Summary
Interleukin-23 is a recently identified member of the IL-12 family of heterodimeric cytokines that modulate subpopulations of T helper cells, and both IL-12 and IL-23 are attractive targets for therapy of autoimmune diseases. IL-23 is a binary complex of a four-helix bundle cytokine (p19) and a soluble class I cytokine receptor p40. IL-12 and IL-23 share p40 as an α-receptor subunit, yet show only 15% sequence homology between their four-helix cytokines p19 and p35, respectively, and signal through different combinations of shared receptors. In order to elucidate the structural basis of p40 sharing, we have determined a 2.3Å crystal structure of IL-23 for comparison to the previously determined structure of IL-12. The docking mode of p19 to p40 is altered compared to p35, deviating by a ‘tilt’ and ‘roll’ that results in an altered footprint of p40 on the A and D helices of the respective cytokines. Binding of p19 to p40 is mediated primarily by an Arginine residue on helix D of p19 that forms an extensive charge and hydrogen-bonding network with residues at the base of the pocket on p40. This ‘Arginine pocket’ is lined with an inner shell of hydrophobic interactions that are ringed by an outer shell of polar interactions. Comparative analysis indicates that the IL-23 and IL-12 complexes ‘mimic’ the network of interactions constituting the central Arginine pocket despite p19 and p35 having limited sequence homology. The majority of the structural epitopes in the two complexes are composed of unique p19 and p35 pair-wise contacts with common residues on p40. Thus, while the critical hotspot is maintained in the two complexes, the majority of the interfaces are structurally distinct and, therefore, provide a basis for the therapeutic targeting of IL-12 versus IL-23 heterodimer formation despite their use of a common receptor subunit.
Keywords: IL-23, IL-12, cytokine, receptor, crystallography
Introduction
The interactions of four-helix bundle cytokines with their class I cytokine receptors control many aspects of cellular development, differentiation, and proliferation across the immune, nervous and hematopoietic systems. Engagement of cytokines by their receptors results in receptor homo- or heterodimerization, leading to the activation of intracellular Jak/Stat signaling cascades1. Four-helix bundle cytokines have a stereotypical up-up-down-down helical topology for both short and long chain members of the family2. Cytokine receptors also have several conserved features, such as the presence of a ‘cytokine binding homology region’ (CHR) that is characterized by tandem Fibronectin-type III domains (FnIII) containing a hallmark pattern of disulfide bonds, and a WSXWS motif in the second of the FnIII domains3. The prototypical long chain cytokine, human Growth Hormone (hGH) was shown to homodimerize its receptors using a ‘site I/site II’ paradigm through the sides of its helices, and that the receptors engaged the cytokine through inter-strand loops at the junction of the two FnIII domains4,5. While this basic building block has been found in structures of all cytokine receptor complexes, the gp130, or IL-6/IL-12 family of cytokines has evolved an additional receptor binding epitope, termed site III, that requires the presence of a top-mounted Ig-domain on the receptors to homo or heterodimerize signaling complexes in the ‘Tall’ receptor family (e.g. gp130, LIF-R, IL-12, etc.)6,7. Recently, several orphan members of the IL-12 class of cytokines, IL-23 and IL-27, have been paired with their cognate receptors, and their biological activities clarified8,9. In particular, IL-23 has generated a great deal of excitement with regards to its potential role in immune modulation of different subpopulations of T helper cells in concert with IL-1210,11.
IL-12 and IL-23 are heterodimeric cytokines that, unlike typical four-helix cytokines that are secreted alone, are secreted from dendritic cells and macrophages as disulfide-linked complexes between the helical cytokines p35 and p19, respectively, and a shared binding protein termed p4011,12,13,14. Both p35 and p19 have sequence homology to IL-6 and G-CSF, marking them as members of the gp130-class of long-chain cytokines, and the p40 subunit is similar in structure to typical class I cytokine receptors such as the non-signaling alpha receptors for IL-6 and CNTF15. In essence, IL-12 and IL-23 represent cytokines constitutively associated with a soluble α-receptor subunit. While many cytokines exist as naturally ‘shed’ soluble complexes with their α-receptors16, IL-12 and IL-23 are unique in that they are secreted as binary complexes.
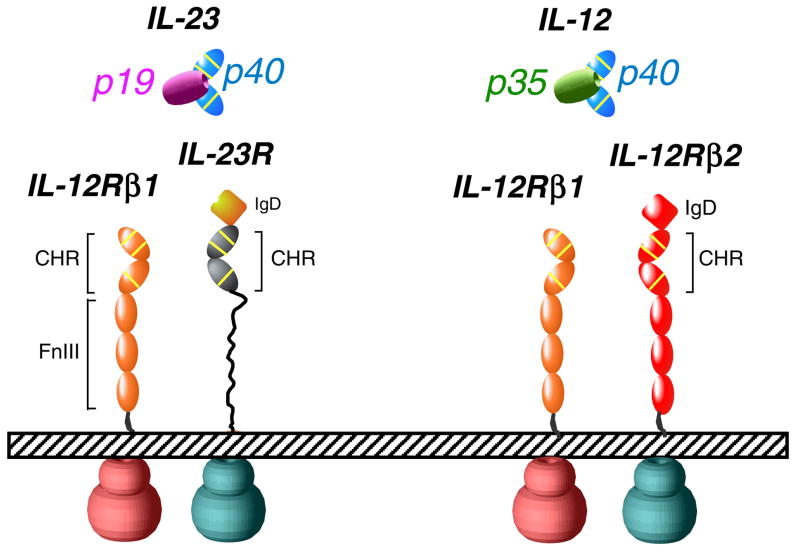
With regards to receptor activation, while IL-12 and IL-23 share p40, they signal through different heterodimeric cell surface complexes involving receptors homologous to gp130. IL-12 (p35/p40) signals through a heterodimer consisting of IL-12Rβ1 and IL-12Rβ217, whereas IL-23 (p19/p40) signals through a heterodimer consisting of IL-12Rβ1 and the IL-12Rβ2-like receptor called IL-23R8. The architecture of the IL-12 and IL-23 receptor signaling complexes can be predicted to recapitulate features of the site I/II/III paradigm based on the organizing principles seen for other members of the tall receptor family such as gp130/IL-6, GCSF/GCSF-R, and LIF/LIF-R7,18. IL-23 also activates the same spectrum of Janus kinase (Jak)/signal transducers and activators of transcription (Stat) signaling molecules as IL-12: Jak2, Tyk2, and Stat1, Stat3, Stat4, and Stat58.
