Abstract
The past two decades have witnessed an explosion in our understanding of pancreatic cancer, and it is now clear that pancreatic cancer is a disease of inherited (germ-line) and somatic gene mutations. The genes mutated in pancreatic cancer include KRAS2, p16/CDKN2A, TP53, and SMAD4/DPC4, and these are accompanied by a substantial compendium of genomic and transcriptomic alterations that facilitate cell cycle deregulation, cell survival, invasion, and metastases. Pancreatic cancers do not arise de novo, and three distinct precursor lesions have been identified. Experimental models of pancreatic cancer have been developed in genetically engineered mice, which recapitulate the multistep progression of the cognate human disease. Although the putative cell of origin for pancreatic cancer remains elusive, minor populations of cells with stem-like properties have been identified that appear responsible for tumor initiation, metastases, and resistance of pancreatic cancer to conventional therapies.
Keywords: pancreas, carcinoma, adenocarcinoma, precursor, genetics, stem cell, mouse model, targeted therapy
EPIDEMIOLOGY, RISK FACTORS, AND DEMOGRAPHICS
Pancreatic cancer is one of the deadliest of all of the solid malignancies. The five-year survival rate is only 4% (1). The American Cancer Society estimates that 37,170 Americans will be diagnosed with pancreatic cancer in the year 2007 and 33,370 will die from it, making pancreatic cancer the fourth leading cause of cancer death (1). In the United States, the age-adjusted incidence of pancreatic cancer is higher in blacks (14.9 cases per 100,000) than in whites (11.1 cases per 100,000), and it is higher in men (12.8 cases per 100,000) than in women (10.0 cases per 100,000) (http://seer.cancer.gov/).
A number of risk factors have been identified (2). Pancreatic cancer is predominantly a disease of the elderly. Pancreatic cancer is rare before the age of 40, and the median age at diagnosis is 73 years (http://seer.cancer.gov/). Cigarette smoking is by far the leading preventable cause of pancreatic cancer (3, 4). Cigarette smoking doubles the risk of pancreatic cancer (Relative Risk = 2), and it is believed that as many as one in four cases of pancreatic cancer may be attributable to smoking (2). Other established risk factors include diets high in meats and fat, low serum folate levels, obesity, long-standing diabetes mellitus, and chronic pancreatitis (2, 5–7). A well-publicized study linking coffee consumption with pancreatic cancer had methodological flaws, and more recent studies did not demonstrate any significant associations (2).
One of the more interesting risk factors for pancreatic cancer is a family history of this disease. Individuals with a strong family history of pancreatic cancer have a significantly increased risk of developing the disease themselves. For example, individuals with a first-degree relative with pancreatic cancer have a 2.3-fold increased risk of developing the malignancy (8). Klein et al. (9) have shown that this risk increases with the number of first-degree relatives one has with pancreatic cancer, and that the simplest model to explain this aggregation in families is an autosomal dominant inheritance of a rare allele. As summarized in Table 1, a number of germ-line genetic alterations have been associated with an increased risk of pancreatic cancer (10–19). These germ-line genetic alterations provide insight into the pathogenesis of pancreatic cancer.
Table 1.
Genes associated with an increased risk of pancreatic cancer
| Individual | Gene | Relative risk | Risk by age 70 | Cancer morphology | Other cancers | Reference(s) |
|---|---|---|---|---|---|---|
| No history | None | 1 | 0.5% | NS | None | |
| Breast cancer | BRCA2 | 3.5–10X | 5% | NS | Breast, ovary, and prostate | (10, 11) |
| BRCA1 | 2X | 1% | Breast cancer with basaloid features | Breast, ovary, and prostate | (12) | |
| FAMMM | P16 (CDKN2A) | 20–34X | 10%–17% | NS | Melanoma | (14, 185) |
| Familial pancreatic cancer (3 FDR) | Unknown | 32X | 16% | NS | Unknown | (9) |
| Familial pancreatitis | PRSS1 | 50–80X | 25%–40% | Pancreatic cancers in the background of severe diffuse chronic pancreatitis | None | (17) |
| Peutz-Jeghers | STK11/LKB1 | 132X | 30%–60% | NS | Gastroesophageal, small bowel, colorectal, and breast | (13, 16) |
| HNPCC | hMLH1, hMSH2, others | Unknown | < 5% | Medullary and colloid phenotypes | Colorectal, endometrial, stomach, ovarian, ureter and renal pelvis, biliary tract, and brain | (18, 186) |
| Young-age-onset pancreatic cancer | FANC-C and FANC-G | Unknown | Unknown | NS | Unknown | (187, 188) |
3 FDR, 3 or more first-degree relatives with pancreatic cancer; FAMMM, familial atypical multiple mole melanoma syndrome; HNPCC, hereditary nonpolyposis colorectal cancer syndrome; NS, nonspecific.
Several features of the genetic syndromes associated with the familial clustering of pancreatic cancer deserve special note (20, 21). First, the penetrance of cancer in the gene carriers is incomplete. For example, some individuals with germ-line BRCA2 gene mutations do not have a strong family history of cancer (11). Goggins et al. (11) studied a series of 41 apparently sporadic pancreatic cancer patients and four (9.8%) of the 41 patients harbored a germ-line BRCA2 gene mutation. Only one of the four had a family history of breast cancer and none had a family history of pancreatic cancer. Clearly, the absence of a strong family history of cancer cannot be used to rule out a germ-line mutation.
Second, individuals can be identified, tested for one of these mutations, and counseled on their pancreatic cancer risk. For example, the Peutz-Jeghers syndrome is characterized by melanocytic macules on the lips and buccal mucosa, and hamartomatous polyps of the gastrointestinal tract (13). Individuals with these stigmata of the Peutz-Jeghers syndrome can be clinically tested for germ-line mutations in the STK11/LKB1 gene. Those found to carry a mutation may then benefit from screening for asymptomatic pancreatic neoplasia (22).
Third, most of these germ-line mutations, with the exception of those in the PRSS1 gene, are also associated with an increased risk of extrapancreatic malignancies. For example, the risk of melanoma in p16/CDKN2A gene mutation carriers is approximately 28% (95% confidence interval = 18% to 40%) by age 80 (23). These extrapancreatic malignancies can also be screened for and, in the case of melanoma, preventative measures such as avoiding sun exposure can also be initiated.
Fourth, pancreatic cancers with microsatellite instability, the hallmark of DNA mismatch repair defects such as those produced by mutations in the hMLH1 and hMSH2 genes, often have a distinctive medullary histology (18, 24). Medullary carcinomas are poorly differentiated; they have expanding borders and a prominent syncytial growth pattern (18, 24). When a pancreatic cancer with this morphology is identified, the morphologic appearance of the cancer can be used to suggest that the patient may benefit from genetic counseling and possibly genetic testing.
Fifth, these germ-line genetic abnormalities may have therapeutic implications (20, 21). For example, the protein product of the BRCA2 gene interacts with the protein products of the Fanconi anemia complementation genes (the FANC genes) to promote homologous recombination (25). As discussed subsequently in further detail, van der Heijden et al. (26, 27) have shown that pancreatic cancers with BRCA2 or other proximal FANC gene mutations are particularly sensitive to Mitomycin C and radiation—therapies known to cause DNA cross-linking and double strand breaks, respectively.
Finally, because of founder mutations, some of these genetic syndromes are more common in certain ethnic groups. For example, 1% of the Ashkenazi Jewish population carries the 6174delT BRCA2 gene mutation (10). Knowledge of these associations can help guide genetic testing.
PATHOLOGY
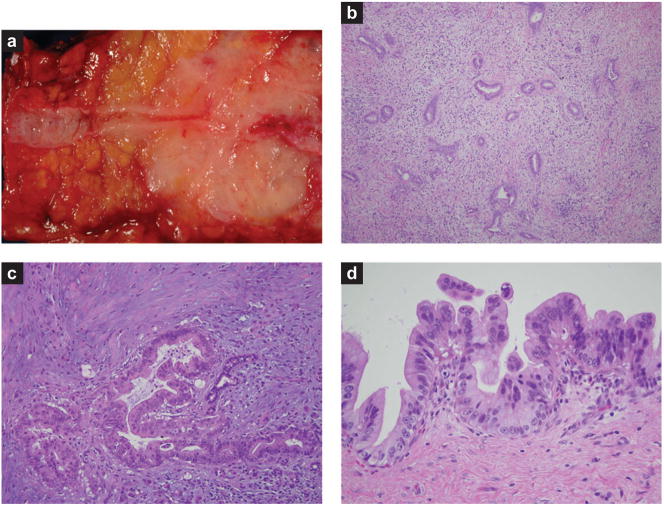
Most cancers of the pancreas are infiltrating ductal adenocarcinomas, and we use the terms pancreatic cancer and infiltrating ductal adenocarcinoma of the pancreas synonymously (21). Pancreatic cancer grossly produces a firm, highly sclerotic mass (Figure 1a). The edges of these cancers are poorly defined, with long tongues of carcinoma extending well beyond the main tumor (21). At the light-microscopic level, pancreatic cancer is composed of an infiltrating gland-forming neoplastic epithelium with an intense desmoplastic reaction (Figure 1b) (21). This desmoplastic reaction is usually so intense that only a minority of the cells in the mass formed by pancreatic cancer are neoplastic cells (Figure 1c). As one might expect from their gross appearance, pancreatic cancers are extremely infiltrative neoplasms. Vascular and perineural invasion are present in the majority of surgically resected cancers, and metastases to regional lymph nodes, the liver, and distant sites are all too common (21).
Figure 1.
Pathology of pancreatic adenocarcinoma and its precursor lesions. (a) Gross photograph of an infiltrating adenocarcinoma. Note the dramatic narrowing of the pancreatic duct associated with the poorly defined white neoplasm. (b) Low-power photomicrograph of an infiltrating adenocarcinoma. Note the haphazard arrangement of the glands and the associated non-neoplastic desmoplastic stroma. (c) High-power photomicrograph of an infiltrating adenocarcinoma. Note the desmoplastic stroma and the marked pleomorphism in the cancer relative to the trapped non-neoplastic duct. (d) High-grade pancreatic intraepithelial neoplasia.
The majority of pancreatic cancers express immunohistochemically detectable Cytokeratin (Cytokeratins 7, 8, 13, 18, and 19), carcinoembryonic antigen, carbohydrate antigen 19–9 (CA19–9), B72.3 (TAG-72), CA 125, and DUPAN 2 (21). Most pancreatic cancers also express a number of mucins, including MUC1, MUC3, MUC4, and MUC5AC (21, 28). More recently described markers include claudin 4 and 18, sea urchin fascin homolog, several of the S-100 proteins (including S-100A4, S-100A6, and S-100P), and the glycosylphosphatidyl-inositol-anchored proteins mesothelin and prostate stem cell antigen (29–32).
A number of histologic variants of pancreatic cancer have been described. These include adenosquamous carcinoma, colloid carcinoma, hepatoid carcinoma, medullary carcinoma, signet-ring cell carcinoma, undifferentiated carcinoma, and undifferentiated carcinoma with osteoclast-like giant cells (21). These are important to recognize because some have distinct pathogenesis (medullary carcinoma), some have a significantly better prognosis (colloid, medullary), and some have a worse prognosis (adenosquamous carcinoma, undifferentiated carcinoma) than infiltrating ductal adenocarcinoma.
PRECURSOR LESIONS
Histologically distinct precursor lesions have been described in the pancreas (33, 34). These precursor lesions include pancreatic intraepithelial neoplasia (PanIN), the intraductal papillary mucinous neoplasm (IPMN), and the mucinous cystic neoplasm (MCN) (35). Lives can be saved if these noninvasive lesions can be detected and treated before they progress to an invasive carcinoma.
