Abstract
Histones are the basic protein components of nucleosomes. They are among the most conserved proteins and are subject to a plethora of post-translational modifications. Specific histone residues are important in establishing chromatin structure, regulating gene expression and silencing, and responding to DNA damage. Here we present HistoneHits, a database of phenotypes for systematic collections of histone mutants. This database combines assay results (phenotypes) with information about sequences, structures, post-translational modifications, and evolutionary conservation. The web interface presents the information through dynamic tables and figures. It calculates the availability of data for specific mutants and for nucleosome surfaces. The database currently includes 42 assays on 677 mutants multiply covering 405 of the 498 residues across yeast histones H3, H4, H2A, and H2B. We also provide an interface with an extensible controlled vocabulary for research groups to submit new data. Preliminary analyses confirm that mutations at highly conserved residues and modifiable residues are more likely to generate phenotypes. Buried residues and residues on the lateral surface tend to generate more phenotypes, while tail residues generate significantly fewer phenotypes than other residues. Yeast mutants are cross referenced with known human histone variants, identifying a position where a yeast mutant causes loss of ribosomal silencing and a human variant increases breast cancer susceptibility. All data sets are freely available for download.
In eukaryotic cells, nucleic acids are packaged with proteins to form a condensed structure. The basic unit of packing is the DNA–protein complex known as nucleosome, comprising a core protein complex wrapped by ∼146 nucleic acid base pairs. The major components of the nucleosome complex are consistent among eukaryotic species, consisting of two copies each of histones H2A, H2B, H3, and H4. This structure plays a key function in DNA packing and gene expression regulation (Smith 1991).
Histone proteins are among the most conserved proteins in eukaryotic cells. Amino acid sequence identities between yeast (Saccharomyces cerevisiae) and human (Homo sapiens) histones are 92% (H4), 90% (H3), 71% (H2A), and 63% (H2B). Residues in the core domains of histones are more highly conserved. This degree of sequence conservation reflects the important roles of histone residues in the nucleosome.
Histone genes other than H4 have had family expansions in higher organisms. Histones H1 and H2A have the largest families, while histones H2B and H3 have only limited numbers of variants. These histone variants provide functional heterogeneity (Brown 2001), such as transcriptional activation/repression, and heterochromatin barriers (Kamakaka and Biggins 2005). For example, a yeast H2A variant, H2A.Z, antagonizes silencing at telomeres and prevents the spread of heterochromatin (Meneghini et al. 2003; Raisner et al. 2005). An H3-like variant, CENPA, replaces the ordinary H3 subunit in the histone octomers in the centromere of eukaryotes (Smith 2002).
Histone residues are heavily modified by post-translational modifications such as acetylation, methylation, and phosphorylation. These modifications enable many functions of the chromosome and mark specific regions of DNA (Iizuka and Smith 2003; Millar and Grunstein 2006). For example, H3 K56 acetylations facilitate gene expression in yeast by recruiting the SWI/SNF nucleosome remodeling complex (Xu et al. 2005) and hypoacetylation of H4 tails mark the silenced heterochromatin by allowing H4 to interact with Sir3 (Grunstein 1998).
The vital and dynamic roles of histones have motivated studies that mutate specific histone residues and determine their phenotypes, as can be done quite easily in Saccharomyces cerevisiae (Smith and Santisteban 1998). Assays have identified H3 and H4 residues that affect rDNA, telomeric, transcriptional, and HM (silent mating locus) silencing (Park et al. 2002; Smith et al. 2002; Thompson et al. 2003; Duina and Winston 2004; Tompa and Madhani 2007). Certain modifiable residues in H3 and H4 have important roles in transcriptional silencing and DNA damage response (Hyland et al. 2005). Studies of the nucleosome SIN and LRS domains have found residues that affect Sin– phenotypes, ribosomal silencing, chromatin folding, and heterochromatin formation (Fry et al. 2006). Systematic screens using a collection of 320 point mutants of histone core residues have identified residues responsible for suppression of Ty (Spt– phenotypes), transcription elongation defects due to 6-azauracil (6AU) sensitivity, and sensitivity to DNA damage agents hydroxyurea (HU) and methyl methanesulfonate (MMS) (Matsubara et al. 2007). Screens using alanine substitutions of all histone core residues have identified essential residues and residues required for H3K4 methylation (Nakanishi et al. 2008).