Biologically, it first seemed likely that these two cytokines would have redundant roles in immune homeostasis, namely T helper (Th)1-type responses that are important for cell-mediated antimicrobial and cytotoxic activities, based on their shared use of p40 as a subunit. However, it was quickly shown that their functions are non-redundant11. Whereas IL-12 drives the typical Th1 responses such as interferon-γ (IFN-γ) production, IL-23 does not influence the development of Th1 cells, but instead drives the development of an alternate CD4+ T cell population, now termed Th17 cells, that are notable for their production of pro-inflammatory IL-17 cytokines19. Several studies have shown that IL-23 regulation of autoreactive Th17 cells plays a critical role in the development of chronic autoimmune disorders20,21,22. Recently, IL-23 has also been shown to possess tumour-promoting proinflammatory activity, and that IL-23 blockade can render tumours susceptible to infiltration by IL-12-induced cytotoxic T cells23. These studies have led to considerable interest in the possibility of therapeutic IL-23 blockade for treatment of autoimmune disorders and, potentially, cancer.
Because IL-12 and IL-23 share their p40 subunit, it is important to delineate common versus cytokine-specific protein-protein interactions to serve as guideposts for the potential development of IL-12 versus IL-23-specific antagonists. Here we present a structural analysis of IL-23 that represents an important benchmark in elucidating the molecular basis of p40 subunit sharing with IL-12, and in understanding the assembly of their respective signaling complexes based on the organizing principles derived from other members of gp130/IL-6/IL-12 family.
Results
We expressed IL-23 by co-infection of insect HiFive cells with recombinant baculoviruses carrying cDNAs for p19 and p40. The complex purified by gel filtration as a single peak of disulfide-linked dimer (data not shown), confirming the integrity of the IL-23 heterodimer as shown for IL-1215. IL-23 was crystallized and the structure determined to a resolution of 2.3Å by molecular replacement using the coordinates of p4015, and p19 was then built from the partial phases in order to eliminate model bias that would be introduced by using p35 as a starting model. During the expression, Asn-linked glycans were deglycosylated with EndoH in order to debulk the complex of carbohydrate moieties, and the surface lysines were methylated in order to grow large, well-ordered crystals. The deglycosylated and methylated IL-23 behaved identically to unmodified IL-23 during purification, while crystal diffraction was markedly improved (~4.5 Å to 2.3 Å).
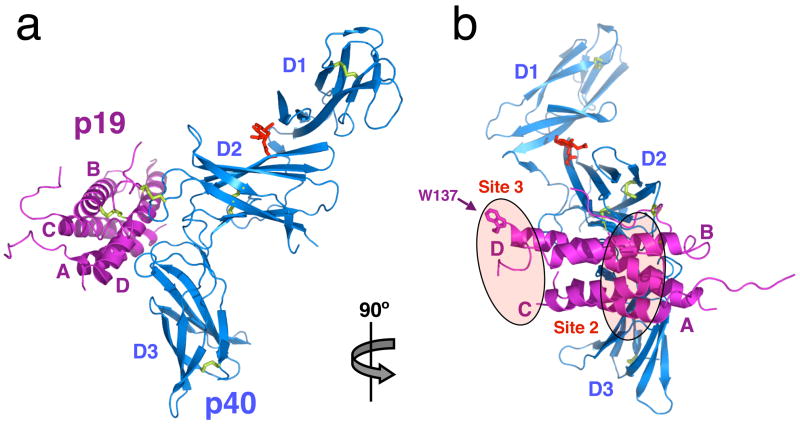
The overall structure of IL-23 is typical of class I cytokine receptor complexes and strongly resembles the p35/p40 heterodimer structure of IL-12 (discussed below) (Figure 2). The p40 subunit, as previously seen, is composed of three domains (D1, D2 and D3) of approximate dimensions 100 × 45 × 25 Å, and superimposes with the previous human p40 structure with an overall root mean square deviation (RMSD) of 1.3Å. The D1 domain is an S-type Ig-fold in which strand A is swapped from the three-strand sheet so that it now joins the four-strand sheet. The D1 domain interacts with the D2 domain in an unusual fashion in which the long axis of the domain is displaced from co-linearity with the D2 domain, such that it engages the top of D2 in a nearly orthogonal orientation (Figure 2b), as opposed to the more conventional end-to-end packing of tandem Ig domains, as seen for the D2 and D3 domains. The D2 and D3 domains represent the canonical CHR found in all class I cytokine receptors, with the conserved disulfide bonds in D2 (C109–120 and C148–C171). We observed one GlcNAc moiety emanating from Asn200 on the D2 domain that packs against the D1 domain, but at a location distant from the cytokine binding site (Figure 2). The remaining consensus Asn-linked glycosylation sites in p40 are unmodified or the glycans disordered in the structure. The D3 domain contains the conserved WSXWS motif (WSEWA in p40) on the G strand. This consensus sequence is a part of a β-bulge and π-cation stacking motif in which side chains from the F strand (R287 and Q289) stack between aromatic residues on the WSEWA motif.
Fig. 2.
Structure of IL-23. (a) Side view of the IL-23 crystal structure. p19 helices A to D are colored in magenta, and p40 domains 1 to 3 colored in blue. The single N-acetylglucosamine residue attached to Asn200 on p40 is shown in red. Disulfide linkages are colored in yellow. (b) Face-on view of IL-23. Receptor interaction sites 2 and 3 are highlighted in ovals, and the key site 3-interacting sidechain of Trp137 is displayed.
