86.5%) were positive for the integrase gene and amplified gene cassette regions by PCR with primers for class 1 integron 5′ and 3′ CS regions. Most cassette-positive isolates contained either 1 (n = 61, 68%) or 2 (n = 26, 29%) integrons (Appendix Table). Three isolates contained >3 integrons. Although 16 different integrons were found in this collection, 19 distinct integron profiles could be identified because of multiple integrons in some isolates (Appendix Table). Six integrons contained only 1 gene cassette: aacA4, aadA2, aadB, blaPSE1, dfrA7, or dfrA15. Seven integrons contained 2 or 3 gene cassettes (listed in order of cassette occurrence in the individual integron): aadB/catB3, aac3A-Id/aadA7, blaOXA30/aadA1, dfrA12/orfF/aadA2, dfrA1/orfC, dfrA1/aadA1, and tnpA/dfrA7. A 4.0 kb integron containing the cassettes arr2/cmlA5/blaOXA10/aadA1was found alone or in combination with an integron containing the single gene cassette aacA4. Two other unique large integrons were found in this collection: a 5.8-kb integron with the gene cassettes qacH/dfrA17/ereA/aadA2/cmlA/aadA1 and a 6.0-kb integron with cassettes aac(6')-IIc/ereA2/IS1247/aac3/arr/ereA2.
A total of 121 class 1 integrons were identified. Of these, 91 were fully sequenced (Appendix Table). With 2 exceptions, genetic drift in gene cassette sequences was not observed. One integron containing the single cassette aadA2 showed 1 base difference from other aadA2 gene cassettes found in this study (a T→C transition at position 39 of the gene cassette, resulting in a synonymous change). One blaOXA30 gene cassette exhibited a point mutation (A→G) at position 31, also a synonymous change. All other gene cassettes within the study showed 100% nucleotide identity to GenBank reported cassettes, except for the dfrA17 gene cassette identified in the unique 5.8-kb integron in the Stanley isolates from Taiwan (Appendix Table). This cassette showed 91% sequence identity to a gene cassette found in uncultured bacteria (GenBank accession no. FM179325) and in E. coli (GenBank accession no. EU687490).
Most gene cassettes found in this study confer resistance to the aminoglycosides (aadA1, aadA2, aadA7, aadB, aacA4, aac, aac3A-Id, aac(6')-IIc), trimethoprim (dfrA1, dfrA7, dfrA12, dfrA15, dfrA17), and β-lactams (blaPSE1, blaOXA10, blaOXA30). Other resistance cassettes included those coding for chloramphenicol resistance (cmlA, cmlA5, catB3), erythromycin resistance (ereA2), rifampin resistance (arr2), and resistance to quaternary ammonium compounds (qacH). Because resistance to the aminoglycoside streptomycin and the β-lactam ampicillin were selection criteria for isolates in this study, the predominance of cassettes encoding resistance to those antimicrobial drugs is not unexpected.
Phenotypic resistance to chloramphenicol was also a selection criterion, but only 3 integrons contained gene cassettes for this resistance. None of the integrons identified in this study carried genes coding for tetracycline resistance, although phenotypic resistance to this antimicrobial drug was also an inclusion criterion. Because the SGI1 contains genes for chloramphenicol and tetracycline resistance, which are not located within integrons, tetracycline- and chloramphenicol-resistant isolates that are SGI1 positive most likely contain these genes. Alternatively, resistance to chloramphenicol and tetracycline may be encoded elsewhere on the chromosome or on structures other than integrons in isolates that are not positive for the SGI1.
The SGI1 was identified in 17 isolates (19%) (Appendix Table). Twelve of the SGI1-positive isolates were serovar Typhimurium, belonged to ST 19, and showed the integron pattern (blaPSE1, 1.0 kb and aadA2, 1.2 kb) characteristic of serotype Typhimurium phage type DT 104. An additional Typhimurium ST 19 isolate containing 4 integrons, including blaPSE1 and aadA2, was also positive for SGI1. Four serotype Albany isolates ST 292 were positive for a variant SGI1, which includes the integrons dfrA1/orfC and blaPSE1 (15). This result indicates chromosomal location of these integrons in these isolates.
