Abstract
The p53 transcription factor is a key tumor suppressor and a central regulator of the stress response. To ensure a robust and precise response to cellular signals, p53 gene expression must be tightly regulated from the transcriptional to the post-translational levels. Computational predictions suggest that several microRNAs are involved in the post-transcriptional regulation of p53. Here we demonstrate that miR-125b, a brain-enriched microRNA, is a bona fide negative regulator of p53 in both zebrafish and humans. miR-125b-mediated down-regulation of p53 is strictly dependent on the binding of miR-125b to a microRNA response element in the 3′ untranslated region of p53 mRNA. Overexpression of miR-125b represses the endogenous level of p53 protein and suppresses apoptosis in human neuroblastoma cells and human lung fibroblast cells. In contrast, knockdown of miR-125b elevates the level of p53 protein and induces apoptosis in human lung fibroblasts and in the zebrafish brain. This phenotype can be rescued significantly by either an ablation of endogenous p53 function or ectopic expression of miR-125b in zebrafish. Interestingly, miR-125b is down-regulated when zebrafish embryos are treated with γ-irradiation or camptothecin, corresponding to the rapid increase in p53 protein in response to DNA damage. Ectopic expression of miR-125b suppresses the increase of p53 and stress-induced apoptosis. Together, our study demonstrates that miR-125b is an important negative regulator of p53 and p53-induced apoptosis during development and during the stress response.
Keywords: MicroRNA, p53, development, human, zebrafish
Recent studies have highlighted the importance of microRNAs (miRNAs), an abundant class of small noncoding RNAs, in many biological processes (Bartel 2004). miRNAs are generated in two steps: First, a hairpin precursor is processed from miRNA primary transcripts by the Drosha complex; and second, the precursor is cleaved by the Dicer complex to become a mature duplex miRNA (Bartel 2004). One strand of the mature miRNA is loaded into the RNA-induced silencing complex (RISC) and brought to its target mRNAs (Bartel 2004). miRNAs often bind to the 3′ untranslated region (UTR) of mRNAs with imperfect complementarity (Bartel 2004). They regulate gene expression by inhibiting translation or repressing stability of the targets (Bartel 2004). Inactivation of miRNA biogenesis by the loss of Dicer leads to severe defects in zebrafish embryos and in mouse prenatal lethality (Bernstein et al. 2003; Giraldez et al. 2005). The role of miRNAs during development has also been notably featured in the self-renewal and differentiation of embryonic stem cells and the formation of various embryonic tissues (He and Hannon 2004; Foshay and Gallicano 2007; Tay et al. 2008).
miRNAs are also implicated in tumorigenesis. Differences in miRNA expression profiles define the signatures of various cancers, and miRNA dysregulation can lead to all the hallmarks of cancer (Garzon et al. 2006; Zhang et al. 2007). miRNAs are known to be both regulators and targets of oncogenes and tumor suppressor genes (Garzon et al. 2006). For example, the let-7 miRNA family regulates the Ras oncogenes (Johnson et al. 2005). The miR-34 family is a key downstream effector of the p53 tumor suppressor (Bommer et al. 2007; Chang et al. 2007; He et al. 2007; Tarasov et al. 2007). miR-125b, a highly conserved homolog of lin-4 essential for the temporal control of post-embryonic differentiation in Caenorhabditis elegans (Olsen and Ambros 1999), was also recently found to be elevated in several types of cancers (Nelson et al. 2006; Bloomston et al. 2007; Shi et al. 2007; Bousquet et al. 2008). Further understanding of the expression and the function of miRNAs in cancers would provide new approaches for diagnosis and therapies against these diseases.
The p53 tumor suppressor is an important transcription factor that safeguards the cell against tumorigenesis and regulates multiple cellular processes (Foulkes 2007). During development, p53 activity is maintained at a very low level (Almog and Rotter 1997). Activation of p53 in response to DNA damage or oncogene activation leads to cell cycle arrest or apoptosis (Foulkes 2007). Inactivation of the p53 pathway is often a key event in tumorigenesis, found in ∼50% of human cancers (Harris 1996). The expression and activity of p53 are monitored by many layers of regulation, mainly at the post-translational level by ubiquitin ligases such as Mdm2 and Mdm4 (Foulkes 2007). Indeed, changes in the protein level of p53 were not observed to correlate with the transcriptional activity of the p53 gene during embryogenesis or differentiation, indicating that post-transcriptional regulation is likely to be involved (Dony et al. 1985; Klinken et al. 1988; Tchang et al. 1993). Although an antisense RNA is implicated in the maturation of p53 pre-mRNA, there is no evidence that it modulates p53 expression (Khochbin and Lawrence 1989).
Here, we report for the first time a miRNA that directly regulates p53. We identified miR-125b as a negative regulator of p53 in both humans and zebrafish. miR-125b binds directly to the 3′ UTR of human and zebrafish p53 mRNAs, and represses p53 protein levels in a manner dependent on its binding site in the p53 3′ UTRs. miR-125b-mediated regulation of p53 is critical for modulating apoptosis in human cells and in zebrafish embryos during development and during the stress response. Our report provides new insights into the function of miR-125b during development and suggests a role in tumorigenesis for this miRNA.
Results
miR-125b binds to the 3′ UTR of human and zebrafish p53 mRNAs
To examine the possibility of p53 regulation by miRNAs, we searched for potential miRNA-binding sites in the p53 mRNA by computational analysis. Searches by TargetScan (Lewis et al. 2005) and miRBase Target (Yoon and De 2006) yielded two different lists of miRNAs. Most of the predicted binding sites (miRNA response elements, MREs) of these miRNAs in the 3′ UTR of p53 are poorly conserved. Only one specific miRNA, miR-125b, targeted both human and zebrafish p53 when the predictions were compared across distant species (Fig. 1A). The putative MREs of miR-125b were also found in the 3′ UTRs of p53 in other vertebrates (Supplemental Fig. 1A). This suggests that miR-125b is likely to be an important regulator of p53.
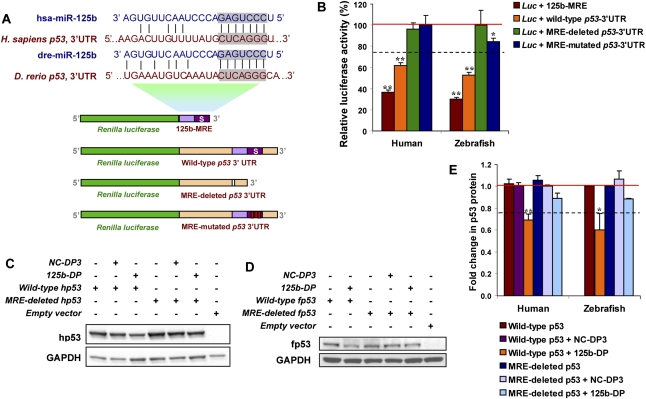
Figure 1.
miR-125b binds to the 3′ UTR of zebrafish and human p53 mRNAs. (A) Outline of luciferase reporter assay for validating the interaction of miR-125b with the 3′ UTR of p53: The MREs of miR-125b in the 3′ UTR of human and zebrafish p53 mRNA were predicted by TargetScan and miRBase Target. Shaded texts indicate the “seed” regions. Each predicted MRE or the whole p53 3′ UTR was inserted into a psiCheck2 vector, immediately downstream from the Renilla luciferase gene. In mutant reporter constructs, the MRE was deleted or a three-mismatch mutation was introduced into the seed region. Each luciferase construct was cotransfected with negative control duplex 1 (NC-DP1) or miR-125b duplex (125b-DP) into HEK-293T cells, and luciferase readings were obtained 48 h after transfection. (B) Repression of luciferase activity due to the interaction between miR-125b and the predicted MREs in the luciferase-MRE or in the luciferase–p53–3′ UTR constructs. Repression was abolished when the MRE was deleted or mutated. Every Renilla luciferase reading was normalized to that of the control firefly luciferase. The luciferase activities of 125b-DP-transfected cells were presented as percentages relative to the level of luciferase in the NC-DP1-transfected cells (this control luciferase level is considered as 100% and is represented by the solid red line). The values represent average ± SEM (n ≥ 6). The dashed line represents the threshold of luciferase activity (75%), suppression of luciferase level below which indicates positive binding. Two-tail t-test results are indicated by (*) P < 0.05 and (**) P < 0.01, relative to the NC-DP1-transfected controls. (C) Overexpression of miR-125b down-regulates the wild-type but not the MRE-deleted human p53 (hp53) in H1299 cells: pCDNA3.1+ vector containing the full-length human p53 cDNA sequence with or without the miR-125b MRE was transfected alone or cotransfected with negative control duplex 3 (NC-DP3) or 125b-DP into H1299 cells. The level of human p53 was analyzed by Western blots 2 d after transfection. (D) Overexpression of miR-125b down-regulates the wild-type but not the MRE-deleted zebrafish p53 (fp53) in H1299 cells: pCDNA3.1+ vector containing the full-length zebrafish p53 cDNA sequence with or without the MRE of miR-125b was transfected alone or cotransfected with NC-DP3 or 125b-DP into H1299 cells. The level of zebrafish p53 was analyzed by Western blots 2 d after transfection. (E) p53 protein level was quantified from the Western blot bands in C and D, normalized to GAPDH level, and presented as fold change ± SEM (n ≥ 3) relative to the p53 level of p53-only-transfected cells (solid red line). The dashed line represents the threshold of suppression (0.75-fold) corresponding to threshold set in the luciferase reporter assay. Two-tail t-test results are indicated by (*) P < 0.05 and (**) P < 0.01, relative to the p53-only-transfected controls.