A recent systematic screen used a systematic library of synthetic histone H3 and H4 mutants consisting of 486 alleles (Dai et al. 2008). Each H3 and H4 residue was mutated to alanine, with existing alanines mutated to serine. Charged residues were also mutated to neutral amino acids and amino acids with opposite charge. Modifiable residues were replaced with amino acids mimicking both modified and unmodified states. Additional mutants included N-terminal tail deletions and multiple mutations. All mutants were tested in 14 assays grouped into six phenotype categories, including lethality, temperature sensitivity, DNA damage response, transcriptional elongation, transcriptional silencing, and response to microtubule disruption.
These studies together provide valuable insights into the functional roles of histone residues, but joint analysis has been difficult due to disparate data formats and the lack of a combined data collection. Here we present a database, HistoneHits (http://histonehits.org), as a resource that integrates sequence, structure, post-translational modification, and phenotypic data for genetic screens of histone mutants. We have deposited data from published systematic screens (Park et al. 2002; Hyland et al. 2005; Fry et al. 2006; Matsubara et al. 2007; Dai et al. 2008; Nakanishi et al. 2008). In addition to on-line analysis tools, database contents may also be downloaded for independent analysis. We provide templates for groups to submit new data sets, including both high-throughput screens of global mutant collections and smaller, more focused data sets. This database is being maintained as a resource for the chromatin community.
Results
Data sources
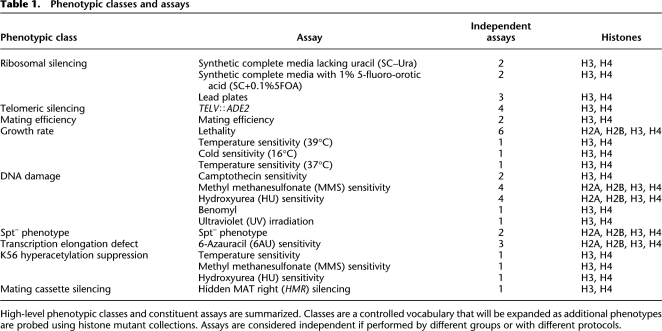
Currently, our database is focused on yeast histones because the power of yeast genetics makes it is easy to connect genotype to phenotype; currently this is more difficult in other organisms, especially those with extensive histone gene duplications. We have deposited data from publications on yeast histone mutagenesis (Park et al. 2002; Hyland et al. 2005; Fry et al. 2006; Matsubara et al. 2007; Dai et al. 2008; Nakanishi et al. 2008) that include several large-scale screens of mutations within yeast histone proteins H2A, H2B, H3, and H4. The database currently has 43 assays grouped in nine phenotypic classes (Table 1).
Table 1.
Phenotypic classes and assays
High-level phenotypic classes and constituent assays are summarized. Classes are a controlled vocabulary that will be expanded as additional phenotypes are probed using histone mutant collections. Assays are considered independent if performed by different groups or with different protocols.
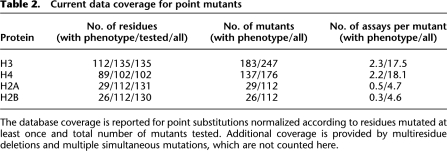
Histones H3 and H4 have the greatest number of phenotypes available for analysis (Table 2). Each residue of histones H3 and H4 has been mutated and tested, with at least one phenotype reported for 83% of H3 and 87.2% of H4 residues. Residues of histones H2A and H2B have been mutated to alanine (Matsubara et al. 2007; Nakanishi et al. 2008). The viable mutants have been tested in several assays. Phenotypes for 26% of H2A and 23% of H2B residues have been reported in at least one assay.
Table 2.
Current data coverage for point mutants
The database coverage is reported for point substitutions normalized according to residues mutated at least once and total number of mutants tested. Additional coverage is provided by multiresidue deletions and multiple simultaneous mutations, which are not counted here.
Templates for uploading new data sets can be prepared through an on-line wizard. This template is designed to eliminate errors in data entry and to accurately capture standardized metadata, including information about strains and plasmids. To ensure data quality, all submissions are reviewed before being added to the database.