Genetic Relatedness of Integron-bearing Nontyphoidal S. enterica
The 90 integron-containing S. enterica isolates represented 17 different STs. Thirty-three (37%) Typhimurium isolates belonged to ST 19 or to STs that differ from 19 at only 1 locus. These isolates contained 10 different class 1 integrons, which combined to create 12 integron profiles (Appendix Table). Eleven (12%) isolates of serovar Isangi belonged to ST 216 or to STs that differ from 216 at 1 or 2 loci. These isolates contained 3 integrons and 4 integron profiles. The remaining isolates represented diverse STs, which differed from each other at 6 or 7 of the MLST loci.
Integron Distribution Across Nontyphoidal S. enterica Genetic Lineages
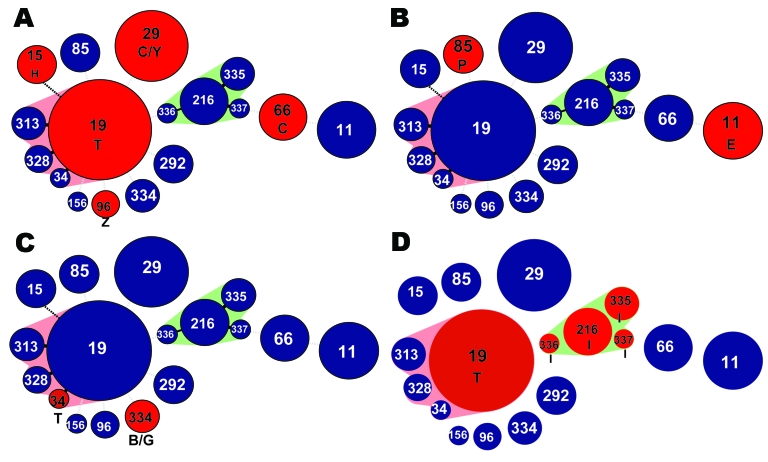
This study identified 5 class 1 integrons that were distributed across different genetic lineages, supporting the role of horizontal gene transfer in the dissemination of antimicrobial drug resistance in nontyphoidal S. enterica. The class 1 integron dfrA12/orfF/aadA2, which confers resistance to trimethoprim and streptomycin/spectinomycin, was identified in S. enterica from 5 different serotypes belonging to 5 STs (Figure, panel A). This integron was geographically widespread, being found in isolates from Europe, the United States, Taiwan, the Philippines, and South Africa. The integron containing the single trimethoprim-encoding dfrA7 cassette was present in isolates from 2 serotypes, 2 STs and 3 different areas (Figure, panel B). The integron dfrA1/aadA1 was found in isolates from 3 serotypes, 2 STs, and 2 areas (Figure, panel C).
Figure.
Minimum spanning trees depicting integron distribution across Salmonella enterica genetic lineages. A) dfrA12/orfF/aadA2; B) dfrA7; C) dfrA1/aadA1; D) arr2/blaOXA30/cmlA5/aadA2. Circles represent unique sequence types (STs). Red circles represent the STs that carried the integron involved in horizontal gene transfer. Numbers in circles represent the ST. Circle size reflects number of isolates in each ST. Pink and green shading indicates closely related groups of isolates. Letters refer to serotypes: B, Brandenburg; C, Cholerasuis; E, Enteriditis; H, Heidelberg; G, Goettingen; I, Isangi; P, Paratyphi A; Z, Schwarzengrund; Y, Stanley; T, Typhimurium. Geographic sources of isolates are as follows: Panel A: ST66, serotype C, Taiwan; ST29, serotype C, Taiwan; ST29, serotype Y, Taiwan; ST96, serotype Z, Denmark and Taiwan; ST19, serotype T, US Centers for Disease Control and Prevention and South Africa; ST15, serotype H, Philippines; Panel B: ST11, serotype E, Uganda and South Africa; ST85, serotype P, Denmark; Panel C: ST334, serotype G, Spain; ST334, serotype B, Spain; ST34, serotype T, Germany; Panel D: ST19, serotype T, South Africa; ST216, ST335, ST336, and ST 337, serotype I, South Africa.