The four-helix bundle p19 is topologically similar to other canonical long chain four-helix cytokines (Figure 2a and 2b), which are so far the only known protein structures to exhibit an up-up-down-down helix topology. As is typical in cytokine structures, the helices are well-ordered, while several inter-helical loops (residues 29–47 (A–B loop), 92–99 (B–C loop), and 123–137 (C–D loop)) lack defined electron density, which likely indicates flexibility of these regions. A comparison of p19 with all four-helix bundle cytokine structures shows the closest similarity with Interleukin-624 (Dali Z-score of 12.7 with RMSD of 2.5Å for 117 overlapping residues). p19 has ~15% sequence identity with p35, and superposition of 107 overlapping residues (out of 133) yields an RMSD of 2.3Å (Dali Z-score of 10.5). Thus, we speculate that even though p19 and p35 both engage p40, they are not simply paralogs of one another, but may have undergone convergent evolution to engage p40 from distinct four-helix bundle precursors. The mature p19 polypeptide sequence is shorter than mature p35 by 27 residues, which is partially manifested in the structure by the truncation of the p19 A, C and D helices by two, one and two helical turns, respectively, compared to p35. A major deviation between the p19 and p35 structures is seen at the C-terminal end of the A–B loop in p35, which forms an 11-amino acid disulfide-bonded loop (missing in p19) that forms two turns of an α-helix and forms numerous interactions with p40.
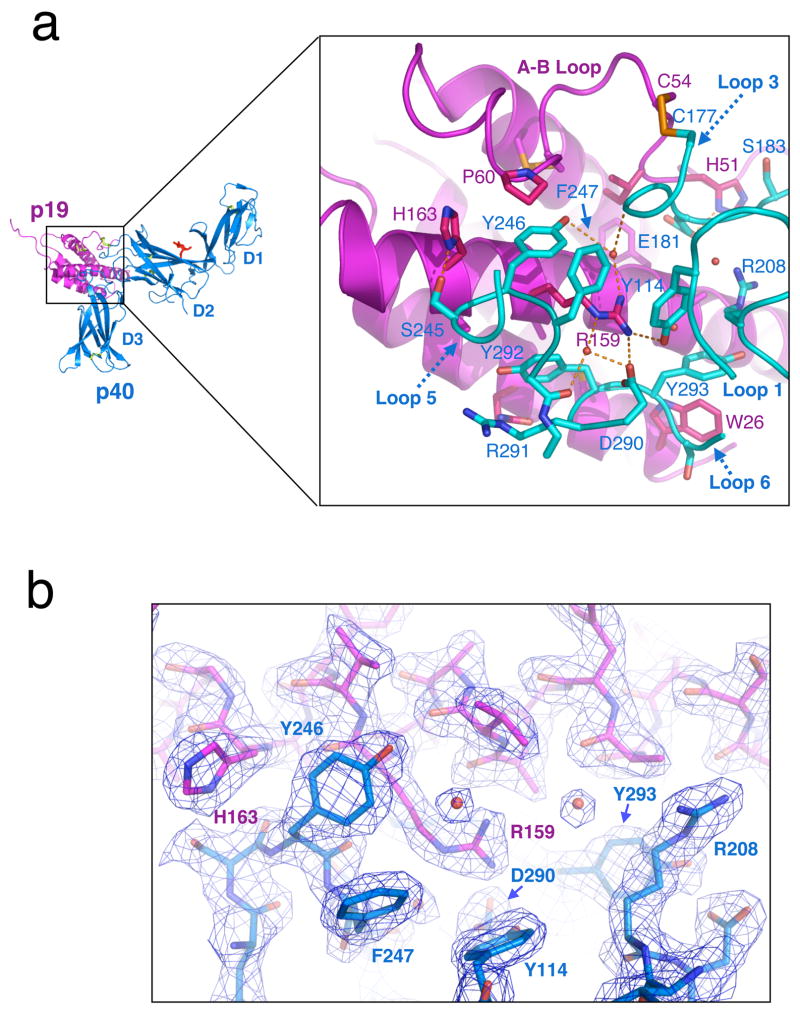
In the IL-23 interface, p40 loops 1 and 3 from the D2 domain, and loops 5 and 6 from D3 interact with the p19 A and D helices, as well as second half of the long AB-loop (Figure 3a). The inter-subunit disulfide bond unique to the IL-12/23 cytokines is formed at the top edge of the interface between p19 residue Cys54 on the AB-loop to p40 Cys177. This disulfide bond is peripheral to the main structural epitope between p19 and p40, and in the case of IL-12, was shown not to be necessary for the p35/p40 interaction15. There is a large receptor-cytokine interface involving 18 residues of p19 and 14 residues of p40 that buries 1730Å2 of surface area, with 830Å2 buried on p19 and 900Å2 buried on p40.
Fig. 3.
Structural anatomy of the p19–p40 interface. (a) Close-up view of the secondary structure and amino acid contacts between p19 and p40. (b) Cut away view of the ‘Arginine pocket’ 2Fo-Fc electron density contoured at 1.5 σ. Several well-ordered waters are visible at the interface, stabilizing the interaction of p40 pocket residues with Arg159.
The p40 binding site forms a ‘volcano-like’ surface with Asp 290 at the base of the crater, which is lined with hydrophobic residues such as Tyr246, Phe247, Tyr292, Tyr293 (Figure 3a). A peripheral group of both apolar and hydrogen bonding residues (e.g. Arg208, Ser245, Glu181, Arg291) form a ring of ‘peaks’ surrounding the crater. In the heart of the interface, p19 extends Arg159 from its helix D into the crater such that its guanidium group forms an extensive network of interactions including a salt bridge to p40 residue Asp290 and a hydrogen bond to Tyr114. The ε-nitrogen of the p19 Arg159 guanidinium group also forms water-mediated hydrogen bonds to Tyr246 and Asp290 (Figure 3a). These waters also hydrogen bond to the main chains of Ala179 of loop 3 and Ser248 of loop 5, respectively. There are several other ordered water molecules visible at the periphery of the interface that appear to stabilize the p40 interaction (Figure 3b). In p40, the aromatic residues lining the crater of the Arginine pocket form multiple van der Waals interactions with backbone and side chain atoms on helices A and D of p19 that surround the centrally protruding Arg159 (Figure 3a). The principal interactions involve p40 Tyr292 packing against the p19 D-helix main chain and forming hydrophobic contacts with p19 Ala155 and 158, and p40 Tyr293 interacting with p19 D-helix Ala152 and A-helix Trp26. The outermost shell of the interface exhibits several additional side-chain specific hydrogen bonds from p40 residues Glu181 and Ser245, which interact with p19 residues His51 from the p19 AB-loop, and His163, respectively (Figure 3a). Collectively, the p19/p40 interface appears to have three shells of interactions: a charged center at the base of the crater, hydrophobic contacts lining the crater, and hydrogen bonded peaks at the interface periphery. Thus, rather than the more conventional interface hotspot where a hydrophobic center is surrounded by polar periphery25,26, IL-23 has a polar center that appears insulated by a ‘gasket’ of hydrophobic interactions.