Five-point grading system for assay results
To assist comparison between qualitative, semi-quantitative, and quantitative phenotypes, the database includes an internal representation of each assay result using a five-point semi-quantitative scale: –2, –1, 0, 1, and 2. In this scheme zero is defined as indistinguishable from the wild-type phenotype (Supplemental Fig. 1). Assay results can be restricted to only the positive or negative side or extreme values of the scale when appropriate. The database also records whether the direction of an assay matches the phenotypic category, or whether it is reversed as in a counterscreen.
Appropriate conversion of assay results into this five-point scale is required for data integrity. The data submission wizard and online tutorial provide examples to help users score assays on the five-point scale, essentially converting traditional scores such as “+” and “++” to +1 and +2, and “–” and “– –” to –1 and –2 (Supplemental Fig. 1). For assays generating numeric values, users may submit the original assay results and appropriate break points for the different categories. All conversions are reviewed carefully before incorporation in the database. The database also saves the original assay results as real numbers and/or text descriptions to ensure data integrity and permit recalculation (database schema, Supplemental Fig. 2).
Global analysis
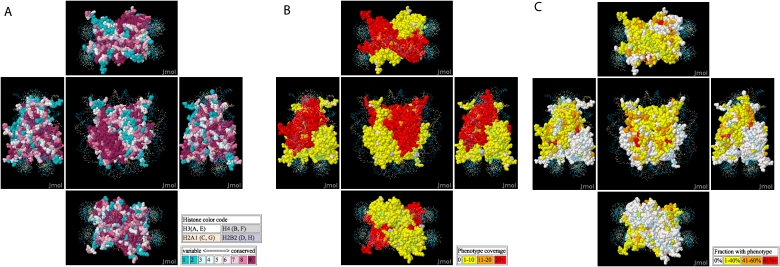
Global analyses show the current progress in assessing histone function by mutagenesis. Residues required for function are expected to be evolutionarily conserved (Fig. 1A), and conservation scores are taken from ConSurf (Landau et al. 2005). Conserved regions can be compared with the depth of coverage of assays for mutants at each position (Fig. 1B) and the fraction of mutants yielding a non-wild-type phenotype (Fig. 1C).
Figure 1.
A nucleosome crystal structure is colored according to evolutionary conservation scores (A), number of mutant/assay combinations tested (B), and the fraction of mutant/assay combinations giving a non-wild-type phenotype (C).
As expected, residues that are involved in nucleosome functions are usually more conserved than other residues. Mutations of the most conserved residues are 2.8 ± 0.5-fold more likely to give phenotypes than mutations of the least conserved residues. The probability that a mutation at a position generates a phenotype increases significantly with evolutionary conservation (Fig. 1A,C; logistic regression P-value < 3 × 10−10).
Geographical domain enrichment
The residues of histone proteins can be roughly partitioned into four major geographical domains: buried, disk surface (protein surface that does not contact DNA), lateral (protein surface that contacts DNA), and tail (unstructured in the crystal) (Dai et al. 2008). Residues in different geographical domains have significantly different probabilities of generating phenotypes (P-value < 10−16; see Methods). Buried and lateral residues generate more phenotypes than residues in the other domains (P-values = 0.0066 and 1.1 × 10−5 after multiple-testing correction, odds ratios = 1.3 and 1.4; see Methods); and tail residues generate significantly fewer phenotypes (P-value = 1.6 × 10−15, odds ratio = 0.48; see Methods). Residues on the disk surface do not show significant differences relative to other residues.
In terms of phenotypic classes, we find that tail residues are significantly underrepresented in the telomeric silencing, growth rate, DNA damage response, transcriptional elongation defect and mating cassette silencing assays. Lateral residues are overrepresented in the rDNA silencing, telomeric silencing, growth rate, DNA damage response, Spt– phenotype, and transcriptional elongation defect assays. Buried residues are overrepresented in the growth rate and mating cassette silencing assays. Geographical domain analysis is provided automatically on-line as part of the search results.