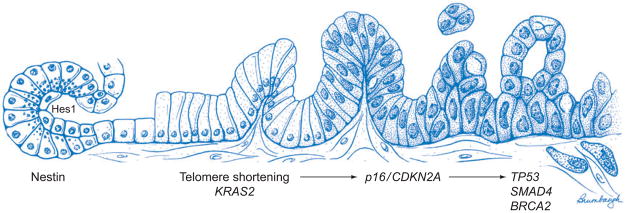
PanINs are microscopic lesions in the smaller (<5 mm) pancreatic ducts (33, 34). PanINs can be papillary or flat, and they are composed of columnar to cuboidal cells with varying amounts of mucin. PanINs are subclassified into PanIN-1, PanIN-2, and PanIN-3 lesions, depending upon the degree of cytologic and architectural atypia (Figure 1 d) (33, 34). PanINs are often present in the pancreatic parenchyma adjacent to infiltrating adenocarcinomas, and several case reports have documented patients with PanINs who later developed an infiltrating pancreatic cancer (36). As illustrated in Figure 2, molecular analyses of PanINs have shown that PanINs harbor many of the same genetic alterations as are found in infiltrating pancreatic cancer (20, 35). Activating point mutations in codon 12 of the KRAS2 gene typically occur in early low-grade PanIN lesions (PanIN-1), whereas inactivating mutations in the p16/CDKN2A gene occur in intermediate lesions (PanIN-2), and inactivating mutations in SMAD4, TP53, and BRCA2 occur in late lesions (PanIN-3) (35). Telomere shortening is also an early event, occurring in PanIN-1 lesions, and telomere shortening may contribute to the accumulation of chromosomal abnormalities in PanINs (37). As discussed below, understanding of the genetics of PanINs was crucial for the development of genetically engineered mouse models of pancreatic cancer, and it forms a basis for early detection strategies.
Figure 2.
Genetic progression model of pancreatic adenocarcinoma. The progression from histologically normal epithelium to low-grade pancreatic intraepithelial neoplasia (PanIN), to high-grade PanIN, to invasive carcinoma (left to right) is associated with the accumulation of specific genetic alterations. On the basis of their temporal appearance in this progression model, the molecular abnormalities can be classified as early (KRAS2 mutation, telomere shortening), intermediate (p16/CDKN2A loss), or late (mutations of DPC4/SMAD4, TP53, BRCA2). These signature genomic alterations are accompanied by a multitude of expression abnormalities that are not illustrated (see text for details). Note that this progression model is specific for PanINs, and other recognized precursor lesions of adenocarcinoma (intraductal papillary mucinous neoplasm and mucinous cystic neoplasm) likely harbor a distinct compendium of genetic alterations in their path to invasive cancer. The state of the field does not definitively establish the cellular origin for PanIN and pancreatic cancer. On the basis of genetically engineered models, two candidates have emerged as the most likely cell of origin for ductal neoplasia in the pancreas: facultative progenitors within the acinar compartment and centroacinar cells. Both competing hypotheses are illustrated to the far left of the figure to underscore the current uncertainty in the literature. Rigorous lineage-tracing experiments have confirmed the ability of differentiated acinar cells to undergo acinar-to-ductal transdifferentiation, via generation of nestin-positive intermediates. This process, known as acinar-to-ductal metaplasia, is a frequent accompaniment of pancreatic neoplasia in experimental models, and is also observed in the setting of human pancreatic cancers. The second putative source of neoplastic ductal cells is centroacinar cells, which are found at the junction of acini and ducts. These are the only cell types in the mature exocrine pancreas with retained Notch activation, as determined via persistent nuclear Hes1 expression. Loss of regulatory signals (e.g., inactivation of PTEN) that lead to uncontrolled centroacinar cell expansion culminates in pancreatic adenocarcinoma. In addition to these putative candidates, this model does not exclude the possibility of a rare, as yet undefined, pancreatic stem cell as the source of ductal neoplasia. See Cellular Origins of Pancreatic Cancer for further details.
IPMNs are mucin-producing epithelial neoplasms that arise within the main pancreatic duct or one of its branches and that often, although not always, have a papillary architecture (21, 34). IPMNs with an intestinal phenotype express the mucin MUC2, whereas most PanINs express MUC1 (21, 34). The SMAD4 gene is inactivated in only a small minority of IPMNs (38), whereas loss of the STK11/LKB1 gene product is significantly more common (39). Because they are larger lesions, screening for IPMNs may be possible using currently available imaging technology (22, 40).
The third histologically distinct precursor lesion in the pancreas is the MCN (21). The vast majority of MCNs arise in women. In contrast to IPMNs, MCNs do not communicate with the larger pancreatic ducts, and, in contrast to PanINs and IPMNs, MCNs have a distinctive ovarian-type stroma (21). The genetic alterations in MCN have not been extensively studied, but it appears that they can have many of the same abnormalities found in infiltrating ductal adenocarcinomas of the pancreas, but at a lower frequency (35).
It is clear that some noninvasive IPMNs and some MCNs of the pancreas progress to invasive adenocarcinoma over time (35). Patients with noninvasive IPMNs and MCNs are on average 3–5 years younger than patients with the corresponding invasive lesion (21). This latter observation suggests a significant window of opportunity during which these precursor lesions can be treated and the patient cured before an invasive cancer develops.
MOLECULAR GENETICS OF PANCREATIC CANCER
One of the most important paradigms to emerge from more than two decades of research in pancreatic cancer has been the understanding that pancreatic cancer is a disease of inherited and somatic mutations. A compendium of signature mutations defines pancreatic adenocarcinoma and differentiates this malignancy from other neoplasms of the pancreas.
KRAS2 and Other Oncogenes
Activating mutations of the KRAS2 oncogene are possibly the single most common genetic abnormality in pancreatic cancer, present in ~90%–95% of cases (41). KRAS2 encodes a member of the RAS family of guanosine triphosphate (GTP)-binding proteins that mediate a range of cellular functions, including proliferation, cell survival, cytoskeletal remodeling, and motility, among others. A variety of stimuli, such as binding of growth factor ligands to the cognate growth factor receptor, results in signal transduction via intermediary proteins that culminate in the activation of the Kras protein. The active protein is bound to GTP, and inactivation occurs through guanosine-triphosphatase-activating proteins, which promote GTP hydrolysis to the diphosphate GDP and attenuate Kras signaling. Activating mutations impair the intrinsic GTPase activity of the KRAS2 gene product, resulting in a protein that is constitutively active in intracellular signal transduction (42). The spectrum of KRAS2 gene mutations in pancreatic cancer is limited principally to alterations of codon 12, with occasional mutations of codons 13 or 61 (41). Mutations of the KRAS2 gene are one of the earliest genetic abnormalities observed in the progression model of pancreatic cancer, demonstrable in approximately 36%, 44%, and 87% of cancer-associated PanIN-1A, PanIN-1B, and PanIN-2/3 lesions (35). PanIN lesions and an adenocarcinoma within the same pancreas may harbor different KRAS2 gene mutations, suggesting that some precursors evolve as independent clones from the one that eventually progresses to the invasive cancer (43). The high frequency of KRAS2 gene mutations in human PanINs supports its role as an initiating event for pancreatic cancer formation, and this has been reiterated in several recent animal models, where expression of mutant Kras is a prerequisite for the development of ductal preneoplasia and cancer (see below) (44–48). In addition to its role in pancreatic cancer initiation, constitutive RAS signaling appears to be required for pancreatic cancer maintenance as well (49).
Activated Kras engages a number of downstream effector pathways, including RAF–mitogen-activated protein kinase (RAF-MAPK), phosphoinositide-3-kinase (PI3K), and RalGDS pathways. Several lines of evidence point toward a critical role for RAF-MAPK signaling in pancreatic carcinogenesis. One-third of pancreatic cancers with wild-type KRAS2 harbor mutations of the BRAF oncogene, thereby resulting in the activation of RAF-MAPK signaling even in the absence of KRAS2 mutations (50). Second abrogation of RAK-MAPK signaling with small molecule inhibitors, or with antisense inhibition of an essential transducer of RAS signaling to RAF, the kinase suppressor of ras 1 (KSR1), results in growth inhibition of pancreatic cancer xenografts (51). The PI3K-AKT pathway is an essential cell survival pathway, with diverse roles in tumorigenesis across multiple solid malignancies. The PI3K-AKT pathway is constitutively activated in most pancreatic cancers, and targeting this pathway with small molecule inhibitors or genetic strategies results in growth inhibition in vitro and in vivo (52). Although RAS certainly contributes to PI3K-AKT signaling in pancreatic cancer, independent genomic events can also activate this pathway, including amplification of the AKT2 gene on chromosome 19q (~10%–15% of pancreatic cancers) (53) and activating mutations of the PIK3CA gene in a subset of IPMNs (54). Recently, Ral A, a downstream moiety in the RalGDS effector pathway, has emerged as another key player in RAS-mediated transformation, as knockdown of Ral A in pancreatic cancer cells essentially abolishes their tumorigenic phenotype (55). Whether these signaling moieties can be utilized as therapeutic targets remains to be determined.
Additional candidates that contribute to oncogenesis have been identified (56–58). For example, the gene encoding the oncogenic transcription factor C-Myc on chromosome 8q is amplified in 20%–30% of pancreatic cancers, and CMYC transcripts are overexpressed in nearly all cases (56). Other oncogenes that are targets of amplification in pancreatic cancer include MYB (chromosome 6q), AIB1/NCOA3 (chromosome 20q), and EGFR (chromosome 7p) (56–58). A number of recurrent chromosomal amplifications have also been identified in the high-resolution copy number experiments (e.g., localized amplicons on chromosomes 18q and 7q), wherein the putative targets are under investigation or simply unknown (56–58). Elucidation of these oncogenic events will undoubtedly provide additional insights into the biology and therapy of pancreatic cancer.
Tumor Suppressor Genes (p16/CDKN2A, TP53, and DPC4/SMAD4)
The gene p16/CDKN2A, also known as INK4A, is the most commonly inactivated tumor suppressor gene in pancreatic cancers (59). The encoded protein belongs to the cyclin-dependent kinase (CDK) inhibitor family and inhibits cell cycle progression through the G1-S checkpoint mediated by CDKs such as CDK4 and -6. Loss of p16/CDKN2A function is seen in approximately 90% of pancreatic cancers. This loss occurs by several different mechanisms, including homozygous deletion (40%), intragenic mutation with loss of the second allele (40%), and epigenetic silencing by promoter methylation (10%–15%) (20).
Inactivation of the TP53 gene on chromosome 17p is present in approximately 50%–75% of pancreatic cancers, and inactivation of the TP53 gene almost always occurs via intra-genic mutation combined with loss of the second allele (60). The p53 protein has a number of important functions in the cell, including regulation of the G1-S cell cycle checkpoint, maintenance of G2-M arrest, and the induction of apoptosis. Loss of p53 function allows cells to survive and divide despite the presence of damaged DNA, leading to the accumulation of additional genetic abnormalities (61).
The deleted in pancreatic carcinoma 4 gene, or DPC4 (also known as SMAD4), on chromosome 18q21 is inactivated in approximately 55% of pancreatic cancers, either by homozygous deletion (30%) or by intragenic mutations and loss of the second allele (25%) (62). The Smad4 protein plays a critical role in signaling through the transforming growth factor-β (TGF-β) pathway. The TGF-β pathway is activated when the TGF-β protein binds to specific cell surface receptors. This triggers an intracellular cascade that results in phosphorylation and nuclear localization of the Smad transcription factors Smad 2/3, complexed with Smad4. The TGF-β pathway has profound growth-inhibitory effects by regulating the expression of specific target genes; therefore, loss of Smad4 in pancreatic cancer cells, which abrogates Smad-dependent TGF-β signaling, provides a selective growth advantage (63). Immunohistochemical labeling for Smad4 protein expression mirrors DPC4/SMAD4 gene status in pancreatic cancers with rare exceptions (64). Loss of Smad4 labeling is seen only at the stage of PanIN-3 lesions, whereas virtually all noninvasive IPMNs and MCNs retain expression (38, 65, 66). Inactivation of DPC4/SMAD4 is uncommon in nonductal neoplasms of the pancreas (21) and is rare in most extrapancreatic malignancies (67). Therefore, immunolabeling for loss of Smad4 is a convenient ancillary diagnostic marker in clinical specimens, including suspected metastases from an occult pancreatic primary.
