Abstract
The Bladder Cancer-Associated Protein gene (BLCAP; previously BC10) is a tumour suppressor that limits cell proliferation and stimulates apoptosis. BLCAP protein or message are downregulated or absent in a variety of human cancers. In mouse and human, the first intron of Blcap/BLCAP contains the distinct Neuronatin (Nnat/NNAT) gene. Nnat is an imprinted gene that is exclusively expressed from the paternally inherited allele. Previous studies found no evidence for imprinting of Blcap in mouse or human. Here we show that Blcap is imprinted in mouse and human brain, but not in other mouse tissues. Moreover, Blcap produces multiple distinct transcripts that exhibit reciprocal allele-specific expression in both mouse and human. We propose that the tissue-specific imprinting of Blcap is due to the particularly high transcriptional activity of Nnat in brain, as has been suggested previously for the similarly organized and imprinted murine Commd1/U2af1-rs1 locus. For Commd1/U2af1-rs1, we show that it too produces distinct transcript variants with reciprocal allele-specific expression. The imprinted expression of BLCAP and its interplay with NNAT at the transcriptional level may be relevant to human carcinogenesis.
INTRODUCTION
The Bladder Cancer-Associated Protein gene (BLCAP; previously BC10) was first identified in a differential gene expression study where it was found to be specifically silenced in invasive human bladder transitional cell carcinoma (1). Loss of BLCAP expression has since also been observed in renal-cell carcinoma (2) and cervical cancer (3). Over-expression of BLCAP in HeLa and human tongue carcinoma cells stimulates apoptosis (3,4). Relative to other tissues, the Novartis gene expression atlas (SymAtlas) shows BLCAP mRNA levels to be high in brain, the pituitary and the immune system (5) (Affymetrix probe set 201032_at).
BLCAP is translated in vitro into a ∼9.8 kDa protein of 87 amino acids whose function is unknown but which putatively includes two trans-membrane domains, cytoplasmic domains at the N and C terminals, a phosphorylation site and may bind DNA (6) (UniProt/SwissProt: P62952). At the amino acid level, BLCAP is identical in human, mouse (P62951), orangutan (Q5R692), rat (P62950), cat (P62953) and cattle (P62954).
Human BLCAP is located on Chr 20q11.2 and is annotated in RefSeq (NM_006698) as having two exons, with the open reading frame confined to the last exon. An exceptional feature of the locus is the existence of a distinct gene, Neuronatin (NNAT), within the intron of BLCAP. Distinct CpG islands (CGI) overlap the transcription start sites (TSSs) of both NNAT and BLCAP, with an additional CGI in the body of NNAT. NNAT is imprinted and paternally expressed in fetal brain (7). The same study found ∼50% of CpGs in a region encompassing the NNAT CGIs to be methylated in brain and other tissues, suggestive of differential, maternal allele-specific methylation. However, no evidence was found for imprinted expression of BLCAP in fetal brain or other tissues, and the BLCAP CGI was unmethylated in all investigated tissues (7).
The orthologous murine Blcap/Nnat locus on Chr 2 largely recapitulates the genomic organization and expression profile of human BLCAP/NNAT. The intron harbours Nnat whose imprinting and paternal-specific expression was first observed in mouse embryo and newborn brain (8). Blcap mRNA levels are relatively high in brain and the pituitary, but not the immune system (SymAtlas probe set gnf1m03525_a_at). In addition, Blcap expression is high in adipose tissue, oocytes and zygotes, for which SymAtlas contains no human data. The promoters of Nnat and Blcap are associated with CGIs that are smaller than in human. In embryos, the Nnat CGI and gene body are methylated only on the maternal allele (9). The Blcap CGI is unmethylated, and expression assays in androgenetic, parthenogenetic and uniparentally disomic embryos suggested that Blcap is not imprinted (10).
Here, we show that murine and human Blcap generate multiple transcripts that are imprinted in the brain where expression occurs preferentially from either the maternal or the paternal allele in a transcript-specific manner. For the mouse, we quantify the relative contributions of representative transcripts to the mRNA pool and demonstrate that the regulation of the locus depends on maternal germline methylation. We argue that past studies found no evidence of Blcap imprinting due to the tissue-specific nature of Blcap imprinting (7,10). We compare the locus with other imprinted domains where the gene carrying the primary imprint mark is located in the intron of a gene that also exhibits parent-of-origin-specific expression (11,12). Based on similarities with these other loci, we propose that the high level of Nnat transcription in brain relative to other tissues contributes to the brain-specific imprinted expression pattern of Blcap. Our findings shed new light on the epigenetic regulation of a tumour suppressor gene and have implications for its deregulation during carcinogenesis.
RESULTS
Multiple Blcap transcripts are generated from distinct promoters in human and mouse
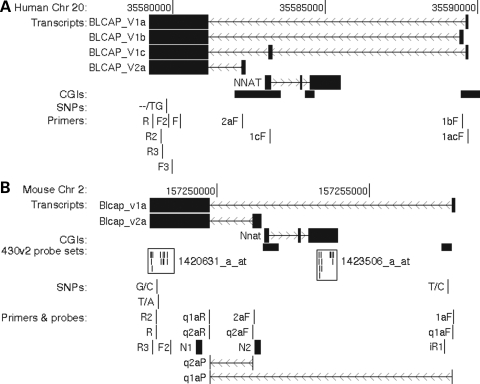
Several mouse (e.g. CJ174807) and human expressed sequence tags (ESTs; e.g. DA770824, DA366527, DA277117) suggested that additional Blcap TSSs exist in both species. In human, ESTs also indicated an alternatively spliced third internal BLCAP exon overlapping the NNAT coding sequence (e.g. DA377434). We have identified at least four distinct human and two distinct mouse Blcap transcripts (Fig. 1). They fall into two categories, depending on whether their TSSs are downstream or upstream of Nnat. We refer to Blcap transcripts with TSSs downstream of Nnat collectively as Blcap_v1 and denote specific transcripts with a one letter suffix. Analogously, Blcap_v2 stands for isoforms with TSSs upstream of Nnat.
Figure 1.