Mutations affecting post-translational modifications
Many histone residues are targets of post-translational modifications (PTMs). These modifications directly affect nucleosome dynamics or recruit enzyme complexes required for various functions (Kouzarides 2007). Post-translational modifications may provide kinetic proofreading to reduce the error rate of gene activation (Blossey and Schiessel 2008). We searched for methylation, acetylation, phosphorylation, and other histone PTMs using UniProt (Wu et al. 2006) and using Textpresso with the spatial relationship ontological category followed by manual curation (Muller et al. 2004). The database currently reports 50 modifications on 31 histone residues.
Modification sites have been specifically targeted by mutation studies. The database contains 40 mutants on 13 methylation sites, 51 mutants on 21 acetylation sites, and 14 mutants on 7 phosphorylation sites. Modifiable residues are more likely to have phenotypes than other residues (P-value = 10−10, odds ratio = 1.7). Residues that are subject to methylation and acetylation have significantly more phenotypes (P-values = 3 × 10−10 and 1 × 10−4, odds ratios = 1.6 and 1.4) than other residues.
When assays are grouped into phenotypic classes, modifiable residues are more likely to have rDNA silencing phenotypes (P-value = 8 × 10−5 after multiple-testing correction, odds ratio = 3.3). Modifiable residues overall are less likely to have growth defects, including lethality, slow growth, and temperature sensitivities (P-value = 0.02, odds ratio = 0.28). This may be because the PTM sites are enriched in tail regions (22 out of 34 PTM sites are in histone tails), which do not alter the structure of the histone core and do not cause serious nucleosome defects. Thus, we restricted attention to the tail and then compared PTM sites with non-PTM sites. In the tail region, the PTM sites are 3.9 times more likely to have phenotypes than the non-PTM sites (P-value = 3.7 × 10−14), especially rDNA silencing phenotypes (P-value = 0.017 corrected for multiple testing, odds ratio = 3.1). Histone modifications are known to regulate rRNA expression (Chen and Pikaard 1997).
Comparison with human sequence variations
Phenotypes of yeast mutants are relevant to understanding the function of human disease-related variants at homologous sites. We cross-referenced yeast mutants with non-synonymous human sequence substitution variants from dbSNP (Sherry et al. 2001) and UniProt (Supplemental Table 1; Wu et al. 2006). Wild-type sequences of human histone proteins were aligned with yeast orthologs using ClustalW (Larkin et al. 2007), providing additional information about geographical domains (Dai et al. 2008) and PTMs for each residue. No domains or PTMs are overrepresented in the coding single nucleotide polymorphism (SNP) residues (multinomial χ2 test for domains; binomial test for PTMs).
Two human histone variants are associated with breast cancer, VAR_036206 on H4 (E63Q) and VAR_035802 on H2A (A52T) (Sjoblom et al. 2006). While the yeast mutant corresponding to VAR_036206 (yeast H4 E63Q) does not have any phenotype in the assays, the E63A mutant at the same position exhibits loss of silencing in a ribosomal silencing assay (lead plate). Disruption of ribosomal synthesis affects cell growth and may increase cancer susceptibility (Ruggero and Pandolfi 2003). Yeast mutants corresponding to VAR_035802 (H2A T53) have not yet been tested. The H4 E63 variant is on the disk surface and H2A T53 is buried (Supplemental Fig. 3).
Ten histone SNPs have been genotyped for frequencies in human populations (Supplemental Table 1). Seven of the corresponding positions have been tested in at least one yeast assay, and three of these show at least one phenotype. While allele frequencies are available for some histone SNPs, the data are so far too limited to test for a correlation between human allele frequency and yeast mutant phenotypes.
Searching the database
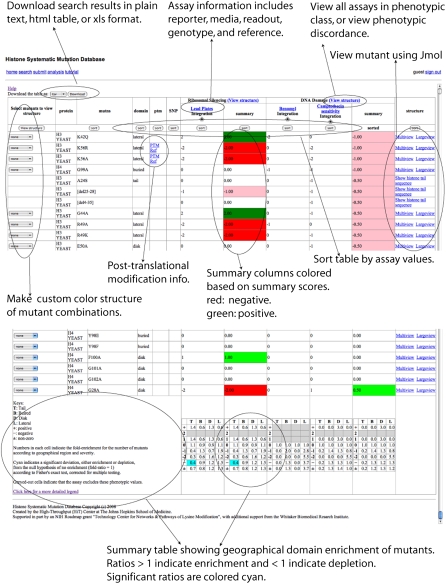
The database is searchable by histone proteins, mutated positions, mutated residue types, mutated PTM sites, phenotypic classes, and constituent assays (Fig. 2). The search results provide links to visualize each mutation on the histone nucleosome crystal structure, or a representation of the tail for mutations in this unstructured region (Fig. 3; see Visualization section below). The results table includes the protein name, the mutated position and geographical domain, and the original and mutated residues. Known PTMs and SNPs, linked to literature references, are provided when these are available. Each column is sortable.