We validated the binding of miR-125b to the 3′ UTR of human and zebrafish p53 using a luciferase reporter assay (Fig. 1A,B). Ectopic expression of miR-125b by transfection of miR-125b duplex into HEK-293T cells suppresses by ∼60% (P < 0.01) the activity of a Renilla luciferase construct containing the miR-125b MREs of human or zebrafish p53 at its 3′ end (Fig. 1B). Similarly, the activity of a luciferase construct containing the entire 3′ UTR of human or zebrafish p53 was suppressed ∼40%–50% (P < 0.01) by ectopic miR-125b (Fig. 1B). Suppression of luciferase activity was abolished when the miR-125b-MREs were deleted from the p53 3′ UTR, and when a 3-base mismatch mutation was introduced into the seed region (Fig. 1B). These data indicate that the predicted MREs are critical for the direct and specific binding of miR-125b to the p53 mRNA.
To confirm the binding of miR-125b to human and zebrafish p53 in vitro, we ectopically expressed the full-length human/zebrafish p53 cDNA in p53-null H1299 cells. Consistent with the luciferase reporter assays, overexpression of miR-125b in H1299 cells significantly repressed both human and zebrafish wild-type p53 protein (30%–40%, P < 0.05) but not when the MREs were deleted from the 3′ UTRs of p53 mRNAs (Fig. 1C–E).
In addition, we validated the specificity and the efficacy of the miR-125b duplex (125b-DP) and the miR-125b antisense oligonucleotide (125b-AS) that were used in our assays. We transfected into HEK-293T cells a luciferase reporter containing a perfect complementary MRE of miR-125b to provide a site with high binding affinity for the miRNA. Indeed, cotransfection of the cells with 125b-DP suppresses the luciferase activity by ∼95%, while no suppression was observed with any of the control miRNA duplexes (negative control duplex, NC-DP1/2/3) (Supplemental Fig. 1B). The luciferase activity suppression by 125b-DP was abrogated when 125b-AS was cotransfected; this effect was not observed with the same concentration of a negative control miRNA antisense oligonucleotide (Supplemental Fig. 1B). Therefore, 125b-DP and 125b-AS are effective and specific for modulating the level of miR-125b. The transfection efficiency and the effect of these oligonucleotides on miR-125b levels were also verified by quantitative RT–PCR (qRT–PCR) in each cell type. For example, in H1299 cells, we found that the transfection of 125b-DP increased the level of miR-125b by ∼25-fold in comparison with the endogenous miR-125b level (Supplemental Fig. 1C).
miR-125b represses endogenous p53 and p53-induced apoptosis in human neuroblastoma cells
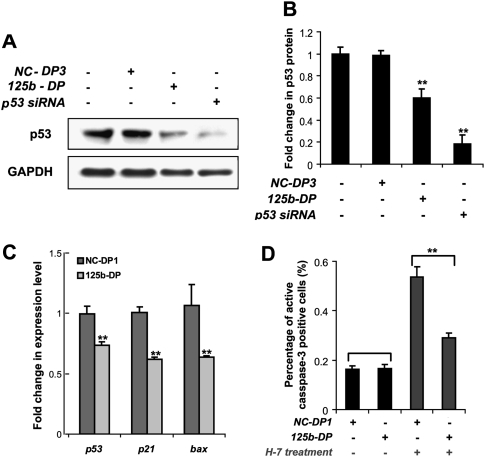
To investigate the regulation of endogenous human p53 by miR-125b, we used the neuroblastoma cell line SH-SY5Y, which is known to express wild-type p53 (Vogan et al. 1993). The endogenous level of miR-125b in undifferentiated SH-SY5Y is relatively low, and transfection of miR-125b duplex brought miR-125b level up by ∼27-fold (Supplemental Fig. 2A). Ectopic expression of miR-125b reduced the level of p53 protein in SH-SY5Y cells by ∼40% (P < 0.01) (Fig. 2A,B). The level of p53 mRNA was also reduced by 125b-DP transfection although the fold change was smaller than that of p53 protein (Fig. 2C). The expression of p21 and bax, the two main targets of p53, also dropped significantly after 125b-DP transfection (Fig. 2C).
Figure 2.
miR-125b represses the endogenous p53 expression and suppresses p53-induced apoptosis in human neuroblastoma SH-SY5Y cells. (A) The endogenous p53 protein level in SH-SY5Y cells 2 d after a transfection with mock (water), negative control duplex 3 (NC-DP3), miR-125b duplex (125b-DP), or p53 siRNA. (B) The p53 protein level was quantified from the Western blot bands in A, normalized to the GAPDH level, and presented as fold change ± SEM (n ≥ 3) relative to the p53 level of mock-transfected cells. (C) The mRNA expression levels of p53, p21, and bax in SH-SY5Y cells 2 d after transfection with NC-DP1 or 125b-DP. The expression was quantified by real-time PCR, normalized to the expression of β-actin, and presented as fold change ± SEM (n ≥ 4) relative to that in the cells transfected with NC-DP1. (D) The percentage of SH-SY5Y cells positive for active caspase-3 was quantified by the Cellomics high-content screening system 2 d after a transfection with NC-DP1 or with 125b-DP. The 10 μM H-7 treatment was applied 24 h before fixing. The values represent average ± SEM (n ≥ 3). For each replicate, 20 images (including at least 10,000 cells) were analyzed. In all panels, two-tail t-test results are indicated by (*) P < 0.05 and (**) P < 0.01, relative to the mock-transfected or NC-DP-transfected controls.
Induction of p53 often leads to apoptosis (Almog and Rotter 1997). However, in neuroblastoma cells, p53 protein is mainly localized to the cytoplasm, so the endogenous activity of nuclear p53 is usually insufficient to modulate apoptosis (Moll et al. 1996). Thus, we predicted that ectopic expression of miR-125b in SH-SY5Y cells will only suppress apoptosis when the p53 pathway is fully activated by an exposure to the drug 1-(5-isoquinolinyl sulfonyl)-2-methyl piperazine (H-7). Exposure to H-7 leads to an increased import of p53 into the nucleus, where p53 becomes active and induces apoptosis (Ronca et al. 1997). Indeed, ectopic expression of miR-125b significantly suppressed H-7-induced apoptosis, but did not affect apoptosis in the untreated SH-SY5Y cells, as quantified by the staining of active-caspase-3 (Fig. 2D).
miR-125b represses endogenous p53 and apoptosis in primary human lung fibroblasts
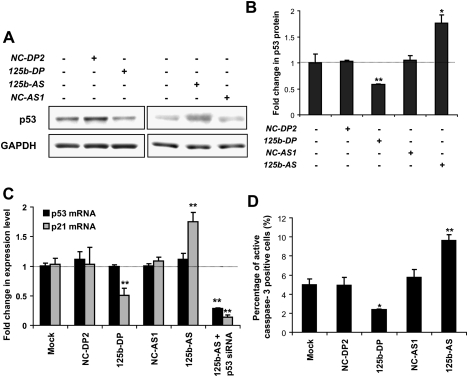
To further demonstrate the repression of human p53 by miR-125b in a physiological context, we examined this regulation in primary human lung fibroblasts that were cultured from normal fetal lungs to homogeneity. The level of miR-125b expression in human lung fibroblasts is relatively high. We were able to knock down the endogenous miR-125b by ∼24-fold with 125b-AS or overexpress miR-125b by ∼26-fold with 125b-DP (Supplemental Fig. 2B). Consistently, overexpression of miR-125b repressed p53 protein levels, while knockdown of miR-125b elevated p53 levels significantly (Fig. 3A,B). The expression of p21 mRNA, a main target of p53, in human lung fibroblasts was also modulated by miR-125b in the same fashion as p53 protein (Fig. 3C). Here, the effect of miR-125b on p21 mRNA level was solely dependent on p53 expression since knockdown of p53 by a siRNA was able to rescue the increase in p21 expression caused by the 125b-AS (Fig. 3C). In addition, 125b-DP represses p21 expression in a dose-dependent manner, with significant suppression still observable at a concentration as low as 10 nM of 125b-DP (Supplemental Fig. 3A). The level of p53 mRNA in human lung fibroblasts, however, was not affected by the changes in miR-125b expression (Fig. 3C). This suggests that miR-125b inhibits the translation of p53 but does not modulate the stability of p53 mRNA in these cells. In addition, miR-125b knockdown led to a substantial increase in apoptotic cells, as quantified by active-caspase-3 staining, while miR-125b overexpression had the opposed effect (Fig. 3D). These data demonstrate that miR-125b expression is both necessary and sufficient for maintaining the physiological levels and the activity of p53 in human lung fibroblasts.