Genomic organization and features of the human BLCAP (A) and murine Blcap (B) loci. Each panel shows a base pair scale, the likely exon–intron structure of transcripts and the positions of CpG islands (CGIs), single nucleotide polymorphisms (SNPs) and PCR primers referred to in the text. Base pair coordinates are with respect to the human Mar 2006 v36.1 and the mouse Feb 2006 v36.0 NCBI genome builds. (A) and (B) are drawn to the same scale, each displaying a genomic region of ∼13 kb. Transcript structures were inferred from ESTs and the results of the PCR, sequencing and northern blot experiments described in the text. Transcripts are shown with filled boxes delineating exons and thin lines with chevrons denoting introns and direction of transcription. (B) also displays the locations of mouse expression microarray (Affymetrix 430v2), TaqMan quantitative real-time PCR and northern probes. The TaqMan probes are intron-spanning, as indicated. Microarray probes are displayed as small vertical lines, and a frame indicates the probes belonging to the same probe set. Primer and probe sequences, and experimental conditions are listed in Supplementary Material, Table S1. The figure was generated with the UCSC genome browser (40).
Microarray measurements suggest preferential maternal expression of murine Blcap
We measured gene expression in whole head samples of newborn mice with maternal and paternal uniparental duplication (UpDp) of distal Chr 2 (13), which includes the Blcap locus. Thus, both homologous copies of the Blcap locus are of either the maternal or the paternal epigenotype. Two microarrays containing probe sets specific to Blcap and Nnat (Fig. 1B) were hybridized with a MatDp(dist2) sample and a PatDp(dist2) sample, respectively. The Blcap probe set detected ∼2.4-fold more Blcap expression in MatDp(dist2) than in PatDp(dist2) material. The Nnat probe set, as expected, essentially failed to detect Nnat message in the MatDp(dist2) sample, while registering high Nnat levels in PatDp(dist2) newborn head (MatDp(dist2)/PatDp(dist2) = ∼0.006).
SNP-based allele-specific direct sequencing and quantitative real-time PCR confirm preferential maternal expression of Blcap_v1a in mouse brain
A T/A SNP between the C57BL/6J (B6) and Mus mus castaneus (cast) mouse strains was identified in the last Blcap exon (Fig. 1B). It was used to determine the allelic origin of Blcap_v1a expression in newborn and adult brain samples of B6 × cast (B×C) and cast × B6 (C×B) inter-subspecific hybrid offspring. Blcap_v1a was specifically amplified from cDNA and directly sequenced. The maternal allele always dominated the sequencing traces, consistent with preferential maternal expression of Blcap_v1a (Fig. 2A). For further confirmation and to more accurately quantify the contribution of the parental alleles, we measured Blcap_v1a expression in MatDp(dist2) and PatDp(dist2) newborn brain samples relative to a matched non-UpDp control sample by TaqMan quantitative real-time PCR (qRT–PCR). We observed ∼2.2-fold more Blcap_v1a expression in MatDp(dist2) than in the control, while PatDp(dist2) brain retained ∼25% of the expression level in the control (Fig. 3A). This corresponds to ∼8.8-fold more Blcap_v1a transcript in MatDp(dist2) than in PatDp(dist2) brain, a larger fold-change than observed via microarray in whole head samples.
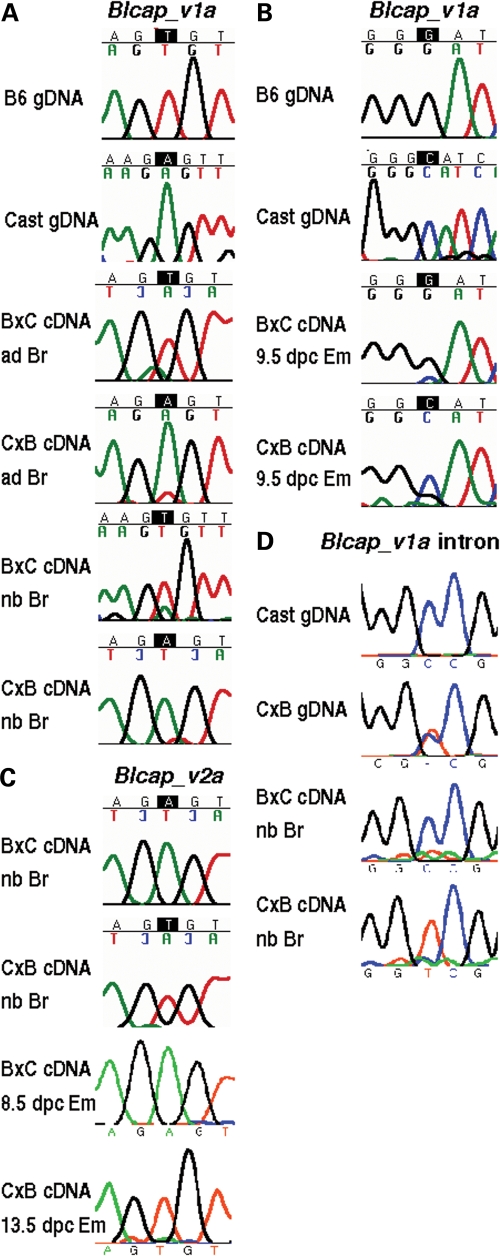
Figure 2.
Allele-specific sequencing results for murine Blcap. (A) The two top-most rows of sequencing traces labelled gDNA (genomic DNA) show the B6 (T) and cast (A) genotypes at the site of the T/A SNP between the two strains (Fig. 1B). The other rows contain the traces obtained from amplifying and sequencing Blcap_v1a using adult (ad) and newborn (nb) brain (Br) cDNA samples of B6 × cast (B×C) and cast × B6 (C×B) inter-subspecific hybrids (denoted in dam × sire order). In both adult and newborn brain and for both directions of the cross, the dominant peak in the trace at the site of the SNP corresponds to the maternal allele, indicating Blcap_v1a expression from preferentially the maternal allele. (B) A G/C SNP between B6 and cast (top gDNA traces and Fig. 1B) was used to determine the allelic origin of Blcap_v1a expression in 9.5 dpc embryo (Em). The maternal allele dominated the sequence traces for the B×C and C×B hybrid embryo samples, indicating preferential maternal expression. (C) The same T/A SNP as in (A) was used to test for allele-specific expression of Blcap_v2a (Fig. 1B). In B×C and C×B newborn brain and whole embryo, Blcap_v2a was expressed from exclusively the paternal allele, in contrast to Blcap_v1a. (D) Allele-specific sequencing of transcripts in the first intron of Blcap_v1a using a T/C SNP between the B6 and cast strains (top gDNA traces and Fig. 1B). Transcripts amplified from B×C and C×B newborn (nb) brain (Br) were predominantly expressed from the paternal allele.