Several tumor suppressor genes are inactivated in smaller proportions (5%–10%) of pancreatic cancers, including the serine-threonine kinase LKB1/STK11 (chromosome 19p) (16) and the TGF-β/activin signaling pathway receptors such as TGFBR1 (chromosome 9q), TGFBR2 (chromosome 3p) (68), and ACVR1B (chromosome 12q) (69). The gene MKK4 on chromosome 17p, which encodes for a stress-activated protein kinase, is preferentially inactivated in subsets of pancreatic cancer metastases, suggesting that the protein product may function as a metastases suppressor (70). In addition to classic oncogenes and tumor suppressor genes, several so-called caretaker or genome maintenance genes are inactivated in pancreatic cancer (61). These genes do not directly influence cell growth and proliferation, but rather prevent the accumulation of DNA damage and maintain genomic fidelity. We have already discussed the role of such caretaker genes—specifically the DNA mismatch repair genes hMLH1 and hMSH2, BRCA2, and related Fanconi anemia (FANC) genes—in the pathogenesis of familial pancreatic cancer (see above). However, somatic inactivation of these genes is also observed in pancreatic cancer outside the familial context.
Telomere Length Abnormalities
Telomeres are hexameric TTAGGG repeats at the ends of chromosome arms that confer stability to chromosomes during cell division and prevent the ends from becoming sticky (71). Loss of telomeres renders chromosome ends highly recombinogenic, with the formation of end-to-end fusions that result in chromosome breakage during anaphase (72). In fact, lagging chromosomal material in dividing cancer cells is often visible as anaphase bridges under the microscope. The continuous cycles of breakage-fusion-bridge that form between chromosomes result in regions of amplifications and deletions within the daughter cell genome. A recent study using an in situ fluorescent probe to evaluate telomere length demonstrated that telomere attrition is one of the earliest demonstrable genetic aberrations in pancreatic cancer, with >90% of even the low-grade PanIN lesions demonstrating marked shortening of telomeres, as compared with normal ductal epithelium (37).
The likely secondary consequence of telomere erosion in PanIN lesions is the creation of an environ that is permissive to acquisition of chromosomal rearrangements. Indirect evidence from concurrent allelotype and mutation analysis of microdissected PanIN lesions implies this to be true, as allelic deletions often precede point mutations at selected tumor suppressor gene loci (73). In most instances, cells harboring this degree of genomic instability are eliminated through activation of p53. However, chromosomal rearrangements likely persist in cells with TP53 gene mutations (as observed in higher-grade PanIN lesions), and these cells will then quickly accrue further genomic alterations (71). Paradoxically, telomerase reactivation, observed in the vast majority of invasive pancreatic cancers (74), is postulated to attenuate ongoing genomic instability and stabilize the neoplastic cells against potentially catastrophic rearrangements.
Epigenetic Abnormalities
It is now evident that epigenetic abnormalities are extremely common in cancers, and these abnormalities provide an alternative mechanism of transcriptional silencing (75). Epigenetic abnormalities in cancer predominantly encompass methylation of CG dinucleotides (CpG islands) in the 5′ regulatory region of tumor suppressor genes, which abrogates RNA polymerase from binding and initiating transcription. In cancers, there is preferential methylation of the gene promoter in the neoplastic cells, but not in the corresponding normal cells within the tissue of origin.
Epigenetic silencing is frequently observed in pancreatic cancers and tends to involve genes that function in tumor suppression and/or critical homeostatic pathways (e.g., p16/CDKN2A, E-cadherin, retinoic acid beta, SOCS-1, TSLC1, and reprimo, among others) (76). Epigenetic silencing is unlikely to be a bystander effect, but has tangible functional consequences on the biology of neoplastic pancreatic cells. For example, the gene reelin (RELN), whose product is involved in regulating neuronal migration, is frequently methylated in pancreatic cancer. Downregulation of RELN transcripts in pancreatic cancer cells confers upon these cells properties of increased motility, invasiveness, and clonogenicity, whereas re-expression of RELN in cell lines with epigenetic silencing of this gene inhibits their migration (77). Similarly, aberrant methylation at certain gene promoters (e.g., reprimo, whose gene product is associated with p53-induced G2-M arrest) is associated with increased genetic instability and adverse prognosis in established pancreatic cancer (78). A subset of genes that undergoes epigenetic modification in pancreatic cancer is also silenced by promoter methylation in PanIN, IPMN, and MCN, the three precursor lesions of adenocarcinoma (76, 79–81). This finding suggests that epigenetic abnormalities are an early event in the multistep progression of pancreatic neoplasia.
One important confounding factor gleaned from these studies has been the age-related methylation at multiple gene promoters observed in normal duodenal mucosa (82). Pure pancreatic juice free of contamination by small intestinal secretions is therefore needed to specifically identify differentially methylated DNA shed into the pancreatic duct by pancreatic cancer cells. As further discussed in the section on translational studies, quantitative analysis of promoter methylation in a limited panel of genes has emerged as a promising new biomarker strategy, with demonstrable superiority to previous qualitative analyses (82).
Although there has been extensive work on the role of promoter hypermethylation in the pathogenesis of cancers, recent data suggest that promoter hypomethylation in candidate genes may also be important in the development and progression of cancer. Genes demonstrating preferential hypomethylation in pancreatic cancers are typically overexpressed in the cancers versus non-neoplastic pancreas (e.g., Maspin, S100A4, mesothelin, prostate stem cell antigen, and claudin4), suggesting a putative oncogenic role for these proteins (83). This has been functionally demonstrated in the case of VAV1, a Rho–guanine exchange factor in the RAS superfamily, which is overexpressed in pancreatic cancers as a consequence of promoter demethylation (84). Even in the presence of oncogenic KRAS2 mutations, downregulation of VAV1 by RNA-mediated interference abrogates the transformed phenotype of pancreatic cancer cell lines, whereas overexpression of the Vav1 protein in primary cancers is associated with an adverse prognosis compared with Vav1-negative cases.
Expression Abnormalities, Including Aberrant MicroRNA Expression in Pancreatic Cancer
Global expression platforms, such as cDNA and oligonucleotide microarrays, and serial analysis of gene expression have helped elucidate large numbers of differentially expressed transcripts in pancreatic cancer and their precursor lesions (29, 85–89). Multiple insights have been gained into the biology of pancreatic cancer and translational applications thereof from these global expression studies.
First, expression profiling of noninvasive lesions has helped elucidate the role of signaling pathways, including Hedgehog (Hh) and Notch, that play a role in pancreatic cancer initiation (see below) (86, 90). Second, comparison of expression profiles between noninvasive precursor lesions and the accompanying adenocarcinoma has identified transcripts such as S100A4, claudin4, CXCR4, and mesothelin that likely play a role in the invasion process (88). Third, these studies have identified an important role for host stromatumor cell interactions in the pathogenesis of pancreatic cancer. Specifically, these studies have helped define that on the basis of distinct expression profiles, “bulk” tumors can be divided into four architectural compartments: the neoplastic epithelium itself, juxtatumoral stroma (i.e., stroma immediately adjacent to the neoplastic cells), panstroma (i.e., the rest of the stroma beyond the juxtatumoral location), and angioendothelium (i.e., the tumor vasculature) (91). Finally, global expression profiling has identified numerous promising diagnostic, imaging, and therapeutic candidates with immediate translation relevance in pancreatic cancer. The example of mesothelin highlights the versatile applications that can be generated from discovery of a single over-expressed gene. Mesothelin, which encodes for a GPI-anchored cell surface protein, is upregulated almost ubiquitously in pancreatic adenocarcinomas, is expressed only infrequently in noninvasive precursor lesions, and is essentially absent in non-neoplastic pancreatic tissues (32, 92). Immunolabeling for mesothelin is useful as an ancillary marker for the diagnosis of pancreatic adenocarcinoma in equivocal biopsy or cytology specimens (93). Because mesothelin is also secreted, detection of elevated serum levels has been proposed as a potential biomarker for pancreatic cancer (94). Mesothelin also appears to be a potent immunogen, and development of a T cell immune response to mesothelin epitopes is a reliable indicator of response to a pancreatic cancer whole cell vaccine in patients (95). Efforts are under way to develop a recombinant mesothelin vaccine and thus eliminate the need for a whole cell vaccine with its attendant nonspecific immunogenic effects. Lastly, humanized antimesothelin monoclonal antibodies and antimesothelin-conjugated Pseudomonas immunotoxins have been developed as a therapeutic strategy in pancreatic cancer, and early phase clinical studies are under way with these agents (96). Thus, global expression studies have identified promising candidates that have the potential to impact all aspects of pancreatic cancer care in the clinic.
MicroRNAs (miRNAs) are a highly conserved family of 18–24-nucleotide RNA molecules that regulate the stability or translational efficiency of complementary target mRNAs (97). In the past five years, ~400 mammalian miRNAs have been identified, and recent evidence suggests the existence of many more that have yet to be characterized. In Caenorhabditis elegans and Drosophila, miRNAs have been demonstrated to regulate cellular differentiation, proliferation, and apoptosis. These functions have led to speculation that miRNAs may participate in tumorigenesis in humans. Indeed, multiple reports of altered miRNA expression in human cancers have been published (98, 99). Furthermore, miRNAs are enriched at sites in the human genome that are often targets of amplification, loss of heterozygosity, or chromosomal breakpoints in neoplastic cells. These initial studies suggest that abnormal expression of miRNAs may be a fundamental feature of cancer cells. There is also evidence that suggests that miRNA deregulation plays a causative role in certain neoplasms. For example, in one study, C. elegans let-7 and its human homolog were shown to regulate the oncogene RAS (100). Human let-7 shows diminished expression in lung cancers, whereas RAS is overexpressed. Moreover, overexpression of let-7 in lung cancer cell lines decreases growth rates. In another study, a group of miRNAs located on human chromosome 13, known as the mir-17 cluster, was shown to accelerate Myc-mediated lymphomagenesis in a mouse model; this is the first established oncogenic miRNA cluster and has been denoted as OncomiR-1 (101). Not unexpectedly, several recent studies showed that miRNA expression is deregulated in pancreatic cancers. A miRNA signature of pancreatic cancer has been elucidated, and it includes the upregulation of miR-21, miR-155, miR-221, and miR-222 (102–104). It is unclear which of these miRNAs plays a pathogenic role in pancreatic cancer and at what stage of tumorigenesis. In addition to enhancing our understanding of the biology of pancreatic cancer, these aberrantly expressed miRNAs might be useful as potential therapeutic targets, with the recent availability of in vivo miRNA knockdown strategies (antagomirs) (105).
Developmental Signaling Pathways: Hedgehog and Notch
The mammalian Hh signaling pathway comprises three secreted ligands—Sonic, Desert, and Indian hedgehog (Shh, Dhh, Ihh)—which bind to and inactivate the 12-transmembrane Patched (PTCH) receptor (106). This, in turn, releases the 7-transmembrane Smoothened (SMO) receptor from its inhibition by PTCH and allows SMO to inhibit Suppressor of Fused (SUFU). In the absence of signaling within this pathway, SUFU enables cytoplasmic sequestration of the Hh transcription factor glioma-associated oncogene homolog 1 (GLI), and inhibition of SUFU results in nuclear translocation of GLI, with transcriptional activation of multiple Hh target genes. Of note, PTCH is itself an Hh target, thus creating a negative feedback loop and regulating physiologic Hh levels. The Hh signaling pathway not only plays critical and diverse roles in developmental patterning, but is also being increasingly recognized as a key component of mature tissue homeostasis. Specifically, sustained Hh signaling within the stem cell niche of various tissues appears to be a prerequisite for maintaining stem cell number and function (107). In a wide range of epithelial sites, injury leads to transient expansion of an Hh-dependent stem cell population that is critical in accomplishing tissue regeneration. Given these significant roles for Hh signaling in the tissue-specific stem cell niche, it is not surprising that deregulation of this pathway has been reported in multiple human cancers, including pancreatic cancer.