Figure 3.
miR-125b represses the endogenous p53 expression and suppresses apoptosis in human lung fibroblast cells. (A) The endogenous p53 level in human lung fibroblast cells 2 d after transfection with mock (water), negative control duplex 2 (NC-DP2), or miR-125b duplex (125b-DP); and 1 d after transfection with mock, negative control antisense 1 (NC-AS1), or miR-125b antisense (125b-AS). (B) The p53 protein level was quantified from the Western blot bands in A, normalized to the GAPDH level and presented as fold change ± SEM (n ≥ 3) relative to the p53 level of mock-transfected cells (dotted line). (C) The levels of p53 mRNA and p21 mRNA in human lung fibroblast cells 2 d after transfection with mock, NC-DP2, 125b-DP, or NC-AS1,125b-AS, or a cotransfection of 125b-AS and p53 siRNA. The expression was quantified by real-time PCR, normalized to the expression of β-actin and presented as fold change ± SEM (n ≥ 4) relative to that in the mock-transfected cells (dotted line). (D) The percentage of human lung fibroblast cells positive for active caspase-3, 2 d after transfection with mock, NC-DP2, 125b-DP, NC-AS1, or 125b-AS was quantified by the Cellomics high-content screening system. The values represent average ± SEM (n ≥ 3). For each replicate, 20 images (including at least 10,000 cells) were analyzed. In all panels, two-tail t-test results are indicated by (*) P < 0.05 and (**) P < 0.01, relative to the mock-transfected controls.
Spatio-temporal expression of miR-125b during zebrafish embryogenesis
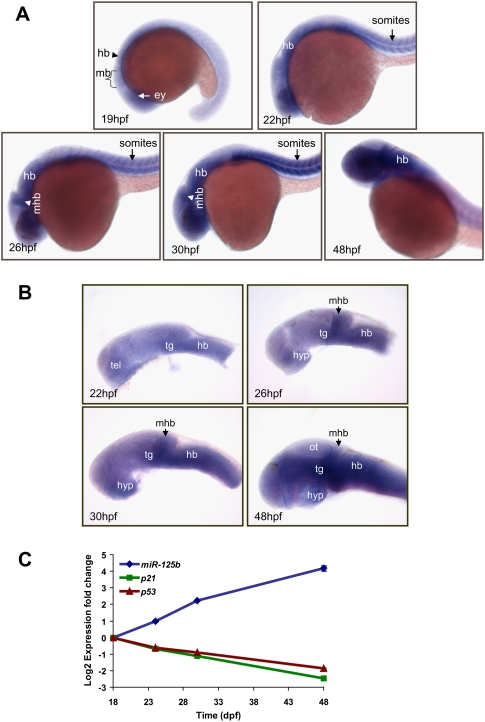
To examine if miR-125b expression is inversely correlated to p53 expression spatio-temporally during development, we analyzed miR-125b expression at different stages of zebrafish embryogenesis. Expression of miR-125b was first detected at 19 h post-fertilization (hpf) by whole mount in situ hybridization (Fig. 4A). miR-125b was present in the whole embryo with enrichment in the brain, the eyes, and the somites at different stages (Fig. 4A,B). The expression pattern in the brain is consistent with previously published data (Wienholds et al. 2005). However, no enriched expression was detected in the spinal cord. Instead, we found a pronounced miR-125b expression in the somites between 22 and 30 hpf (Fig. 4A). By 22 hpf, miR-125b expression was enriched in the eyes, the somites, the telencephalon, and the midbrain with stronger expression in the tegmentum and hindbrain (Fig. 4A,B). Between 26 and 30 hpf, miR-125b was strongly expressed in the hypothalamus, the tegmentum, the midbrain–hindbrain boundary, and the hindbrain (Fig. 4B). miR-125b expression continues to increase in the brain such that the optic tectum became the only region with weak miR-125b expression by 48 hpf (Fig. 4A,B).
Figure 4.
Spatio-temporal expression of miR-125b during zebrafish embryogenesis. (A) Whole-mount in situ hybridization of miR-125b in zebrafish embryos at 19 hpf, 22 hpf, 26 hpf, 30 hpf, and 48 hpf. Side view of the whole body excluding the tail is shown. (B) Side view of zebrafish brain, in situ hybridization with miR-125b at 22 hpf, 26 hpf, 30 hpf, and 48 hpf. In A and B, each image shows the expression pattern of miR-125b in a representative embryo. The same pattern was observed in all 20 embryos examined at each developmental stage. (ey) Eye; (hb) hindbrain; (hyp) hypothalamus; (mhb) midbrain–hindbrain boundary; (ot) optic tectum; (tel) telencephalon; (tg) tegmentum. (C) The expression pattern of miR-125b, p53, and p21 during zebrafish development: Transcript levels were quantified by real-time PCR, normalized to internal controls (18S or β-actin), and presented as log2 fold change ± SEM (n ≥ 4) relative to the expression at 18 hpf.
Analysis of miR-125b expression in whole embryo lysate by qRT–PCR showed that the expression initiated at 18 hpf and increased exponentially from 18 to 48 hpf (Fig. 4C). Interestingly, p53 and p21 expression was inversely correlated with miR-125b up-regulation over time (Fig. 4C). We also compared the spatio-temporal expression pattern of miR-125b (our in situ hybridization analysis) with the expression pattern of p53 mRNA (Yamaguchi et al. 2008). p53 and miR-125b were observed to be coexpressed in the brain and the eyes at ∼24 hpf. In the brain, miR-125b expression increases steadily from 24 to 48 hpf, while p53 expression decreases gradually during that same period. In the somites, miR-125b expression is enriched from 22 to 30 hpf, while p53 expression is not observed. Western blots also showed that p53 protein can be detected at 18 hpf and decreases to undetectable levels by 48 hpf (data not shown). The inverse correlation between miR-125b and p53 expression/activity supports our hypothesis that p53 is down-regulated by miR-125b during zebrafish embryogenesis.
Loss of miR-125b leads to severe defects in zebrafish embryos
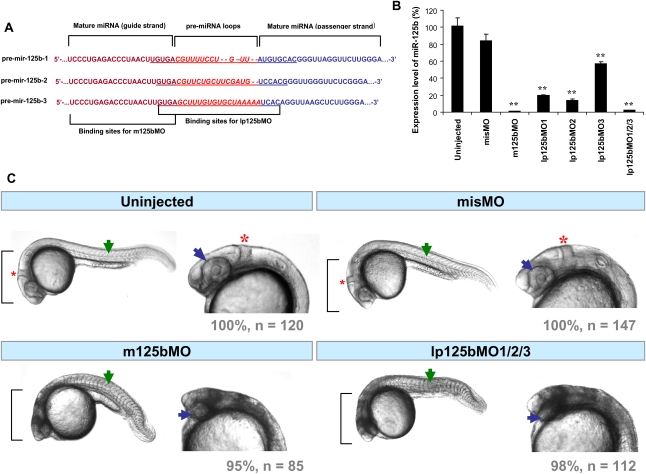
To probe for the function of miR-125b in zebrafish, we synthesized four different morpholinos against miR-125b (Fig. 5A): one (m125bMO) targeting the mature guide strand, and three (lp125bMOs) targeting the precursors. In zebrafish, mature miR-125b is derived from three different precursor isoforms with sequence differences in the loop region (Fig. 5A); lp125bMOs were designed to bind to each of these loops. According to Kloosterman et al. (2007), binding of the morpholinos to the loop regions of miRNA precursors is able to block processing of the miRNAs, hence down-regulating the mature miRNA level. Near-complete knockdown of mature miR-125b was observed with m125bMO and also with a combination of the three lp125bMOs (Fig. 5B). Individual lp125bMOs also suppressed the expression of miR-125b, albeit incompletely (Fig. 5B). As a control, injection of a morpholino (misMO) with five mismatches different from m125bMO did not cause in any significant change in miR-125b expression (Fig. 5B).
Figure 5.
Loss of miR-125b in zebrafish embryos. (A) Design of morpholinos targeting either the guide strand of mature miR-125b (m125b) or the loop regions of pre-mir-125b (lp125b). Three different lp125b morpholinos (lp125bMO1/2/3) were designed for the three isoforms of pre-mir-125b. (B) qRT–PCR elucidating the effects of miR-125b morpholinos on the endogenous level of zebrafish miR-125b at 24 hpf. One-cell-stage embryos were injected with m125bMO or lp125bMO1/2/3 (individually or together). A morpholino (misMO) with 5 nt different from m125bMO was used as control. Total RNA was obtained from the embryos at 24 hpf. All the expression values were normalized to 18S RNA levels and presented as average percentage ± SEM (n ≥ 4) relative to the expression values in uninjected controls. Two-tail t-test results are indicated by (**) P < 0.01, relative to the uninjected control. (C) Loss-of-function morphology at 24 hpf: Morphants typically exhibit severe cell death in the brain (brackets), absence of the midbrain–hindbrain boundary (*), smaller eyes (blue arrows), and deformed somites (green arrows). Each control/morphant embryo is shown with a lateral view of the whole body and a magnified view of the head. The total number of embryos (n) in each treatment and the percentage of embryos having the same phenotype as in the representative picture are indicated below each image.
