Abstract
Introduction
Pancreatic ductal adenocarcinoma (i.e., pancreatic cancer) is an almost universally lethal disease. The identification of precursor lesions of pancreatic cancer provides an opportunity for early detection and potential therapeutic intervention before the development of invasive cancer.
Discussion
It is now established that pancreatic cancers do not arise de novo but rather exhibit a sequential histological and genetic progression of precursor lesions culminating in frank, invasive neoplasia. Pancreatic intraepithelial neoplasia (PanIN) is the most common non-invasive precursor lesion of pancreatic cancer. The development of a consensus nomenclature scheme for PanINs has facilitated research into pancreatic cancer precursors and enabled standardization of results across institutions.
Conclusion
PanINs harbor many of the molecular alterations observed in invasive pancreatic cancer, confirming their status as true non-invasive precursor lesions. Recently developed genetically engineered mouse models of pancreatic cancer also demonstrate the stepwise PanIN progression model, underscoring the commonalities in pancreatic neoplasia between mouse and man.
Keywords: Pancreatic cancer, Pancreatic intraepithelial neoplasia, PanIN, Genetics
Introduction
Pancreatic cancer is a disease with a dismal prognosis. In the United States, approximately 33,000 patients are diagnosed with pancreatic cancer annually, and nearly an equal number will die from their malignancy. Worldwide pancreatic cancer causes an estimated 213,000 deaths each year [1]. For all stages combined, the 1-year survival rate is around 20%, and the overall 5-year survival rate is only 4% despite the availability of improved surgical and medical avenues [2, 3].
The high mortality rate for pancreatic cancer is primarily because of the advanced stage at which the neoplasm is diagnosed and because there are no sensitive and specific tools to detect the disease at an earlier stage. More than 80% of the patients with pancreatic cancer have locally advanced or distant metastastic disease at the time of diagnosis, rendering their malignancies surgically inoperable. Currently, surgical resection remains the only curative treatment. Studies from high-volume centers with optimal staging report up to a 15–20% 5-year survival rate in patients undergoing surgical resection [4, 5]. Even if pancreatic cancer is diagnosed early and surgical resection with curative intent is performed, nearly all patients develop local recurrence and/or distant metastases after surgery and eventually succumb to the debilitating effects of metastatic growth [6]. Unfortunately, conventional therapeutic modalities like chemo-radiation have had minimal impact, and the long-term survival of patients with pancreatic cancer has not improved in the last five decades [7, 8].
Improved patient survival has been achieved in a variety of epithelial neoplasms (e.g., colorectal, lung, breast, cervix, and prostate cancer), largely because of identification of cancers at their primary anatomic sites at an early, often pre-invasive stage [9, 10]. At this moment, however, there is no equivalent of a “Pap smear” or a “PSA test” for pancreatic cancer, which can conveniently detect early neoplasia. Nevertheless, it is now recognized that, analogous to other epithelial cancers, pancreatic cancers do not arise de novo but rather undergo a stepwise progression through histologically well-defined non-invasive precursor lesions, culminating in frank, invasive neoplasia. Although putative precursor lesions of pancreatic cancer were first documented over a century ago [11], it was only in the latter half of the last century that multiple lines of evidence began to coalesce, associating invasive pancreatic cancer with these lesions. For example, meticulous autopsy studies confirmed that the prevalence of what are now recognized as precursor lesions increased with age, thus paralleling the frequency of invasive pancreatic cancer. Similarly, most surgically resected pancreata harboring invasive cancer also tend to demonstrate non-invasive intra-ductular lesions in the surrounding parenchyma, suggesting an etiologic association [12–14]. Most importantly, careful molecular analyses over the last 10 years have unequivocally demonstrated that these precursor lesions share many of the underlying genetic alterations observed in the infiltrating cancer, underscoring their precursor status [15–17].
By the late 1990s, over 70 different terminologies were in use to describe these non-invasive ductal lesions, leading to considerable difficulties in comparing inter-institutional studies. Therefore, there was a dire need for the establishment of an international nomenclature scheme for precursor lesions of pancreatic adenocarcinomas. In 1999, the National Cancer Institute hosted a Pancreatic Cancer Think Tank at Park City, Utah, from which meeting emerged a consensus nomenclature scheme for precursor lesions of pancreatic cancer. The “Pancreatic Intraepithelial Neoplasia” (PanIN) scheme for classifying these lesions, first proposed by Klimstra and Longnecker, has since become a gold standard at academic centers worldwide [18, 19].
Histology
The detailed histopathological grading of PanIN lesions and their distinction from other neoplastic and non-neoplastic conditions in the pancreas have been described elsewhere [18, 19]. The reader is also directed to a freely accessible “teaching site” on the World Wide Web for this purpose, located at http://pathology.jhu.edu/pancreas_panin. Briefly, PanINs are microscopic lesions in the smaller (less than 5 mm) pancreatic ducts. PanINs can be papillary or flat, and they are composed of columnar to cuboidal cells with varying amounts of mucin. PanINs are classified into a fourtier classification, including PanIN-1A, PanIN-1B (low-grade PanINs), PanIN-2 (intermediate grade PanINs), PanIN-3 (high-grade PanIN), reflecting a progressive increase in histologic grade culminating in invasive neoplasia. The lowest grade PanIN lesions can be flat (1A) or papillary (1B) but are characterized by absence of nuclear atypia and retained nuclear polarity. PanIN-2 lesions are architecturally slightly more complex than PanIN-1 lesions, and they have more nuclear changes including loss of nuclear polarity, nuclear crowding, variation in nuclear size (pleomorphism), nuclear hyperchromasia, and nuclear pseudostratification. Mitoses are rarely seen. In contrast, PanIN-3 lesions, also referred to as “carcinoma-in-situ”, demonstrate widespread loss of polarity, nuclear atypia, and frequent mitoses. However, as a pre-invasive lesion, PanIN-3 is still contained within the basement membrane [18, 19]. As discussed above, PanINs are often present in the pancreatic parenchyma adjacent to infiltrating adenocarcinomas, and several case reports have documented patients with high-grade PanINs in the remnant pancreas who later developed an infiltrating pancreatic cancer [15]. In summary, just as there is a progression in the colorectum from adenoma, to adenoma with dysplasia, to invasive cancer, so too is there histologic and genetic progressions from PanIN-1, to PanIN-2, to PanIN-3, to invasive ductal adenocarcinoma in the pancreas [20].