In the vast majority of epithelial malignancies with aberrant Hh activation, constitutive signaling is a consequence of Hh ligand overexpression, rather than a mutational event in one of the pathway components (108). Several lines of evidence implicate the Hh signaling pathway as one of the initiating events in pancreatic cancer. First, Hh ligand is overexpressed in two precursor lesions of pancreatic adenocarcinoma, PanINs and IPMNs, as well as in chronic pancreatitis, a known risk factor for pancreatic cancer (109). Second, the expression profile of immortalized human pancreatic ductal cells selected for stable GLI overexpression closely resembles that of microdissected human PanIN lesions, consistent with the Hh pathway regulating a distinct transcriptional program in these precursor lesions (86). Finally, in recently developed animal models, Hh signaling cooperates with oncogenic Kras in the development of extensive PanIN lesions in mice (see below) (110). Sustained Hh signaling also appears to be critical in the maintenance of pancreatic cancers, as blockade of the Hh pathway in vivo with the small molecule inhibitor cyclopamine results in modest growth inhibition of subcutaneous pancreatic cancer xenografts (109); a more profound effect is observed in spontaneously metastasizing orthotopic xenograft models, where cyclopamine abrogates pancreatic cancer metastases disproportionate to its effects on the primary cancer (111). These findings are consistent with the role of the Hh pathway in regulating metastases, and they have important therapeutic implications for the clinical translation of this class of agents (see below).
The Notch pathway is another pathway critical in developmental patterning and in cell fate determination (112). The Notch pathway in mammals comprises membrane-bound Notch ligands (Jagged and Delta-like) that interact with Notch receptors 1–4 on neighboring cells. The resulting intramembrane proteolysis of the Notch receptor by tumor necrosis factor α-converting enzyme and γ-secretase enzyme allows nuclear translocation of the Notch intracellular domain (NICD). In the nucleus, NICD displaces a transcriptional corepressor complex bound to the CBF1/RBP-Jκ protein, and the NICD-CBF1 association activates transcription of a family of basic helix-loop-helix (bHLH) genes, including Hes1, Hey1, and HeyL. The translated bHLH proteins mediate Notch pathway effects, principal among which is the ability to restrain differentiation and maintain cells in an undifferentiated, precursor state. For further discussion on the Notch signaling pathway, the reader is referred to the review by Aster et al. (112a) in this volume of the Annual Review of Pathology: Mechanisms of Disease.
Notch signaling plays a seminal role in pancreatic development by maintaining an appropriate pool of precursor cells for exocrine differentiation and restricting premature differentiation along the endocrine lineage (113). In the mature pancreas, Notch signaling is active principally in the centroacinar cells (CACs), located at the junction of acini and ducts (see Cellular Origins of Pancreatic Cancer) (90). Expression of Notch receptor, ligands, and gene targets is elevated in PanIN lesions as well as in invasive cancer (90). Moreover, expression of the Notch gene target Hes1 is seen in precursor lesions arising in mouse models of pancreatic cancer (44), suggesting a role for this pathway in pancreatic cancer initiation (see Animal Models of Pancreatic Cancer). Blockade of Notch signaling in pancreatic cancer cell lines, with RNA interference or pharmacologic agents, leads to growth inhibition and apoptosis, underscoring the importance of this pathway as a new therapeutic target in established cancer (114).
ANIMAL MODELS OF PANCREATIC CANCER
Although the pancreas was one of the first organs wherein tissue-specific transgene expression was achieved (115, 116), almost two decades passed before the development of an animal model that faithfully recapitulated the cognate human disease. These early genetically engineered mice (GEM) utilized acinarspecific promoters such as elastase to drive oncogene expression in the mouse pancreas, which, not surprisingly, yielded neoplasms of predominantly acinar histogenesis (116, 117). A major breakthrough was achieved in 2003 with the development of a GEM that expressed an oncogenic KrasG12D allele from its endogenous promoter (and thus, at physiologic levels) in the developing pancreas, through Cre-mediated recombination driven by Pdx1 (or Ptf1/p48) regulatory elements (44).
Pdx1 is a homeodomain protein critical for early pancreatic development, and both differentiated exocrine and endocrine cell types in the mature pancreas arise from a Pdx1-expressing progenitor population (118). Ptf1/p48 is a marker of early pancreatic specification, and its expression persists in the exocrine acinar compartment postnatally (119). The Pdx1-Cre; LSL-KrasG12D mice develop the entire compendium of precursor ductal lesions (murine PanIN or mPanIN) observed in the human pancreatic cancer progression model, with a minor proportion (~10%) developing invasive, metastatic adenocarcinomas after a long latency of several months (44); a near identical phenotype is observed in the Ptf1/p48-Cre; LSL-KrasG12D mice. The low frequency of progression to invasive adenocarcinoma in these GEM is likely a consequence of Kras-induced senescence in the preinvasive lesions, underscoring the need for cooperating mutations in the Ink4a-Rb or Arf-p53 checkpoints to develop invasive neoplasia (120). This, in fact, is the case because engineering mice that misexpress oncogenic Kras in the pancreas in combination with a conditional Ink4a/Arf or p53-null allele (45, 47), or a conditional knock-in mutant p53R172H (121), develop lethal, metastatic pancreatic cancers with complete penetrance and shorter latency (weeks to few months, based on the genotype). Several caveats have emerged from the generation of these genetically engineered mouse models:
GEM that harbor either pancreas-specific deletion of Ink4a/Arf or p53, in the absence of conditional KrasG12D expression, do not demonstrate an obvious pancreatic phenotype (44, 45, 47). Thus, mutant Kras appears to be the initiator genetic lesion, and the additional genetic hits serve as accelerators of disease progression (122). As a corollary, Ink4a, Arf, and p53 appear to be dispensable for normal pancreatic development because Cre-mediated deletion under Pdx1 or Ptf1/p48 regulatory elements abrogate the corresponding transcripts in all differentiated exocrine and endocrine cell types.
Expectedly, there is an increasing latency for the development of invasive neoplasia, if the second conditional allele with KrasG12D is heterozygous for Ink4a/Arf or p53, versus homozygous null (47); in the former instances, the invasive cancer usually demonstrates loss of the retained wild-type allele. Furthermore, murine pancreatic cancers that arise in the backdrop of conditional homozygous alleles (i.e., with a rapidly progressive course) have locally advanced lethal tumor burden accompanied by micrometastatic disease, whereas cancers arising with longer latencies in the compound heterozygous mice typically develop gross systemic metastases (44, 45, 47). It is postulated that the longer latency permits emergence of subclones with more aggressive metastatic potential (123).
The choice of cooperating mutation with KrasG12D has a bearing on the histologic spectrum of the invasive cancer; thus, GEM with conditional Ink4a deletions are biased toward developing poorly differentiated neoplasms, including those with sarcomatous or anaplastic histology (something uncommonly observed in human pancreatic cancer), whereas a mutant p53 backdrop usually lends itself to well- or moderately differentiated adenocarcinomas (44, 45, 47). The presence or absence of an intact TGF-β signaling pathway also has an impact on tumor histology, as discussed subsequently.
Recently, three models of pancreatic neoplasia have been described wherein the integrity of the TGF-β signaling pathway is compromised by either conditional deletion of the Smad4 allele (46, 124) or the type II TGFβ receptor (Tgfbr2) (48). These new models have provided fascinating insights into the role of TGF-β signaling in pancreatic neoplasia. First, it appears that conditional deletion of Tgfbr2 or Smad4 is not sufficient to induce either mPanIN lesions or invasive cancer, underscoring the importance of multiple genetic hits (46, 48, 124). Second, deletion of either Tgfbr2 or Smad4 cooperates with mutant Kras in inducing pancreatic neoplasia, with shorter latencies than observed in Pdx1-Cre; KrasG12D GEM alone. There is, however, one crucial distinction: Although GEM with pancreas-specific loss of Tgfbr2 and mutant Kras expression develop well-differentiated adenocarcinomas with 100% penetrance and lethality (48), the combination of Smad4 loss with mutant Kras yields pancreata that harbor cystic neoplasms, either IPMNs (46) or MCNs (124), respectively. The basis for this attenuated phenotype in the setting of Smad4 loss is unclear, but it might reflect a more pervasive abrogation of signal transduction downstream of the deleted Tgfbr2 receptor. Third, consistent with the role of TGF-β signaling in promoting epithelial-to-mesenchymal transition (125), loss of Smad4 is associated with decreased evidence of epithelial-to-mesenchymal transition in the murine pancreatic cancers. Thus, pancreatic cancers that arise in Smad4 wild-type, Ink4a/Arf-null, KrasG12D mice demonstrate a high proportion of poorly differentiated carcinomas, whereas Ink4a/Arf-null, KrasG12D mice that also harbor conditional Smad4 deletions typically develop well- to moderately differentiated adenocarcinomas expressing epithelial markers, such as Cytokeratin 19 and E-cadherin (46).
In addition to expanding our understanding of the basic biology of pancreatic cancer, the recently developed compendium of GEM is a treasure trove for performing studies on early detection, molecular imaging, tumor immunology, and experimental therapeutics (126). Many of the oncogenic pathways and genes upregulated in human pancreatic cancers (EGFR, Hh, Notch, cyclooxygenase-2, matrix metalloproteinases, etc.) are overexpressed in the preinvasive lesions and/or the associated invasive cancer (44, 46, 47, 121). Thus, there is an opportunity to explore chemoprevention and treatment strategies in a biologically relevant preclinical model. How these mouse hospitals will compare to the more traditional human tumor xenograft models remains to be seen, but there is little doubt that their emergence has garnered tremendous interest in the scientific community to study pancreatic cancer.
CELLULAR ORIGINS OF PANCREATIC CANCER
The cell of origin of human pancreatic cancer is not defined; however, significant progress has been made in defining specific populations within the cancer that are selectively able to initiate new tumors when transplanted into mice. Although there are significant differences between normal stem cells and these tumor-initiating cells, the tumor-initiating cells are often termed cancer stem cells.
Emerging evidence from multiple solid cancers supports the existence of cancer stem cells (127), which were first identified within hematopoeitic malignancies (128). Normal stem cells undergo asymmetric cell division, with one daughter cell retaining self-renewal capacity and the other differentiating into a transit-amplifying cell, with limited proliferative potential; it is postulated that cancer stem cells divide in an identical manner. On the basis of tumor initiation in heterotransplantation experiments, cancer stem cells typically comprise 0.1%–1% of cells in a given cancer (127). A series of elegant studies in solid cancers (breast cancer, gliomas, colon cancer, etc.) has confirmed the existence of self-renewing tumor-initiating subpopulations, which can be enriched on the basis of a variable expression of a panel of cell surface markers, such as CD133, CD24, CD44 (129–131). Li and colleagues (132) have recently isolated a tumor-initiating population from human pancreatic cancer, which is characterized by a surface CD44+, CD24+ ESA+ immunophenotype. These cells, comprising <1% of the bulk population, had a 100-fold increased tumorigenic potential compared with nontumorigenic cancer cells, with 50% of animals injected with as few as 100 CD44+CD24+ESA+ cells, forming tumors that were histologically indistinguishable from the human tumors from which they originated (132).