Isolates of ST 216, 335, 336, and 337 (serovar Isangi) and ST 19 (serovar Typhimurium) from South Africa contained an identical 4.0 kb integron not previously reported in S. enterica, with the cassettes arr2/cmlA5/blaOXA10/aadA1 (Figure, panel D). In some serovar Typhimurium ST 19 isolates, this integron was found in combination with the aacA4 integron. The presence of a unique integron in genetically unrelated isolates from the same area indicates that this integron may have been horizontally transferred.
An integron containing the single gene cassette aacA4 was found alone in isolates of serovar Isangi with ST 335 from Uganda and South Africa. This integron was also found in serovar Typhimurium isolates with ST 19, both alone and in combination with the integron arr2/cmlA5/blaOXA10/aadA1. These isolates belong to different genetic lineages, in that they differ at all 7 MLST loci. The occurrence of the same integron in these genetically unrelated isolates further supports a role for horizontal gene transfer in the dissemination of antimicrobial drug resistance in nontyphoidal S. enterica.
Evidence for Clonal Expansion of Integron-mediated MDR in S. enterica
The serotype Typhimurium phage type DT104 integron pattern, with 2 class 1 integrons of sizes 1.0 and 1.2 kb and bearing the gene cassettes blaPSE1 and aadA2, was found in 12 (13%) serovar Typhimurium isolates of ST 19 (Table 4) and in 1 additional serovar Typhimurium ST 19 isolate, which also contained 2 other integrons. This result is consistent with the hypothesis that a common ancestor has undergone clonal expansion in a number of areas.
Table 4. Evidence of clonal expansion among isolates from a global collection of Salmonella enterica isolates*.
| Integron profile | No. isolates | Serotype | Sequence type | Source |
|---|---|---|---|---|
|
blaPSE1, aadA2 |
4 | Typhimurium | 19 | United States/ACHD |
| 2 | Typhimurium | 19 | Spain | |
| 3 | Typhimurium | 19 | Italy | |
| 3 |
Typhimurium |
19 |
South Africa |
|
|
blaOXA30/aadA1
|
1 | Typhimurium | 328 | Philippines |
| 2 | Typhimurium | 19, 328 | Taiwan | |
| 1 |
Typhimurium |
313 |
South Africa |
|
|
aadB/catB3
|
1 | Typhimurium | 19 | Taiwan |
| 1 |
Typhimurium |
328 |
Philippines |
|
|
dfrA12/orfF/aadA2
|
1 | Schwarzengrund | 96 | Taiwan |
| 1 |
Schwarzengrund |
96 |
Denmark |
|
|
dfrA12/orfF/aadA2
|
1 | Typhimurium | 19 | United States/CDC |
| 2 |
Typhimurium |
19 |
South Africa |
|
|
aacA4
|
1 | Isangi | 335 | South Africa |
| 1 |
Isangi |
335 |
Uganda |
|
| dfrA7 | 6 | Enteritidis | 11 | Uganda |
| 2 | Enteritidis | 11 | South Africa |
*ACHD, Allegheny County Health Department; CDC, Centers for Disease Control and Prevention.
Serovar Typhimurium isolates of ST 19 from Taiwan and ST 328 from the Philippines contained 2 class 1 integrons with the cassettes aadB/catB3 and blaOXA30/aadA1. The integron blaOXA30/aadA1 was also found alone in an isolate of serovar Typhimurium ST 328 from Taiwan and in combination with an integron containing the cassette aadB in an isolate of serovar Typhimurium ST 313 from South Africa. ST 313 and ST 328 are closely related to ST 19, differing from it at only 1 locus (ST 313 differs from ST 19 at the MLST locus sucA, ST 328 and ST 19 differ at MLST locus aroC; see Appendix Table). Therefore, these isolates are all closely related, and their integrons may represent an instance of clonal expansion.