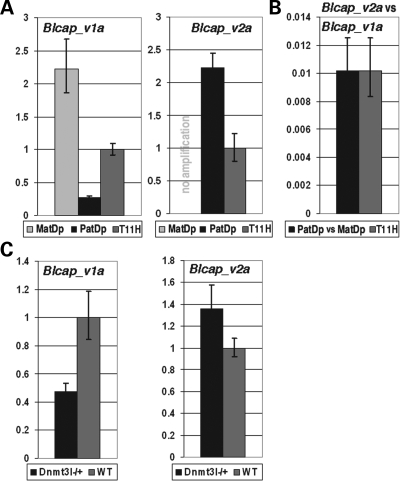
Figure 3.
TaqMan quantitative real-time PCR (qRT–PCR) measurements of Blcap_v1a and Blcap_v2a. Error bars indicate the 95% confidence interval. (A) Blcap_v1a and Blcap_v2a levels in MatDp(dist2) and PatDp(dist2) newborn (nb) brain relative to a matched non-UpDp control (T11H). Blcap_v1a was ∼2.2 times more abundant in MatDp(dist2) than in control nb brain, and achieved only ∼25% of the control expression level in PatDp(dist2) nb brain, which corresponds to a MatDp(dist2)/PatDp(dist2) expression ratio of ∼8.8. This reflects the preferential maternal expression of Blcap_v1a. Blcap_v2a was not present in MatDp(dist2) nb brain (no amplification), and ∼2-fold more abundant in PatDp(dist2) than in non-UpDp nb brain, consistent with paternal-only expression. (B) Left bar: amount of Blcap_v2a in PatDp(dist2) nb brain relative to Blcap_v1a in MatDp(dist2) nb brain. Right bar: amount of Blcap_v2a in non-UpDp nb brain relative to Blcap_v1a in the same sample. In both cases, Blcap_v2a reached only ∼1% of the Blcap_v1a transcript level, i.e. in nb brain, Blcap_v1a is ∼100-fold more abundant than Blcap_v2a. (C) Quantification of Blcap_v1a and Blcap_v2a levels in 8.5 dpc Dnmt3l−/+ embryos relative to wild-type (WT) litter-mates. Expression of Blcap_v1a was reduced to ∼50% of WT level in Dnmt3l−/+ embryos, while Blcap_v2a increased in abundance by ∼40%. This is consistent with the notion that the maternal allele-specific methylation of the Nnat promoter CGI is established during oogenesis and that methylation of this CGI positively regulates Blcap_v1a expression, while it inhibits Blcap_v2a expression.
Preferential maternal Blcap_v1a expression is present in embryo, but not in placenta or adult murine tissues other than brain
A second, G/C SNP between B6 and cast (Fig. 1B) was used for allele-specific sequencing assays of cDNA derived from whole embryo at 9.5 days post coitum (dpc), 13.5 dpc placenta and testis, heart, lung, liver and skeletal muscle tissue of adult mice. Preferential maternal Blcap_v1a expression was only observed in embryo (Fig. 2B and Supplementary Material, Fig. S1), likely due to the brain component of this RNA sample. Biallelic expression of Blcap_v1a in tissues other than brain likely contributed to the reduced fold-change observed via microarray in whole head samples in comparison to the brain samples assayed by qRT–PCR.
Blcap_v2a originating upstream of Nnat is, like Nnat, only expressed from the paternal allele
EST evidence for an alternative Blcap TSS upstream of the Nnat promoter prompted us to determine whether this transcript isoform was imprinted (Blcap_v2a in Fig. 1B). We amplified a product with intron boundaries identical to EST CJ174807 including almost the entire length of the last Blcap exon. Sequencing showed that in newborn brain and whole embryo, Blcap_v2a was exclusively expressed from the paternal allele (Fig. 2C). This was confirmed by TaqMan qRT–PCR: amplification failed in MatDp(dist2) newborn brain due to a lack of Blcap_v2a transcripts, while an approximately 2-fold increase of Blcap_v2a was observed in PatDp(dist2) newborn brain relative to the non-UpDp control (Fig. 3A).
Since the microarray probe set for Blcap is located in the last exon that is shared by Blcap_v1a and Blcap_v2a, the microarray measurements likely reflect the cumulative levels of both isoforms. Nevertheless, the microarray detected a ∼2.4-fold change in MatDp(dist2) relative to PatDp(dist2), suggesting that the paternally expressed Blcap_v2a is much less abundant than the predominantly maternally expressed Blcap_v1a. Based on our previous qRT–PCR measurements in newborn brain, Blcap_v1a is ∼100-fold more abundant than Blcap_v2a (Fig. 3B). Hence, Blcap_v2a is unlikely to have significantly contributed to the microarray measurements so that the small MatDp(dist2)/PatDp(dist2) ratio in relation to the qRT–PCR results is most likely due to the brain-specific imprinting of Blcap.
Transcript analysis by northern hybridization confirms findings for Blcap_v1a and Blcap_v2a
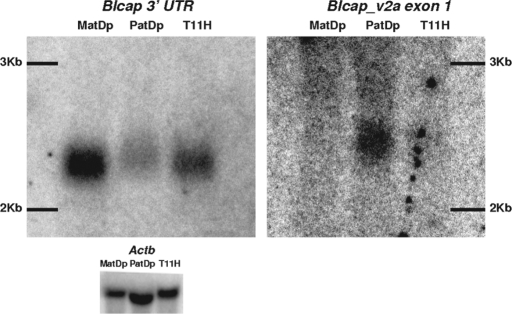
Two northern blots with newborn brain RNA of MatDp(dist2), PatDp(dist2) and non-UpDp mice were hybridized with a probe to the last Blcap exon and a probe to the first exon of Blcap_v2a. The former generated a single band in all three lanes (Fig. 4), which corresponds to the known size of Blcap_v1a (2079 bp), the more abundant of the two Blcap transcripts. The lack of additional distinct bands is evidence for the absence of additional major Blcap transcripts. However, the last Blcap exon contributes >87% to the length of the two known transcripts so that additional transcripts with small alternative first exons may have been subsumed in the one band. The band was most intense in the MatDp(dist2) and least intense in the PatDp(dist2) lane, even though the RNA load of the PatDp(dist2) lane was higher (Fig. 4). This indicates that in newborn brain, transcripts sharing the last Blcap exon cumulatively originate most often from the maternal allele, consistent with the microarray measurements in whole newborn head.
Figure 4.