It is important to note that PanINs are the most common, albeit not the only, recognized precursor lesions for pancreatic cancer. Two “macroscopic” precursor lesions (so called because they present typically as radiologically detectable cysts in the pancreas [21]) are intraductal papillary mucinous neoplasm and mucinous cystic neoplasms (MCNs). Intraductal papillary mucinous neoplasms (IPMNs) are mucin-producing epithelial neoplasms, which arise within the main pancreatic duct or one of its branches, and that often, although not always, have a papillary architecture [19, 22]. By definition, IPMNs involve the larger pancreatic ducts. Those that involve the main pancreatic ducts are designated “main duct type”, while those that involve the secondary branches of the main pancreatic duct are designated “branch duct type” [18, 19, 23]. Two features characterize MCNs at the light microscopic level. First, the cysts are lined by columnar, mucin-containing epithelium. Second, the underlying stroma has the appearance of ovarian stroma, and in fact, expresses hormonal receptors like estrogen and progesterone [24, 25]. Similar to PanINs, the cystic precursor lesions also demonstrate a multi-step histological and genetic progression to invasive neoplasia but will not be discussed within the scope of the current review.
As discussed above, the strongest evidence establishing the precursor lesional status for PanINs has been derived from comparative molecular analyses with invasive pancreatic cancer. Herein, we discuss some of the most common seminal alterations that are seen in PanIN lesions and likely contribute to the stepwise genetic progression model of pancreatic cancer.
Oncogene mutations in PanIN lesions
Oncogenes can be activated through a variety of mechanisms including point mutations within the gene and amplification of the gene itself. A growing numbers of oncogenes have been identified that are targeted in pancreatic cancer. The most common activating point mutation involves the KRAS oncogene, on chromosome 12p, in over 90% of pancreatic ductal adenocarcinomas [26, 27]. This is the highest fraction of RAS alteration found in any human tumor type. Frequent mutation sites involve codons 12, 13, and 61, but in pancreatic ductal cancers, the majority occur in codon 12 [28]. The KRAS family proteins encode small GTP-binding cytoplasmic proteins and regulate cell-cycle progression via the mitogen-activated protein kinase and AKT cascades [29]. Activating mutations impair the intrinsic GTPase activity of the KRAS gene product, resulting in a protein that is constitutively active in intracellular signal transduction [30]. Mutations of the KRAS gene are one of the earliest genetic abnormalities observed in the progression model of pancreatic cancer, demonstrable in approximately 36%, 44%, and 87% of cancer-associated PanIN-1A, PanIN-1B, and PanIN-2/3 lesions, respectively [31]. The frequency of KRAS gene mutations is somewhat lower (~10%) in PanIN lesions arising in the backdrop of chronic pancreatitis [32]. Of note is given that PanIN lesions and an adenocarcinoma within the same pancreas may harbor different KRAS gene mutations, suggesting that some precursors evolve as independent clones from the one that eventually progress to the invasive cancer [33]. The high frequency of KRAS gene mutations in human PanINs supports its role as an initiating event for pancreatic cancer formation. This fact has been reiterated in several recent animal models (see discussion below) where expression of mutant Kras is a prerequisite for the development of ductal pre-neoplasia and cancer [34, 35]. In addition to its role in pancreatic cancer initiation, constitutive RAS signaling appears to be required for pancreatic cancer maintenance as well [36].
Tumor-suppressor gene mutations in PanIN lesions
Tumor-suppressor genes are genes that promote tumor growth when inactivated. Tumor-suppressor genes are recessive, which means that two copies need to be mutated for loss of function, and they can be inactivated by a variety of mechanisms: first, by an intragenic mutation in one allele (copy of a gene) coupled with loss of the second allele; second, by deletion of both alleles (homozygous deletion); and third, by hypermethylation of the promoter of the gene, thus silencing gene expression. In sporadic cancers, these alterations are both somatic mutations acquired during life, while patients with inherited forms of cancer inherit one mutant allele in the germline, while the second allele is somatically mutated in the cancer cells. Three tumor-suppressor genes, p16INK4A/CDKN2A, TP53, and DPC4/SMAD4/MADH4, are inactivated in a significant proportion of PanINs, mirroring their relative frequencies of loss of function in invasive adenocarcinomas.
The p16INK4A/CDKN2A gene, located on the short arm of chromosome 9 (9p), is one of the most frequently inactivated tumor-suppressor genes in pancreatic cancer [37]. Remarkably, virtually all pancreatic carcinomas have loss of p16INK4A/CDKN2A function in 40% of pancreatic cancer through homozygous deletion, in 40% by an intragenic mutation coupled with loss of the second allele, and in 15% by hypermethylation of the p16INK4A/CDKN2A gene promoter [37, 38]. The p16INK4A/CDKN2A gene encodes the cell-cycle checkpoint protein p16, which binds to the cyclin-dependent kinases Cdk4 and Cdk6, thereby inhibiting binding of cyclin D1, resulting in G1-S cell-cycle arrest [39]. Loss of p16INK4A/CDKN2A results in inappropriate phosphorylation of retinoblastoma (Rb)-1, thereby facilitating progression of the cell cycle through the G1/S transition [40]. Thus, the p16/Rb pathway is inactivated in virtually all pancreatic cancers, leading to an inappropriate progression through the G1 phase of the cell cycle. Loss of p16 expression is also seen in cancer-associated PanINs, with 30% of PanIN-1A and PanIN-1B, 55% of PanIN-2, and 71% of PanIN-3 lesions, demonstrating loss of nuclear p16 protein expression [41]. In contrast, loss of p16 expression is less frequently observed in PanIN lesions arising in the backdrop of chronic pancreatitis (respectively, 0%, 11%, 16%, and 40% for PanIN-1A, PanIN-1B, PanIN-2, and PanIN-3) [42].
The TP53 tumor-suppressor gene on chromosome 17p encodes for the p53 protein [43, 44]. The p53 protein has a number of important functions in the cell, including regulation of the G1/S cell-cycle checkpoint, maintenance of G2/M arrest, and the induction of apoptosis. The TP53 gene is inactivated in 55–75% of pancreatic cancers, almost always by an intragenic mutation in one allele coupled with loss of the second allele [44]. The loss of TP53 means that two critical controls of cell number (cell division and cell death) are deregulated in the majority of pancreatic cancers. By immunohistochemistry, p53 accumulation is usually seen in the advanced PanIN-3 lesions, which is consistent with TP53 gene mutations being a late genetic event in pancreatic cancer progression [45, 46].