Although the class 1 integron dfrA12/orfF/aadA2 appears to have been circulated through horizontal gene transfer, this integron has also spread through clonal expansion of nontyphoidal S. enterica. This integron appeared in isolates of serovar Schwarzengrund ST 96 in Taiwan and Denmark and in serovar Typhimurium isolates of ST 19 in the United States and South Africa. Similarly, integrons with the single gene cassettes aacA4 and dfrA7 also exhibited both horizontal gene transfer and clonal expansion. The aacA4 integron was found in serovar Isangi isolates of ST 335 in Uganda and South Africa. The dfrA7 integron was found in serovar Enteritidis isolates of ST 11, also in Uganda and South Africa.
Discussion
To better understand the dissemination of integron-mediated antimicrobial drug resistance, this study characterized the class 1 integrons and genetic lineages associated with 90 multidrug-resistant isolates obtained from a global collection of nontyphoidal S. enterica. Integrons found in this collection were diverse in size, gene cassette combination and distribution, and presented evidence for roles of clonal expansion and horizontal gene transfer.
Horizontal gene transfer is an important factor in the dissemination of antimicrobial drug–resistance genes, particularly when those genes are associated with mobile elements such as plasmids, transposons, and integrons (10,11,14). In this study, the widespread distribution of the dfrA12/orfF/aadA2 integron among several STs and across several distinct regions is an example of horizontal gene transfer, as is the presence of the integrons dfrA7, dfrA1/aadA1, and arr2/cmlA5/blaOXA10/aadA1 in different genetic backgrounds (when assessed by MLST). These integrons are potentially capable of transmitting drug resistance to other S. enterica isolates or to other bacteria.
Integrons are widely distributed among bacterial species. The integron found in the greatest number of different genetic lineages in this study, dfrA12/orfF/aadA2, has been previously reported in a number of species, including E. coli (GenBank accession no. AF335108, unpub. data), Serratia marcesens (Genbank accession no. AF284063, unpub. data), and Salmonella (25). The integron dfrA1/aadA1 has been documented in E. coli from Turkey (26) and cited in E. coli in GenBank entries from India (GenBank accession no. EF417897, unpub. data) and Kenya (GenBank accession no. EF417897, unpub. data), in Klebsiella pneumonia from Poland (GenBank accession no. AY007807, unpub. data), and in S. enterica (GenBank accession no. AM746675, unpub. data). Transfer of integrons between different bacterial species has been documented in the clinical setting, which poses a serious threat to containment of nosocomial infections (27). The existence of identical integrons in different types of bacteria and the ability of these integrons to be transferred in vivo indicates that many bacteria acquire integrons from a common pool. This is an important consideration in efforts to halt the development and spread of antimicrobial drug resistance.
In this study, clonal expansion of S. enterica appears to be responsible for a large fraction of the dissemination of drug resistance integrons. Although several integrons demonstrated evidence of clonal expansion, including aacA4, aadB/catB3, blaOXA30/aadA1, dfrA7, and dfrA12/orfF/aadA2, the strongest evidence is presented by the prevalence and geographic ubiquity of the serotype Typhimurium DT104 pattern clone, in which the resistance-encoding integrons (blaPSE1 and aadA2) are chromosomally integrated. These integrons are still mobilizable (28) but chromosomal location may make them more likely to be disseminated through clonal expansion than through horizontal gene transfer, particularly in the absence of antimicrobial selective pressure.
Our study assessed mechanisms of dissemination of integrons in a collection that is more genetically and geographically diverse than is typical for studies of integrons in S. enterica. MLST, used for assessment of genetic relatedness of isolates in this study, is better suited to analysis of global populations than other commonly-used methods, such as pulsed-field gel electrophoresis. In addition, this study focused on the relative contributions of clonal expansion and horizontal gene transfer to the dissemination of class 1 integron borne genes coding for antimicrobial drug resistance, which has not previously been well explored.