Detection of Blcap_v1a and Blcap_v2a in MatDp(dist2), PatDp(dist2) and non-UpDp (T11H) newborn brain RNA by northern hybridization. The signal obtained with an Actb probe is shown as a loading control (bottom). Left: Hybridization signal produced by the probe designed to the last Blcap exon (N1 in Fig. 1B). A single band was observed in all three lanes where the relative band intensity was highest in the MatDp(dist2) lane and lowest in the PatDp(dist2) lane. Band position and relative intensities were consistent with the known size of the major Blcap transcript Blcap_v1a (2079 bp) and overall preferential expression of Blcap from the maternal allele in newborn brain. Right: Results obtained with a probe specific to the first exon of Blcap_v2a (N2 in Fig. 1B). No band was present in the MatDp(dist2) lane. A single band was visible in the PatDp(dist2) lane. At the corresponding position in the non-UpDp lane, a very faint band was present. This indicates paternal-only expression of Blcap_v2a. The band was slightly shifted upward relative to the band obtained with the N1 probe, as is expected of the slightly larger Blcap_v2a (2258 bp). The exposure times necessary to visualize the results for Blcap_v2a were considerably longer than for the N1 probe, which is consistent with the low abundance of Blcap_v2a relative to Blcap_v1a as determined by qRT–PCR.
The probe specific for Blcap_v2a generated an obvious band in only the PatDp(dist2) lane at a position consistent with the slightly larger size of Blcap_v2a (2258 bp) relative to Blcap_v1a. At the same position, a very faint band could be seen in the non-UpDp lane, but there was no signal in the MatDp(dist2) lane, indicating paternal-only expression.
Blcap expression is regulated by maternal germline methylation
In the mouse, the DNA methyltransferase 3-like (Dnmt3l) protein is required during oogenesis to establish the maternal-specific methylation marks that in the offspring, control the parent-of-origin-specific expression of most imprinted genes (14,15). In Dnmt3l−/+ offspring of Dnmt3l−/− dams and wild-type sires, the imprinting control regions (ICRs) that normally carry a maternal methylation mark are unmethylated on both alleles, leading to the misregulation of developmentally crucial imprinted genes and death by 9.5 dpc (14). Our previous microarray measurements of gene expression in Dnmt3l−/+ 8.5 dpc embryos versus wild-type litter mates showed a decrease of Blcap expression to ∼70% of wild-type levels and an increase in Nnat expression by ∼40% (16). Measurements of Blcap_v1a and Blcap_v2a by qRT–PCR showed a reduction of Blcap_v1a in Dnmt3l−/+ embryos to ∼50% of wild-type levels, whereas Blcap_v2a levels increased by ∼40% (Fig. 3C).
In embryo, the Nnat promoter (including a BssHII restriction site ∼50 bp upstream of the TSS) and gene body were previously shown to be methylated on only the maternal allele (9). In human, differential methylation of NNAT has been observed even farther upstream, to within the last exon of BLCAP (7). The above changes in Blcap expression levels in Dnmt3l−/+ embryos are consistent with the notion that this methylation mark is established during oogenesis and that in the offspring, it acts as the ICR of the Blcap locus. To investigate this further, we performed a bisulfite mutagenesis methylation assay of the Blcap_v2a and Nnat promoter region (Supplementary Material, Fig. S2). Overall, we observed a loss of methylation in Dnmt3l−/+ relative to wild-type samples across the entire region, suggesting that its methylation significantly depends on the activity of Dnmt3l during oogenesis.
Human orthologues of Blcap_v1a and Blcap_v2a are imprinted in fetal brain
The genomic organization of the human BLCAP locus is similar to murine Blcap, including alternative BLCAP isoforms (Fig. 1A). BLCAP_V1a, b and c are orthologous to Blcap_v1a in the sense that their TSSs are downstream of NNAT. Analogously, BLCAP_V2a upstream of NNAT is orthologous to Blcap_v2a. To determine whether this similarity extends to the imprinting status of these BLCAP isoforms, we performed allele-specific sequencing assays in brain samples of human fetuses that were heterozygous for a TGn repeat copy number polymorphism (rs_11474161) in the last BLCAP exon where n = 7 or n = 8, i.e., n differed between the parental alleles by one.
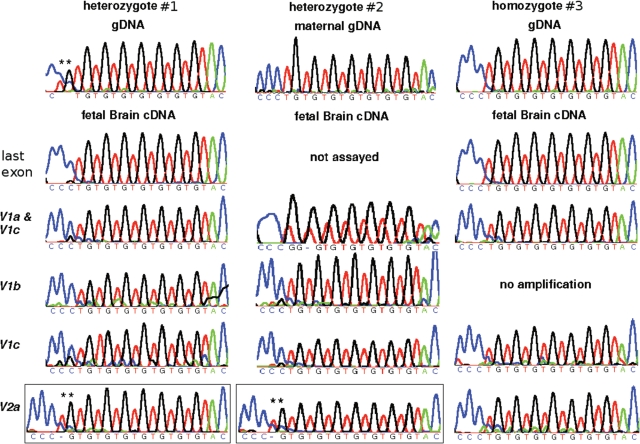
In fetus no. 1, BLCAP_V1a, b and c were predominantly expressed from the n = 7 allele, while BLCAP_V2a transcripts originated from the n = 8 allele, indicating reciprocal allele-specific expression (Fig. 5). Primers designed to the last BLCAP exon, thought to be shared by all isoforms, amplified only the n = 7 allele, suggesting that the BLCAP_V1 isoforms transcribed from the n = 7 allele are overall more abundant than isoforms like BLCAP_V2a that are expressed from the n = 8 allele. This recapitulates our observations on the relative expression levels of Blcap_v1a and Blcap_v2a in the mouse. For fetus no. 1, no parental sample was available, precluding parent-of-origin determination. For a second fetus (no. 2), maternal genomic DNA (gDNA) was available and the maternal genotype was n = 7 (Fig. 5). Predominantly the maternal n = 7 allele of BLCAP_V1a, b and c was present in brain cDNA of fetus no. 2, as opposed to BLCAP_V2a which originated exclusively from the paternal n = 8 allele (Fig. 5). These data indicate that the parent-of-origin-specific expression profile of murine Blcap is conserved in human, including the relatively minor contribution of paternal transcripts like BLCAP_V2a to the overall BLCAP transcript pool.
Figure 5.