Figure 2.
The database can be searched through a web interface with a flexible combination of keys, including proteins, mutated positions, mutated PTM sites, original amino acids, assays, and phenotypic classes.
Figure 3.
The search result page integrates assay results with sequence, structural, PTM, and SNP information.
Results are provided for each assay, together with a summary score for each phenotypic class. Each assay name is linked to its metadata: a description of the assay, a summary of the assay result distribution, and an interface to visualize all the mutants in the assay. A “view structure” link for each phenotype category generates an interface to visualize all mutants for assays within the category. Residues may be color coded by assay results, or according to the concordance and discordance across multiple assays. The bottom row of the table summarizes enrichment or depletion of non-wild-type phenotypes according to geographical domain (see Geographical Domain Enrichment section).
Visualization
The nucleosome is represented by a yeast crystal structure (White et al. 2001), downloaded as 1ID3 from PDB (Berman et al. 2000). Two views of the structure, multiview (the default) and largeview, are provided. Multiview (Fig. 1; Supplemental Fig. 4) shows a small structure in the center and four side structures from the top, bottom, left and right. The largeview lacks the side views, but has a much larger en face view of the structure.
Multiview and largeview share the same toolbox. Users can spin the structure, zoom in and out, switch between spacefill and ribbon views and select specific histone chains to display. The visualization is implemented using Jmol (http://www.jmol.org/), and Jmol commands are available through the Jmol menu and console by clicking on the word “Jmol” in the lower right corner of the structure display area.
For each assay, a combined view shows all the tested deletions and multiple mutations (Supplemental Fig. 5). This view can be found in the assay description page. Mutants are colored green, light green, gray, pink, and red for 2, 1, 0, –1, and –2, respectively. Multiple mutations to different amino acids are represented with different symbols.
Analysis of phenotypic discrepancies and assay concordance
Assays may have conflicting results for the same histone mutant. Phenotypic discordance may be due to differences in the strain background, differences in the use of plasmids or DNA integration to introduce the mutant sequence, or differences in the assay protocol. The database provides a fast method to identify and visualize discordant assay results. As an example, two recent lethality assays (Dai et al. 2008; Nakanishi et al. 2008) tested 215 overlapping mutants for growth defects and lethality. Of the 215 mutants tested, 186 are viable in both assays, and 14 mutants are lethal in both. The remaining 18 mutants are lethal in only one of the two assays (Supplemental Table 2). These discrepancies can be visualized using the “visualize structure” link in the phenotype class header of the results (Fig. 3). The discrepant mutants are primarily located on the lateral surface, which interacts with DNA molecules (Supplemental Fig. 4, top view).
The same method can be adapted to identify residues that give concordant or discordant phenotypes for related assays. Silencing is a relevant example because the mechanisms for silencing can depend on the transcript. Some silencing mutants either enhance or suppress silencing for all transcripts, possibly by affecting histone-DNA interactions. Other mutants are specific to certain classes of transcripts, or even affect silencing in opposite directions. Visualization permits identification of histone surface patches responsible for generic and specific silencing phenotypes (Supplemental Fig. 6).
Data submission
Users are encouraged to submit their data directly to our database. The data can be as complex as multiple large scale screens done with a systematic collection of mutants or as simple as a single list of mutants isolated by traditional means. The database includes a web interface that generates a spreadsheet template that can be filled and uploaded (Supplemental Fig. 7). This template has the description of the assay and several examples of how to enter assay results.
In order to generate this template, the web interface collects reference information (PubMed ID or prepublication information), protein name, strain, plasmid, and assay description. The assay description includes the name, phenotypic class, readout, format, and reporter. The direction, indicating whether an assay is a screen or a counterscreen, is also recorded. Most of these fields are selected from controlled vocabularies provided to a user. New terms are readily added following a review.
