Abstract
We performed a genome-wide siRNA screen in mouse embryonic stem (ES) cells to identify genes essential for self-renewal, and found 148 genes whose down-regulation caused differentiation. Many of the identified genes function in gene regulation and/or development, and are highly expressed in ES cells and embryonic tissues. We further identified target genes of two transcription regulators Cnot3 and Trim28. We discovered that Cnot3 and Trim28 co-occupy many putative gene promoters with c-Myc and Zfx, but not other pluripotency-associated transcription factors. They form a unique module in the self-renewal transcription network, separate from the core module formed by Nanog, Oct4, and Sox2. The transcriptional targets of this module are enriched for genes involved in cell cycle, cell death, and cancer. This supports the idea that regulatory networks controlling self-renewal in stem cells may also be active in certain cancers and may represent novel anti-cancer targets. Our screen has implicated over 100 new genes in ES cell self-renewal, and illustrates the power of RNAi and forward genetics for the systematic study of self-renewal.
Keywords: ES cells, self-renewal, RNAi, genetic screen
Embryonic stem cells (ES cells) are pluripotent cells derived from the inner cell mass of the developing embryo. They can be cultured continuously in their pluripotent state, and can also be induced to differentiate into cell types from all three germ layers (for review, see Keller 2005). A number of signal transduction pathways have been shown to be important to ES cell self-renewal, including the LIF, BMP, MAPK, and Wnt pathways (Niwa et al. 1998; Burdon et al. 1999; Ying et al. 2003, 2008; Sato et al. 2004). At the same time, self-renewal is also controlled by pluripotency-associated transcription factors. Nanog, Sox2, and Oct4 form the core of the self-renewal transcription network. They physically interact with each other and form large protein complexes (Wang et al. 2006). They are also transcriptionally interconnected and co-occupy promoters of numerous target genes (Boyer et al. 2005; Loh et al. 2006; Chen et al. 2008; Kim et al. 2008).
Several approaches have been used recently to improve our understanding of ES cell self-renewal. First, novel self-renewal genes were identified by expression profiling for genes highly expressed in ES cells followed by functional genetic studies (Ivanova et al. 2006; Zhang et al. 2006). Second, a protein network involved in self-renewal was discovered by biochemical purification of proteins interacting with known pluripotency factors (Wang et al. 2006; Liang et al. 2008). Finally, the target genes of several self-renewal transcription factors have been identified by chromatin immunoprecipitation (ChIP) in ES cells (Boyer et al. 2005; Bernstein et al. 2006; Loh et al. 2006; Mikkelsen et al. 2007; Chen et al. 2008; Kim et al. 2008; Meissner et al. 2008). Despite these efforts, however, the picture of self-renewal still remains largely incomplete, as these studies have relied primarily on bootstrapping strategies to expand the network of self-renewal genes using previously known factors.
Results
Genome-wide siRNA screen
While previous studies have identified several self-renewal genes and pathways, it is likely that novel classes of important self-renewal genes exist. Therefore, we chose to execute a functional analysis to systematically explore the mechanism of self-renewal. We carried out a genome-wide RNAi screen in mouse ES cells to identify genes that are required for the maintenance of self-renewal. We employed the Oct4GiP reporter ES cell line for the screen, which expresses the green fluorescent protein (GFP) under the control of mouse Oct4 gene regulatory elements (Ying et al. 2002). GFP expression in this cell line correlates faithfully with the ES cell identity, and can thus be used to quantitatively monitor the self-renewal and differentiation status of the cells (Ying and Smith 2003). To detect changes in GFP fluorescence, we used fluorescence-activated cell sorting (FACS) analysis for its superior sensitivity. As a proof of principle, we confirmed that knockdown of known self-renewal genes such as Oct4 or Nanog induces differentiation of the Oct4GiP cells, and validated that the extent of differentiation after siRNA transfections can be readily measured by the percentage of GFP-negative cells (percent of differentiation) by FACS (Supplemental Fig. S1).