Another commonly inactivated tumor-suppressor gene in pancreatic cancer is DPC4, also known as SMAD4/MADH4. DPC4 is a tumor-suppressor gene on chromosome 18q and is one of the most commonly inactivated genes in pancreatic ductal adenocarcinoma, detected in approximately 55% of the cases. Inactivation occurs either through homozygous deletion, in approximately 30%, or loss of one allele coupled with an intragenic mutation in the second allele in approximately 25% [47–49]. The DPC4 gene codes for the protein Smad4, and Smad4 plays a critical role in signaling through the transforming growth factor type β (TGF-β) pathway. The TGF-β pathway is activated when the TGF-β proteins bind to specific cell surface receptors. This triggers an intracellular cascade that results in the nuclear localization of Smad4. Once in the nucleus, Smad4 has growth controlling effects by regulating the expression of specific target genes [29, 50]. Therefore, loss of DPC4 and, thus loss of Smad4 protein, interferes with the intracellular signaling cascades downstream from TGF-β and activin, resulting in decreased growth inhibition via loss of pro-apoptotic signaling or inappropriate G1/S transition [51, 52]. Immunohistochemical labeling for Smad4 protein expression mirrors DPC4/SMAD4/MADH4 gene status with rare exceptions, and like TP53, loss of Smad4 expression is a late genetic event in pancreatic carcinoma progression. Smad4 expression is intact in PanIN-1 and PanIN-2 lesions, but loss of Smad4 expression is observed in 31–41% of PanIN-3 lesions [48].
Genome-maintenance genes mutations in PanIN lesions
Genome-maintenance genes are those that function to identify and repair damage to DNA. They do not directly influence cell growth and proliferation but rather prevent the accumulation of DNA damage and maintain genomic fidelity. When a genome-maintenance gene is inactivated, DNA damage is not repaired efficiently, and DNA mutations accumulate. If these mutations occur in cancer-associated genes, they can contribute to tumorigenesis [53]. Although gross chromosomal abnormalities are frequent in pancreatic ductal adenocarcinomas, genetic instability also occurs through DNA mismatch repair defects [54]. The DNA mismatch repair genes hMLH1 and hMSH2 are examples of genome-maintenance genes targeted in pancreatic cancer [49]. Their encoded proteins work together to repair small insertions, deletions, and other sequence mismatches in newly replicated DNA. Either by mutation or promoter hypermethylation, one of these genes can be inactivated. As a result, DNA repair is compromised, and mutations accumulate in repetitive tracts, producing alterations known as “microsatellite instability” (MSI). Approximately 4% of pancreatic cancers have MSI, and these cancers have a specific microscopic appearance called “medullary histology”. Medullary histology is characterized by pushing borders, syncytial growth pattern, and lymphocytic infiltrate. Furthermore, MSI is associated with poor differentiation and lack of KRAS and TP53 mutations, and germline mutations of this gene are associated with the human non-polyposis colorectal cancer syndrome [55–57].
Another class of genome-maintenance genes includes the Fanconi anemia family of genes. Fanconi anemia is a hereditary cancer susceptibility disorder, with the occurrence of hematologic abnormalities or acute myelogenous leukemia at an early stage, usually leading to death before the age of 20. Patients who survive into adulthood often develop solid tumors [58]. The genes that mutated in pancreatic cancer include the BRCA2, the FANCC gene, and the FANCG gene [58, 59]. These genes are targeted in a small percentage of pancreatic cancers, namely less than 10%. Of these, BRCA2 appears to be particularly significant, because germline BRCA2 mutations, including a founder germline mutation prevalent in the Ashkenazi Jewish population, result in a predisposition to pancreatic cancer in the affected kindred [60]. In ductal pancreatic cancers 7% to 10% harbor an inactivating intragenic inherited mutation of one copy of the BRCA2 gene accompanied by loss of heterozygosity [61, 62]. Among the three cases of pancreatic cancer with germline mutation of BRCA2, loss of remaining wild-type allele was present in a single PanIN-3 lesion but none in 13 low-grade PanINs, confirming that bi-allelic inactivation of the BRCA2 gene, like the TP53 gene, is a late event in pancreatic cancer [63].
Telomere length abnormalities in PanIN lesions
Telomeres are structures present at the ends of linear chromosomes, comprising hexameric DNA repeat sequences (TTAGGG) in association with telomere-binding proteins. These telomeric repeat sequences prevent fusion between ends of chromosomes, and so we can assume that telomeres serve as sort of protective “caps”. It appears that telomeres become abnormally short very early in the development of pancreatic neoplasia [64]. These shortened telomeres can presumably lead to the abnormal fusion of chromosome ends and in this fashion to chromosome instability, promoting further neoplastic progression in these cells [53]. Such a chromosome fusion leads to so-called anaphase bridges during mitosis [65]. During cellular replication, these anaphase bridges frequently break, generating unstable chromosome ends that are subject to abnormal fusion events and subsequent chromosomal rearrangements [66]. Telomere length abnormalities are one of the earliest event in the pancreatic progression model, with more than 90% of even the lowest grade PanIN lesions demonstrating marked shortening of telomeres as compared with normal ductal epithelium [64]. It is believed that this loss of telomere integrity in PanIN lesions is one of the major causes for the loss of tumor-suppressor genes and the gain of oncogenes described earlier.
Epigenetic abnormalities in PanIN lesions
In addition to genetic changes, we now know that epigenetic abnormalities are a common hallmark of cancers. Epigenetic abnormalities in cancer occur predominantly trough methylation of CG dinucleotides (“CpG islands”) in the promoter region of genes, leading to silencing of transcription [67]. In cancers, there is preferential methylation of the gene promoter in the neoplastic cells but not in the corresponding normal cells within the tissue of origin. Numerous studies have showed promoter hypermethylation of several genes, which have a function in tumor suppression and/or critical homeostatic pathways, to be an important mechanism for gene inactivation in many types of cancer [68, 69]. A recent study of a large number of microdissected PanIN lesions has found that as many as 70% of the earliest PanIN-1A lesions demonstrate evidence of aberrant promoter methylation [70]. In addition to previously documented genes –p16 and proenkephalin, this study found evidence of progressive hypermethylation in NPTX2, SARP2, Reprimo, and LHX1 [70–73]. These results suggest that aberrant CpG island hypermethylation begins in early stages of PanINs, and its prevalence progressively increases during neoplastic progression. The aberrantly methylated genes in PanIN lesions can be detected with methylation-specific PCR, making them potentially attractive for early detection. For that reason, the detection of aberrantly methylated genes in the pancreatic juice of patients with pancreatic carcinoma might be a promising diagnostic strategy [74].