Antimicrobial drug resistance is a serious and increasing problem in S. enterica and in other gram-negative pathogens, such as Pseudomonas aeruginosa, Acinetobacter baumannii, K. pneumoniae and E. coli (29). The genes that code for antimicrobial drug resistance in these pathogens have proven to be remarkably mobile and widely distributed within and between species. The dissemination of integrons bearing antimicrobial drug resistance gene cassettes in Salmonella and other bacteria is a complex process that involves both the horizontal transfer of mobile genetic elements and the expansion of particularly fit clones. The combined effect of these mechanisms is that, if integrons confer an adaptive benefit caused by the presence of antimicrobial drug selective pressure or if clones harboring these integrons have increased fitness caused by other factors, then the integrons may disseminate rapidly both geographically and among diverse species. It is important to understand these mechanisms of transmission to develop methods for surveillance and control of antimicrobial drug resistance.
Supplementary Material
Description of integrons and associated serotypes, MLST results, and countries of origin of 90 isolates with the ACSSuT phenotype used in this study*
Acknowledgments
We thank Stanley M. Reynolds and Nkuchia M. M’ikanatha for assistance with serotyping; and Bruce W. Dixon, Dennis Hansen, Tersia Kruger, Karen H. Keddy, Alessandra Carattoli, Jose Maria Casellas, Wen Chien Ko, John Bates, Miguel Usera,Herman Goossens, Celia C. Carlos, Jean Whichard, Timothy J. Barrett, Fred Angulo, C. Anthony Hart, and Frances Jamieson for providing isolates. The analysis of MLST data in the study depended in part on the website for MLST at the Max-Planck Institute for Infectious Biology, Berlin (http://web.mpiib-berlin.mpg.de), curated by Mark Achtman.
This study was supported by grant R56 AI059385 from the National Institute of Allergy and Infectious Diseases (Bethesda, MD, USA). The isolate collection was assembled with the support of grant R01-C1322404 from the Centers for Disease Control and Prevention, Atlanta, GA, USA.
Biography
Ms Krauland is a doctoral student in the Epidemiology Department at the Graduate School of Public Health, University of Pittsburgh. She has conducted her doctoral research in the Infectious Diseases Epidemiology Research Unit, University of Pittsburgh School of Medicine and Graduate School of Public Health. Her research interests include dissemination of antimicrobial drug resistance, molecular evolution of pathogens, and molecular subtyping methods.
Footnotes
Suggested citation for this article: Krauland MG, March JW, Paterson DL, Harrison LH. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg Infect Dis [serial on the Internet]. 2009 Mar [date cited]. Available from http://www.cdc.gov/EID/content/15/3/388.htm
References
- 1.Hald T, Lo Fo Wong DM, Aarestrup FM. The attribution of human infections with antimicrobial resistant Salmonella bacteria in Denmark to sources of animal origin. Foodborne Pathog Dis. 2007;4:313–26. 10.1089/fpd.2007.0002 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. World Health Organization antimicrobial resistance fact sheet no. 139. April 2003. [cited 2008 Mar 25]. Available from http://www.who.int/mediacentre/factsheets/fs139
- 3.Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, et al. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38(Suppl 3):S127–34. 10.1086/381578 [DOI] [PubMed] [Google Scholar]
- 4.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2004. Atlanta: The Centers; 2007. [Google Scholar]
- 6.Davis MA, Hancock DD, Besser TE. Multiresistant clones of Salmonella enterica: the importance of dissemination. J Lab Clin Med. 2002;140:135–41. 10.1067/mlc.2002.126411 [DOI] [PubMed] [Google Scholar]
- 7.Butaye P, Michael GB, Schwarz S, Barrett TJ, Brisabois A, White DG. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect. 2006;8:1891–7. 10.1016/j.micinf.2005.12.020 [DOI] [PubMed] [Google Scholar]
- 8.Threlfall EJ. Epidemic Salmonella typhimurium DT 104—a truly international multiresistant clone. J Antimicrob Chemother. 2000;46:7–10. 10.1093/jac/46.1.7 [DOI] [PubMed] [Google Scholar]