Human allele-specific sequencing results. For each fetus, the top row contains a trace generated from genomic DNA (gDNA). For fetus no. 1 (left), this trace demonstrates heterozygosity for a TGn repeat copy number polymorphism in the last BLCAP exon (n = 7 for one allele, and n = 8 for the other; asterisks highlight the additional TG dinucleotide). For fetus no. 2 (middle), the maternal gDNA trace shows the maternal genotype to be n = 7. The second and subsequent rows display traces obtained with fetal brain cDNA. In fetus no. 1, only the n = 7 allele is evident after sequencing of PCR products generated with primers designed to the last BLCAP exon, thought to be common to all BLCAP isoforms. Similar results were observed for amplicons specific to isoforms BLCAP_V1a and c, BLCAP_V1b and BLCAP_V1c. This was also the case for fetus no. 2. In contrast, the n = 8 allele was seen upon sequencing BLCAP_V2a products in both fetuses nos 1 and 2 (framed traces; asterisks highlight the additional TG). These data indicate that BLCAP_V2a is predominantly expressed from the paternal allele, while the other isoforms are preferentially expressed from the maternal allele. For comparison, an n = 7 homozygous fetus (no. 3) is shown on the right.
Paternal allele-specific transcripts extending into the first intron of Blcap_v1a
The first intron of Blcap_v1a prior to Nnat contains three AATAAA polyadenylation (polyA) site motifs in the sense orientation relative to Blcap. Two of them are only 25 bp apart, are located within a ∼60 bp region that is evolutionarily conserved among mammals, and coincide with the ends of multiple ESTs and an mRNA that likely correspond to Blcap transcripts (BY664891, CJ320720, AV349270, BB178170, AK038884). However, they may also correspond to anti-sense transcripts generated by Nnat. All these transcripts were recovered from brain tissues or Rathke's pouch, an embryonic precursor of parts of the pituitary.
Allele-specific sequencing assays were performed using a T/C SNP between B6 and cast near the start of the Blcap_v1a intron (Fig. 1B). Multiple, differentially spliced transcripts were amplified from newborn brain. Sequencing of amplicons that covered the SNP position and whose parental origin could thus be determined, revealed that they originated from the paternal allele (Fig. 2D).
The Commd1/U2af1-rs1 locus generates paternal allele-specific transcripts similar to Blcap_v2a
The murine Commd1 (=Murr1)/U2af1-rs1 locus on proximal Chr 11 has been noted previously as being similar in organization to Blcap/Nnat (11). In previous microarray experiments, we compared MatDp(prox11) with PatDp(prox11) tissues, including brain (17). For Commd1/U2af1-rs1, these microarray measurements reflected the known preferential maternal expression of Commd1 and the paternal-only expression of U2af1-rs1 in brain (11,18). However, as with Blcap, we found EST evidence (e.g. BG074660, BE137092, BU582903) for alternative Commd1 transcripts (Commd1_v2a, b, c) originating from within or near the 5′-end of the intronic U2af1-rs1 gene (Supplementary Material, Fig. S3A). Using a G/A SNP between the B6 and cast strains in the first exon of Commd1_v2b, we determined that this isoform is exclusively paternally expressed (Supplementary Material, Fig. S3B). The Commd1 microarray probe sets map to the second and third Commd1 exons that are common to all isoforms (Supplementary Material, Fig. S3A). The fact that they detected preferential maternal expression suggests that paternal Commd1 transcripts like Commd1_v2a are much less abundant than the maternally expressed Commd1_v1a that was assayed in reference (11), similar to the relative abundances of Blcap_v1a and Blcap_v2a.
DISCUSSION
Tissue- and promoter-specific imprinting of Blcap/Blcap
The BLCAP/Blcap locus generates transcripts from distinct promoters downstream (BLCAP_V1/Blcap_v1) and upstream (BLCAP_V2/Blcap_v2) of NNAT/Nnat (Fig. 1). We found Blcap_v1a to be preferentially expressed from maternally inherited alleles in brain but not other murine tissues, indicating brain-specific imprinting (Fig. 2A and Supplementary Material, Fig. S1). Similarly, BLCAP_V1 transcripts are preferentially maternally expressed in human fetal brain (Fig. 5). In contrast, the BLCAP_V2a/Blcap_v2a isoforms are predominantly paternally expressed in brain (Figs 2C and 5). In mouse brain, Blcap_v1a is much more abundant than Blcap_v2a (Fig. 3B), explaining the overall dominance of Blcap transcripts from maternal alleles (microarray data and Fig. 4), which is also evident in human (Fig. 5). The transcriptional profile of BLCAP/Blcap is thus a complex combination of tissue-specific expression, promoters of different efficiencies and tissue- and promoter-specific imprinting.
This complexity is reminiscent of the GRB10/Grb10 locus. In mouse, the ‘major’ Grb10 promoter is active in a range of tissues but only on maternally inherited alleles (19), while distinct brain-specific promoters are utilized on only paternal alleles (20–23). In human, the ‘major’ GRB10 promoter is active on both parental alleles (20,24,25). BLCAP/Blcap thus shares the promoter-specific expression of either predominantly the maternal or the paternal alleles (Figs 2A and C), but in contrast to GRB10/Grb10, this ‘reciprocal’ imprinting pattern occurs in the same tissue and is conserved in human (Fig. 5).
Previously, Blcap expression has been detected by qualitative methods in both parthenogenote and androgenote as well as in MatDp(dist2) and PatDp(dist2) mouse embryos (10). This is consistent with the contribution of non-brain tissues in which we found Blcap_v1a to be expressed biallelically (Supplementary Material, Fig. S1) and the incomplete silencing of the paternal allele of Blcap_v1a even in brain (Figs 2A and 3A). Similarly in human, a qualitative expression assay yielded biallelic results for BLCAP in various fetal tissues including brain (7). The brain-specific result is again probably due to the incomplete suppression of BLCAP_V1 transcription from the paternal allele (Fig. 5).
Transcriptional interference hypothesis
The Blcap/Nnat locus is similar in many respects to the Commd1/U2af1-rs1 locus (11,18). Commd1_v1a is preferentially maternally expressed, particularly in adult brain where the paternally expressed U2af1-rs1 is abundant relative to earlier developmental stages and other tissues (11). In situ hybridizations showed Commd1 and U2af1-rs1 to be co-expressed widely throughout the brain and in the same cell types (11). In addition, paternally expressed anti-sense transcripts overlap the Commd1_v1a promoter region, are most abundant in brain and likely correspond to U2af1-rs1 transcripts (11). Transcription of U2af1-rs1 through the Commd1_v1a promoter was postulated to interfere with Commd1_v1a expression in cis where the degree of interference directly relates to U2af1-rs1 expression levels (11). This transcriptional interference mechanism would explain the imprinted expression profile of Commd1_v1a.