Alterations in apomucin expression in PanIN lesions
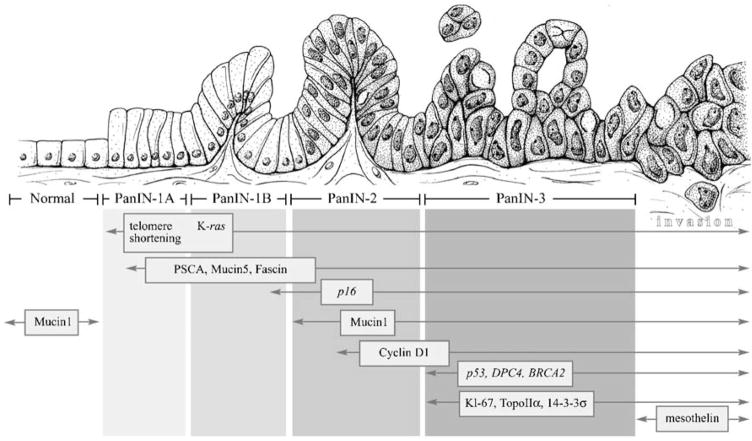
The apomucins MUC1, MUC2, and MUC5 are frequently overexpressed in epithelial cancers, particularly those arising in the gastrointestinal tract and pancreas [75]. MUC1 is expressed in the normal pancreatic ducts and acini and is responsible for the maintenance of lumen formation. MUC1 expression is also often encountered in invasive pancreatic ductal adenocarcinomas [76, 77]. Maitra et al. showed that MUC1 expression was present 43% in PanIN-2 and 85% in PanIN-3 but in only 6% and 5% in PanIN-1A/B. Thus, in the multi-step progression of pancreatic adenocarcinomas, MUC1 expression within normal intra- and interlobular ducts appears to be decreased in the low-grade PanINs (PanIN-1A and 1B). However, MUC1 appears to be subsequently re-expressed in the advanced PanIN lesions, and this expression persists into invasive adenocarcinoma. Of interest, unlike MUC1, the expression of the apomucin MUC2 is uncommon in both normal pancreas and in invasive ductal adenocarcinomas [45, 76]. In contrast, MUC2 expression is commonly seen in IPMNs and their associated invasive colloid carcinomas [78]. These mucins can be used to distinguish PanINs from IPMNs, because PanINs, in contrast to IPMNs with intestinal differentiation, do not express MUC2. Furthermore, MUC5 is similar to MUC1 in that it is also expressed in the majority of invasive ductal adenocarcinomas. In contrast to MUC1, however, MUC5 is not expressed in normal ducts, but its expression is up-regulated even in the earliest PanIN lesions and persists thereafter in the majority of lesions of all histologic grades [45, 79]. These mucins are also potentially detectable by imaging [80], and they may be useful for screening and as therapeutic targets for the treatment of precursor lesions [81, 82] (Fig. 1).
Fig. 1.
A “PanINgram” illustrating some of the molecular alterations that occur during the multi-step progression of pancreatic adenocarcinomas. The molecular abnormalities listed are not comprehensive, and additional alterations are discussed in the text at the appropriate juncture. Adapted from [45]
Aberrant expression of proteins in PanIN lesions
The protein cyclin D1 is a co-factor in the phosphorylation and inactivation of the Rb protein, which plays a central role in cell-cycle regulation [39]. Over-expression of the cyclin D1 protein has been documented in 60–85% of invasive pancreatic adenocarcinoma in imunohistochemistry studies [83, 84]. Cyclin D1 overexpression in pancreatic cancer has been associated with a poor prognosis and decrease in survival [85]. In the development of pancreatic cancer, cyclin D1 overexpression appears to be an intermediate step with nuclear overexpression in 29% of PanIN-2 lesion and 57% of PanIN-3 lesions but no expression in normal pancreatic ducts, PanIN-1A, or PanIN-1B lesions [45].
Cyclooxygenase-2 (COX-2) regulates the metabolism of arachidonic acid into prostaglandins and other pro-inflammatory products. COX-2 has been implicated in tumorigenesis in which metabolites of COX-2 activate a range of signaling pathways, leading to cancer cell proliferation, survival, invasion, and angiogenesis [86]. These processes may be secondary to activation of the MAP kinase signaling pathway and nuclear factor kappa B (NF κB)-mediated signaling [87]. In pancreatic cancer, COX-2 levels are up-regulated, and also in PanIN lesions, COX-2 is expressed. In general, COX-2 follows the trend of expressions, which increases from normal pancreatic ducts to PanIN to adenocarcinoma, with significantly higher expression in PanIN-2/3 compared with PanIN-1A/1B [88]. The appearances of COX-2 in PanIN lesions suggest the possibility of a potential target for chemoprevention using selective COX-2 inhibitors [89].
Certain proteins were first identified as overexpressed in pancreatic cancer based on global expression analyses and subsequent validation in tissue sections. Many of these proteins, not surprisingly, are also overexpressed in precursor lesions. For example, protein prostate stem cell antigen (PSCA) is overexpressed in 30% of PanIN-1 lesions, and respectively 40%, 60%, and 60% in PanIN-2, PanIN-3, and invasive cancer, mandating the classification of PSCA as an early event in the progression model [45]. The patterns of protein expression in PanIN lesions are important, because the proteins expressed in low-grade PanINs may be reasonable chemoprevention targets, while those expressed late (in PanIN-3 lesions) are potential markers for the early detection of pancreatic neoplasia.
Signaling pathways and PanIN lesions
It is known that several embryonic signaling pathways (Notch, Hedgehog, and Wnt pathways) play an important role in multiple tissues during development in utero, and these pathways are for the most part turned off in adult somatic cells, including the exocrine pancreas. Recently, abnormal transcriptional activation of these pathways has been reported in both human and mouse models of pancreatic neoplasia [90–93]. The Notch signaling plays a critical role in maintaining the balance among cell proliferation, differentiation, and apoptosis. Over-expression of Notch pathway receptors (Notch 1–4), ligands (Jagged 1–2), and transcriptional targets (Hes 1) are up-regulated in PanIN lesions and in invasive adenocarcinoma. Notch activation in PanIN lesions appears to be ligand dependent, with Jagged-1 identified by micro-array analysis as one of the significantly overexpressed genes in early PanIN lesions [90, 94].