In the screen, we depleted proteins one at a time in the Oct4GiP cells in 96- or 384-well plates with Dharmacon siGenome library smart pools, and measured cellular differentiation after 4 d. The siRNA library used in the primary screen contained siRNA pools against 16,683 mouse genes. Genes were considered potential hits if the corresponding siRNA smart pools increased the percent of differentiation by two standard deviations from the plate average. We then rescreened the four individual siRNAs of the smart pools against the primary hits in a secondary screen (Fig. 1A). We found that a total of 148 genes scored with one or more siRNAs (Supplemental Table S1). We rediscovered several genes that have been shown previously to be important in ES cell self-renewal, including Il6st, Mbd3, Oct4, Rif1, and Sox2, supporting the validity of our screen. We also identified many genes that have been implicated previously in ES cell functions or embryonic development. For example, Mga and Zfp219 are highly expressed in the inner cell mass (Yoshikawa et al. 2006); Rnf2, Yy1, and Zfp219 are part of a protein network that includes some known self-renewal proteins (Wang et al. 2006; Liang et al. 2008); Cbx1, D630039A03Rik, Eya1, Hist1h3i, and Zfp13 are transcriptional targets of the Nanog–Oct4–Sox2 complex (Loh et al. 2006; Kim et al. 2008). We were also encouraged that the identified genes are functionally important because we found multiple components of previously known protein complexes, such as Trim28–Cbx1, Cpsf1–Cpsf2–Cpsf3–Fip1l1, and Cul3–Nedd8–Rbx1. Gene ontology (GO) analysis showed that the identified genes are enriched for “nucleic acid binding” and “transcription factors,” and many of them function in developmental pathways and gene regulation (Fig. 1B; Supplemental Table S2).
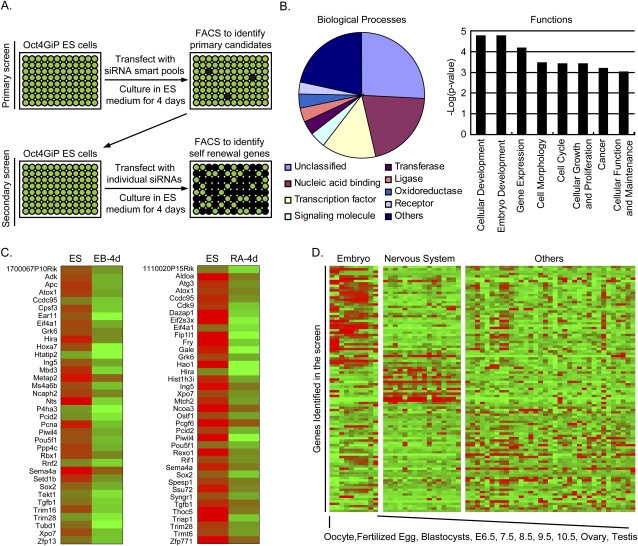
Figure 1.
Genome-wide RNAi screen for self-renewal genes in mouse ES cells. (A) Outline of the screen. (Dark wells) Differentiated cells that are GFP-negative; (green wells) self-renewing cells that are GFP-positive. (B) Functional categorization of candidate self-renewal genes identified from the screen. (Left panel) Molecular functions (Panther Classification System). (Right panel) Biological processes (Ingenuity Pathway Analysis). Only selected categories were shown. For complete lists, see Supplemental Table S2. (C) Expression of candidate self-renewal genes during ES cell differentiation. (Right panel) Differentiation after 4 d in RA. (Left panel) Differentiation after 4 d of EB formation. (D) Relative expression level of candidate self-renewal genes in 61 different tissues. Genes were clustered based on their expression levels in embryonic tissues, nervous system, and other tissues.
Like any RNAi screens, it is conceivable that some of the identified candidates may be false positives due to off-target effects of the siRNAs. Therefore, we further assembled a list of high-confidence candidates based on the following analysis. First, 48 genes were scored by multiple siRNAs in the screen, and are thus less likely caused by off-target effects. Second, we assessed the functional relevance of the identified genes in self-renewal and embryonic development by examining their expression patterns during ES cell differentiation and across mouse tissues from different developmental stages. We reasoned that genes important for self-renewal may participate in developmental switches, and should be highly expressed in ES cells and embryonic tissues and turned off in differentiated cells or adult tissues. Indeed, during ES cell differentiation, 61 of the 128 genes (47%) represented on the Affymetrix arrays were down-regulated after either retinoic acid (RA) treatment or embryoid body (EB) formation (Fig. 1C; Supplemental Table S2). Finally, in 61 mouse tissues, 43 of the 105 genes (41%) whose transcripts were detected had higher expression in embryonic tissues (Fig. 1D; Supplemental Table S2). In total, 104 of the 148 genes were either scored by more than one siRNA or highly expressed in ES cells or embryonic tissues. They were selected as high-confidence candidates and are most likely important for self-renewal.