Severe developmental defects were observed in the miR-125b morphants, where the most apparent phenotype was the accumulation of dead cells in the brain (Fig. 5C). This phenotype was observed by 24 hpf in almost all embryos microinjected with m125bMO or with lp125bMO1/2/3 (Fig. 5C; Table 1). Other morphological defects upon miR-125b knockdown include smaller eyes, a missing midbrain–hindbrain boundary, and deformities in the somites. These data are consistent with the expression pattern of miR-125b and demonstrate its importance in zebrafish development.
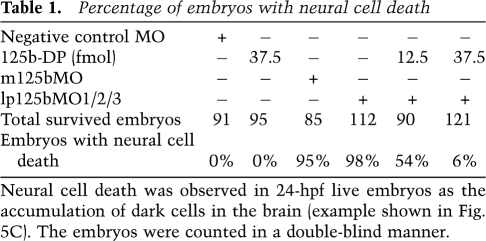
Table 1.
Percentage of embryos with neural cell death
Neural cell death was observed in 24-hpf live embryos as the accumulation of dark cells in the brain (example shown in Fig. 5C). The embryos were counted in a double-blind manner.
Loss of miR-125b increases p53 and p53-dependent apoptosis in zebrafish
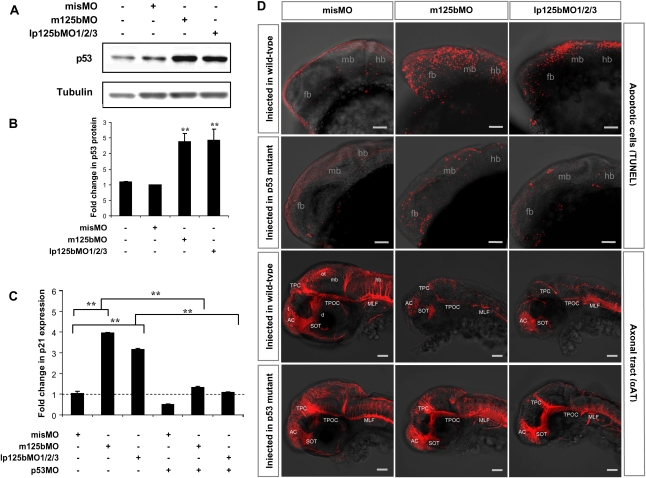
As a result of miR-125b knockdown by injection of m125bMO or lp125bMO1/2/3 into one-cell-stage embryos, the endogenous level of p53 protein was elevated in zebrafish embryos at 24 hpf (Fig. 6A,B). p21 was also up-regulated in both types of morphants (Fig. 6C). When the morphants were coinjected with a morpholino blocking translation of p53, p21 expression was restored to wild-type levels, indicating that the up-regulation of p21 by miR-125b required p53 (Fig. 6C).
Figure 6.
Loss of miR-125b elevates p53 and triggers p53-dependent apoptosis in zebrafish embryos. (A) Elevation of p53 protein caused by loss of miR-125b in zebrafish embryos: Embryos were injected with misMO, m125bMO, or lp125bMO1/2/3. Western blotting was performed at 24 hpf. (B) The p53 protein level was quantified from the Western blot bands in A, normalized to tubulin level, and presented as fold change ± SEM (n ≥ 3) relative to the p53 level in the misMO-injected embryos. Two-tail t-test results are indicated by (**) P < 0.01, relative to the misMO-injected control. (C) qRT–PCR of p21 transcripts at 24 hpf in embryos injected with different combinations of morpholinos. p53MO indicates a morpholino blocking translation of p53. The values were normalized to the expression level of β-actin and represented as average fold change ± SEM (n ≥ 4) relative to the expression level in misMO-injected embryos (dashed line). Two-tail t-test results are indicated as (**) P < 0.01. (D) TUNEL assay for detecting apoptotic cells (visualized as red spots) in the 24-hpf brains and acetylated tubulin staining (αAT) marking mature neurons and axonal tracts in the 48-hpf brains of wild-type and p53M214K mutant embryos microinjected with misMO, m125bMO, or lp125bMO1/2/3. Each image is a projection of multiple optical slides obtained from a representative embryo. Three embryos were observed for each condition for the TUNEL assay, and five were observed for each condition in the αAT staining. All of them had a similar phenotype as the representative images. (AC) Anterior commissure; (d) diencephalon; (fb) forebrain; (hb) hindbrain; (mb) midbrain; (MLF) medial longitudinal fasciculus; (ot) optic tectum; (SOT) supraoptic tract; (TPC) tract of posterior commissure; (TPOC) tract of postoptic commissure; (t) telencephalon. Bar, 50 μm.
At 24 hpf, an increase in terminal dUTP nick end labeling (TUNEL)-positive apoptotic cells was observed in the midbrain and hindbrain domains of both m125bMO- and lp125bMO1/2/3-injected embryos (Fig. 6D). Enhanced apoptosis was observed in the m125bMO morphants only from 18 hpf, consistent with the stage when miR-125b expression was first detected (Supplemental Fig. 5). Apoptosis reached a peak at 24 hpf, when the brain defects were the most severe (Supplemental Fig. 5). Apoptosis decreased gradually by 30 hpf (Supplemental Fig. 5), but the hatched larvae were still defective, with distorted heads and abnormal behaviors.
We then asked whether the cell death phenotype in miR-125b morphants was caused by the elevation in p53 protein. To ablate p53 function, we used the zebrafish p53M214K mutant, which is defective in p53 activity but still undergoes normal embryogenesis (Berghmans et al. 2005). Remarkably, knockdown of miR-125b, whether by m125bMO or by lp125bMO1/2/3, had no observable effects on brain apoptosis (Fig. 6D). Similar effect was observed with the coinjection of p53 MO and miR-125b MOs (Supplemental Fig. 6). Defects in the midbrain–hindbrain boundary and the somites of miR-125b morphants were also rescued in p53M214K mutants or by coinjection of p53 MO (Supplemental Fig. 6). Additionally, miR-125b morphants exhibited severe defects in axonal pathfinding, as observed by anti-acetylated tubulin immunostaining (Fig. 6D). Most major primary axonal tracts were markedly reduced in the miR-125b morphants, but they were rescued substantially by the loss of p53-mediated apoptosis in the p53M214K mutant (Fig. 6D). Taken together, these data demonstrate that the excessive p53 activity in miR-125b morphants is responsible for the abnormal increase in apoptosis and most of the observed morphological defects. Therefore, the p53 pathway is likely to be the major target that mediates the function of miR-125b during the early development of zebrafish.
Synthetic miR-125b duplex rescues apoptosis in miR-125b morphants by restoring the normal level of p53
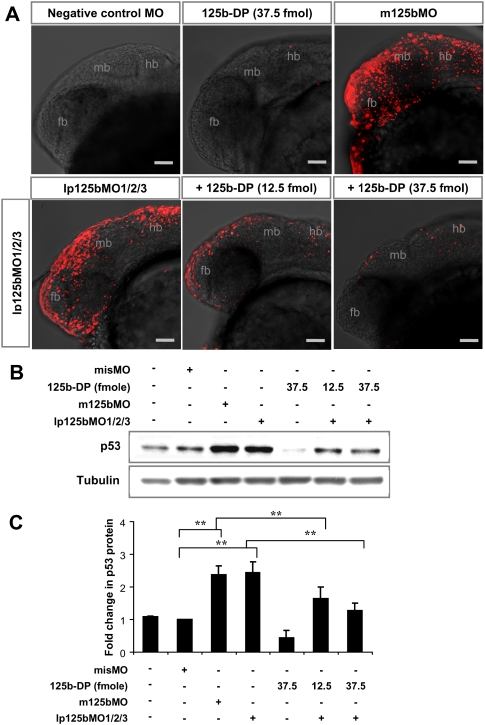
In order to address the specificity of miR-125b morpholinos, we attempted to rescue the morphant phenotype with coinjection of synthetic miR-125b duplex into lp125bMO1/2/3 morphants. Indeed, the number of apoptotic cells in the morphants was reduced significantly by miR-125b duplex in a dose-dependent manner (Fig. 7A). The efficiency of rescue was quantified by the number of embryos with visible dead cells in the live brain at 24 hpf. Both m125bMO and lp125bMO1/2/3 injection caused more than 90% of embryos to show neural cell death, whereas coinjection of the miR-125b duplex reduced this number to only 6% (Table 1). This rescue of apoptosis corresponds to the reduction in p53 protein by miR-125b duplex (Fig. 7B,C). Importantly, since this duplex does not bind to the lp125bMOs, rescue of the lp125bMO1/2/3 morphants demonstrates that synthetic miR-125b duplex produced mature miR-125b that replenished the endogenous miR-125b to repress p53 and down-regulate apoptosis.
Figure 7.