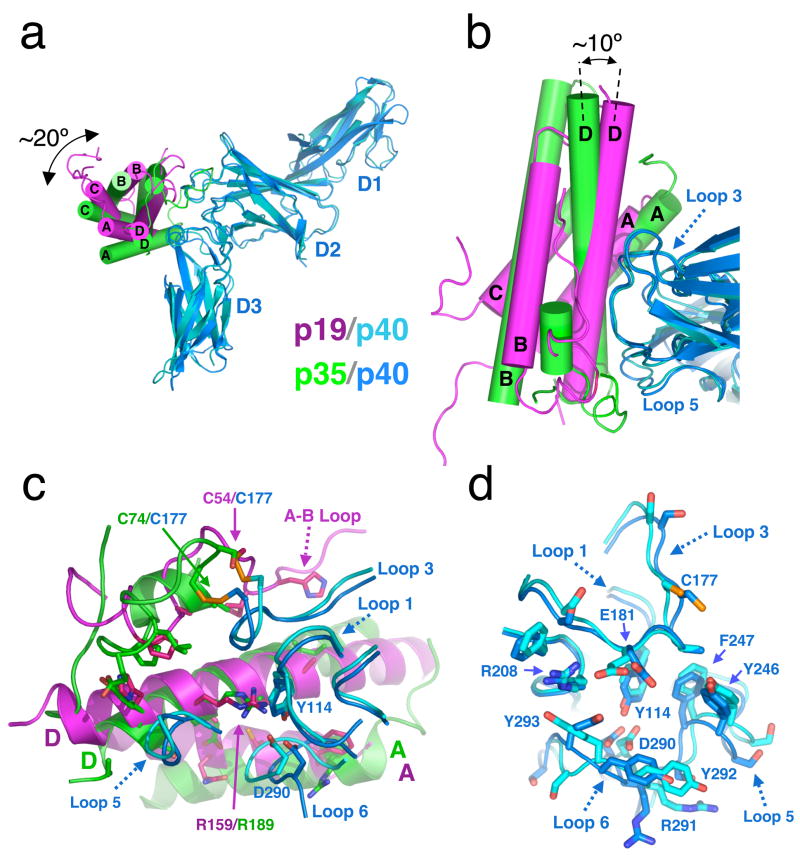
Although both p19 and p35 primarily use their A and D helices to engage p40, there are substantial positional changes with respect to the docking modes on p40. When the p40 molecules from both complexes are superimposed and the complexes are viewed looking down the barrels of the p19 and p35 four-helix bundles, it is clear that p19 is rotated clockwise towards the D2 domain of p40 by approximately 20 degrees (Figure 4a). When viewed from the top, p19 is also tilted towards p40 by approximately 10 degrees compared to p35 (Figure 4b). The overall result of the rotation and tilt of p19 with respect to p35 is that p19 has more intimate interaction between the N- and C-terminal ends of its D and A helices with p40 than p35, and p35 has more intimate interactions with p40 at the C and N-terminal ends of its D and A helices, respectively (Figure 4b). In this latter region of the IL-12 interface, p40 loops 3 and 5 interdigitate into a ‘corner’ of p35 formed by an 11-amino acid disulfide bonded loop C-terminal to Cys74 – this mini-loop is missing in p19. Thus, while p35 is less complementary with p40 at one end relative to p19, the rotation and tilt of the four-helix bundle results in improved complementarity at the opposite end.
Fig. 4.
Comparative analysis of IL-23 and IL-12. (a) p19 (pink) is ‘rolled’ toward the D2 of p40 (blue) by ~20° and (b) tilted by ~10° when compared to p35 (green). (c) Overlay of the p19–p40 and p35–p40 interaction interfaces when p40 is structurally superimposed. Note the intact side chain positioning of p19 Asp159 and p40 ‘Arginine pocket’ residues. All contact residues on p19 and p35 are drawn as sticks. (d) Comparison of the four-helix bundle interacting loops of p40 from the two structures (IL-23 p40 in cyan and IL-12 p40 in blue) reveals only slight distortions in the positioning of the loops when bound to their respective four-helix cytokines.
In comparison to IL-1215, IL-23 has utilized the same network of charge and hydrogen bonds involving p40 residues Asp290 and Tyr114 at the base of the crater by structurally ‘mimicking’ the Arg189 on the D-helix of p35 with its own p19 D-helix Arg at position 159 (Figure 4c). While a mutational analysis of interface residues was carried out for IL-12 to assess the energetic importance of p35/p40 contacts in complex assembly15, such data does not currently exist for IL-23. However, instructive comparisons can be made for obviously analogous structural contacts. In IL-12, residues that make up the hydrogen-bonding network involving the central Arg on the cytokine helix D, p40 residues Asp290 and Tyr114, are energetically critical as their mutation abrogates complex formation. We suggest that these residues in IL-23 are likely to be similarly energetically important. Given the vanishing sequence identity between p19 and p35, conservation of this core ‘hotspot’ in both complexes was unexpected. The positional correspondence of these residues (Figure 4c) and their interactions in the face of highly divergent sequence and docking positions on p40 further suggests convergent structural evolution from distinct precursors to recapitulate the features of this ‘Arginine’ pocket in both heterodimers. The hydrogen bonds in the Arginine pocket in the two complexes are not exactly similar, most likely due to a slight shift in backbone position of the p19 Arg159 versus p35 Arg189, which affects the side chain location and geometry of the resultant bonding network (Figure 4c). Further direct comparisons of the role of bound water molecules in the respective interfaces is complicated by the differing resolutions of the structures (IL-23 2.3Å versus IL-12 2.8Å). Nevertheless, it is clear that the interactions that form the Arginine pocket are structurally conserved, and therefore likely to be important for heterodimer formation in both cytokines.