Validation of selected self-renewal genes
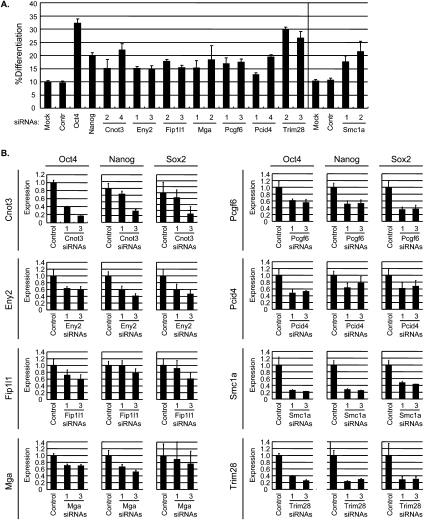
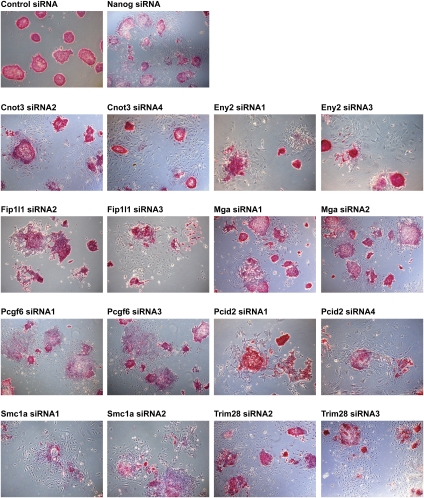
Among the high-confidence candidates, we selected eight genes (Cnot3, Eny2, Fip1l1, Mga, Pcgf6, Pcid2, Smc1a, and Trim28) to further validate their functions in ES cell self-renewal. We used two different siRNAs against each to minimize possible off-target effects, and verified that the siRNAs we used knocked down their corresponding genes efficiently by quantitative PCRs (qPCRs) (Supplemental Fig. S2). To prove that these genes are important for self-renewal, we used four different assays to measure the self-renewal and differentiation state of ES cells after depleting proteins encoded by each of the eight genes. First, we performed reporter assays with Oct4GiP cells. Depleting proteins encoded by any of the eight genes caused a clear increase in the percentage of differentiated cells, as judged by a reduction in GFP fluorescence (Fig. 2A). Second, we performed qPCRs on endogenous ES cell markers Oct4, Nanog, and Sox2. We found that depletion of the candidate proteins resulted in a decrease in the expression of Oct4, Nanog, and Sox2 to various extents (Fig. 2B), suggesting that self-renewal was indeed compromised. Third, we performed alkaline phosphatase staining on cells transfected with siRNAs against the candidate genes. Undifferentiated ES cells express high levels of alkaline phosphatase while differentiated cells do not. We found that candidate gene siRNA-transfected cells formed colonies with reduced alkaline phosphatase activity and an obviously altered differentiation morphology (Fig. 3). We also repeated the alkaline phosphatase staining assays in a different ES cell line (CCE), and observed similar phenotypes (Supplemental Fig. S3). Finally, we performed immunofluorescence staining of SSEA-1, a specific marker for undifferentiated mouse ES cells. We found that cells transfected with siRNAs against the candidate genes expressed much lower levels of SSEA-1 (Supplemental Fig. S4). Based on the results of these assays, we conclude that Cnot3, Eny2, Fip1l1, Mga, Pcgf6, Pcid2, Smc1a, and Trim28 all play important roles in self-renewal.
Figure 2.
Validation of selected self-renewal genes identified in the screen. (A) Oct4GiP reporter assays after knockdown of candidate self-renewal genes. Oct4GiP ES cells were transfected with siRNAs against candidate self-renewal genes (two different siRNAs for each gene). Percent of differentiation (% Differentiation) was defined by percentage of GFP-negative cells, and was measured 96 h after transfection by FACS. (B) Knockdown of candidate self-renewal genes causes down-regulation of ES cell marker genes. ES cells were transfected with siRNAs against candidate genes and cells were collected 96 h after transfection. Level of Oct4, Nanog, and Sox transcripts were measured by qPCRs from cDNAs prepared from the transfected cells.
Figure 3.
Morphological changes and loss of alkaline phosphatase staining after the silencing of candidate genes. ES Oct4GiP cells were transfected with siRNAs against candidate self-renewal genes and replated at low density on the second day. Cells were cultured in ES cell medium, and alkaline phosphatase staining was performed 5 d after plating.