Synthetic miR-125b rescues apoptosis in miR-125b morphants. (A) TUNEL assay for detecting apoptotic cells in the 24-hpf brains: Embryos were injected with a standard negative control morpholino, miR-125b duplex (125b-DP), m125bMO, or lp125bMO1/2/3. Two different concentrations of 125b-DP (12.5 fmol and 37.5 fmol per injection) were used to rescue the embryos injected with lp125bMO1/2/3. Each image is a projection of multiple optical slides from a representative embryo. Three embryos were observed for each condition, and all of them had a similar phenotype as the representative images. (fb) Forebrain; (hb) hindbrain; (mb) midbrain. Bar, 50 μm. (B) Regulation of p53 protein in the morphants and the rescued embryos. Western blotting was performed at 24 hpf. (C) p53 protein level was quantified from the Western blot bands in B, normalized to tubulin level, and presented as fold change ± SEM (n ≥ 3) relative to the p53 level in the embryos injected with misMO. Two-tail t-test results are indicated by (**) P < 0.01.
Stress-induced p53 and apoptosis are repressed by ectopic miR-125b
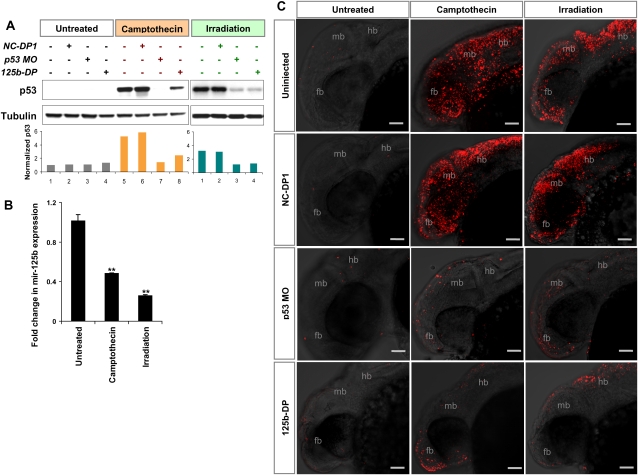
To further elucidate the role of miR-125b in zebrafish development, we examined the ability of miR-125b to suppress p53 during the stress response in zebrafish embryos. p53 can be induced quickly by agents that cause DNA damage, leading to cell cycle arrest and apoptosis (Kuerbitz et al. 1992; Langheinrich et al. 2002). To induce DNA damage, we irradiated zebrafish embryos with 40 Gy of γ-rays or treated them with 500 nM camptothecin for 8 h. As expected, the p53 protein increases dramatically after both treatments in wild-type embryos (Fig. 8A). Interestingly, both treatments resulted in a significant drop in miR-125b expression (Fig. 8B), suggesting that the down-regulation of miR-125b allows a smooth up-regulation of p53 in this stress response pathway.
Figure 8.
Overexpression of miR-125b rescues stress-induced apoptosis. (A) Regulation of p53 protein in zebrafish embryos injected with negative control duplex 1 (NC-DP1), p53 morpholino (p53 MO), or miR-125b duplex (125b-DP). At 24 hpf, uninjected and injected embryos were treated with 500 nM camptothecin for 8 h or subjected to 40 Gy of γ-irradiation. Protein lysate from the two treatments with two sets of untreated control were loaded on two separate gels. The bar chart presents quantification of p53 Western blot band intensity, normalized to tubulin levels, and presented as fold change relative to the uninjected untreated control of each blot. (B) Regulation of miR-125b in uninjected embryos or those treated with 500 nM camptothecin for 8 h or subjected to 40 Gy of γ-irradiation, normalized to 18S RNA level, and presented as average fold change relative to untreated control ± SEM (n ≥ 6). Two-tail t-test results are indicated as (**) P < 0.01, relative to the untreated control. (C) Staining of apoptotic cells in embryos uninjected or injected with NC-DP1, p53 MO, or 125b-DP, treated with 500 nM camptothecin for 8 h or with 40 Gy of γ-irradiation. Embryos were fixed at 32 hpf and subjected to TUNEL assay. Each image is a projection of multiple optical slides from a representative embryo. Three embryos were observed for each condition, and all of them had a similar phenotype as in the representative image. (fb) Forebrain; (hb) hindbrain; (mb) midbrain. Bar, 50 μm.
To test whether an ectopic expression of miR-125b can reduce the extent of the DNA damage stress response, we exposed miR-125b-duplex-injected embryos to γ-irradiation or camptothecin treatment. As anticipated, the level of p53 protein in the treated embryos was reduced significantly by miR-125b duplex (Fig. 8A). Staining of apoptotic cells in the embryonic brain further demonstrated that the severe apoptosis induced by γ-irradiation or camptothecin was rescued significantly by the injection of miR-125b duplex (Fig. 8C). In fact, the rescue effects of miR-125b duplex during the DNA damage response were nearly as great as the effects of p53 knockdown via a morpholino (Fig. 8C). A negative control duplex had no effect (Fig. 8C).
Discussion
The function of miR-125b in regulating p53 and p53-dependent-apoptosis
Our data establish miR-125b as a bona fide negative regulator of p53. We validated this interaction in two vertebrate species, humans and zebrafish, suggesting that this interaction is an essential negative regulatory element of the p53 pathway. The direct interaction between miR-125b and p53 mRNA was elucidated by several lines of evidence: (1) The 3′ UTR of both human and zebrafish mRNAs contain a putative binding site (the MRE) for miR-125b with significant seed match. (2) miR-125b suppresses the activity of a luciferase reporter fused with the 3′ UTR of human/zebrafish p53 mRNA in a MRE-dependent manner. (3) miR-125b represses the ectopic expression of human/zebrafish wild-type p53 cDNA but not the MRE-deleted p53 cDNA in a p53-null background (H1299 cells). Our report is the first to identify a miRNA that directly regulates p53.
Moreover, we showed that the negative regulation of p53 by miR-125b is physiologically relevant to cell function and embryonic development. Ectopic expression of miR-125b is able to repress p53 protein modestly but significantly in human fetal lung fibroblasts and human neuroblastoma SH-SY5Y cells. An increase of miR-125b also represses apoptosis, whereas knockdown of miR-125b increases the level of p53 protein and apoptosis in both in human cells and during zebrafish embryogenesis. In particular, zebrafish embryos displayed a specific neural cell death phenotype upon miR-125b knockdown. These data are consistent with a prior report showing that Drosophila mutants lacking functional miR-125b and let-7 exhibit increased cell death during pupal development (Caygill and Johnston 2008). Therefore, miR-125b may well be a conserved survival factor that directly or indirectly keeps the level of p53 low during development to support normal tissue growth.
A reduction in miR-125b level is also important for the in vivo p53-dependent stress response. We demonstrated that miR-125b was down-regulated in zebrafish embryos just 8 h after γ-irradiation or camptothecin treatment, while p53 protein level was elevated dramatically. Furthermore, the DNA damage stress-induced p53 and apoptosis in zebrafish embryos was repressed by miR-125b ectopic expression. Similarly, we also observed a down-regulation in miR-125b in human neuroblastoma SH-SY5Y cells after a 24-h treatment with etoposide, a topoisomerase inhibitor known to induce DNA damage and activate p53 (Supplemental Fig. 3B,C). Our data suggest that the down-regulation of miR-125b in response to the stress of DNA damage is conserved in both zebrafish and humans. It would be interesting to further characterize the mechanism of miR-125b down-regulation in response to DNA damage.
Specificity versus off-target effects
We demonstrated that miR-125b acts as a direct negative regulator of p53 in human and zebrafish. The induction of p53 and the increase in apoptosis is an expected phenotype of miR-125b knockdown. However, the dramatic effects of the miR-125b morpholinos in zebrafish raised concerns of a possible off-target effect.
Morpholinos are a useful tool for loss-of-function studies in zebrafish; however, their frequent off-target effect is a main concern for functional analysis (Ekker and Larson 2001). Particularly, increase in neural cell death by 24 hpf has been considered a nonspecific effect of 15%–20% of the morpholinos used in zebrafish (Ekker and Larson 2001). Recently, Robu et al. (2007) demonstrated that this effect is mediated through the p53 pathway since the increase in apoptosis is associated with p53 activation and can be completely reversed by coinjection of p53 morpholino. The specific mechanism by which p53 is activated by mistargeting of morpholinos was not explained (Robu et al. 2007). The question is whether activation of p53 and increase in neural cell death is always an off-target effect. There are many examples showing that the overexpression of p53 or knockdown of p53’s negative regulators can lead to the same phenotype in zebrafish (Langheinrich et al. 2002; Campbell et al. 2006; Ghiselli 2006; Bretaud et al. 2007). Robu et al. (2007) suggested that even if the morpholino targets a known regulator of p53, it can also have some off-target effect, as in the case of a Mdm2 morpholino. They believe that a morphant phenotype can be considered as on-target only if it can be rescued by overexpression of the targeted gene (Robu et al. 2007). This approach has been used previously to identify new regulators of p53 in zebrafish (Ghiselli 2006).
In order to address the specificity of miR-125b morpholinos, we followed two approaches. First, two different sets of morpholinos were used to knock down miR-125b: the m125bMO (targets the mature miR-125b) and the three lp125bMOs (target the loop region of the three pre-mir-125b isoforms). The sequence of m125bMO overlaps with the lp125bMOs by only 3–4 nucleotides (nt) (Fig. 5A). Thus, the probability of all of these morpholinos having the same off-target effect is very low (2%–4%). Second, and importantly, we were able to rescue miR-125b morphants specifically with synthetic miR-125b duplex, as suggested by Robu et al. (2007).