Aberrant activation of the Hedgehog signaling pathway has been reported in PanINs and pancreatic cancer, as well as in genetically engineered murine models (see discussion below) of PanIN [91, 92]. Global transcriptional profiling of human PanINs revealed up-regulation of extra-pancreatic foregut markers including pepsinogen C, MUC6, Sox-2, KLF4, and TFF1 as a consequence of overexpression of Gli1, a downstream mediator of Hedgehog signaling. Furthermore, activation of the Hedgehog pathway in a human pancreatic ductal epithelial cell line resulted in a similar up-regulation of foregut markers seen in the early PanIN lesions [95]. It is interesting to note that the aberrantly expressed markers of foregut are not present in normal ductal epithelium.
Activation of the Wnt signaling pathways usually occurs via activating mutations of β-catenin or loss-of-function mutations of the APC tumor-suppressor gene; either event leads to stabilization and nuclear translocation of β-catenin and transcription of Wnt target genes [96]. Several studies demonstrated that Wnt pathway mutations are rare in pancreatic ductal adenocarcinoma, although they are frequently observed in non-ductal tumors (e.g., solid pseudopapillary tumors, pancreatoblastomas, and acinar cell carcinomas) [97, 98]. In PanIN lesions, nuclear β-catenin expression is a rare event, and this reiterates the existence of two distinct, genetically divergent pathways of neoplasia in the pancreas: one resulting in the more common, conventional ductal adenocarcinoma and the other resulting in the less common non-ductal neoplasms [45].
Mouse models
Since the development of genetically engineered mouse models with pancreatic cancer, our understanding of the genetics of human PanINs and invasive pancreatic cancer has improved a lot. A major breakthrough was achieved in 2003, when Hingorani and colleagues developed a mouse model with pancreatic neoplasia that expressed an oncogenic KRASG12D allele from its endogenous promoter through Cre-mediated recombinant driven by Pdx1 regulatory elements [35]. Pdx1 is involved in early pancreatic cell fate determination. Pdx1 expression is critical in pancreatic development, and homozygous deletion of Pdx1 causes pancreatic agenesis [99]. The Pdx1-Cre, LSL-KrasG12D mice develop the entire histologic compendium of murine PanIN (mPanIN) lesions observed in the cognate human disease, and in a subset of mice, develop invasive pancreatic carcinomas as well. Although expression of mutant Kras itself is not enough for developing invasive cancer, it is sufficient to initiate PanINs. The fact that these animals developed PanIN lesions before they developed invasive cancer has helped to validate the hypothesis that PanINs can progress to invasive cancer. However, when engineering mice that mis-express oncogenic Kras in the pancreas were combined with bi-allelic INK4a/Arf deletion or an oncogenic Trp53R172H allele, these mice developed aggressive, metastatic pancreatic cancers, with complete penetrance and shorter latency. On the other hand, abrogation of either INK4a/Arf or TP53 signaling alone in the absence of oncogenic Kras does not lead to the development of pancreatic carcinomas or associated precursor lesions, underscoring the crucial importance of Kras signaling in initiating the cascade of events, which result in pancreatic carcinogenesis [34, 100, 101]. Of interest, the mPanIN lesions in the various LSL-KrasG12D mice not only demonstrate the morphological spectrum of human PanIN lesions but they also carry many of the alterations described above, such as overexpression of Notch, Hedgehog, and COX-2 [35, 101]. These mouse models have significantly facilitated defining the role of these genes in the progression of pancreatic neoplasia.
Mouse models can also be used to examine the role of other medical conditions and environmental factors in the development of pancreatic cancer [102, 103]. For example, Guerra et al. reported that when Kras mutations are created in adult mice, these genetically engineered mice do not develop lesions or pancreatic cancer. However, if these mice are challenged with a mild form of pancreatitis, they will develop the full spectrum of PanINs and invasive pancreatic carcinoma. This study provides an excellent example of how genetics and environmental factors interplay in the development of pancreatic cancer, especially when we translate these studies into human observations [103, 104].
At last, mouse models are potentially useful tools to explore pre-clinical diagnostic and therapeutic strategies for pancreatic neoplasia. As already mentioned, these mouse models recapitulate not only the morphology of the cognate human disease but also many of the signaling pathways like Notch, Hedgehog, and COX-2 [35, 101]. Thus, there is a unique opportunity to explore chemoprevention and treatment strategies in a biologically relevant pre-clinical model.
Conclusion
Putative precursor lesions of pancreatic cancer were documented over a century ago. However, it took many decades to define the various histological types of precursor lesions in the pancreas and to credential these lesions as true precursors to invasive adenocarcinoma. Nevertheless, the detailed mechanisms involved in the initiation and progression of these precursor lesions remain to be elucidated. An improved understanding of the pathogenesis of PanIN lesions will enable us to develop better tools for primary and secondary prevention of pancreatic cancer, as well as improve existing tools for early diagnosis.
Acknowledgments
The authors thank the NCI R01 CA113669, The Sol Goldman Pancreatic Cancer Research Center, and the Michael Rolfe Foundation for Pancreatic Cancer Research.