LIF–Stat3, BMP–Smads, and Erk pathways are known to be important in self-renewal (Niwa et al. 1998; Burdon et al. 1999; Ying et al. 2003, 2008; Sato et al. 2004). We found no significant changes in the expression or phosphorylation of Stat3, Smad proteins, and Erk kinases after siRNA treatment for seven of the eight genes (Supplemental Fig. S5), except for Smc1a. However, the Smc1a transfected cells were largely differentiated as judged by their morphology, and the differentiation may have contributed to the changes observed in the Western blots. Therefore, these eight genes are unlikely to regulate self-renewal through the regulation of Stat3, Smad1, or Erk in the LIF, BMP, and MAPK pathways, and are likely to regulate self-renewal through novel pathways.
ChIP–chip of Cnot3 and Trim28
Among the eight genes studied, we selected two transcription regulators, Cnot3 and Trim28, for further investigation because they scored with multiple siRNAs in the screen and had relatively strong phenotypes. Reduction of the expression of Cnot3 or Trim28 by four different siRNAs all resulted in increases in the percentage of differentiated cells in the reporter assay, and all the siRNAs were also confirmed to knock down their target genes (Supplemental Fig. S6). Furthermore, silencing these two genes also up-regulated the transcription of differentiation markers (Supplemental Fig. S7). Thus, both Cnot3 and Trim28 are important for the maintenance of self-renewal.
Cnot3 belongs to the Ccr4–Not complex that regulates gene expression both transcriptionally and post-transcriptionally (Collart and Timmers 2004). Other components of the Ccr4–Not complex have been shown to be important for embryonic development in both Drosophila (Zaessinger et al. 2006) and Caenorhabditis elegans (Molin and Puisieux 2005). In our screen, we identified only Cnot3 but not other components of the Ccr4–Not complex. Therefore, Cnot3 may have functions independent of the Ccr4–Not complex in ES cells. Trim28 is essential for mouse embryonic development (Cammas et al. 2000) and silencing of murine leukemia virus in ES cells (Wolf and Goff 2007), and it interacts with the proteins encoded by several pluripotency genes, including Nanog, Rex1, and Dax1, although the physiological significance of these interactions is not known (Wang et al. 2006). Trim28 also interacts with heterochromatin protein HP1, and the interaction is important for heterochromatin-mediated gene silencing (Ryan et al. 1999) and endoderm differentiation (Cammas et al. 2004). Recently, Trim28 was also found to be important for self-renewal (Fazzio et al. 2008). Consistent with these findings, we identified both Trim28 and an HP1 protein that interacts with it in our screen: Cbx1 (HP1β). It is possible that Trim28 and Cbx1 regulate genes that are important for self-renewal through modification of chromatin structures.
We found that both Cnot3 and Trim28 are highly expressed in ES cells and embryonic tissues and down-regulated during ES cell differentiation (Supplemental Fig. S8). To explore the pathways regulated by Cnot3 and Trim28, we sought to identify the genes whose promoter regions are bound by these factors. We employed the recently developed biotin-mediated ChIP (Biotin-ChIP) system (Kim et al. 2008) and determined the binding sites for Cnot3 and Trim28 in promoter regions of mouse ES cells (Supplemental Fig. S9). We identified 1669 sites (corresponding to 1547 genes) that were occupied by Cnot3, and 3331 sites (corresponding to 3073 genes) that were occupied by Trim28 (Fig. 4A; Supplemental Table S3). To verify the Biotin-ChIP results, we performed qPCRs on 20 Cnot3- or Trim28-binding sites and confirmed the binding (Supplemental Fig. S10A,B). For Trim28, we also confirmed some of the binding sites identified by the Biotin-ChIP method with ChIP assays using an antibody against the endogenous Trim28 protein (Supplemental Fig. S10C). From the Biotin-ChIP analysis, we found that the vast majority of the Cnot3- and Trim28-binding sites were in close proximity to transcription start sites (TSSs) in the mouse genome (Fig. 4B), consistent with the idea that they may regulate the transcription of these genes. From the sequences bound by Cnot3 and Trim28, we deduced their consensus binding motifs to be CGGCXGCG and GCCGCGXX, respectively (Fig. 4C). Interestingly, both Cnot3 and Trim28 bound to the Cnot3 promoter region (Supplemental Table S3; Supplemental Fig. S10), and may therefore regulate Cnot3 expression in ES cells to maintain self-renewal. Trim28 also occupies the promoter regions of many other pluripotency genes, including Nanog, Sox2, Tcf3, Il6st, and Lefty2, and thus may play a central role in the self-renewal network.