Here, we have shown that specifically in brain, Blcap_v1a, like Commd1_v1a, is preferentially expressed from the maternal allele. Nnat, like U2af1-rs1, is highly expressed in brain compared to other tissues. Nnat expression is particularly low in the murine tissues in which we found Blcap_v1a to be biallelically expressed (Fig. 2B). A NeuroBlast (26) search, based on the in situ hybridization data for thousands of genes that compose the Allen Brain Atlas (27), found Blcap to have the most similar expression pattern in brain relative to Nnat (Supplementary Material, Table S2). This suggests that in brain, Nnat and Blcap are co-expressed in the same cell types. Our data on paternal allele-specific transcripts in the first intron of Blcap_v1a may reflect Nnat transcripts extending through the Blcap_v1a promoter region (Fig. 2D). Thus, the transcriptional interference mechanism proposed by Wang et al. (11) for Commd1/U2af1-rs1 may also be acting at the Blcap/Nnat locus.
Alternative polyadenylation hypothesis
For the H13/Mcts2 imprinted locus, we recently demonstrated parent-of-origin-specific H13 polyA site selection (12). An H13 intron contains the paternally expressed gene Mcts2. Alternative H13 polyA sites upstream of Mcts2 are preferentially utilized on the paternal allele. On the maternal allele, H13 transcription proceeds through the inactive Mcts2 to downstream polyA sites. The ICR of the locus is a CGI overlapping the Mcts2 promoter that carries a maternal germ-line methylation mark. The organization of the H13/Mcts2 locus thus is similar to Blcap/Nnat. The paternal allele-specific transcripts overlapping the first exon and extending into the intron of Blcap_v1a (Fig. 2D), instead of being generated by Nnat, may reflect the use of an alternative Blcap polyA site on specifically paternal alleles and preferentially in brain. This assumes that high transcriptional activity of Nnat can reduce the utilization of the canonical Blcap polyA site in favour of the alternative site. Like transcriptional interference, alternative polyA could explain the observed brain-specific preferential maternal expression of Blcap_v1a.
Possible implications of BLCAP/NNAT imprinting for carcinogenesis
Expression studies in various cancers have consistently found BLCAP to be absent or down-regulated relative to normal or less severe controls (Supplementary Material, Table S3). Reducing BLCAP expression by RNAi in osteosarcoma cells has been reported to increase cell growth and colony formation, alters the cell cycle and promotes cell proliferation (28). In contrast, over-expression of BLCAP in HeLa and human tongue cancer cells has been shown to stimulate apoptosis (3,4).
Similar studies of NNAT include observations of down- as well as up-regulation in different cancers (Supplementary Material, Table S3). Several of these investigations assessed the DNA methylation state of the NNAT promoter and/or the effect of DNA de-methylation agents on the expression of NNAT, but none of the reports about elevated NNAT expression in cancer included information on its methylation state. De-methylation and reactivation of maternal NNAT may have occurred in these cases. Global as well as locus-specific hypomethylation, often in introns, has previously been observed in cancer (29,30).
In the context of the transcriptional interference model, this may be relevant to carcinogenesis in tissues like brain where NNAT is highly expressed. Reactivation of maternal NNAT would lead to an overall down-regulation of BLCAP, akin to our results for Blcap in Dnmt3l−/+ mouse embryos (Fig. 3C). So, with respect to BLCAP expression levels, hypomethylation of NNAT would have an effect similar to hypermethylation and silencing of the BLCAP promoter.
NNAT and BLCAP have not been studied in concert in cancer biology. Some studies employed large-scale methods that implicitly generated limited data for both genes (31,32). Care needs to be taken when choosing polymorphisms for allele-specific profiling: BLCAP message is the target of RNA editing, particularly in brain, generating what appear to be A/G SNPs (33,34). We believe that a simultaneous allele-specific profiling of NNAT and BLCAP expression and chromatin modifications in tumours and cell lines would be insightful.
MATERIALS AND METHODS
Mouse samples
Tissues of B6, cast and inter-subspecies hybrids (B×C and C×B) were dissected, snap-frozen and stored at −80°C. The MatDp(dist2), PatDp(dist2) and non-UpDp control newborn mice were generated as described in reference (13). Briefly, these mice are the offspring of heterozygotes for the T(2:9)11H reciprocal translocation that have inherited the distal part of both homologous copies of Chr 2 from either the mother, MatDp(dist2), or the father, PatDp(dist2), due to a non-disjunction event. The non-UpDp controls were carriers of the T(2:9)11H translocation with an otherwise normal, that is, biparental and balanced genome. The generation of the 8.5 dpc Dnmt3l−/+ embryos and normal litter mates is described in reference (14). RNA and DNA were extracted with the Qiagen RNeasy mini (Cat. No. 74104) and DNeasy tissue kits (Cat. No. 69504) following manufacturer protocols. cDNA was made with the Invitrogen Superscript First-Strand Synthesis System (Cat. No. 12371-019). Between 1 and 5 µg of total RNA, quantified with an Agilent 2100 BioAnalyzer, were used in a standard 50 µl oligo dT-primed reaction following the manufacturer protocol.
Human samples
The brain samples of fetuses 1 and 3 were obtained from the MRC-Wellcome Human Developmental Biology Resource. RNA and DNA were extracted with the Qiagen RNeasy mini (Cat. No. 74104) and DNeasy tissue kits (Cat. No. 69504) following manufacturer protocols. cDNA was synthesized using the Invitrogen Superscript First-Strand Synthesis System (Cat. No. 12371-019). For fetus F46, brain cDNA and maternal genomic DNA samples were collected under the guidelines of the Hammersmith and Queen Charlotte's and Chelsea Hospitals Trust Research Ethics Committee (Registration No. 2005/6028). Informed consent was collected from all subjects.
Expression microarrays
Two Affymetrix MOE 430 v2 expression microarrays were hybridized with probes produced from total RNA samples of the whole heads of a MatDp(dist2) and a PatDp(dist2) newborn mouse, respectively. Probe generation, array hybridization and analysis are described in references (16) and (17). The data can be viewed using WAMIDEX (https://atlas.genetics.kcl.ac.uk), the web interface to our collection of imprinting-related microarray data (16). The raw array data are available from GEO (GSE11789).
PCR
All reactions were performed using Abgene Reddymix (Cat. No. AB-0575/LD/A). Unless noted otherwise, total reaction volume was 10 µl, including 1 µl cDNA template and 0.5 µl mix of 20 µm forward and reverse primer. Primer sequences and cycling conditions are listed in Supplementary Material, Table S1. Primers were designed using Primer3Plus (35). For fetus F46, two rounds of PCR with nested primers were necessary to obtain enough product from brain cDNA for sequencing. Two microlitre of the first round PCR reaction were used as template in the second round.