The identification of pancreatic cancer tumor-initiating cells is an important milestone for two reasons: First, it is well documented that tumor-initiating cells in other malignancies demonstrate significantly greater resistance to conventional cytotoxic agents and to radiation than the differentiated bulk population (133, 134). This has enormous implications for therapy of pancreatic cancer, as most strategies are currently based on reducing tumor volume rather than eradicating the cells responsible for tumor initiation and maintenance (135). Using combined therapeutic approaches that target both the tumor-initiating cells and the differentiated bulk population is most likely to eradicate disease recurrence and achieve a lasting cure (111). Second, identification of pancreatic tumor-initiating cells provides an opportunity to elucidate the putative cell of origin for pancreatic cancer, on the premise that pancreatic tumor-initiating cells are the most proximate derivative of this elusive cell type. Compelling evidence suggests that tumor-initiating cells arise from either resident normal tissue stem cells or a progenitor cell that acquires capacity for self-renewal through dysregulation of critical self-renewal pathways (136, 137); the subsequent acquisition of oncogenic mutations permits niche-independent, cell-autonomous expansion of these deregulated stem/progenitor cells, culminating in neoplasia (138).
In parallel with these advances using human pancreatic cancer samples, experimental animal models continue to provide insights into the cellular origins of experimentally induced pancreatic cancer. Of note, the expression of a mutant Kras gene driven by CK19 regulatory elements in GEM, which targets transgene expression to the mature ductal epithelium, yields a minimal phenotype, characterized by periductal lymphocytic infiltration and an absence of mPanINs or neoplasia (139). This suggests that differentiated ductal cells are unlikely to be the proximate cell of origin for pancreatic adenocarcinoma, and supports a stem/progenitor cell origin, as implicated in the discussion above. By contrast, targeting mutant Kras to the entire pancreatic anlage by Pdx1 regulatory elements results exclusively in ductal neoplasms, without acinar or endocrine lesions (44, 121). Thus, even if differentiated ductal cells can be excluded as the source of mPanINs and adenocarcinoma, the putative cell of origin is such that it responds to mutant Kras expression by differentiating along a ductal lineage. Unlike the marrow and neural tissues, a discrete pancreatic stem cell with multilineage differentiation potential has not been isolated (132).
If tumor-initiating cells represent the neoplastic transformation of resident tissue stem or progenitor cells, where do the latter reside in the pancreas? Two potential candidates with at least limited epithelial plasticity have emerged in the exocrine pancreas, which fulfill some of the criteria for stemness (see also Figure 2) (140). The first is the CAC, located at the junction of acini and ducts. CACs are the only cell type with retained Notch activation (as assessed by nuclear Hes1 expression) in the mature exocrine pancreas (90). As stated above, Notch signaling in the developing pancreas represses differentiation, and sustained activation of this pathway in CACs suggests persistence of a precursor-like transcriptional program (90, 113). Further evidence is derived from studies in mice with conditional deletion of the PTEN protein phosphatase gene in the pancreas; these mice develop prominent ductal metaplasia resulting from expansion of Hes1-expressing CACs (141). A minority of Pten-deficient mice develop pancreatic adenocarcinoma, bolstering a role for CACs in exocrine neoplasia (141).
The second contender for the cell of origin for pancreatic neoplasia is more provocative, in that differentiated acinar cells are themselves considered to harbor facultative progenitor properties. Unlike dedicated stem cells, facultative progenitors acquire a precursor phenotype under specific circumstances, such as organ injury and regeneration (113). For example, in caerulein-induced chemical pancreatitis, surviving acinar cells can repress the terminal exocrine transcriptional program and undergo a process of dedifferentiation, expressing markers of undifferentiated pancreatic progenitor cells such as Pdx1 and Hes1 (142). Under conditions of growth factor stimulation (e.g., activated EGFR signaling), acinar cells undergo a process of acinar-to-ductal metaplasia (ADM), characterized by the abnormal formation of tubular structures (i.e., tubular complexes) with both ductal and acinar differentiation (143). Rigorous lineage-tracing experiments in reporter mice have confirmed ADM to be a true transdifferentiation process; in other words, ductal elements within foci of ADM are tagged, confirming their acinar histogenesis (144). ADM occurs through the generation of nestin-positive intermediates (113). Nestin is an intermediate filament that marks precursor cells at many tissue sites, including the pancreas (145), and its expression in cells of acinar lineage reiterates the ability of differentiated acinar cells to acquire a progenitor phenotype. One of the strongest lines of evidence supporting the role of the acinar compartment in pancreatic neoplasia is derived from a recent study by Guerra and colleagues (146), wherein targeting an oncogenic KrasG12D allele to elastase-expressing acinar cells during development results in the formation of mPanINs and adenocarcinomas. The mPanINs arise in the backdrop of ADM and demonstrate focal expression of acinar markers. In this model of conditional KrasG12D expression, activating the oncogenic allele in adult pancreata did not yield mPanINs, unless coexistent chronic caerulein-induced injury was superimposed (146). Thus, injury-induced expansion of acinar facultative progenitors that respond to an oncogenic KrasG12D allele might be a prerequisite for the development of mPanINs and subsequent adenocarcinoma in the adult pancreas; this study also provides a rationale for the established association between longstanding chronic pancreatitis (or some rare forms of hereditary pancreatitis) and the risk for pancreatic adenocarcinoma in humans (2).
TRANSLATIONAL STUDIES IN PANCREATIC CANCER
The past two decades have seen advances in our fundamental understanding of the nature of pancreatic cancer, from the characterization of precursor lesions to the identification of an ever increasing compendium of molecular abnormalities, and the development of relevant animal models. Unfortunately, despite these advances, pancreatic cancer remains a disease of near uniform lethality. Therefore, much attention has now focused on translational studies in pancreatic cancer, that is, those that can directly impact patient care and outcome. Although a comprehensive cataloging of these efforts would be beyond the scope of this review, we highlight selected examples of bench-to-bedside translation, particularly those in the areas of early detection and therapy.
Early Detection of Pancreatic Cancer
Pancreatic cancer, once invasive, is for the most part incurable. Therefore, the best chance for a cure currently rests in early detection of this neoplasm before it invades (20). The most widely utilized tumor marker for pancreatic cancer in the clinic is the sialylated Lewisa blood group antigen CA19–9. Although the sensitivity of CA19–9 as a tumor marker in patients presenting with symptoms suspected to be due to pancreatic cancer is ~80%, this value is considerably diminished (~55%) in small, resectable cancers (<3 cm) (147). Furthermore, in high-risk, asymptomatic individuals whose pancreata harbor preinvasive lesions (IPMNs, high-grade PanINs), serum CA19–9 is often normal, thus largely precluding its role as a screening tool on the lines of a prostate-specific antigen test for the pancreas (22, 40). The development of tumor markers that can supplant serum CA19–9 in the diagnosis of asymptomatic pancreatic neoplasms, and particularly high-grade noninvasive lesions, is thus a priority.
Global gene expression studies of pancreatic cancers have yielded a plethora of differentially expressed genes, whose secreted gene products have shown considerable potential as new serum markers for pancreatic cancer. A promising candidate to emerge from these studies has been macrophage inhibitory cytokine 1 (MIC1) (148), identified by serial analysis of gene expression and oligonucleotide microarrays as differentially overexpressed in invasive pancreatic cancers (149) as well as in IPMNs (150). Elevated serum MIC1 antigen levels significantly outperformed CA19–9 and other tumor markers in distinguishing patients with resectable pancreatic cancers from healthy controls (150, 151). Other tumor markers whose discovery has been facilitated by global gene expression profiling strategies include the gene products of osteopontin (152), tissue inhibitor of metalloproteinase-1 (153), and mesothelin genes (32), among others; preliminary studies in cohorts of pancreatic cancer and control patients have confirmed the potential utility of some of these markers in the diagnosis of early-stage (i.e., resectable) pancreatic cancers (151–153). In addition to gene expression profiling, ongoing efforts at large-scale proteomic analyses of pancreatic cancers have also facilitated the discovery of candidate biomarkers (154, 155). Along these lines, mass spectrometric analysis of the pancreatic cancer secretome is a novel strategy for biomarker discovery because secreted proteins, rather than the whole cell proteome, is the subset most likely to yield promising circulating tumor markers (156). Quantitative secretome analysis has identified several secreted protein candidates (e.g., perlecan, CD9, and the fibronectin receptor), which await validation in clinical material (156). One important caveat that has emerged from these studies has been the realization that there is unlikely to be a single magic bullet in the serum that will provide the sensitivity and specificity needed to screen asymptomatic individuals, and therefore, any screening assay for pancreatic cancer will likely incorporate two or more tumor markers.
In addition to serum, pancreatic juice is also a bona fide clinical source for developing biomarkers of pancreatic cancer (157, 158). Pancreatic juice, being a more proximate surrogate of the ductal epithelium than serum, would expectedly have an enriched fraction of tumor markers, unadulterated by serum components (147). As an example, mutant KRAS2 is readily detected in pancreatic juice but can be identified in the peripheral circulation only at the stage of advanced, inoperable pancreatic cancers (159). Unlike serum, however, pancreatic juice is obtained during the course of an invasive endoscopic procedure, and therefore, juice-based biomarkers cannot be utilized as a screening modality in the general population. The assays developed in the context of pancreatic juice are best applicable to high-risk individuals suspected of harboring early-stage pancreatic cancers or high-grade noninvasive lesions (22, 40).
Aberrantly methylated genes may also serve as markers of pancreatic cancer. For example, quantitative methylation-specific polymerase chain reaction for assessment of promoter methylation of a panel of genes in pancreatic juice better predicts for pancreatic cancer than conventional methylation-specific polymerase chain reaction, with 100% specificity if >1% methylation at two or more gene promoters is utilized as an inclusion criterion (82). Along the same lines, recent advances in quantitative assays allow for the measurement of KRAS2 gene mutation load in pancreatic juice using an ultrasensitive ligation-based amplification strategy (160) and for the identification of differentially expressed transcripts by microarrays and real-time polymerase chain reaction (161). All of these quantitative techniques are currently in the experimental realm, and their performance in larger-scale clinical cohorts needs to be elucidated, either independently or as an adjunct to radiology and cytologic examination.
Developing Improved Pancreatic Cancer Therapies
The clinical reality is that the majority of patients present with locally advanced or distant metastatic, surgically inoperable disease. Therefore, the development of more potent therapies for advanced pancreatic cancer mandates great urgency. Gemcitabine is the first line of therapy for advanced pancreatic cancer (162) and is increasingly being used as adjuvant treatment in patients with resectable disease (163). The pivotal Phase III trial, which demonstrated the superior efficacy of gemcitabine over the then standard of care 5-fluorouracil, reported an enhancement of median survival from 4.4 to only 5.6 months in patients with advanced pancreatic cancer (164). The EGFR tyrosine kinase inhibitor erlotinib (Tarceva®) is the first targeted agent approved by the Food and Drug Administration for pancreatic cancer (165); unfortunately, the combination of erlotinib and gemcitabine improves survival by less than a month compared with gemcitabine alone in patients with advanced disease. Clearly, there is considerable room for improvements in pancreatic cancer therapy. A large number of investigational agents are being developed for application to pancreatic cancer, targeting growth factor, angiogenic, and other signaling pathways upregulated in this malignancy (166); the discussion of these is beyond the scope of this review. We discuss two specific paradigms illustrating how the knowledge gleaned from pancreatic cancer genetics is serving as a road map for identifying new therapeutic agents against this malignancy.
Synthetic lethal screening was developed in the context of mapping genetic interactions in Drosophila and in yeast, wherein the mutation of two functionally related genes in parallel is lethal to the organism, whereas mutation of any one by itself does not affect the organism’s viability. Hartwell and colleagues (167) were the first to suggest that this paradigm be extrapolated to human cancer therapy, such that an intrinsic genetic or biochemical defect specific to the neoplastic cells is capable of inducing cell death when combined with an extrinsic insult (i.e., therapy) that is synthetic lethal to the former. Non-neoplastic cells, by virtue of lacking this specific genetic or biochemical defect, would bypass the effects of synthetic lethality, providing a broad therapeutic window for cancer-specific cytotoxicity (168). An example of synthetic lethality in the context of pancreatic cancers encompasses the ~5%–7% of cases that harbor loss-of-function mutations in the BRCA2 gene or other members of the Fanconi anemia complex (e.g., FANCC and FANCG), which play a critical role in homologous recombination repair (26). Neoplastic cells lacking BRCA2 (or FANC family) gene function are defective in homologous recombination repair and are rendered selectively sensitive to the effects of DNA cross-linking agents such as Mitomycin C, and to γ-irradiation. In the minority of pancreatic cancers harboring defects in homologous recombination repair, Mitomycin C provides a potential therapeutic window not accorded by standard therapies such as gemcitabine (27, 169).