In the first approach, our data showed that knockdown of miR-125b by either m125bMO or lp125bMOs resulted in the same phenotype: up-regulation of p53 protein and increase in neural cell death at 24 hpf (Figs. 5C, 6A–D). In fact, the severity of the phenotype was dependent on the dose and the efficacy of the morpholinos. For m125bMO or for the combination of lp125bMO1/2/3, injection of 0.25 pmol of morpholino had no effect; 0.5 pmol of morpholino induced a mild neural cell death in ∼80% of injected embryos; and 0.75 pmol of morpholino produced severe neural cell death in ∼95% of the embryos. m125bMO reduced the level of mature miR-125b to almost zero (Fig. 5B), and the apoptotic phenotype was the most severe in embryos injected with this morpholino. Injection of all three lp125bMOs resulted in a comparable effect to that of m125bMO because this coinjection would inhibit the processing of all pre-mir-125b isoforms. Injection of each lp125bMO individually resulted in an incomplete knockdown of miR-125b (Fig. 5B) and led to a mild neural cell death. In particular, the level of mature miR-125b was reduced more by lp125bMO1 and lp125bMO2 than by lp125bMO3, probably due to the lower expression of pre-mir-125b-3 in the embryos. As a consequence, injection of lp125bMO1 or lp125bMO2 resulted in more cell death than that of lp125bMO3. In addition, to test whether sequence specificity was important for knockdown of miR-125b, we designed a control morpholino that had the same length and GC content as the m125bMO but contained five mismatches. This mismatched morpholino (misMO) was not able to knock down miR-125b (Fig. 5B). Moreover, embryos injected with misMO exhibited no difference in morphology from those of uninjected controls (Fig. 5C).
The specificity of knockdown was further demonstrated by the second approach: Overexpression of miR-125b by injection of synthetic miR-125b duplex rescued the effect of lp125bMO1/2/3 in a dose-dependent manner. Specifically, injection of 37.5 fmol of the miR-125b duplex reduced the level of p53 protein and the number of apoptotic cells significantly (Fig. 7A–C). With this dose, the percentage of embryos with neural cell death dropped from 98% in the lp125bMO1/2/3 morphant population, to 6% in the rescued embryos in which lp125bMO1/2/3 was coinjected with the miR-125b duplex (Table 1). Because lp125bMO1/2/3 can only bind to the loop regions of mir-125b precursors and block processing of endogenous miRNA precursors, it cannot interact with the synthetic miR-125b duplex. Moreover, lp125bMO1/2/3 was injected at a 20- to 30-fold (750:37.5 to 750:12.5) higher dose than the miR-125b duplex; hence, the rescue could not be due to a titration of the morpholino. Instead, this result implies that the mature miR-125b processed from the injected synthetic duplex was able to replenish endogenous miR-125b, and thus repress p53 and down-regulate apoptosis in the embryos.
These two lines of evidence strongly suggest that the miR-125b morpholinos used in our experiments were on-target. Additionally, the phenotype of the miR-125b morphants is also explained by the temporal and spatial expression pattern of miR-125b. Defects in the miR-125b morphants appear precisely in the regions where miR-125b is normally strongly expressed; e.g., the brain, the eyes, and the somites (cf. Figs. 5C and 4A). The onset of increased apoptosis in the morphants is also correlated to the stage (18 hpf) when miR-125b begins to express in the embryos (cf. Supplemental Fig. 5 and Fig. 4C).
Moreover, the expression of miR-125b was up-regulated during development, while the activity of p53 was decreasing (Fig. 4C). In contrast, miR-125b was down-regulated in response to DNA damage, while p53 was up-regulated under the same condition (Fig. 8). The inverse correlation between miR-125b expression and p53 strengthens our conclusion that miR-125b is a physiological regulator of p53 in zebrafish.
Conservation of miR-125b targets in the p53 network
Besides p53, miR-125b may also target other components of the p53 network. Sinha et al. (2008) suggested that miR-125b targets seven genes that, with the exception of bak1, are upstream regulators of p53. The mRNAs of these genes all contain putative binding sites for miR-125b in their 3′ UTRs. One of these targets, bak1 mRNA, has been shown to bind to miR-125b in human prostate cancer cell lines (Shi et al. 2007). We compared the putative binding sites of the seven targets across a number of vertebrates and found that each site is broadly (although not strictly) conserved among vertebrates (Supplemental Table 3). Each species has a binding site for miR-125b in the sequence of at least one of the seven targets. Supporting these findings, we demonstrated that these seven genes in the p53 network were indeed down-regulated by miR-125b ectopic expression in the human neuroblastoma SH-SY5Y cells and/or in mouse fibroblast Swiss-3T3 cells (Supplemental Fig. 4C). Hence, these genes are likely to be targets of endogenous miR-125b in both human and mouse. Furthermore, miR-125b ectopic expression also down-regulated p53 mRNA as well as the p53 targets, p21 and bax mRNAs in mouse fibroblast Swiss-3T3 cells. Since the binding site for miR-125b in the 3′ UTR of p53 mRNA is not conserved in mouse, the down-regulation of p53 expression by miR-125b may be mediated indirectly by the down-regulation of the genes upstream of p53 in mouse Swiss-3T3 cells. Although further experiments are required to validate the direct targets of miR-125b in the p53 network, our analysis strongly suggests that the p53 network, as a whole, is a broadly conserved target of miR-125b regulation in vertebrates.
Most miRNAs and their mRNA-binding motifs (the seed regions) are strictly conserved across species, but their targets are less well-conserved (Chan et al. 2005; Rajewsky 2006). The loss/gain during evolution of an individual mRNA target may make very little impact on the function of a miRNA with multiple targets (Chen and Rajewsky 2006). This corroborates our finding that miR-125b targets multiple genes in the p53 network, where the redundancy of these targets allows for their relatively neutral loss/gain across various species. While not every target is strictly conserved, the overall regulation of p53 activity by miR-125b may still be conserved during evolution via one or another component of the p53 network.
Other targets and other functions of miR-125b
Genes in the p53 network are most probably not the only targets of miR-125b. We observed several defects in zebrafish embryos after ectopic expression of miR-125b, including a delay in growth, rounding of the body, thickening of yolk extension, and loss of brain ventricles and of the midbrain–hindbrain boundary (data not shown). These phenotypic effects are not observed in p53-deficient zebrafish. In addition, we also found that ectopic expression of miR-125b promotes neuronal differentiation in human neural progenitors and neuroblastoma cells (M.T.N. Le, H. Xie, B. Zhou, P.H. Chia, M. Um, G. Udolph, H. Yang, B. Lim, and H.F. Lodish, in prep.). This phenotype was not recapitulated by knockdown of p53. Additional targets that mediate the function of miR-125b in zebrafish and in human neural cells remain to be identified.
The implication of miR-125b in tumorigenesis
Recent reports suggest that miR-125b acts as a tumor suppressor in several types of cancers but as an oncogene in other types. The expression of miR-125b is down-regulated in ovarian carcinoma and thyroid carcinoma (Nelson et al. 2006; Volinia et al. 2006; Iorio et al. 2007; Nam et al. 2008) but elevated in pancreatic cancer, oligodendroglial tumors, prostate cancer, myelodysplastic syndromes (MDS), and acute myeloid leukemia (AML) (Nelson et al. 2006; Bloomston et al. 2007; Shi et al. 2007; Bousquet et al. 2008). Particularly in MDS and AML patients, a t(2:11)(p21:q23) translocation with the breakpoint mapped near the genomic location of mir-125b-1 locus leads to a sixfold to 90-fold up-regulation of miR-125b (Bousquet et al. 2008). In addition, miR-125b was shown to suppress cell cycling in hepatocellular carcinomas (Li et al. 2008) but to promote proliferation of prostate cancer cells (Shi et al. 2007). Our study supports the notion that miR-125b acts as an oncogene by negatively regulating p53 and suppressing p53-dependent apoptosis. It would be interesting to further examine how p53 activity is affected by the elevated level of miR-125b in prostate cancer and in AML and whether miR-125b-p53 dysregulation represents a new mechanism for cellular transformation in certain types of cancer.
Materials and methods
Cloning and mutagenesis of the luciferase reporters and the p53 expression constructs
The MREs or the whole 3′ UTR of p53 were cloned into the psiCHECK-2 vector (Promega), between the XhoI and NotI sites, immediately 3′ downstream from the Renilla luciferase gene. The top (sense) and bottom (antisense) strands of each MRE were designed to contain XhoI and NotI sites, respectively (Supplemental Table 1). They were synthesized, annealed, and ligated into the psiCheck-2 vector. The 3′ UTR of human p53 was amplified from the total cDNA of SH-SY5Y cells by a nested PCR and inserted into TOPO PCR2.1 (Invitrogen). Subsequently, the UTR was released from the TOPO vector by XhoI and NotI and ligated into the psiCheck2 vector. The 3′ UTR of zebrafish p53 was amplified from total cDNA of 18-hpf zebrafish embryos, digested with XhoI/NotI, and ligated directly into the psiCheck2 vector. The full-length cDNA encoding human and zebrafish p53 was cloned into a pcDNA3.1+ vector, in between the EcoRI and XhoI sites, downstream from the CMV promoter. The human p53 cDNA was PCR-amplified from the total cDNA of SH-SY5Y cells. The zebrafish p53 cDNA was PCR-amplified from the total cDNA of 18hpf zebrafish embryos, using fp53Fe/ fp53Rx primers.