The surrounding interactions in the IL-23 and IL-12 interfaces, however, are almost entirely different (Table 2), which was to be expected given the limited sequence identity between p19 and p35. Comparison of the two interfaces reveals that, with the exception of the central Arginine residue discussed above, there are no other conserved pairwise interactions (Table 2). Many of the same p40 residues are buried within the different interfaces (Figure 5a), and serve similar structural roles in both complexes, but through interactions with distinct constellations of amino acids on the p19 and p35 surfaces (Figure 5b). For instance in the interface ‘outer shell’, p40 residue Glu181, which was shown in IL-12 to be energetically-critical for heterodimer formation15, forms a hydrogen bond with Arg183 in p35 versus His51 in p19. Ser245 of p40, whose mutation to Ala had a modest effect in the IL-12 system, hydrogen bonds to the amide nitrogen of Tyr193 in p35 versus the side chain of His163 in p19. In the IL-23 complex, p40 forms several more interactions with the p19 AB-loop near the Cys54-Cys177 interchain disulfide linkage (Figure 4c), which may be a result of the rotation of p19 towards p40 relative to p35. In IL-12, the AB-loop is largely disordered with the exception of residues C-terminal to the p35 Cys74, and this disorder may be a result of the gap between p35 and p40 above the D helix, formed by the relative rotation of p35 away from p40. Collectively, p40 displays an ability to engage two different cytokine surfaces through a combination of shared (Arg pocket) and distinct interactions (outer shells).
Table 2.
Shared and distinct contacts between p40 and the four helix bundles p19 and p35
| p40 residue | p19 residue | vdW* | H-bonds** | p35 residue | vdW* | H-bonds** |
|---|---|---|---|---|---|---|
| Tyr 114 | Arg 159 | 1 | 1 | Arg 189 | 4 | 1 |
| Ser 175 | 0 | 0 | Ser 73 | 3 | 0 | |
| Ala 176 | 0 | 0 | Ser 73 | 2 | 0 | |
| Cys 177 | Cys 54 | 4 | 0 | Ser 73 | 6 | 0 |
| Cys 74 | ||||||
| Pro 178 | 0 | 0 | Cys 74 | 1 | 0 | |
| Ala 179 | Asp 59 | 2 | 0 | Val 60 | 3 | 0 |
| Val 156 | Leu 68 | |||||
| Ala 180 | Ile 52 | 2 | 0 | 0 | 0 | |
| Glu 181 | His 51 | 18 | 3 | Ile 182 | 11 | 3 |
| Ile 52 | Arg 183 | |||||
| Phe 153 | Thr 186 | |||||
| Ser 183 | His 51 | 1 | 0 | 0 | 0 | |
| Arg 208 | Ala 152 | 4 | 0 | Ile 182 | 3 | 0 |
| Thr 242 | 0 | 0 | Leu 68 | 1 | 0 | |
| Pro 243 | His 163 | 1 | 0 | Glu 67 | 4 | 0 |
| Ser 245 | Ala 162 | 6 | 1 | Ser 192 | 11 | 1 |
| His 163 | Tyr 193 | |||||
| Thr 167 | Ala 196 | |||||
| Tyr 246 | Cys 58 | 15 | 1 | Pro 65 | 9 | 1 |
| Pro 60 | Arg 189 | |||||
| Arg 159 | ||||||
| His 163 | ||||||
| Phe 247 | 0 | 0 | Arg 189 | 2 | 0 | |
| Ser 248 | 0 | 0 | Ser 192 | 1 | 0 | |
| Gln 289 | 0 | 0 | Arg 34 | 2 | 0 | |
| Asp 290 | Arg 159 | 1 | 2 | Arg 34 | 2 | 1 |
| Arg 291 | 0 | 0 | Arg 34 | 2 | 0 | |
| Tyr 292 | Gln 19 | 17 | 0 | Arg 34 | 16 | 2 |
| Cys 22 | Arg 181 | |||||
| Ala 155 | Asp 188 | |||||
| Ala 158 | Arg 189 | |||||
| Arg 159 | ||||||
| Ala 162 | ||||||
| Tyr 293 | Trp 26 | 6 | 0 | Arg 34 | 15 | 1 |
| Ala 152 | Arg 181 | |||||
| Ile 182 | ||||||
| Val 185 | ||||||
| Ser 294 | Trp 26 | 8 | 0 | Arg 34 | 1 | 1 |
| Ser 295 | 0 | 0 | Arg 34 | 1 | 0 |
van der Waals cutoff distance of 3.9Å
hydrogen bond cutoff distance of 3.5Å
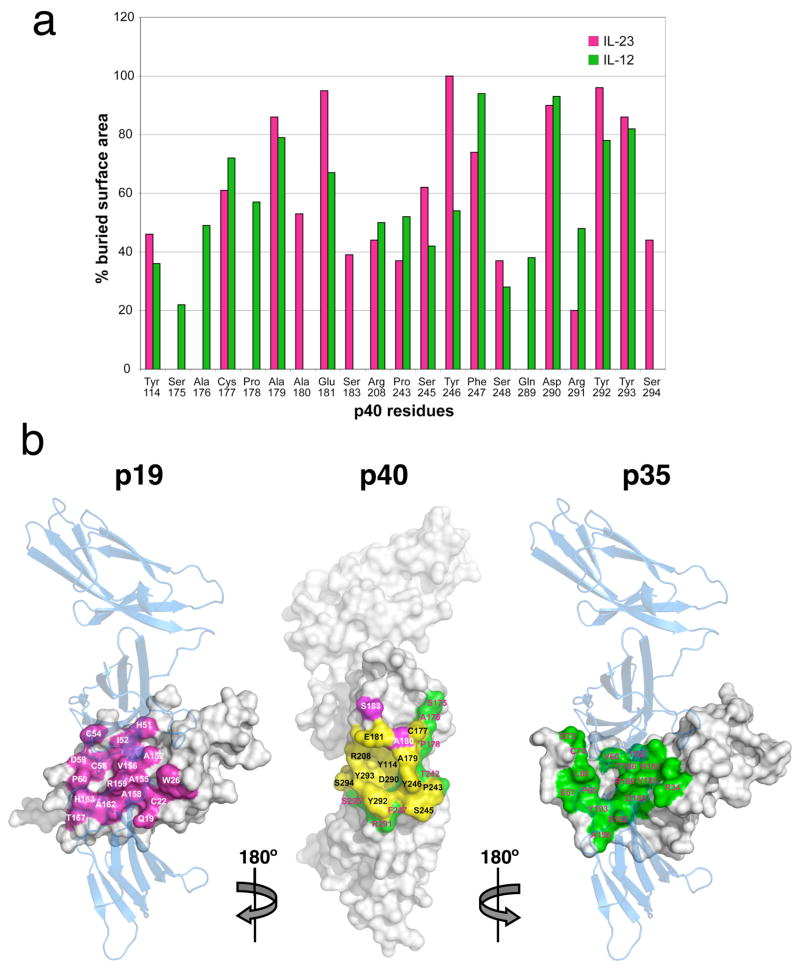
Fig. 5.