References
- 1.Parker JF, Florell SR, Alexander A, DiSario JA, Shami PJ, Leachman SA. Pancreatic carcinoma surveillance in patients with familial melanoma. Arch Dermatol. 2003;139:1019–1025. doi: 10.1001/archderm.139.8.1019. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society. Cancer facts & figures 2007. American Cancer Society; Atlanta: 2007. pp. 1–52. [Google Scholar]
- 4.Sohn TA, Yeo CJ, Cameron JL, Koniaris L, Kaushal S, Abrams RA, Sauter PK, Coleman J, Hruban RH, Lillemoe KD. Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg. 2000;4:567–579. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- 5.Winter JM, Cameron JL, Campbell KA, Arnold MA, Chang DC, Coleman J, Hodgin MB, Sauter PK, Hruban RH, Riall TS, Schulick RD, Choti MA, Lillemoe KD, Yeo CJ. 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg. 2006;10:1199–1210. doi: 10.1016/j.gassur.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Li DH, Xie KP, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 7.Isozaki H, Okajima K, Morita S, Takeda Y, Ishibashi T, Tanimura M, Hara H, Niki M, Akimoto H, Kobayashi M. Review of pancreatic cancer. Bull Osaka Med Coll. 1990;36:1–11. [PubMed] [Google Scholar]
- 8.Matsuno S, Egawa S, Arai K. Trends in treatment for pancreatic cancer. J Hepatobiliary Pancreat Surg. 2001;8:544–548. doi: 10.1007/s005340100023. [DOI] [PubMed] [Google Scholar]
- 9.Negm RS, Verma M, Srivastava S. The promise of biomarkers in cancer screening and detection. Trends Mol Med. 2002;8:288–293. doi: 10.1016/s1471-4914(02)02353-5. [DOI] [PubMed] [Google Scholar]
- 10.Smith RA, Cokkinides V, Eyre HJ. American Cancer Society guidelines for the early detection of cancer, 2006. CA Cancer J Clin. 2006;56:11–25. doi: 10.3322/canjclin.56.1.11. [DOI] [PubMed] [Google Scholar]
- 11.Hulst SPL. Information on the genesis of the adenocarcinoma and carcinoma of the pancreas. Virchows Archiv fur Pathologische Anatomie und Physiologie und fur Klinische Medizin. 1905;180:288–316. [Google Scholar]
- 12.Cubilla AL, Fitzgerald PJ. Morphological lesions associated with human primary invasive nonendocrine pancreas cancer. Cancer Res. 1976;36:2690–2698. [PubMed] [Google Scholar]
- 13.Andea A, Sarkar F, Adsay VN. Clinicopathological correlates of pancreatic intraepithelial neoplasia: a comparative analysis of 82 cases with and 152 cases without pancreatic ductal adenocarcinoma. Mod Pathol. 2003;16:996–1006. doi: 10.1097/01.MP.0000087422.24733.62. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz AM, Henson DE. Familial and sporadic pancreatic carcinoma, epidemiologic concordance. Am J Surg Pathol. 2007;31:645–646. doi: 10.1097/PAS.0b013e31802d6d42. [DOI] [PubMed] [Google Scholar]
- 15.Brat DJ, Lillemoe KD, Yeo CJ, Warfield PB, Hruban RH. Progression of pancreatic intraductal neoplasias to infiltrating adenocarcinoma of the pancreas. Am J Surg Pathol. 1998;22:163–169. doi: 10.1097/00000478-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Brockie E, Anand A, Albores-Saavedra J. Progression of atypical ductal hyperplasia/carcinoma in situ of the pancreas to invasive adenocarcinoma. Ann Diagn Pathol. 1998;2:286–292. doi: 10.1016/s1092-9134(98)80020-8. [DOI] [PubMed] [Google Scholar]
- 17.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 18.Hruban RH, Adsay NV, Bores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Kloppel G, Longnecker DS, Luttges J, Offerhaus GJ. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Hruban RH, Takaori K, Klimstra DS, Adsay NV, Albores-Saavedra J, Biankin AV, Biankin SA, Compton C, Fukushima N, Furukawa T, Goggins M, Kato Y, Kloppel G, Longnecker DS, Luttges J, Maitra A, Offerhaus GJ, Shimizu M, Yonezawa S. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–987. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 20.Hruban RH, Fukushima N. Pancreatic adenocarcinoma: update on the surgical pathology of carcinomas of ductal origin and PanINs. Mod Pathol. 2007;20(Suppl 1):S61–S70. doi: 10.1038/modpathol.3800685. [DOI] [PubMed] [Google Scholar]
- 21.Winter JM, Cameron JL, Lillemoe KD, Campbell KA, Chang D, Riall TS, Coleman J, Sauter PK, Canto M, Hruban RH, Schulick RD, Choti MA, Yeo CJ. Periampullary and pancreatic incidentaloma: a single institution’s experience with an increasingly common diagnosis. Ann Surg. 2006;243:673–680. doi: 10.1097/01.sla.0000216763.27673.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz-Winnenthal FH, Zgraggen K, Volk C, Schmied BM, Buchler MW. Intraductal papillary mucinous tumors of the pancreas. Curr Gastroenterol Rep. 2003;5:133–140. doi: 10.1007/s11894-003-0082-y. [DOI] [PubMed] [Google Scholar]
- 23.Longnecker DS, Adsay NV, Fernandez-del Castillo C, Hruban RH, Kasugai T, Klimstra DS, Kloppel G, Luttges J, Memoli VA, Tosteson TD, Yanagisawa A, Wilentz R, Zamboni G. Histopathological diagnosis of pancreatic intraepithelial neoplasia and intraductal papillary-mucinous neoplasms: interobserver agreement. Pancreas. 2005;31:344–349. doi: 10.1097/01.mpa.0000186245.35716.18. [DOI] [PubMed] [Google Scholar]
- 24.Fukushima N, Mukai K. ‘Ovarian-type’ stroma of pancreatic mucinous cystic tumor expresses smooth muscle phenotype. Pathol Int. 1997;47:806–808. doi: 10.1111/j.1440-1827.1997.tb04462.x. [DOI] [PubMed] [Google Scholar]
- 25.Hruban RH, Klimstra DS, Pitman MB. Atlas of tumor pathology. Tumors of the pancreas. AFIP, Armed Forces Institute of Pathology; Washington, DC: 2007. [Google Scholar]
- 26.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 27.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, Kensler TW, Bose KK, Cameron JL, Bos JL. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 28.Deramaudt T, Rustgi AK. Mutant KRAS in the initiation of pancreatic cancer. Biochim Biophys Acta. 2005;1756:97–101. doi: 10.1016/j.bbcan.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer. 2003;2:15. doi: 10.1186/1476-4598-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hingorani SR, Tuveson DA. Ras redux: rethinking how and where Ras acts. Curr Opin Genet Dev. 2003;13:6–13. doi: 10.1016/s0959-437x(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 31.Lohr M, Kloppel G, Maisonneuve P, Lowenfels AB, Luttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luttges J, Diederichs A, Menke MA, Vogel I, Kremer B, Kloppel G. Ductal lesions in patients with chronic pancreatitis show K-ras mutations in a frequency similar to that in the normal pancreas and lack nuclear immunoreactivity for p53. Cancer. 2000;88:2495–2504. doi: 10.1002/1097-0142(20000601)88:11<2495::aid-cncr10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 33.Laghi L, Orbetegli O, Bianchi P, Zerbi A, Di CV, Boland CR, Malesci A. Common occurrence of multiple K-RAS mutations in pancreatic cancers with associated precursor lesions and in biliary cancers. Oncogene. 2002;21:4301–4306. doi: 10.1038/sj.onc.1205533. [DOI] [PubMed] [Google Scholar]