DNA sequencing
2.5 µl of PCR reaction was treated with 1 µl USB Exosap-IT (Cat. No. 78200) in a 15 µl reaction to inactivate primers and unincorporated nucleotides. Depending on the concentration of the PCR product as estimated by gel electrophoresis using 1.2% agarose, 2.5 or 5 µl of the Exosap reaction were used as template in a 10 µl sequencing reaction together with 0.5–1.0 µl BigDye and 2 µl 5× sequencing buffer from the ABI BigDye v3.1 kit (Cat. No. 4337455) and 0.4 µl 20 µm primer. Primer sequences and cycling conditions are listed in Supplementary Material, Table S1. The products were precipitated with 300 mm sodium acetate in ethanol, collected by centrifugation, washed with 70% ethanol, resuspended and denatured in formamide and sequenced on an ABI 3730xl sequencer. Sequence reads were analysed with the Staden package (http://staden.sourceforge.net/) (36).
qRT–PCR
Custom TaqMan gene expression assays (primer-probe mixes) for Blcap_v1a and v2a were ordered from ABI (Cat. No. 4331348). One microlitre of cDNA template was diluted in 3.5 µl water and added to 0.5 µl primer-probe mix in 5 µl ABI TaqMan Gene Expression Master Mix (Cat. No. 4370048). The reactions were performed on an ABI 7900ht Lightcycler and analysed using the ΔΔCt method with Actb as the endogenous control (37). The confidence intervals shown in Figure 3 are based on four replicate reactions per transcript and template. Primer and probe sequences and cycling conditions are listed in Supplementary Material, Table S1.
Northern hybridization
Two blots were generated in parallel from 10 µg per well of total RNA extracted from single MatDp(dist2), PatDp(dist2) or non-UpDp control newborn brains. Size estimates are based on a DNA ladder run in parallel. PCR primers and conditions for the generation of the templates for the ∼200 bp probes N1 and N2 shown in Figure 1B are given in Supplementary Material, Table S1. Twenty microlitre of template PCR product was purified by gel electrophoresis in 1.2% low melting point agarose and gel-extracted with the Qiagen QiaEx II kit (Cat. No. 20021). Five microlitre (∼25 ng) of purified template was used to make αdCT 32P-labelled probe with the NEBlot kit (Cat. No. N1500S) in a standard 50 µl reaction, which was purified with a Roche mini Quick Spin DNA Column (Cat. No. 11814419001). The probe in 10 ml Sigma PerfectHyb solution (Cat. No. H7033-125ML) was incubated with the respective blot (pre-hybridized with 10 ml PerfectHyb for 2hrs) at 65°C overnight. Blots were washed at 65°C for 20 min in 2% SSC, 0.5% SDS and for 40 min in 0.5% SSC, 0.5% SDS. The blot probed with N1 (last Blcap exon) was exposed to phosphor screen for 15 h. The exposure time for the N2 blot (Blcap_v2a exon 1) was 84 h. The phosphor screen was scanned with an Amersham Typhoon 9200 Variable Mode Imager at 50 µm resolution.
Bisulfite mutagenesis DNA methylation assay
Genomic DNA extracted from 7.75 dpc Dnmt3l−/+ and wild-type visceral yolk sac (VYS) tissue was bisulfite-converted using the Zymo Research EZ DNA Methylation-Direct Kit (Cat. No. D5020) according to the manufacturer's protocol (∼500 ng input DNA per reaction). Genomic DNA of 8.5 dpc Dnmt3l−/+ embryos was converted as described (12). Four microlitre of converted DNA were used for each PCR with Thermoprime Taq DNA polymerase (Thermo Scientific Cat. No. AB-0301) according to the manufacturer's protocol, adjusted for a 20 µl reaction volume and the addition of 4 µl 5M Betaine (Sigma Cat. No. B0300). The products were directly sequenced as well as cloned and sequenced using the pGEM-T Easy vector (Promega Cat. No. A1360) and DH5α competent cells (Invitrogen). Primers and cycling conditions are provided in Supplementary Material, Table S1. Primers were designed using BiSearch (38). ‘Beads-on-a-string’ diagrams were produced with BiQ Analyzer (39).
SUPPLEMENTARY MATERIAL
FUNDING
The Wellcome Trust to R.J.O.; Biotechnology and Biological Sciences Research Council to R.J.O.; European Molecular Biology Organisation to R.S. Funding to pay the Open Access charges was provided by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Colin Beechey and Jo Peters for the MatDp(dist2) and PatDp(dist2) material, and Deborah Bourc'his and Timothy Bestor for the Dnmt3l−/+ material.
Conflict of Interest statement. The authors declare that there are no conflicts of interest with regard to the work presented in this article.