Comparison of shared and distinct contact surfaces in the IL-23 and IL-12 complexes. (a) Histogram of buried surface area contributed by each p40 residue involved in p19 and/or p35 interaction. (b) Surface representation of p40 (middle panel) with residues interacting with p19 (pink), p35 (green), or p19 and p35 (yellow) highlighted. The p40-interacting surfaces of p19 (left panel in pink) and p35 (right panel in green) are shown with a transparent p40 overlay to demonstrate orientation.
The cross-reactivity of p40 is not due to large-scale conformational changes in the binding site. Superposition of p40 from both heterodimers reveals that the backbone conformations of the p40 loops are generally similar, although several of the loops are slightly distorted (Figure 4d). We interpret this as structural accommodation of the different p19 and p35 cytokine surfaces by somewhat flexible, solvent exposed p40 loops as has been previously reported for receptor-cytokine interactions in the gp130 and IL-4/13 receptor systems26,27, rather than an ‘induced fit’ binding mechanism in which distinct loop arrangements accommodate the divergent surfaces. Strikingly, the center of the Arginine pocket, including residues Asp290, Tyr114, and Phe247 of p40 have preserved their side chain and main chain positions in the two complexes (Figure 4d). These residues, including Phe247, are energetically essential for formation of the IL-12 complex15. The main conformational deviations seen in the p40 binding site are localized to loop 6 of the p40 D3 domain, where Arg291, Tyr292, and Tyr293 assume different side chain positions. It therefore appears that p40 has preserved the structural context of most of the energetically critical residues used in the IL-12 heterodimer that are also in contact with p19, further supporting their conserved role in IL-23.
To further investigate the basis for the cross-reactivity of p40 for p19 and p35, we analyzed the inter-chain contacts and buried surface area for each p40 residue that makes contact with either p19 or p35 (Table 2 and Figure 5a). Clearly, p40 uses the same binding surface to contact both p19 and p35 (Figure 5a). However, when analyzed in terms of inter-atomic contacts (Table 2 and Figure 5b), of the 23 total p40 residues that make contact with either cytokine, we find that twelve (~1/2) of these residues are shared contact residues. Nine of the remaining residues only contact p35, while two only contact p19. The bulk of this disparity reflects the added contact area provided by the A–B loop extension found in p35. The two amino acids that exclusively contact p19 are found at the C-terminal end of loop 3 in p40 and interact with p19 in the ordered section of the A–B loop around the interchain disulfide at Cys54 (discussed earlier). This data is represented graphically in Figure 5b, with shared residues on the surface representation of p40 (middle panel) colored yellow and p19 and p35-exclusive residues colored pink and green, respectively.
Discussion
As both IL-12 and IL-23 are attractive therapeutic targets for various autoimmune diseases, one goal is to develop small molecule inhibitors of heterodimer formation15. Small molecule antagonism of cytokine-receptor interactions remains a daunting, but important, challenge given how important many cytokine-receptor interactions are in human health and disease. While antibodies can be highly effective antagonists, production costs are high and special storage and handling is required. Despite the immense difficulties in producing small molecule antagonists of protein-protein interactions, one notable exception has been the four-helix cytokine Interleukin-2 (IL-2), for which a panel of high affinity small molecules have been created that antagonize its association with its α-receptor28. Comparison of the IL-2/drug and IL-2/IL-2Rα complexes revealed that the drug utilizes a similar binding epitope on the cytokine as the receptor29. Further, an alanine scanning study showed that the drug uses the same energetic hotspots on IL-2 as the receptor, despite inducing conformational changes on the cytokine surface30. Given this encouraging proof of concept, small molecule targeting of cytokines remains an active effort in the pharmaceutical industry.
Due to the different activities of IL-12 and IL-23 on Th1 and Th17 responses, respectively, the ultimate therapeutic goal would likely be to target one cytokine specifically in order to modulate these responses independently. The shared use of p40 by these cytokines would presumably make it difficult to develop drugs against p40 that target one heterodimer versus the other. Our comparative structural analysis of the p19 and p35 interfaces with p40 reveals that the majority of the helical cytokine surfaces involved in p40 interactions are structurally unique and cross-reactivity of a drug targeted against p19 or p35 would be unlikely. On the other hand, the majority of p40 residues contacting p19 also contact p35, rendering p40 a more difficult target for design of a cytokine-specific antagonist. However, the p40 binding surfaces are not entirely redundant, and in fact several residues are used specifically in p19 or p35 interactions (Table 2, and Figure 5b). For example, p40 residues Ala180 and Ser183 only contact p19, whereas approximately nine p40 resides are used only for p35 contact. Most of these latter residues are on the p40 loops 5 and 6, which have extensive contact with the p35 disulfide-bonded mini-loop that is not present in p19. Therefore we conclude there are sufficient structural differences in these interfaces that, in principle, a small molecule or antibody could selectively interfere with formation of IL-12 or IL-23 by targeting either the cytokines or p40.