A second example of synthetic lethality involves the methylthioadenosine phosphorylase (MTAP) gene, whose gene product is the critical enzyme in the salvage pathway of purine synthesis. The MTAP gene is located on chromosome 9p21, approximately 100 kb telomeric to the p16/CDKN2A gene, which undergoes frequent homozygous deletion in pancreatic cancer and, less commonly, in high-grade noninvasive lesions (170, 171). In approximately half of these cases with p16/CDKN2A homozygous deletion, the MTAP gene is codeleted, thereby blocking the salvage pathway of purine synthesis in the affected pancreatic cancer cells (170, 171). In these instances, purine synthesis in the neoplastic cells is essentially entirely dependent upon the de novo pathway, and therefore, blockade of de novo purine synthesis by systemic inhibitors (e.g., L-alanosine, an analog of L-aspartic acid) can effectively terminate ongoing purine synthesis in the MTAP-null cancer cells (172). In contrast, non-neoplastic cells, which retain MTAP gene function, will escape synthetic lethality and continue to synthesize purines via the intact salvage pathway. A convenient immunohistochemical assay for Mtap protein can help identify which cancers harbor MTAP gene deletions and thus enable appropriate patient selection (170, 171).
Another loss-of-function mutation in pancreatic cancer that has recently been utilized as a substrate for synthetic lethal screening is DPC4/SMAD4, which is inactivated in ~55% of pancreatic cancers (62). A large-scale screening of ~20,000 compounds identified a single agent that is selectively toxic to cells lacking DPC4/SMAD4 function, with minimal impact at comparable dosages on cells with retained gene function (173); this compound currently awaits further evaluation in pancreatic cancer.
Although synthetic lethal targeting generally involves a loss-of-function mutation specific to the cancer cells, a second related paradigm that is increasingly being used to develop targeted therapies involves aberrant gene activation and has been termed oncogene addiction. This term, first coined by Bernard Weinstein, describes a phenomenon of acquired dependence by the cancer cells on the proliferation and survival signals exerted by mutated, constitutively activated oncogenes (174). Oncogene addiction predicts that acute inhibition of an aberrantly activated oncogene will profoundly impact growth and survival in neoplastic cells bearing the mutant gene, but that these effects will be mitigated in cells with the wild-type allele (168). The best known examples of oncogene addiction result from mutations in genes encoding receptor and nonreceptor protein kinases, such as EGFR, ERBB2, BRAF, and KIT (CD117), among others (175–178). Neoplasms that are relatively resistant to conventional cytotoxic agents, yet harbor kinase gene mutations, often respond dramatically to small molecule inhibitors or monoclonal antibodies directed against the mutant oncoprotein (179). The ability to selectively target critical oncogenic mutations in subsets of cancers has given new impetus to the concept of individualized therapy (177, 180).
In the context of pancreatic cancers, the obvious oncogenic target would be mutant KRAS2 or its gene product, which is constitutively activated in 90% of cases and is considered a critical initiating event for this malignancy (122). Unfortunately, farnesyltransferase inhibitors, which interfere with critical posttranslational modification of RAS proteins, have been largely ineffective in clinical trials (181). Although the reason for this is not entirely clear, it is believed to be a result of compensatory increase in the activity of geranylgeranyltransferase enzymes, which bypass the farnesyltransferase-inhibitor-induced blockade of farnesyltransferase (182).
As our knowledge of the pancreatic cancer genome has expanded, additional mutant oncogenes have emerged as candidates for targeted therapies. For example, EGFR is mutated in a small subset (~3%) of pancreatic cancers, and patients bearing one of these tumors might benefit from tyrosine kinase inhibitors or antibodies that antagonize the mutant receptor (183). Recently, activating PI3KCA (PI3K catalytic, alpha) gene mutations were reported in ~10% of pancreatic neoplasms, particularly IPMNs and associated colloid cancers, and these cancers are likely susceptible to small molecule inhibitors of the protein kinase (54).
Efforts are under way to perform large-scale sequencing of pancreatic cancers to identify the entire gamut of germ-line and somatic coding sequence alterations. It is certain that these analyses will elucidate new oncogenic mutations, which in turn will be the substrate for targeted therapy in subsets of pancreatic cancers. In the future, individualized therapy of pancreatic cancers is likely to become the standard of care, with the choice of first-line agents dictated by the genetic alterations in an individual neoplasm. The availability of high-throughput mutation detection platforms that can garner information on hundreds of cancer-associated mutations in a single run will greatly facilitate the clinical applicability of genomic data to treatment protocol design (184).
SUMMARY POINTS
Pancreatic cancer runs in some families and a number of genes responsible for this aggregation (BRCA2, BRCA1, CDKN2A/p16, STK11/LKB1, DNA mismatch repair genes) have been discovered.
A number of histologically distinct variants of pancreatic cancer with distinct clinical features have been described, of which ductal adenocarcinoma is by far the most common and most lethal subtype.
Three noninvasive precursor lesions of invasive pancreatic cancer have been described, namely PanIN, IPMN, and MCN. These represent an opportunity to cure pancreatic neoplasia.
A genetic progression model of pancreatic cancer has been developed, which incorporates genomic, transcriptomic, and proteomic abnormalities implicated in the development of this malignancy. Specifically, alterations observed in the early (KRAS2 mutations, telomere shortening), intermediate (p16/CDKN2A loss), and later (DPC4, TP53, and BRCA2 mutations) stages of disease have been identified.
Genetically engineered mouse models of pancreatic cancer have been developed, which recapitulate the entire spectrum of histologic progression observed in the cognate human disease. In these models, mutant Kras2 expression directed to the exocrine pancreas appears to be the prerequisite for developing ductal lesions.
Pancreatic cancer “stem cells” have been identified, raising the possibility of developing tumor-initiating cell-targeted therapies.
Understanding the genetic basis of pancreatic cancer has been instrumental in developing rational early-detection assays and improved therapeutic strategies. Sequencing of the pancreatic cancer genome is on the horizon and will enormously facilitate safer and more efficacious targeted therapies for this uniformly lethal disease.
RELATED RESOURCES
The Johns Hopkins Pancreatic Cancer Web site. http://pathology.jhu.edu/pancreas/
The National Familial Pancreas Tumor Registry. http://pathology2.jhu.edu/pancreas/nfptr.cfm
Pancreatic Cancer in the Ashkenazi Jewish Population. http://pathology2.jhu.edu/pancreas/ashkenazi jewish ancestry.cfm
Pancreatic Cancer in African Americans. http://pathology2.jhu.edu/pancreas/African Americans.cfm
Listing of Pancreatic Cancer Web sites. http://pathology2.jhu.edu/pancreas/othersites.cfm
Acknowledgments
Funding to the authors was provided by the Sol Goldman Pancreatic Cancer Research Center (A.M. and R.H.H.), the Michael Rolfe Foundation for Pancreatic Cancer Research (A.M.), the Lustgarten Foundation for Pancreatic Cancer Research (A.M.), and the National Institutes of Health (grant number P50CA062924 to R.H.H. and R01CA113669 to A.M.). We thank Jennifer Parsons-Brumbaugh for preparing the illustrations.
Footnotes
DISCLOSURE STATEMENT
Dr. Hruban has the potential to receive milestone payments and royalties from Cerus Corporation as a result of the mesothelin invention.
LITERATURE CITED
- 1.Am. Cancer Soc. Cancer Facts and Figures. Atlanta, GA: Am. Cancer Soc; 2007. [Google Scholar]
- 2.Lowenfels AB, Maisonneuve P. Epidemiology and risk factors for pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:197–209. doi: 10.1016/j.bpg.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Stolzenberg-Solomon RZ, Pietinen P, Barrett MJ, Taylor PR, Virtamo J, Albanes D. Dietary and other methyl-group availability factors and pancreatic cancer risk in a cohort of male smokers. Am J Epidemiol. 2001;153:680–87. doi: 10.1093/aje/153.7.680. [DOI] [PubMed] [Google Scholar]
- 4.Villeneuve PJ, Johnson KC, Hanley AJ, Mao Y. Alcohol, tobacco and coffee consumption and the risk of pancreatic cancer: results from the Canadian Enhanced Surveillance System case-control project. Canadian Cancer Registries Epidemiology Research Group. Eur J Cancer Prev. 2000;9:49–58. doi: 10.1097/00008469-200002000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Michaud DS, Liu S, Giovannucci E, Willett WC, Colditz GA, Fuchs CS. Dietary sugar, glycemic load, and pancreatic cancer risk in a prospective study. J Natl Cancer Inst. 2002;94:1293–300. doi: 10.1093/jnci/94.17.1293. [DOI] [PubMed] [Google Scholar]
- 6.Michaud DS, Giovannucci E, Willett WC, Colditz GA, Stampfer MJ, Fuchs CS. Physical activity, obesity, height, and the risk of pancreatic cancer. JAMA. 2001;286:921–29. doi: 10.1001/jama.286.8.921. [DOI] [PubMed] [Google Scholar]
- 7.Everhart J, Wright D. Diabetes mellitus as a risk factor for pancreatic cancer. A meta-analysis. JAMA. 1995;273:1605–9. [PubMed] [Google Scholar]
- 8.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, et al. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–38. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 10.Ozcelik H, Schmocker B, Di Nicola N, Shi XH, Langer B, et al. elT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat Genet. 6174d;16:17–18. doi: 10.1038/ng0597-17. [DOI] [PubMed] [Google Scholar]
- 11.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–64. [PubMed] [Google Scholar]
- 12.Lynch HT, Deters CA, Snyder CL, Lynch JF, Villeneuve P, et al. BRCA1 and pancreatic cancer: Pedigree findings and their causal relationships. Cancer Genet Cytogenet. 2005;158:119–25. doi: 10.1016/j.cancergencyto.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 13.Giardiello FM, Brensinger JD, Tersmette AC, Goodman SN, Petersen GM, et al. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–53. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- 14.Bartsch DK, Sina-Frey M, Lang S, Wild A, Gerdes B, et al. CDKN2A germline mutations in familial pancreatic cancer. Ann Surg. 2002;236:730–37. doi: 10.1097/00000658-200212000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de vos tot Nederveen Cappel WH, Offerhaus GJ, van Puijenbroek M, Caspers E, Gruis NA, et al. Pancreatic carcinoma in carriers of a specific 19 base pair deletion of CDKN2A/p16 (p16-leiden) Clin Cancer Res. 2003;9:3598–605. [PubMed] [Google Scholar]
- 16.Su GH, Hruban RH, Bansal RK, Bova GS, Tang DJ, et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154:1835–40. doi: 10.1016/S0002-9440(10)65440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowenfels AB, Maisonneuve P, DiMagno EP, Elitsur Y, Gates LK, Jr, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–46. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 18.Goggins M, Offerhaus GJ, Hilgers W, Griffin CA, Shekher M, et al. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+ Am J Pathol. 1998;152:1501–7. [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen GM, de Andrade M, Goggins M, Hruban RH, Bondy M, et al. Pancreatic cancer genetic epidemiology consortium. Cancer Epidemiol Biomarkers Prev. 2006;15:704–10. doi: 10.1158/1055-9965.EPI-05-0734. [DOI] [PubMed] [Google Scholar]
- 20.Maitra A, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:211–26. doi: 10.1016/j.bpg.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Hruban RH, Klimstra DS, Pitman MB. Tumors of the Pancreas. Washington, DC: Armed Forces Inst. Pathol; 2006. [Google Scholar]
- 22.Canto MI, Goggins M, Hruban RH, Petersen GM, Giardiello FM, et al. Screening for early pancreatic neoplasia in high-risk individuals: a prospective controlled study. Clin Gastroenterol Hepatol. 2006;4:766–81. doi: 10.1016/j.cgh.2006.02.005. quiz 665. [DOI] [PubMed] [Google Scholar]
- 23.Begg CB, Orlow I, Hummer AJ, Armstrong BK, Kricker A, et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst. 2005;97:1507–15. doi: 10.1093/jnci/dji312. [DOI] [PubMed] [Google Scholar]
- 24.Wilentz RE, Goggins M, Redston M, Marcus VA, Adsay NV, et al. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: a newly described and characterized entity. Am J Pathol. 2000;156:1641–51. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 26.van der Heijden MS, Brody JR, Gallmeier E, Cunningham SC, Dezentje DA, et al. Functional defects in the Fanconi anemia pathway in pancreatic cancer cells. Am J Pathol. 2004;165:651–57. doi: 10.1016/S0002-9440(10)63329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11:7508–15. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 28.Adsay NV, Basturk O, Cheng JD, Andea AA. Ductal neoplasia of the pancreas: nosologic, clinicopathologic, and biologic aspects. Semin Radiat Oncol. 2005;15:254–64. doi: 10.1016/j.semradonc.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, et al. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–22. [PubMed] [Google Scholar]