Deletion or mutation of the miR-125b MRE in the p53 construct was performed using the QuickChange site-directed mutagenesis kit (Promega) according to the manufacturer's instructions. The sequences of all primers are provided in Supplemental Table 1.
Luciferase reporter assay
miRNA duplexes including negative control duplex 1 and 2 (NC-DP1/2, negative control PremiR #1 and #2), negative control duplex 3 (NC-DP3, miR-7 PremiR), and miR-125b duplex (125b-DP, miR-125b PremiR); miRNA antisense oligonucleotide including negative control antisense 1 (NC-AS1, negative control AntimiR 1) and miR-125b antisense (125b-AS, miR-125b AntimiR) were purchased from Ambion and dissolved in water. miR-7 duplex was used as a negative control in our experiments because no seed match of miR-7 of can be found in the 3′ UTR of human and zebrafish p53 mRNAs. Ten nanograms of each psiCHECK-2 construct were cotransfected with 10 nM miRNA duplexes or 100 nM miRNA antisenses into HEK-293T cells in a 96-well plate using lipofectamin-2000 (Invitrogen). After 48 h, the cell extract was obtained; firefly and Renilla luciferase activities were measured with the Dual-Luciferase reporter system (Promega) according to the manufacturer's instructions.
Cell culture, transfection, and drug treatments
Human HEK-293T cells, human neuroblastoma SH-SY5Y cells, p53-null human lung carcinoma H1299 cells, mouse Swiss-3T3 cells, and human lung fibroblast cells were maintained in DMEM or RPMI media, supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Invitrogen). H1299 cells, SH-SY5Y cells, Swiss-3T3 cells, and human lung fibroblast cells were transfected in suspension with 4 × 105 cells per well in six-well plates using lipofectamin-2000 (Invitrogen). Plasmids (human/zebrafish wild-type or mutant p53 constructs) were transfected into H1299 cells at a final concentration of 0.5 μg/mL. miRNA duplexes and antisense oligonucleotides were transfected at a final concentration of 80 nM and 100 nM, respectively (unless otherwise stated). p53 siRNA (Dharmacon) was transfected at 60 nM final concentration.
H-7 and etoposide (Sigma) were dissolved in water and dimethyl sulfoxide (DMSO), respectively. SH-SY5Y cells (untransfected or 24 h after transfection with miRNA duplexes) were treated with 10 μM H-7 or 10 μM etoposide for 24 h. Control cells were treated with water or DMSO, respectively.
Active caspase-3 assay
Two days after transfection, SH-SY5Y cells and human lung fibroblast cells were fixed with 4% paraformaldehyde and treated with cold methanol for 10 min at −20°C. After 1-h blocking with 0.3% Triton X-100 and 3% goat serum in phosphate-buffered saline (PBS), the cells were incubated with anti-active-caspase-3 antibody (BD Biosciences) overnight at 4°C and then incubated with Alexa Fluor 568 goat anti-rabbit secondary antibody (Invitrogen) and Hoechst (Invitrogen). Fluorescent images of the cells were collected and analyzed by the Cellomics high content screening system.
Whole mount in situ hybridization
Whole mount in situ hybridizations with double-Dig-labeled miR-125b miRCURY LNA probe (Exiqon) on zebrafish embryos were performed essentially as described (Wienholds et al. 2005). Modifications to the protocol include an incubation of the 19-, 22-, and 24-hpf embryos for 30 sec and of the 30-hpf embryos for 1 min with PCR-grade proteinase K (Roche) after fixing. The hybridization mix was prepared by adding 20 pmol of miR-125b doubled-labeled LNA probe to every 1 mL of hybridization solution. The hybridization temperature used was 20°C below the melting temperature of the miR-125b LNA probe. Optimal signal-to-noise ratio during color development was obtained by washing the embryos with 5× Tris-buffered saline containing 0.1% Tween 20 (TBST buffer) between color reactions. This cycle of washing with 5× TBST followed by color development was repeated thrice. As a control, the same protocol was used for double-Dig-labeled miR-7 miRCURY LNA probe (Exiqon) on zebrafish embryos, and no signal was observed before 48 hpf, consistent with the prior report (Wienholds et al. 2005).
Microinjection in zebrafish embryos
Wild-type and p53M214K mutant zebrafish were maintained by standard protocols (Brand et al. 2002). All injections were carried out at the one- to four-cell stage with 2 nL of solution into each embryo. In the knockdown experiments, miR-125b morpholinos were injected at 0.75 pmol per embryo (lp125bMMO1/2/3 indicates the coinjection of three lp125bMOs, 0.25 pmol each); p53 morpholino was coinjected at 1 pmol per embryo; miR-125b duplex was injected at 12.5 fmol or 37.5 fmol per embryo. In the stress response experiments, the embryos were injected with 37.5 fmol per embryo of NC-DP1 or 125b-DP or with 0.5 pmol per embryo p53 MO. Sequences of all morpholinos (GeneTools) are shown in Supplemental Table 2.
qRT–PCR
RNA was extracted from cells or zebrafish embryos using Trizol reagent (Invitrogen) and subsequently column-purified with RNeasy kits (Qiagen). For qRT–PCR of miR-125b, 100 ng of total RNA was reverse-transcribed and subjected to Taqman miRNA assay (Applied Biosystems). For qRT–PCR of mRNAs, cDNA synthesis was performed with 1 μg of total RNA using the High Capacity cDNA Archive Kit (Applied Biosystems). Subsequently, human p53 and zebrafish 18S expression was analyzed by TaqMan assay; the expression of all other genes was analyzed by SYBR assay (Applied Biosystems) following the manufacturer's protocol.
Western blot assay
Cells were lysed in RIPA buffer (Pierce). Zebrafish embryos were dechorionated, deyolked, and homogenized in T-PER reagent (Thermo Fisher Scientific) containing protease inhibitor (Roche). Protein was separated by a 10% polyacrylamide gel and transferred to a methanol-activated PVDF membrane (GE Healthcare). The membrane was blocked for 1 h in PBST containing 7.5% milk and subsequently probed with 0.5 μg/mL anti-p53 antibody (Santa Cruz Biotechnologies) or 1.4 μg/mL Zfp53-9.1 antibody (Lee et al. 2008), anti-Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (Abcam), or 1 μg/mL anti-α-tubulin antibody (Sigma Aldrich) overnight at 4°C. After 1-h incubation with goat-anti-mouse HRP-conjugated secondary antibody (Santa Cruz Biotechnologies), the protein level was detected with luminol reagent (Santa Cruz Biotechnologies). Intensity of the protein bands was quantified using ImageJ.
TUNEL assay
Embryos were dechorionated and fixed in 2% paraformaldehyde (Fluka) overnight at 4°C. They were then dehydrated in methanol (50%, 75%, 95%, 100% series) and incubated with cold acetone for 10 min at −20°C. After permeabilization in PBST containing fresh 0.1% sodium citrate for 15 min, they were assayed using the in situ cell death detection kit TMR red (Roche) according to the manufacturer's instructions. High-resolution images were obtained by confocal microscope.
Immunostaining
Embryos were dechorionated and fixed in 4% paraformaldehyde overnight at 4°C. After three washes with PBDT (PBS containing 2% BSA, 1% DMSO and 0.5% Triton X-100), the embryos were incubated with cold acetone for 20 min at −20°C followed by three additional PBDT washes. Subsequently, the embryos were blocked with 1× blocking buffer (Roche) for 1 h, then incubated with mouse anti-acetylated tubulin monoclonal antibody, 1:200 (Sigma) overnight at 4°C. The embryos were washed extensively in PBDT (30 min, six times) and incubated with Alexa Fluor 568 goat-anti-mouse IgG antibody, 1:200 (Molecular Probe) for 4 h. After five washes in PBDT (30 min each), the embryos were refixed in 4% paraformaldehyde overnight at 4°C.
Image acquisition and microscope settings
Fluorescent images of the TUNEL assays and the acetylated tubulin staining were obtained with an LSM510 confocal laser-scanning microscope (Carl Zeiss Vision GmbH). A bright-field image was acquired at the same time as the fluorescent image. Projection of image stacks was made by the Zeiss image browser. Images were then imported into Adobe Photoshop for cropping, resizing, and orientation. Contrast and brightness were adjusted equally for all images of the same figure.
Images of live embryos were obtained by an SZX12 stereomicroscope (Olympus) and a MagnaFIRE SP camera (Olympus). The embryos were mounted in 3% methyl-cellulose. Images were acquired with a 65× objective, at a resolution of 1280 × 1024, with ∼100-msec exposure and 8-bit depth at room temperature. The image set of each embryo was combined, resized, cropped, and oriented using Adobe Photoshop.