- 34.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 36.Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3:413–423. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- 37.Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 38.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, Baylin SB, Kern SE, Herman JG. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 39.Sherr CJ. Cell cycle control and cancer. Harvey Lect. 2000;96:73–92. [PubMed] [Google Scholar]
- 40.Sellers WR, Rodgers JW, Kaelin WG., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci U S A. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilentz RE, Geradts J, Maynard R, Offerhaus GJ, Kang M, Goggins M, Yeo CJ, Kern SE, Hruban RH. Inactivation of the p16 (INK4A) tumor-suppressor gene in pancreatic duct lesions: loss of intranuclear expression. Cancer Res. 1998;58:4740–4744. [PubMed] [Google Scholar]
- 42.Rosty C, Geradts J, Sato N, Wilentz RE, Roberts H, Sohn T, Cameron JL, Yeo CJ, Hruban RH, Goggins M. p16 Inactivation in pancreatic intraepithelial neoplasias (PanINs) arising in patients with chronic pancreatitis. Am J Surg Pathol. 2003;27:1495–1501. doi: 10.1097/00000478-200312000-00001. [DOI] [PubMed] [Google Scholar]
- 43.Kern SE. p53: tumor suppression through control of the cell cycle. Gastroenterology. 1994;106:1708–1711. doi: 10.1016/0016-5085(94)90431-6. [DOI] [PubMed] [Google Scholar]
- 44.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, Kern SE. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–3033. [PubMed] [Google Scholar]
- 45.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, Cameron JL, Yeo CJ, Hruban RH. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 46.Moore PS, Sipos B, Orlandini S, Sorio C, Real FX, Lemoine NR, Gress T, Bassi C, Kloppel G, Kalthoff H, Ungefroren H, Lohr M, Scarpa A. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 47.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 48.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–2006. [PubMed] [Google Scholar]
- 49.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, Sohn TA, Yeo CJ, Kern SE, Hruban RH. Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol. 2000;156:37–43. doi: 10.1016/S0002-9440(10)64703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schutte M, Hruban RH, Hedrick L, Cho KR, Nadasdy GM, Weinstein CL, Bova GS, Isaacs WB, Cairns P, Nawroz H, Sidransky D, Casero RA, Jr, Meltzer PS, Hahn SA, Kern SE. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 51.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 52.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 53.Maitra A, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:211–226. doi: 10.1016/j.bpg.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Han HJ, Yanagisawa A, Kato Y, Park JG, Nakamura Y. Genetic instability in pancreatic cancer and poorly differentiated type of gastric cancer. Cancer Res. 1993;53:5087–5089. [PubMed] [Google Scholar]
- 55.Goggins M, Offerhaus GJ, Hilgers W, Griffin CA, Shekher M, Tang D, Sohn TA, Yeo CJ, Kern SE, Hruban RH. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+ Am J Pathol. 1998;152:1501–1507. [PMC free article] [PubMed] [Google Scholar]
- 56.Wilentz RE, Goggins M, Redston M, Marcus VA, Adsay NV, Sohn TA, Kadkol SS, Yeo CJ, Choti M, Zahurak M, Johnson K, Tascilar M, Offerhaus GJ, Hruban RH, Kern SE. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: a newly described and characterized entity. Am J Pathol. 2000;156:1641–1651. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto H, Itoh F, Nakamura H, Fukushima H, Sasaki S, Perucho M, Imai K. Genetic and clinical features of human pancreatic ductal adenocarcinomas with widespread microsatellite instability. Cancer Res. 2001;61:3139–3144. [PubMed] [Google Scholar]
- 58.van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–2588. [PubMed] [Google Scholar]
- 59.van der Heijden MS, Brody JR, Gallmeier E, Cunningham SC, Dezentje DA, Shen D, Hruban RH, Kern SE. Functional defects in the Fanconi anemia pathway in pancreatic cancer cells. Am J Pathol. 2004;165:651–657. doi: 10.1016/S0002-9440(10)63329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer J. 2001;7:266–273. [PubMed] [Google Scholar]
- 61.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, Kern SE. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 62.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, Campra D, Grutzmann R, Rehder H, Rothmund M, Schmiegel W, Neoptolemos JP, Bartsch DK. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 63.Goggins M, Hruban RH, Kern SE. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am J Pathol. 2000;156:1767–1771. doi: 10.1016/S0002-9440(10)65047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, Cameron JL, Offerhaus GJ, Hicks JL, Wilentz RE, Goggins MG, De Marzo AM, Hruban RH, Maitra A. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal CP, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci U S A. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Hagan RC, Chang S, Maser RS, Mohan R, Artandi SE, Chin L, DePinho RA. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 67.Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16(Spec1):R88–R95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
- 68.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 69.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 70.Sato N, Fukushima N, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2007 doi: 10.1038/MODPATHOL.3800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 72.Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, Yeo CJ, Hruban RH, Goggins M. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato N, Goggins M. The role of epigenetic alterations in pancreatic cancer. J Hepatobiliary Pancreat Surg. 2006;13:286–295. doi: 10.1007/s00534-005-1057-1. [DOI] [PubMed] [Google Scholar]
- 74.Goggins M. Identifying molecular markers for the early detection of pancreatic neoplasia. Semin Oncol. 2007;34:303–310. doi: 10.1053/j.seminoncol.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luttges J, Zamboni G, Longnecker D, Kloppel G. The immunohistochemical mucin expression pattern distinguishes different types of intraductal papillary mucinous neoplasms of the pancreas and determines their relationship to mucinous noncystic carcinoma and ductal adenocarcinoma. Am J Surg Pathol. 2001;25:942–948. doi: 10.1097/00000478-200107000-00014. [DOI] [PubMed] [Google Scholar]