The IL-23 heterodimer engages the signaling receptors IL-12Rβ1 and IL-23R in order to induce a cellular response via the JAK/STAT pathway (Figure 1). Previous structural studies in the gp130 system provide an architectural template to model the IL-23 quaternary complex 6,26,31. Cytokines of the gp130 family possess a ‘site III’ at the tip of the cytokine that engages an Ig-domain on one of the signaling receptors, in addition to the canonical sites I and II on each side of the four-helix bundle. In the p19 structure, the loop connecting the C-D helices contains a Tryptophan (Trp137) residue that is a signature hallmark of the site III interaction (Figure 2b), as is also seen in members of the gp130-cytokine family 6,31. For IL-23, the p19/p40 heterodimer represents the site I complex. Given that IL-12Rβ1 does not possess an Ig-domain, the only possible site it could engage on IL-23 would be site II (Figure 2b), binding the A and B helices of p19 in an orientation similar to the manner in which IL-6 engages gp130. This would leave the tip of p19 available to engage the Ig-domain of IL-23R to form a quaternary signaling complex. By analogy, the p35/p40 heterodimer would also engage IL-2Rα1 at site II, and recruit the Ig-domain containing IL-12Rβ2 to site III to similarly form a quaternary complex. Future structural studies will clarify the assembly pathways and molecular architecture of the IL-23 and IL-12 signaling complexes, and will likely reveal additional targets of opportunity for structure-based design of antagonists.
Fig. 1.
Schematic representation of IL-23 and IL-12 signaling complexes. IgD stands for “Ig-like domain”, while CHR denotes “cytokine-binding homology region” and FnIII denotes “Fibronectin-type III domain”. Yellow lines denote two conserved disulfide linkages and the conserved WSXWS motif.
Materials and Methods
Protein expression and purification
Recombinant IL-23 used for crystallization was expressed using the baculovirus system. High-titer baculovirus stocks were prepared by transfection and amplification in Spodoptera frugiperda (SF9) cells cultured at 28° C in SF900 II media (Invitrogen). Protein expression was carried out in Trichopulsia ni (Hi-Five, Invitrogen) cells growing in suspension in Insect Xpress media (Lonza).
Human p19 (residues 20–189) and p40 (residues 23–328) cDNAs were cloned into the insect cell expression vector pACSG2 (BD Biosciences) in frame with an N-terminal gp67 leader sequence and C-terminal hexa-histidine tag. To generate glycan-minimized IL-23, kifunensine-treated Hi-Five cells were co-infected with p19 and p40 viruses along with virus carrying the endoglycosidase-H (endoH) cDNA, and protein was allowed to express for 48 h. The p19/p40 IL-23 heterocomplex was captured from supernatant using Ni-agarose (Qiagen) with yields approaching 20 mg/L. Ni-purified IL-23 was diluted to 1 mg/mL, subjected to reductive methylation32, concentrated, and purified by FPLC on a Superdex 200 16/60 size exclusion column (GE Healthcare) equilibrated in 10 mM Hepes, pH 7.0, 150 mM NaCl. Peak fractions were pooled and concentrated for crystallization.
Crystallization and data collection
Initial crystallization trials yielded small triangular plate-like crystals from the Peg-Ion crystallization screen (Hampton Research) using the sitting drop method at 25°C. Ultimately, diffraction-quality crystals were grown in sitting drops at 25° C by mixing 0.5 μL of protein (15 mg/mL in 10 mM Hepes-NaOH pH 7.0, 150 mM NaCl) with an equal volume of 0.1 M Hepes-NaOH pH 7.0, 20% PEG-3350, and 0.2 M Potassium Nitrate. Crystals grew to maximum dimensions of 300×300×100 μm in one to two weeks. Crystals were flash frozen in liquid nitrogen using mother liquor containing 25% v/v glycerol as a cryoprotectant, and a 2.3Å data set was collected under cryo-cooled conditions at beamline 11-1 at the Stanford Synchotron Radiation Laboratory. Diffraction data was processed using MOSFLM33 and SCALA34. Data processing statistics can be found in Table 1.
Table 1.
Data collection and refinement statistics for IL-23
| IL-23 native | |
|---|---|
| Data collection | |
| Space group | C 2 |
| Cell dimensions | |
| a, b, c (Å) | 116.1, 60.0, 160.4 |
| α, β, γ (°) | 90, 90.5, 90 |
| Wavelength (Å) | 0.976 |
| Resolution (Å)* | 30-2.30 (2.42-2.30) |
| Rmerge* | 0.073 (0.556) |
| I/σI* | 13.6 (2.5) |
| Completeness (%)* | 99.9 (100.0) |
| Redundancy* | 3.7 (3.7) |
| Refinement | |
| Resolution (Å) | 30-2.30 |
| No. reflections (total/test) | 46921/2502 |
| Rwork/Rfree | 0.228/0.268 |
| No. atoms | 7016 |
| Protein | 6786 |
| Sugar | 28 |
| Water | 202 |
| B-factors | |
| Protein | 46.8 |
| Sugar | 56.6 |
| Water | 40.8 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.527 |
| Ramachandran plot** | |
| Most favorable | 87.1 % |
| Additionally allowed | 12.6 % |
| Generously allowed | 0.0 % |
| Disallowed | 0.2 % |
Highest resolution shell in parentheses
Ramachandran statistics calculated with PROCHECK39
Structure determination and refinement
Initial phase information was obtained with the program PHASER35 using the p40 subunit from the IL-12 structure (PDB ID 1F45) as a molecular replacement search model. After initial refinement of the molecular replacement solution, the alpha helices of p19 were visible and could be built into the electron density map using COOT36. The model was refined by iterative rounds of simulated annealing, positional minimization, and b-factor refinement using CNS37 followed by model adjustment with COOT. The final model went through restrained refinement using REFMAC38 resulting in a final Rwork and Rfree of 22.8% and 26.8%, respectively. Ramachandran analysis by PROCHECK39 indicates 87.1% of residues reside in the most favorable regions, 12.8% in additionally allowed regions, and two residues (K95 in each p40 chain) found in the disallowed region. All structural figures were prepared using PyMol (http://www.pymol.org). Contact and buried surface area were calculated with the CCP4 program suite34 and the Protein Interfaces, Surfaces and Assemblies (PISA) server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html).