Statistical analysis
Two-tail t-tests were used to determine the significance of differences between the treated samples and the controls where values resulted from luciferase reporter assay, qRT–PCR, Western blots, or high content screening. The tests were performed using Microsoft Excel, where the test type is always set to two-sample equal variance.
Acknowledgments
We thank all our colleagues in GIS and Whitehead, especially Adrian Lim, Lingbo Zhang, and Jun-Liang Tay for technical support; Soh Boon Seng and Moonyoung Um for training on cell culture and for providing the human lung fibroblasts and SH-SY5Y cells; Frank McKeon, Chin Yan Lim, Wai-Leong Tam, Sinnakaruppan Mathavan, and Senthil Raja Jayapal for fruitful discussions; and Alejandro De Los Angles for proofreading the manuscript. We also acknowledge our IMCB colleagues Kar-Lai Poon for participating in the double-blind experiment; Jun Chen and Jinrong Peng for the p53M214K mutant zebrafish and camptothecin; Kian-Chung Lee and Sir David Lane for the anti-p53 antibody and the H1299 cells; and Hang Nguyen, Svitlana Korzh, Amanda Goh, and Quo Lin for helpful advice. M.T.N.L. and H.X. were supported by SMA graduate fellowships. C.T., N.S.C., B.L., and V.K. were supported by A-STAR, Singapore. B.L. and H.F.L. were partially supported by SMA grant C-382-641-001-091. B.L. was also supported by NIH grants DK47636 and AI54973. H.F.L. and B.Z. were supported by NIH grant R01 DK068348. M.T.N.L. designed and performed the experiments, analyzed the data, and wrote the paper. C.T. performed in situ hybridization and provided M.T.N.L. with training in zebrafish biology. N.S.C. performed Western blots and assisted with writing the paper. H.X. and B.Z. helped on the preliminary analysis of miRNA expression in SH-SY5Y cells. V.K. supervised the technical aspects of the zebrafish experiments. B.L. and H.F.L. conceived the research and supervised the experimental design and the writing of the paper.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1767609.
Supplemental material is available at http://www.genesdev.org.
References
- Almog N., Rotter V. Involvement of p53 in cell differentiation and development. Biochim. Biophys. Acta. 1997;1333:F1–F27. doi: 10.1016/S0304-419X(97)00012-7. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berghmans S., Murphey R.D., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bloomston M., Frankel W.L., Petrocca F., Volinia S., Alder H., Hagan J.P., Liu C.G., Bhatt D., Taccioli C., Croce C.M. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Bommer G.T., Gerin I., Feng Y., Kaczorowski A.J., Kuick R., Love R.E., Zhai Y., Giordano T.J., Qin Z.S., Moore B.B., et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Bousquet M., Quelen C., Rosati R., Mansat-De Mas V., La Starza R., Bastard C., Lippert E., Talmant P., Lafage-Pochitaloff M., Leroux D., et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M., Granato M., Nusslein-Volhard C. Keeping and raising zebrafish. In: Nusslein-Volhard C., Dahm R., editors. Zebrafish: A pratical approach. Oxford University Press; New York: 2002. pp. 7–37. [Google Scholar]
- Bretaud S., Allen C., Ingham P.W., Bandmann O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson's disease. J. Neurochem. 2007;100:1626–1635. doi: 10.1111/j.1471-4159.2006.04291.x. [DOI] [PubMed] [Google Scholar]
- Campbell W.A., Yang H., Zetterberg H., Baulac S., Sears J.A., Liu T., Wong S.T., Zhong T.P., Xia W. Zebrafish lacking Alzheimer presenilin enhancer 2 (Pen-2) demonstrate excessive p53-dependent apoptosis and neuronal loss. J. Neurochem. 2006;96:1423–1440. doi: 10.1111/j.1471-4159.2006.03648.x. [DOI] [PubMed] [Google Scholar]
- Caygill E.E., Johnston L.A. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Elemento O., Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput. Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Rajewsky N. Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb. Symp. Quant. Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- Dony C., Kessel M., Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985;317:636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- Ekker S.C., Larson J.D. Morphant technology in model developmental systems. Genesis. 2001;30:89–93. doi: 10.1002/gene.1038. [DOI] [PubMed] [Google Scholar]
- Foshay K.M., Gallicano G.I. Small RNAs, big potential: The role of MicroRNAs in stem cell function. Curr. Stem Cell Res. Ther. 2007;2:264–271. doi: 10.2174/157488807782793781. [DOI] [PubMed] [Google Scholar]
- Foulkes W.D. p53—Master and commander. N. Engl. J. Med. 2007;357:2539–2541. doi: 10.1056/NEJMp0707422. [DOI] [PubMed] [Google Scholar]
- Garzon R., Fabbri M., Cimmino A., Calin G.A., Croce C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Ghiselli G. SMC3 knockdown triggers genomic instability and p53-dependent apoptosis in human and zebrafish cells. Mol. Cancer. 2006;5:52. doi: 10.1186/1476-4598-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Harris C.C. The 1995 Walter Hubert Lecture–molecular epidemiology of human cancer: Insights from the mutational analysis of the p53 tumour-suppressor gene. Br. J. Cancer. 1996;73:261–269. doi: 10.1038/bjc.1996.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M.V., Visone R., Di L.G., Donati V., Petrocca F., Casalini P., Taccioli C., Volinia S., Liu C.G., Alder H., et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Khochbin S., Lawrence J.J. An antisense RNA involved in p53 mRNA maturation in murine erythroleukemia cells induced to differentiate. EMBO J. 1989;8:4107–4114. doi: 10.1002/j.1460-2075.1989.tb08595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinken S.P., Holmes K.L., Morse H.C., III, Thorgeirsson S.S. Transcriptional and post-transcriptional regulation of c-myc, c-myb, and p53 during proliferation and differentiation of murine erythroleukemia cells treated with DFMO and DMSO. Exp. Cell Res. 1988;178:185–198. doi: 10.1016/0014-4827(88)90390-4. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerbitz S.J., Plunkett B.S., Walsh W.V., Kastan M.B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc. Natl. Acad. Sci. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langheinrich U., Hennen E., Stott G., Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr. Biol. 2002;12:2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- Lee K.C., Goh W.L., Xu M., Kua N., Lunny D., Wong J.S., Coomber D., Vojtesek B., Lane E.B., Lane D.P. Detection of the p53 response in zebrafish embryos using new monoclonal antibodies. Oncogene. 2008;27:629–640. doi: 10.1038/sj.onc.1210695. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li W., Xie L., He X., Li J., Tu K., Wei L., Wu J., Guo Y., Ma X., Zhang P., et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int. J. Cancer. 2008;123:1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- Moll U.M., Ostermeyer A.G., Haladay R., Winkfield B., Frazier M., Zambetti G. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol. Cell. Biol. 1996;16:1126–1137. doi: 10.1128/mcb.16.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E.J., Yoon H., Kim S.W., Kim H., Kim Y.T., Kim J.H., Kim J.W., Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- Nelson P.T., Baldwin D.A., Kloosterman W.P., Kauppinen S., Plasterk R.H., Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Robu M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., Ekker S.C. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronca F., Chan S.L., Yu V.C. 1-(5-Isoquinolinesulfonyl)-2-methylpiperazine induces apoptosis in human neuroblastoma cells, SH-SY5Y, through a p53-dependent pathway. J. Biol. Chem. 1997;272:4252–4260. doi: 10.1074/jbc.272.7.4252. [DOI] [PubMed] [Google Scholar]
- Shi X.B., Xue L., Yang J., Ma A.H., Zhao J., Xu M., Tepper C.G., Evans C.P., Kung H.J., Vere White R.W. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl. Acad. Sci. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A.U., Kaimal V., Chen J., Jegga A.G. Dissecting microregulation of a master regulatory network. BMC Genomics. 2008;9:88. doi: 10.1186/1471-2164-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A., Meister G., Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Tay Y.M., Tam W.L., Ang Y.S., Gaughwin P.M., Yang H., Wang W., Liu R., George J., Ng H.H., Perera R.J., et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- Tchang F., Gusse M., Soussi T., Mechali M. Stabilization and expression of high levels of p53 during early development in Xenopus laevis. Dev. Biol. 1993;159:163–172. doi: 10.1006/dbio.1993.1230. [DOI] [PubMed] [Google Scholar]
- Vogan K., Bernstein M., Leclerc J.M., Brisson L., Brossard J., Brodeur G.M., Pelletier J., Gros P. Absence of p53 gene mutations in primary neuroblastomas. Cancer Res. 1993;53:5269–5273. [PubMed] [Google Scholar]
- Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Fujimori-Tonou N., Yoshimura Y., Kishi T., Okamoto H., Masai I. Mutation of DNA primase causes extensive apoptosis of retinal neurons through the activation of DNA damage checkpoint and tumor suppressor p53. Development. 2008;135:1247–1257. doi: 10.1242/dev.011015. [DOI] [PubMed] [Google Scholar]
- Yoon S., De M.G. Computational identification of microRNAs and their targets. Birth Defects Res. C Embryo Today. 2006;78:118–128. doi: 10.1002/bdrc.20067. [DOI] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]