Research groups can either submit their assay results in five-point scale, or submit the raw assay results and communicate with database staff to effect the conversion. The database saves the original assay results for consistency checks and possible recalculations.
Discussion
We presented a histone systematic mutation database as a resource for histone mutagenesis analyses (http://histonehits.org). This database provides a controlled but yet extensible vocabulary to describe the assays on histone mutants. Assays are described in terms of name, reporter, format, readout, and strain. Mutations can be integrated into genomic DNA or expressed from a plasmid. Point mutations, multiple mutations, and deletions can all be deposited in the database. Assays are grouped by phenotypic classes that they are designed to probe. Because the assay readouts are all scaled to the five-point grade system, assay results can be easily compared across studies. Researchers have used this database to deposit their data and perform global data analyses (Dai et al. 2008).
We confirmed that the number of phenotypes for a mutant increases with the residue conservation score (Landau et al. 2005). Mutations on modifiable residues are more likely to have phenotypes, namely rDNA silencing phenotype and low mating efficiencies. Analysis by geographical domains indicates that buried and lateral residues have significantly more phenotypes and tail residues have significantly fewer phenotypes.
The database will expand to include more extensive curation of small-scale experiments, mutations in histone variants and in histones from other species such as Xenopus laevis (Ferreira et al. 2007). Mapping mutagenesis studies across histone families may provide valuable insights into histone residue functions.
A second planned enhancement is to include residue-level physical interactions between histones and other proteins. For example, histone chaperone ASF1A (also known as CCG1-interacting factor A [CIA-I]) disrupts the histone H3–H4 tetramer to form a protein complex (Natsume et al. 2007). These physical interactions between histone residues and other proteins are highly relevant to protein function. We will curate such interactions from sources such as the PDB database (Berman et al. 2000). Connecting individual histone residues with other complexes through physical and genetic interactions will be a powerful method for mapping chromatin modification networks.
Methods
Statistical tests for residue geographical domain enrichments
To test whether residues in different geographical domains have different probabilities to generate phenotypes, we fit a logistic regression model with the number of phenotypes and the number of assays on each mutant as response variables and the geographical domains of the mutants as the covariates. The difference of deviance between this model and the null model is identical to the likelihood-ratio test statistic (McCullagh and Nelder 1989), which approximately follows a χ2 distribution with three degrees of freedom (due to the three additional parameters from the four geographical domains). To test whether a specific domain generates more or fewer phenotypes, we fit four logistic regression models, one for each domain versus the rest of the protein, using the same response variables. These P-values were calculated for two-sided sets and corrected for fourfold multiple testing.
To investigate assay-specific domain enrichment, we calculated the fraction of the tested residues for each domain as the number of the tested residues in the domain divided by the total number of the tested residues. We then calculated the fraction of the tested residues in a domain having a certain assay score. The ratio of these reveals the overrepresentation (>1) or underrepresentation (<1) of certain domains having an assay score. We perform this analysis for assay scores from –2 to 2, positive, negative and nonzero scores. To assess the significance of the ratio, we used Fisher's exact test on a 2 by 2 contingency table, corrected for multiple testing.
Assay description
Assay results can depend on factors such as strain and integration method. The metadata for each assay is captured in attributes including the assay name, reporter, format, readout, and strain (Supplemental Table 3). Different assays may share some attributes.
This design also provides a controlled vocabulary for the data submission. Research groups can choose from the existing entries in each attribute to describe their assays. They may, of course, propose new entries if necessary. We will review each new entry before depositing it into the database.
Phenotypic classes
Some assays are designed to probe related phenotypes, such as temperature sensitivity at different temperatures, and DNA damage sensitivity to different drugs. We group these assays through the phenotypic classes that they are designed to probe. For each mutant, we summarize the assay results for each phenotypic class by calculating its mean value. The mutants under each phenotypic class can be visualized on a nucleosome crystal structure based on their assay results, or based on their accordance and discordance across assays.