REFERENCES
- 1.Gromova I., Gromov P., Celis J.E. Identification of true differentially expressed mRNAs in a pair of human bladder transitional cell carcinomas using an improved differential display procedure. Electrophoresis. 1999;20:241–248. doi: 10.1002/(SICI)1522-2683(19990201)20:2<241::AID-ELPS241>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 2.Rae F.K., Stephenson S.A., Nicol D.L., Clements J.A. Novel association of a diverse range of genes with renal cell carcinoma as identified by differential display. Int. J. Cancer. 2000;88:726–732. doi: 10.1002/1097-0215(20001201)88:5<726::aid-ijc7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 3.Zuo Z., Zhao M., Liu J., Gao G., Wu X. Functional analysis of bladder cancer-related protein gene: a putative cervical cancer tumor suppressor gene in cervical carcinoma. Tumour Biol. 2006;27:221–226. doi: 10.1159/000093057. [DOI] [PubMed] [Google Scholar]
- 4.Yao J., Duan L., Fan M., Yuan J., Wu X. Overexpression of BLCAP induces S phase arrest and apoptosis independent of p53 and NF-B in human tongue carcinoma: BLCAP overexpression induces S phase arrest and apoptosis. Mol. Cell. Biochem. 2007;297:81–92. doi: 10.1007/s11010-006-9332-2. [DOI] [PubMed] [Google Scholar]
- 5.Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gromova I., Gromov P., Celis J.E. bc10: a novel human bladder cancer-associated protein with a conserved genomic structure downregulated in invasive cancer. Int. J. Cancer. 2002;98:539–546. doi: 10.1002/ijc.10244. [DOI] [PubMed] [Google Scholar]
- 7.Evans H.K., Wylie A.A., Murphy S.K., Jirtle R.L. The neuronatin gene resides in a ‘micro-imprinted’ domain on human chromosome 20q11.2. Genomics. 2001;77:99–104. doi: 10.1006/geno.2001.6612. [DOI] [PubMed] [Google Scholar]
- 8.Kagitani F., Kuroiwa Y., Wakana S., Shiroishi T., Miyoshi N., Kobayashi S., Nishida M., Kohda T., Kaneko-Ishino T., Ishino F. Peg5/Neuronatin is an imprinted gene located on sub-distal chromosome 2 in the mouse. Nucleic Acids Res. 1997;25:3428–3432. doi: 10.1093/nar/25.17.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikyo N., Williamson C.M., John R.M., Barton S.C., Beechey C.V., Ball S.T., Cattanach B.M., Surani M.A., Peters J. Genetic and functional analysis of Neuronatin in mice with maternal or paternal duplication of distal chromosome 2. Dev. Biol. 1997;190:66–77. doi: 10.1006/dbio.1997.8681. [DOI] [PubMed] [Google Scholar]
- 10.John R.M., Aparicio S.A., Ainscough J.F., Arney K.L., Khosla S., Hawker K., Hilton K.J., Barton S.C., Surani M.A. Imprinted expression of neuronatin from modified BAC transgenes reveals regulation by distinct and distant enhancers. Dev. Biol. 2001;236:387–399. doi: 10.1006/dbio.2001.0327. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Joh K., Masuko S., Yatsuki H., Soejima H., Nabetani A., Beechey C.V., Okinami S., Mukai T. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol. Cell. Biol. 2004;24:270–279. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood A.J., Schulz R., Woodfine K., Koltowska K., Beechey C.V., Peters J., Bourc'his D., Oakey R.J. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattanach B.M., Beechey C.V. Genomic imprinting in the mouse: possible final analysis. In: Reik W., Surani M.A., editors. Genomic Imprinting: Frontiers in Molecular Biology. Vol. 18. New York, Oxford, Tokyo: IRL Press; 1997. pp. 118–145. [Google Scholar]
- 14.Bourc'his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 15.Bourc'his D., Proudhon C. Sexual dimorphism in parental imprint ontogeny and contribution to embryonic development. Mol. Cell. Endocrinol. 2007;282:87–94. doi: 10.1016/j.mce.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Schulz R., Woodfine K., Menheniott T.R., Bourc'his D., Bestor T., Oakey R.J. WAMIDEX: a web atlas of murine genomic imprinting and differential expression. Epigenetics. 2008;3:89–96. doi: 10.4161/epi.3.2.5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz R., Menheniott T.R., Woodfine K., Wood A.J., Choi J.D., Oakey R.J. Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res. 2006;34:e88. doi: 10.1093/nar/gkl461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatada I., Sugama T., Mukai T. A new imprinted gene cloned by a methylation-sensitive genome scanning method. Nucleic Acids Res. 1993;21:5577–5582. doi: 10.1093/nar/21.24.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyoshi N., Kuroiwa Y., Kohda T., Shitara H., Yonekawa H., Kawabe T., Hasegawa H., Barton S.C., Surani M.A., Kaneko-Ishino T., Ishino F. Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russell syndrome gene. Proc. Natl Acad. Sci. USA. 1998;95:1102–1107. doi: 10.1073/pnas.95.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hikichi T., Kohda T., Kaneko-Ishino T., Ishino F. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 2003;31:1398–1406. doi: 10.1093/nar/gkg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnaud P., Monk D., Hitchins M., Gordon E., Dean W., Beechey C.V., Peters J., Craigen W., Preece M., Stanier P., et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 2003;12:1005–1019. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 22.Yamasaki-Ishizaki Y., Kayashima T., Mapendano C.K., Soejima H., Ohta T., Masuzaki H., Kinoshita A., Urano T., Yoshiura K., Matsumoto N., et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 2007;27:732–742. doi: 10.1128/MCB.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanz L.A., Chamberlain S., Sabourin J.C., Henckel A., Magnuson T., Hugnot J.P., Feil R., Arnaud P. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008 doi: 10.1038/emboj.2008.142. Advance online publication on Jul 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blagitko N., Mergenthaler S., Schulz U., Wollmann H.A., Craigen W., Eggermann T., Ropers H.H., Kalscheuer V.M. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum. Mol. Genet. 2000;9:1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- 25.Hitchins M.P., Monk D., Bell G.M., Ali Z., Preece M.A., Stanier P., Moore G.E. Maternal repression of the human GRB10 gene in the developing central nervous system; evaluation of the role for GRB10 in Silver-Russell syndrome. Eur. J. Hum. Genet. 2001;9:82–90. doi: 10.1038/sj.ejhg.5200583. [DOI] [PubMed] [Google Scholar]
- 26.Lau C., Ng L., Thompson C., Pathak S., Kuan L., Jones A., Hawrylycz M. Exploration and visualization of gene expression with neuroanatomy in the adult mouse brain. BMC Bioinformatics. 2008;9:153. doi: 10.1186/1471-2105-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 28.Fan D.G., Fan Q.Y., Zhang H.Z., Zhang Y.H., Liu Y.Y. A prospective genetic therapy study of extremity saving operations: the regulatory function of gene BLCAP in the development of human osteosarcoma. Chin. J. Clin. Rehabil. 2004;8:982–983. [Google Scholar]
- 29.Kanai Y., Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 30.Wilson A.S., Power B.E., Molloy P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Dekel B., Metsuyanim S., Schmidt-Ott K.M., Fridman E., Jacob-Hirsch J., Simon A., Pinthus J., Mor Y., Barasch J., Amariglio N., et al. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 2006;66:6040–6049. doi: 10.1158/0008-5472.CAN-05-4528. [DOI] [PubMed] [Google Scholar]
- 32.Keshet I., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 33.Clutterbuck D.R., Leroy A., O'Connell M.A., Semple C.A. A bioinformatic screen for novel A-I RNA editing sites reveals recoding editing in BC10. Bioinformatics. 2005;21:2590–2595. doi: 10.1093/bioinformatics/bti411. [DOI] [PubMed] [Google Scholar]
- 34.Paz N., Levanon E.Y., Amariglio N., Heimberger A.B., Ram Z., Constantini S., Barbash Z.S., Adamsky K., Safran M., Hirschberg A., et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staden R., Beal K.F., Bonfield J.K. The Staden package, 1998. Methods Mol. Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arányi T., Váradi A., Simon I., Tusnády G.E. The BiSearch web server. BMC Bioinformatics. 2006;7:431. doi: 10.1186/1471-2105-7-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bock C., Reither S., Mikeska T., Paulsen M., Walter J., Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 40.Kent J.W., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.