- 30.Crnogorac-Jurcevic T, Missiaglia E, Blaveri E, Gangeswaran R, Jones M, et al. Molecular alterations in pancreatic carcinoma: Expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J Pathol. 2003;201:63–74. doi: 10.1002/path.1418. [DOI] [PubMed] [Google Scholar]
- 31.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, et al. Discovery of new markers of cancer through serial analysis of gene expression: Prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–24. [PubMed] [Google Scholar]
- 32.Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, et al. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clin Cancer Res. 2001;7:3862–68. [PubMed] [Google Scholar]
- 33.Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–86. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 34.Hruban RH, Takaori K, Klimstra DS, Adsay NV, Albores-Saavedra J, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–87. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 35.Maitra A, Fukushima N, Takaori K, Hruban RH. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12:81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- 36.Brat DJ, Lillemoe KD, Yeo CJ, Warfield PB, Hruban RH. Progression of pancreatic intraductal neoplasias to infiltrating adenocarcinoma of the pancreas. Am J Surg Pathol. 1998;22:163–69. doi: 10.1097/00000478-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 37.van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–47. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iacobuzio-Donahue CA, Klimstra DS, Adsay NV, Wilentz RE, Argani P, et al. Dpc-4 protein is expressed in virtually all human intraductal papillary mucinous neoplasms of the pancreas: comparison with conventional ductal adenocarcinomas. Am J Pathol. 2000;157:755–61. doi: 10.1016/S0002-9440(10)64589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahin F, Maitra A, Argani P, Sato N, Maehara N, et al. Loss of Stk11/Lkb1 expression in pancreatic and biliary neoplasms. Mod Pathol. 2003;16:686–91. doi: 10.1097/01.MP.0000075645.97329.86. [DOI] [PubMed] [Google Scholar]
- 40.Canto MI, Goggins M, Yeo CJ, Griffin C, Axilbund JE, et al. Screening for pancreatic neoplasia in high-risk individuals: an EUS-based approach. Clin Gastroenterol Hepatol. 2004;2:606–21. doi: 10.1016/s1542-3565(04)00244-7. [DOI] [PubMed] [Google Scholar]
- 41.Caldas C, Kern SE. K-ras mutation and pancreatic adenocarcinoma. Int J Pancreatol. 1995;18:1–6. doi: 10.1007/BF02825415. [DOI] [PubMed] [Google Scholar]
- 42.Hingorani SR, Tuveson DA. Ras redux: rethinking how and where Ras acts. Curr Opin Genet Dev. 2003;13:6–13. doi: 10.1016/s0959-437x(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 43.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–43. [PubMed] [Google Scholar]
- 44.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 45.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–46. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-β signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–60. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3:413–23. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- 50.Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003;163:1255–60. doi: 10.1016/S0002-9440(10)63485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing HR, Cordon-Cardo C, Deng X, Tong W, Campodonico L, et al. Pharmacologic inactivation of kinase suppressor of ras-1 abrogates Ras-mediated pancreatic cancer. Nat Med. 2003;9:1266–68. doi: 10.1038/nm927. [DOI] [PubMed] [Google Scholar]
- 52.Altomare DA, Tanno S, De Rienzo A, Klein-Szanto AJ, Skele KL, et al. Frequent activation of AKT2 kinase in human pancreatic carcinomas. J Cell Biochem. 2003;88:470–76. doi: 10.1002/jcb.10287. [DOI] [PubMed] [Google Scholar]
- 53.Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636–41. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schonleben F, Qiu W, Ciau NT, Ho DJ, Li X, et al. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12:3851–55. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–94. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 56.Nowak NJ, Gaile D, Conroy JM, McQuaid D, Cowell J, et al. Genome-wide aberrations in pancreatic adenocarcinoma. Cancer Genet Cytogenet. 2005;161:36–50. doi: 10.1016/j.cancergencyto.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci USA. 2004;101:9067–72. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, et al. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–62. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–30. [PubMed] [Google Scholar]
- 60.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, et al. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–33. [PubMed] [Google Scholar]
- 61.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 62.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–53. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 63.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–21. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 64.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, et al. Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol. 2000;156:37–43. doi: 10.1016/S0002-9440(10)64703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–6. [PubMed] [Google Scholar]
- 66.Iacobuzio-Donahue CA, Wilentz RE, Argani P, Yeo CJ, Cameron JL, et al. Dpc4 protein in mucinous cystic neoplasms of the pancreas: frequent loss of expression in invasive carcinomas suggests a role in genetic progression. Am J Surg Pathol. 2000;24:1544–48. doi: 10.1097/00000478-200011000-00011. [DOI] [PubMed] [Google Scholar]
- 67.Schutte M, Hruban RH, Hedrick L, Cho KR, Nadasdy GM, et al. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–30. [PubMed] [Google Scholar]
- 68.Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor β receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–32. [PubMed] [Google Scholar]
- 69.Su GH, Bansal R, Murphy KM, Montgomery E, Yeo CJ, et al. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci USA. 2001;98:3254–57. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xin W, Yun KJ, Ricci F, Zahurak M, Qiu W, et al. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin Cancer Res. 2004;10:8516–20. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 71.Meeker AK, De Marzo AM. Recent advances in telomere biology: implications for human cancer. Curr Opin Oncol. 2004;16:32–38. doi: 10.1097/00001622-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 72.Gisselsson D. Chromosome instability in cancer: How, when, and why? Adv Cancer Res. 2003;87:1–29. doi: 10.1016/s0065-230x(03)87164-6. [DOI] [PubMed] [Google Scholar]
- 73.Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, et al. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol. 2001;158:1677–83. doi: 10.1016/S0002-9440(10)64123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sato N, Mizumoto K, Nagai E, Tanaka M. Telomerase as a new target for pancreatic cancer treatment. J Hepatobiliary Pancreat Surg. 2002;9:322–27. doi: 10.1007/s005340200036. [DOI] [PubMed] [Google Scholar]
- 75.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 76.Sato N, Goggins M. The role of epigenetic alterations in pancreatic cancer. J Hepatobiliary Pancreat Surg. 2006;13:286–95. doi: 10.1007/s00534-005-1057-1. [DOI] [PubMed] [Google Scholar]
- 77.Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–65. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 78.Sato N, Fukushima N, Matsubayashi H, Iacobuzio-Donahue CA, Yeo CJ, Goggins M. Aberrant methylation of Reprimo correlates with genetic instability and predicts poor prognosis in pancreatic ductal adenocarcinoma. Cancer. 2006;107:251–57. doi: 10.1002/cncr.21977. [DOI] [PubMed] [Google Scholar]
- 79.Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, et al. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–81. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jansen M, Fukushima N, Rosty C, Walter K, Altink R, et al. Aberrant methylation of the 5′ CpG island of TSLC1 is common in pancreatic ductal adenocarcinoma and is first manifest in high-grade PanlNs. Cancer Biol Ther. 2002;1:293–96. doi: 10.4161/cbt.84. [DOI] [PubMed] [Google Scholar]
- 81.Sato N, Ueki T, Fukushima N, Iacobuzio-Donahue CA, Yeo CJ, et al. Aberrant methylation of CpG islands in intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology. 2002;123:365–72. doi: 10.1053/gast.2002.34160. [DOI] [PubMed] [Google Scholar]
- 82.Matsubayashi H, Canto M, Sato N, Klein A, Abe T, et al. DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease. Cancer Res. 2006;66:1208–17. doi: 10.1158/0008-5472.CAN-05-2664. [DOI] [PubMed] [Google Scholar]
- 83.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–66. [PubMed] [Google Scholar]
- 84.Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, Savoy DN, Molina JR, et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 85.Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, et al. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–26. [PubMed] [Google Scholar]
- 86.Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, et al. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619–26. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 87.Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, et al. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol. 2002;160:1745–54. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sato N, Fukushima N, Maitra A, Iacobuzio-Donahue CA, van Heek NT, et al. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am J Pathol. 2004;164:903–14. doi: 10.1016/S0002-9440(10)63178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fukushima N, Sato N, Prasad N, Leach SD, Hruban RH, Goggins M. Characterization of gene expression in mucinous cystic neoplasms of the pancreas using oligonucleotide microarrays. Oncogene. 2004;23:9042–51. doi: 10.1038/sj.onc.1208117. [DOI] [PubMed] [Google Scholar]
- 90.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, et al. Notch mediates TGF α-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–76. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 91.Iacobuzio-Donahue CA, Ryu B, Hruban RH, Kern SE. Exploring the host desmoplastic response to pancreatic carcinoma: gene expression of stromal and neoplastic cells at the site of primary invasion. Am J Pathol. 2002;160:91–99. doi: 10.1016/S0002-9440(10)64353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, et al. Multi-component analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–12. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 93.McCarthy DM, Maitra A, Argani P, Rader AE, Faigel DO, et al. Novel markers of pancreatic adenocarcinoma in fine-needle aspiration: Mesothelin and prostate stem cell antigen labeling increases accuracy in cytologically borderline cases. Appl Immunohistochem Mol Morphol. 2003;11:238–43. doi: 10.1097/00129039-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 94.Corso CD, Stubbs DD, Lee SH, Goggins M, Hruban RH, Hunt WD. Real-time detection of mesothelin in pancreatic cancer cell line supernatant using an acoustic wave immunosensor. Cancer Detect Prev. 2006;30:180–87. doi: 10.1016/j.cdp.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 95.Thomas AM, Santarsiero LM, Lutz ER, Armstrong TD, Chen YC, et al. Mesothelin-specific CD8+ T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med. 2004;200:297–306. doi: 10.1084/jem.20031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hassan R, Bera T, Pastan I. Mesothelin: a new target for immunotherapy. Clin Cancer Res. 2004;10:3937–42. doi: 10.1158/1078-0432.CCR-03-0801. [DOI] [PubMed] [Google Scholar]
- 97.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–80. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 99.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–38. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 100.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 101.Hammond SM. RNAi, microRNAs, and human disease. Cancer Chemother Pharmacol. 2006;58(Suppl 7):63–68. doi: 10.1007/s00280-006-0318-2. [DOI] [PubMed] [Google Scholar]
- 102.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hampton T. MicroRNAs linked to pancreatic cancer. JAMA. 2007;297:937. doi: 10.1001/jama.297.9.937. [DOI] [PubMed] [Google Scholar]
- 104.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 105.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–89. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 106.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–11. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 107.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–54. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 108.Watkins DN, Peacock CD. Hedgehog signalling in foregut malignancy. Biochem Pharmacol. 2004;68:1055–60. doi: 10.1016/j.bcp.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 109.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–56. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–73. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598–608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 112.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Path Mech Dis. 3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leach SD. Epithelial differentiation in pancreatic development and neoplasia: new niches for nestin and Notch. J Clin Gastroenterol. 2005;39:S78–82. doi: 10.1097/01.mcg.0000155547.83901.a3. [DOI] [PubMed] [Google Scholar]