Web interface and database implementation
The web interface is implemented using AJAX (Asynchronous JavaScript and XML) compiled using the Google Web Toolkit version 1.4. The database uses MySQL (Supplemental Fig. 2). This interface communicates with the database through server side scripts, which are implemented in Perl as CGI programs with Perl DBI module and CGI modules. Data sets are exchanged between the web interface and the server side scripts using the JSON format and the Perl JSON module. The Jmol applet provides structural visualizations and the Perl GD module generates images of deletions and multiple mutations.
Acknowledgments
This material is based upon work supported by the National Science Foundation under grant no. NSF CAREER 0546446 to J.S.B. J.S.B. acknowledges additional support from NIH/NIGMS R41GM073492 and the Whitaker Foundation. J.D.B. and J.S.B. acknowledge support from NIH Roadmap Grant No. U54RR020839.
Footnotes
[Supplemental material is available online at www.genome.org. HistoneHits is freely available at http://histonehits.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083402.108.
References
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blossey R., Schiessel H. Kinetic proofreading of gene activation by chromatin remodeling. HFSP J. 2008;2:167–170. doi: 10.2976/1.2909080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.T. Histone variants: Are they functionally heterogeneous? Genome Biol. 2001;2:REVIEWS0006. doi: 10.1186/gb-2001-2-7-reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Pikaard C.S. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes & Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Hyland E.M., Yuan D.S., Huang H., Bader J.S., Boeke J.D. Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A.A., Winston F. Analysis of a mutant histone H3 that perturbs the association of Swi/Snf with chromatin. Mol. Cell. Biol. 2004;24:561–572. doi: 10.1128/MCB.24.2.561-572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira H., Somers J., Webster R., Flaus A., Owen-Hughes T. Histone tails and the H3 αN helix regulate nucleosome mobility and stability. Mol. Cell. Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry C.J., Norris A., Cosgrove M., Boeke J.D., Peterson C.L. The LRS and SIN domains: Two structurally equivalent but functionally distinct nucleosomal surfaces required for transcriptional silencing. Mol. Cell. Biol. 2006;26:9045–9059. doi: 10.1128/MCB.00248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Hyland E.M., Cosgrove M.S., Molina H., Wang D., Pandey A., Cottee R.J., Boeke J.D. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M., Smith M.M. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 2003;13:154–160. doi: 10.1016/s0959-437x(03)00020-0. [DOI] [PubMed] [Google Scholar]
- Kamakaka R.T., Biggins S. Histone variants: Deviants? Genes & Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Sano N., Umehara T., Horikoshi M. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells. 2007;12:13–33. doi: 10.1111/j.1365-2443.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- McCullagh P., Nelder J.A. Generalized linear models. Chapman and Hall; London: 1989. [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Millar C.B., Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Muller H.M., Kenny E.E., Sternberg P.W. Textpresso: An ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S., Sanderson B.W., Delventhal K.M., Bradford W.D., Staehling-Hampton K., Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Park J.H., Cosgrove M.S., Youngman E., Wolberger C., Boeke J.D. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D., Pandolfi P.P. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Smith M.M. Histone structure and function. Curr. Opin. Cell Biol. 1991;3:429–437. doi: 10.1016/0955-0674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- Smith M.M. Centromeres and variant histones: What, where, when and why? Curr. Opin. Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- Smith M.M., Santisteban M.S. Genetic dissection of histone function. Methods. 1998;15:269–281. doi: 10.1006/meth.1998.0631. [DOI] [PubMed] [Google Scholar]
- Smith C.M., Haimberger Z.W., Johnson C.O., Wolf A.J., Gafken P.R., Zhang Z., Parthun M.R., Gottschling D.E. Heritable chromatin structure: Mapping “memory” in histones H3 and H4. Proc. Natl. Acad. Sci. 2002;99(Suppl 4):16454–16461. doi: 10.1073/pnas.182424999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.S., Snow M.L., Giles S., McPherson L.E., Grunstein M. Identification of a functional domain within the essential core of histone H3 that is required for telomeric and HM silencing in Saccharomyces cerevisiae. Genetics. 2003;163:447–452. doi: 10.1093/genetics/163.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa R., Madhani H.D. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics. 2007;175:585–593. doi: 10.1534/genetics.106.067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C.L., Suto R.K., Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Zhang K., Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]