- 9.Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, et al. Evolutionary history of Salmonella typhi. Science. 2006;314:1301–4. 10.1126/science.1134933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli A. Importance of integrons in the diffusion of resistance. Vet Res. 2001;32:243–59. 10.1051/vetres:2001122 [DOI] [PubMed] [Google Scholar]
- 11.Blahna MT, Zalewski CA, Reuer J, Kahlmeter G, Foxman B, Marrs CF. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J Antimicrob Chemother. 2006;57:666–72. 10.1093/jac/dkl020 [DOI] [PubMed] [Google Scholar]
- 12.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis. 2007;13:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carattoli A. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr Issues Mol Biol. 2003;5:113–22. [PubMed] [Google Scholar]
- 14.Carattoli A, Villa L, Pezzella C, Bordi E, Visca P. Expanding drug resistance through integron acquisition by IncFI plasmids of Salmonella enterica Typhimurium. Emerg Infect Dis. 2001;7:444–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. The genetics of Salmonella genomic island 1. Microbes Infect. 2006;8:1915–22. 10.1016/j.micinf.2005.12.028 [DOI] [PubMed] [Google Scholar]
- 16.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, Ginocchio C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–9. 10.1016/0890-8508(92)90002-F [DOI] [PubMed] [Google Scholar]
- 17.Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maguire AJ, Brown DF, Gray JJ, Desselberger U. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob Agents Chemother. 2001;45:1022–9. 10.1128/AAC.45.4.1022-1029.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyd D, Cloeckaert A, Chaslus-Dancla E, Mulvey MR. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob Agents Chemother. 2002;46:1714–22. 10.1128/AAC.46.6.1714-1722.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 21.Torpdahl M, Skov MN, Sandvang D, Baggesen DL. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J Microbiol Methods. 2005;63:173–84. 10.1016/j.mimet.2005.03.006 [DOI] [PubMed] [Google Scholar]
- 22.Falush D, Torpdahl M, Didelot X, Conrad DF, Wilson DJ, Achtman M. Mismatch induced speciation in Salmonella: model and data. Philos Trans R Soc Lond B Biol Sci. 2006;361:2045–53. 10.1098/rstb.2006.1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidgell C, Reichard U, Wain J, Linz B, Torpdahl M, Dougan G, et al. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect Genet Evol. 2002;2:39–45. 10.1016/S1567-1348(02)00089-8 [DOI] [PubMed] [Google Scholar]
- 24.Harbottle H, White DG, McDermott PF, Walker RD, Zhao S. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J Clin Microbiol. 2006;44:2449–57. 10.1128/JCM.00019-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerra B, Soto SM, Arguelles JM, Mendoza MC. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:-]. Antimicrob Agents Chemother. 2001;45:1305–8. 10.1128/AAC.45.4.1305-1308.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozgumus OB, Celik-Sevim E, Alpay-Karaoglu S, Sandalli C, Sevim A. Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in Turkey. J Microbiol. 2007;45:379–87. [PubMed] [Google Scholar]
- 27.Leverstein-van Hall MA, Box AT, Blok HE, Paauw A, Fluit AC, Verhoef J. Evidence of extensive interspecies transfer of integron-mediated antimicrobial resistance genes among multidrug-resistant Enterobacteriaceae in a clinical setting. J Infect Dis. 2002;186:49–56. 10.1086/341078 [DOI] [PubMed] [Google Scholar]
- 28.Boyd DA, Peters GA, Ng L, Mulvey MR. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 2000;189:285–91. 10.1111/j.1574-6968.2000.tb09245.x [DOI] [PubMed] [Google Scholar]
- 29.O’Mahony R, Quinn T, Drudy D, Walsh C, Whyte P, Mattar S, et al. Antimicrobial resistance in nontyphoidal salmonella from food sources in Colombia: evidence for an unusual plasmid-localized class 1 integron in serotypes typhimurium and anatum. Microb Drug Resist. 2006;12:269–77. 10.1089/mdr.2006.12.269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of integrons and associated serotypes, MLST results, and countries of origin of 90 isolates with the ACSSuT phenotype used in this study*