- 76.Terada T, Ohta T, Sasaki M, Nakanuma Y, Kim YS. Expression of MUC apomucins in normal pancreas and pancreatic tumours. J Pathol. 1996;180:160–165. doi: 10.1002/(SICI)1096-9896(199610)180:2<160::AID-PATH625>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 77.Nagata K, Horinouchi M, Saitou M, Higashi M, Nomoto M, Goto M, Yonezawa S. Mucin expression profile in pancreatic cancer and the precursor lesions. J Hepatobiliary Pancreat Surg. 2007;14:243–254. doi: 10.1007/s00534-006-1169-2. [DOI] [PubMed] [Google Scholar]
- 78.Adsay NV, Merati K, Andea A, Sarkar F, Hruban RH, Wilentz RE, Goggins M, Iocobuzio-Donahue C, Longnecker DS, Klimstra DS. The dichotomy in the preinvasive neoplasia to invasive carcinoma sequence in the pancreas: differential expression of MUC1 and MUC2 supports the existence of two separate pathways of carcinogenesis. Mod Pathol. 2002;15:1087–1095. doi: 10.1097/01.MP.0000028647.98725.8B. [DOI] [PubMed] [Google Scholar]
- 79.Kim GE, Bae HI, Park HU, Kuan SF, Crawley SC, Ho JJ, Kim YS. Aberrant expression of MUC5AC and MUC6 gastric mucins and sialyl Tn antigen in intraepithelial neoplasms of the pancreas. Gastroenterology. 2002;123:1052–1060. doi: 10.1053/gast.2002.36018. [DOI] [PubMed] [Google Scholar]
- 80.Foss CA, Fox JJ, Feldmann G, Maitra A, Iacobuzio-Donohue C, Kern SE, Hruban RH, Pomper MG. Radiolabeled anti-claudin 4 and anti-prostate stem cell antigen: initial imaging in experimental models of pancreatic cancer. Mol Imaging. 2007;6:131–139. [PubMed] [Google Scholar]
- 81.Singh AP, Chaturvedi P, Batra SK. Emerging roles of MUC4 in cancer: a novel target for diagnosis and therapy. Cancer Res. 2007;67:433–436. doi: 10.1158/0008-5472.CAN-06-3114. [DOI] [PubMed] [Google Scholar]
- 82.Ohuchida K, Mizumoto K, Yamada D, Fujii K, Ishikawa N, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. Quantitative analysis of MUC1 and MUC5AC mRNA in pancreatic juice for preoperative diagnosis of pancreatic cancer. Int J Cancer. 2006;118:405–411. doi: 10.1002/ijc.21317. [DOI] [PubMed] [Google Scholar]
- 83.Gansauge S, Gansauge F, Ramadani M, Stobbe H, Rau B, Harada N, Beger HG. Overexpression of cyclin D1 in human pancreatic carcinoma is associated with poor prognosis. Cancer Res. 1997;57:1634–1637. [PubMed] [Google Scholar]
- 84.Biankin AV, Kench JG, Morey AL, Lee CS, Biankin SA, Head DR, Hugh TB, Henshall SM, Sutherland RL. Overexpression of p21(WAF1/CIP1) is an early event in the development of pancreatic intraepithelial neoplasia. Cancer Res. 2001;61:8830–8837. [PubMed] [Google Scholar]
- 85.Kornmann M, Ishiwata T, Itakura J, Tangvoranuntakul P, Beger HG, Korc M. Increased cyclin D1 in human pancreatic cancer is associated with decreased postoperative survival. Oncology. 1998;55:363–369. doi: 10.1159/000011879. [DOI] [PubMed] [Google Scholar]
- 86.Sarkar FH, Adsule S, Li Y, Padhye S. Back to the future: COX-2 inhibitors for chemoprevention and cancer therapy. Mini Rev Med Chem. 2007;7:599–608. doi: 10.2174/138955707780859431. [DOI] [PubMed] [Google Scholar]
- 87.Wu KK. Control of cyclooxygenase-2 transcriptional activation by pro-inflammatory mediators. Prostaglandins Leukot Essent Fatty Acids. 2005;72:89–93. doi: 10.1016/j.plefa.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 88.Maitra A, Ashfaq R, Gunn CR, Rahman A, Yeo CJ, Sohn TA, Cameron JL, Hruban RH, Wilentz RE. Cyclooxygenase 2 expression in pancreatic adenocarcinoma and pancreatic intraepithelial neoplasia: an immunohistochemical analysis with automated cellular imaging. Am J Clin Pathol. 2002;118:194–201. doi: 10.1309/TPG4-CK1C-9V8V-8AWC. [DOI] [PubMed] [Google Scholar]
- 89.Sclabas GM, Uwagawa T, Schmidt C, Hess KR, Evans DB, Abbruzzese JL, Chiao PJ. Nuclear factor kappa B activation is a potential target for preventing pancreatic carcinoma by aspirin. Cancer. 2005;103:2485–2490. doi: 10.1002/cncr.21075. [DOI] [PubMed] [Google Scholar]
- 90.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, Hruban RH, Ball DW, Schmid RM, Leach SD. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 91.Berman DM, Karhadkar SS, Maitra A, Montes De OR, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 92.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del CC, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, Malhotra V, Sood N, Midda V, Monga DK, Kokkinakis DM, Monga SP. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kimura K, Satoh K, Kanno A, Hamada S, Hirota M, Endoh M, Masamune A, Shimosegawa T. Activation of Notch signaling in tumorigenesis of experimental pancreatic cancer induced by dimethylbenzanthracene in mice. Cancer Sci. 2007;98:155–162. doi: 10.1111/j.1349-7006.2006.00369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, Hruban RH, Goggins M, Leach SD. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619–1626. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 96.Taipale J, Beachy PA. The Hedgehog and Wnt signaling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 97.Abraham SC, Klimstra DS, Wilentz RE, Yeo CJ, Conlon K, Brennan M, Cameron JL, Wu TT, Hruban RH. Solid-pseudopapillary tumors of the pancreas are genetically distinct from pancreatic ductal adenocarcinomas and almost always harbor beta-catenin mutations. Am J Pathol. 2002;160:1361–1369. doi: 10.1016/s0002-9440(10)62563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abraham SC, Wu TT, Hruban RH, Lee JH, Yeo CJ, Conlon K, Brennan M, Cameron JL, Klimstra DS. Genetic and immunohistochemical analysis of pancreatic acinar cell carcinoma: frequent allelic loss on chromosome 11p and alterations in the APC/beta-catenin pathway. Am J Pathol. 2002;160:953–962. doi: 10.1016/s0002-9440(10)64917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 100.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, DePinho RA. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 102.Archer H, Jura N, Keller J, Jacobson M, Bar-Sagi D. A mouse model of hereditary pancreatitis generated by transgenic expression of R122H trypsinogen. Gastroenterology. 2006;131:1844–1855. doi: 10.1053/j.gastro.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 103.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 104.Lowenfels AB, Maisonneuve P, Dimagno EP, Elitsur Y, Gates LK, Jr, Perrault J, Whitcomb DC. Hereditary pancreatitis and the risk of pancreatic cancer. International hereditary pancreatitis study group. J Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]