- 114.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006;106:2503–13. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- 115.Ornitz DM, Palmiter RD, Hammer RE, Brinster RL, Swift GH, MacDonald RJ. Specific expression of an elastase-human growth hormone fusion gene in pancreatic acinar cells of transgenic mice. Nature. 1985;313:600–2. doi: 10.1038/313600a0. [DOI] [PubMed] [Google Scholar]
- 116.Ornitz DM, Hammer RE, Messing A, Palmiter RD, Brinster RL. Pancreatic neoplasia induced by SV40 T-antigen expression in acinar cells of transgenic mice. Science. 1987;238:188–93. doi: 10.1126/science.2821617. [DOI] [PubMed] [Google Scholar]
- 117.Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL, Lee DC. Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell. 1990;61:1121–35. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- 118.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–9. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 119.Krapp A, Knofler M, Ledermann B, Burki K, Berney C, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–63. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bardeesy N, Sharpless NE. RAS unplugged: negative feedback and oncogene-induced senescence. Cancer Cell. 2006;10:451–53. doi: 10.1016/j.ccr.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 121.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 122.Deramaudt T, Rustgi AK. Mutant KRAS in the initiation of pancreatic cancer. Biochim Biophys Acta. 2005;1756:97–101. doi: 10.1016/j.bbcan.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 123.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 124.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–43. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 125.Zavadil J, Bottinger EP. TGF-β and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 126.Hruban RH, Rustgi AK, Brentnall TA, Tempero MA, Wright CV, Tuveson DA. Pancreatic cancer in mice and man: the Penn Workshop 2004. Cancer Res. 2006;66:14–17. doi: 10.1158/0008-5472.CAN-05-3914. [DOI] [PubMed] [Google Scholar]
- 127.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 128.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–48. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 129.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–88. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–15. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 131.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 132.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–37. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 133.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 134.Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, et al. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103:2332–36. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huff CA, Matsui W, Smith BD, Jones RJ. The paradox of response and survival in cancer therapeutics. Blood. 2006;107:431–34. doi: 10.1182/blood-2005-06-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–37. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 137.Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci USA. 2004;101:4158–63. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–31. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 139.Brembeck FH, Schreiber FS, Deramaudt TB, Craig L, Rhoades B, et al. The mutant K-ras oncogene causes pancreatic periductal lymphocytic infiltration and gastric mucous neck cell hyperplasia in transgenic mice. Cancer Res. 2003;63:2005–9. [PubMed] [Google Scholar]
- 140.Stanger BZ, Dor Y. Dissecting the cellular origins of pancreatic cancer. Cell Cycle. 2006;5:43–46. doi: 10.4161/cc.5.1.2291. [DOI] [PubMed] [Google Scholar]
- 141.Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–95. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 142.Jensen JN, Cameron E, Garay MV, Starkey TW, Gianani R, Jensen J. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–41. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 143.Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 144.Means AL, Meszoely IM, Suzuki K, Miyamoto Y, Rustgi AK, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–76. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 145.Esni F, Stoffers DA, Takeuchi T, Leach SD. Origin of exocrine pancreatic cells from nestin-positive precursors in developing mouse pancreas. Mech Dev. 2004;121:15–25. doi: 10.1016/j.mod.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 146.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 147.Rosty C, Goggins M. Early detection of pancreatic carcinoma. Hematol Oncol Clin N Am. 2002;16:37–52. doi: 10.1016/s0889-8588(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 148.Bauskin AR, Brown DA, Kuffner T, Johnen H, Luo XW, et al. Role of macrophage inhibitory cytokine-1 in tumorigenesis and diagnosis of cancer. Cancer Res. 2006;66:4983–86. doi: 10.1158/0008-5472.CAN-05-4067. [DOI] [PubMed] [Google Scholar]
- 149.Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, Hruban RH, Kern SE. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–26. [PubMed] [Google Scholar]
- 150.Koopmann J, Buckhaults P, Brown DA, Zahurak ML, Sato N, et al. Serum macrophage inhibitory cytokine 1 as a marker of pancreatic and other periampullary cancers. Clin Cancer Res. 2004;10:2386–92. doi: 10.1158/1078-0432.ccr-03-0165. [DOI] [PubMed] [Google Scholar]
- 151.Koopmann J, Rosenzweig CN, Zhang Z, Canto MI, Brown DA, et al. Serum markers in patients with resectable pancreatic adenocarcinoma: macrophage inhibitory cytokine 1 versus CA19–9. Clin Cancer Res. 2006;12:442–46. doi: 10.1158/1078-0432.CCR-05-0564. [DOI] [PubMed] [Google Scholar]
- 152.Koopmann J, Fedarko NS, Jain A, Maitra A, Iacobuzio-Donahue C, et al. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomark Prev. 2004;13:487–91. [PubMed] [Google Scholar]
- 153.Zhou W, Sokoll LJ, Bruzek DJ, Zhang L, Velculescu VE, et al. Identifying markers for pancreatic cancer by gene expression analysis. Cancer Epidemiol Biomark Prev. 1998;7:109–12. [PubMed] [Google Scholar]
- 154.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, et al. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–63. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 155.Chen R, Yi EC, Donohoe S, Pan S, Eng J, et al. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–97. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 156.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–71. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 157.Chen R, Pan S, Cooke K, Moyes KW, Bronner MP, et al. Comparison of pancreas juice proteins from cancer versus pancreatitis using quantitative proteomic analysis. Pancreas. 2007;34:70–79. doi: 10.1097/01.mpa.0000240615.20474.fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, et al. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–55. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 159.Yamada T, Nakamori S, Ohzato H, Oshima S, Aoki T, et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin Cancer Res. 1998;4:1527–32. [PubMed] [Google Scholar]
- 160.Shi C, Eshleman SH, Jones D, Fukushima N, Hua L, et al. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141–47. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 161.Rogers CD, Fukushima N, Sato N, Shi C, Prasad N, et al. Differentiating pancreatic lesions by microarray and QPCR analysis of pancreatic juice RNAs. Cancer Biol Ther. 2006;5:1383–89. doi: 10.4161/cbt.5.10.3323. [DOI] [PubMed] [Google Scholar]
- 162.Ko AH, Tempero MA. Treatment of metastatic pancreatic cancer. J Natl Compr Cancer Netw. 2005;3:627–36. doi: 10.6004/jnccn.2005.0036. [DOI] [PubMed] [Google Scholar]
- 163.Oettle H, Post S, Neuhaus P, Gellert K, Langrehr J, et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–77. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- 164.Burris HA, 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–13. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 165.Moore MJ. Brief communication: a new combination in the treatment of advanced pancreatic cancer. Semin Oncol. 2005;32:5–6. doi: 10.1053/j.seminoncol.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 166.Von Hoff DD. What’s new in pancreatic cancer treatment pipeline? Best Pract Res Clin Gastroenterol. 2006;20:315–26. doi: 10.1016/j.bpg.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 167.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–68. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 168.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 169.Gallmeier E, Kern SE. Targeting Fanconi anemia/BRCA2 pathway defects in cancer: the significance of preclinical pharmacogenomic models. Clin Cancer Res. 2007;13:4–10. doi: 10.1158/1078-0432.CCR-06-1637. [DOI] [PubMed] [Google Scholar]
- 170.Hustinx SR, Hruban RH, Leoni LM, Iacobuzio-Donahue C, Cameron JL, et al. Homozygous deletion of the MTAP gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapy. Cancer Biol Ther. 2005;4:83–86. doi: 10.4161/cbt.4.1.1380. [DOI] [PubMed] [Google Scholar]
- 171.Hustinx SR, Leoni LM, Yeo CJ, Brown PN, Goggins M, et al. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod Pathol. 2005;18:959–63. doi: 10.1038/modpathol.3800377. [DOI] [PubMed] [Google Scholar]
- 172.Karikari CA, Mullendore M, Eshleman JR, Argani P, Leoni LM, et al. Homozygous deletions of methylthioadenosine phosphorylase in human biliary tract cancers. Mol Cancer Ther. 2005;4:1860–66. doi: 10.1158/1535-7163.MCT-05-0103. [DOI] [PubMed] [Google Scholar]
- 173.Wang H, Han H, Von Hoff DD. Identification of an agent selectively targeting DPC4 (deleted in pancreatic cancer locus 4)-deficient pancreatic cancer cells. Cancer Res. 2006;66:9722–30. doi: 10.1158/0008-5472.CAN-05-4602. [DOI] [PubMed] [Google Scholar]
- 174.Weinstein IB. Cancer. Addiction to oncogenes: the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 175.Gazdar AF, Shigematsu H, Herz J, Minna JD. Mutations and addiction to EGFR: the Achilles ‘heal’ of lung cancers? Trends Mol Med. 2004;10:481–86. doi: 10.1016/j.molmed.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 176.Blagosklonny MV. Gefitinib (iressa) in oncogene-addictive cancers and therapy for common cancers. Cancer Biol Ther. 2004;3:436–40. doi: 10.4161/cbt.3.5.984. [DOI] [PubMed] [Google Scholar]
- 177.Davies M, Hennessy B, Mills GB. Point mutations of protein kinases and individualised cancer therapy. Expert Opin Pharmacother. 2006;7:2243–61. doi: 10.1517/14656566.7.16.2243. [DOI] [PubMed] [Google Scholar]
- 178.Rapp UR, Gotz R, Albert S. BuCy RAFs drive cells into MEK addiction. Cancer Cell. 2006;9:9–12. doi: 10.1016/j.ccr.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 179.Haber DA, Bell DW, Sordella R, Kwak EL, Godin-Heymann N, et al. Molecular targeted therapy of lung cancer: EGFR mutations and response to EGFR inhibitors. Cold Spring Harbor Symp Quant Biol. 2005;70:419–26. doi: 10.1101/sqb.2005.70.043. [DOI] [PubMed] [Google Scholar]
- 180.Weinstein IB, Joe AK. Mechanisms of disease: oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–57. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 181.Van Cutsem E, van de Velde H, Karasek P, Oettle H, Vervenne WL, et al. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol. 2004;22:1430–38. doi: 10.1200/JCO.2004.10.112. [DOI] [PubMed] [Google Scholar]
- 182.Lerner EC, Zhang TT, Knowles DB, Qian Y, Hamilton AD, Sebti SM. Inhibition of the prenylation of K-Ras, but not H- or N-Ras, is highly resistant to CAAX peptidomimetics and requires both a farnesyltransferase and a geranylgeranyltransferase I inhibitor in human tumor cell lines. Oncogene. 1997;15:1283–88. doi: 10.1038/sj.onc.1201296. [DOI] [PubMed] [Google Scholar]
- 183.Kwak EL, Jankowski J, Thayer SP, Lauwers GY, Brannigan BW, et al. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin Cancer Res. 2006;12:4283–87. doi: 10.1158/1078-0432.CCR-06-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 185.Parker JF, Florell SR, Alexander A, DiSario JA, Shami PJ, Leachman SA. Pancreatic carcinoma surveillance in patients with familial melanoma. Arch Dermatol. 2003;139:1019–25. doi: 10.1001/archderm.139.8.1019. [DOI] [PubMed] [Google Scholar]
- 186.Yamamoto H, Itoh F, Nakamura H, Fukushima H, Sasaki S, et al. Genetic and clinical features of human pancreatic ductal adenocarcinomas with widespread microsatellite instability. Cancer Res. 2001;61:3139–44. [PubMed] [Google Scholar]
- 187.Couch FJ, Johnson MR, Rabe K, Boardman L, McWilliams R, et al. Germ line Fanconi anemia complementation group C mutations and pancreatic cancer. Cancer Res. 2005;65:383–86. [PubMed] [Google Scholar]
- 188.van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–88. [PubMed] [Google Scholar]