The final model contains two copies of IL-23 in the asymmetric unit, with p40 chain A and p19 chain C making up one copy and p40 chain B and p19 chain D the other copy. p40 chain A consists of amino acids 1–156 and 164–306, with two loops modeled mostly as alanines (residues 99–100, 102–104, 258–260, 262–263). p40 chain B consists of amino acids 1–156, 162–224, 229–258, and 263–305 with one loop modeled mostly as alanines (99–100 and 102–104) and alanines at positions 258 and 263. p19 chain C consists of amino acids 1–29, 47–94, 100–125, and 137–169, with alanines modeled at positions 29 and 47–48. p19 chain D consists of amino acids 1–28, 47–91, 97–122, and 137–170, with 3 histidine residues modeled at the C-terminus. A single N-acetylglucosamine moiety is found in each p40 chain attached to Asp 200. The amino acid numbering scheme used corresponds to the mature protein chain with the signal peptide cleaved, in order to facilitate comparison between IL-12 and IL-23. Additionally, all four chains have 1 to 2 amino acids from the gp67 signal sequence modeled at their N-termini, with the mature protein sequence starting at position 1.
Protein Data Bank accession code
The atomic coordinates and structure factors for IL-23 have been deposited in the Protein Data Bank under accession code 3DUH.
Acknowledgments
We thank Sean Juo for assistance with data collection, structure analysis and helpful discussion of the manuscript. PJL is a Damon Runyon Fellow, supported by the Damon Runyon Cancer Research Foundation (DRG-1928-06). This work was funded by an NIH grant (AI51321) to KCG. KCG is also supported by the Keck Foundation and the Howard Hughes Medical Institute.
References
- 1.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 2.Sprang SR, Bazan JF. Cytokine structural taxonomy and mechanisms of receptor engagement . Curr Opin Struct Biol. 1993;3:815–827. [Google Scholar]
- 3.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci U S A. 1990;87:6934–8. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–12. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 5.Wells JA, de Vos AM. Hematopoietic receptor complexes. Annu Rev Biochem. 1996;65:609–34. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- 6.Chow D, He X, Snow AL, Rose-John S, Garcia KC. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–5. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]
- 7.Boulanger MJ, Garcia KC. Shared cytokine signaling receptors: structural insights from the gp130 system. Adv Protein Chem. 2004;68:107–46. doi: 10.1016/S0065-3233(04)68004-1. [DOI] [PubMed] [Google Scholar]
- 8.Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, To W, Wagner J, O’Farrell AM, McClanahan T, Zurawski S, Hannum C, Gorman D, Rennick DM, Kastelein RA, de Waal Malefyt R, Moore KW. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 9.Pflanz S, Hibbert L, Mattson J, Rosales R, Vaisberg E, Bazan JF, Phillips JH, McClanahan TK, de Waal Malefyt R, Kastelein RA. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J Immunol. 2004;172:2225–31. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- 10.Trinchieri G, Pflanz S, Kastelein R. The IL-12 family of heterodimeric cytokines: New players in the regulation of T-cell responses. Immunity. 2003;19:641–644. doi: 10.1016/s1074-7613(03)00296-6. [DOI] [PubMed] [Google Scholar]
- 11.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annual Review of Immunology. 2007;25:221–42. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 12.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M, Gorman D, Wagner J, Zurawski S, Liu Y, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 13.Gubler U, Chua AO, Schoenhaut DS, Dwyer CM, McComas W, Motyka R, Nabavi N, Wolitzky AG, Quinn PM, Familletti PC. Coexpression of two distinct genes is required to generate secreted bioactive cytotoxic lymphocyte maturation factor. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:4143–7. doi: 10.1073/pnas.88.10.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf SF, Temple PA, Kobayashi M, Young D, Dicig M, Lowe L, Dzialo R, Fitz L, Ferenz C, Hewick RM. Cloning of cDNA for natural killer cell stimulatory factor, a heterodimeric cytokine with multiple biologic effects on T and natural killer cells. Journal of Immunology. 1991;146:3074–81. [PubMed] [Google Scholar]
- 15.Yoon C, Johnston SC, Tang J, Stahl M, Tobin JF, Somers WS. Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12. Embo J. 2000;19:3530–41. doi: 10.1093/emboj/19.14.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briso EM, Dienz O, Rincon M. Cutting Edge: Soluble IL-6R Is Produced by IL-6R Ectodomain Shedding in Activated CD4 T Cells. J Immunol. 2008;180:7102–6. doi: 10.4049/jimmunol.180.11.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Presky DH, Yang H, Minetti LJ, Chua AO, Nabavi N, Wu CY, Gately MK, Gubler U. A functional interleukin 12 receptor complex is composed of two beta-type cytokine receptor subunits. Proc Natl Acad Sci U S A. 1996;93:14002–7. doi: 10.1073/pnas.93.24.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huyton T, Zhang JG, Luo CS, Lou MZ, Hilton DJ, Nicola NA, Garrett TP. An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. Proc Natl Acad Sci U S A. 2007;104:12737–42. doi: 10.1073/pnas.0705577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. Journal of Biological Chemistry. 2003;278:1910–4. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 20.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. Journal of Clinical Investigation. 2006;116:1310–6. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. Journal of Experimental Medicine. 2003;198:1951–7. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. Journal of Experimental Medicine. 2005;201:233–40. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–5. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 24.Somers W, Stahl M, Seehra JS. 1.9 A crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling. EMBO Journal. 1997;16:989–97. doi: 10.1093/emboj/16.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–6. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 26.Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol Cell. 2003;12:577–89. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 27.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–72. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanos CD, Randal M, Wells JA. Potent small-molecule binding to a dynamic hot spot on IL-2. Journal of the American Chemical Society. 2003;125:15280–1. doi: 10.1021/ja0382617. [DOI] [PubMed] [Google Scholar]
- 29.Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. The structure of interleukin-2 complexed with its alpha receptor. Science. 2005;308:1477–80. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- 30.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc Natl Acad Sci U S A. 2006;103:15422–7. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–4. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 32.Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–22. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography. 1992:26. [Google Scholar]
- 34.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–33. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of a protein structure. J Appl Crystallogr. 1993;26:283. [Google Scholar]