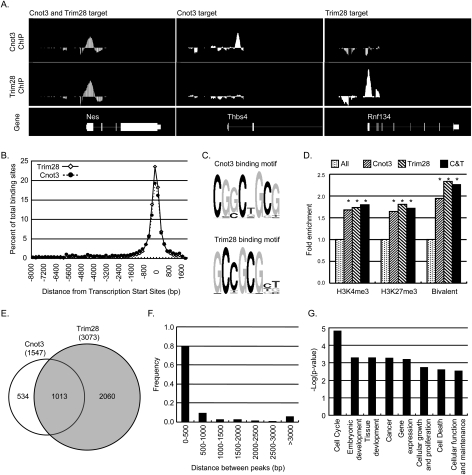
Figure 4.
Identification of Cnot3 and Trim28 target genes. (A) Chromosome view of examples of Cnot3 and Trim28 target genes. (B) Distribution of Cnot3- and Trim28-binding sites from target gene TSSs. (C) Consensus binding motif of Cnot3 and Trim28. (D) H3K4 and H3K27 methylation in promoters of Cnot3 and Trim28 target genes. (Fold enrichment) Percentage of H3 methylation in Cnot3 or Trim28/percentage of H3 methylation in the genome. (Asterisks) P < 10−8, calculated by hypergeometric distribution. (E) Venn diagram of target genes co-occupied by Cnot3 and Trim28. (F) Histogram of distance between Cnot3- and Trim28-binding sites in genes occupied by both. (G) GO analysis (by Ingenuity Pathway Analysis) of Cnot3 and Trim28 target genes. Only selected functional categories were shown. For a complete list, see Supplemental Table S3.
Histone modifications play an important role in controlling gene expression. Histone 3 Lys 4 trimethylation (H3K4me3) is generally associated with active transcription and histone 3 Lys 27 trimethylation (H3K27me3) is typically correlated with repression. A unique feature of ES cells is that a large set of development-related genes are marked by both H3K4me3 and H3K27me3, and the bivalent modification is proposed to poise these genes for activation or repression (Bernstein et al. 2006; Mikkelsen et al. 2007). Inspection of the histone modifications on Cnot3 and Trim28 target promoters revealed that they are highly enriched for H3K4me3, H3K27me3, and bivalent methylations (Fig. 4D), indicating that Cnot3 and Trim28 target genes are dynamically regulated in ES cells and are therefore likely to be important for ES cell function.
Surprisingly, Cnot3 and Trim28 co-occupied 1073 binding sites (corresponding to 1013 target genes, P = 2.2 × 10−16, hypergeometric distribution), and their binding sites are in close proximity to each other (Fig. 4E,F). However, we were not able to detect physical interactions between the Cnot3 and Trim28 proteins by affinity purification (data not shown). GO analysis indicated that the common targets of Cnot3 and Trim28 are enriched for genes involved in cell cycle, development, cell growth, cell death, and gene expression (Fig. 4G; Supplemental Table S3). In addition, they are also enriched for H3K4me3 and H3K27me3 modifications (Fig. 4D). These observations suggest that Cnot3 and Trim28 work cooperatively to sustain self-renewal through regulation of a large set of target loci.
A unique module in the self-renewal transcription network formed by Cnot3, Trim28, c-Myc, and Zfx
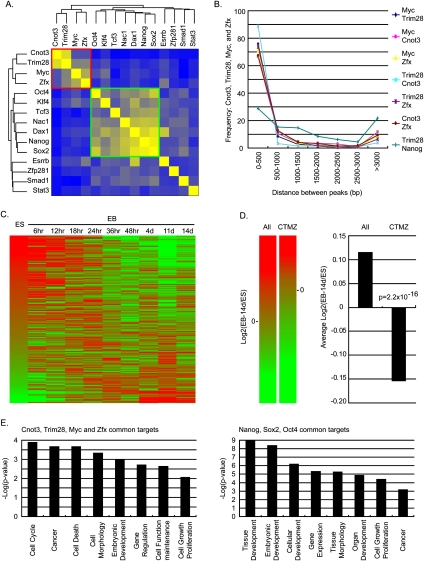
Recently, target genes of many pluripotency-related transcription factors have been determined in ES cells, leading to the discovery of transcription networks regulating self-renewal (Chen et al. 2008; Kim et al. 2008). To determine how Cnot3 and Trim28 fit in these transcription networks, we compared their transcriptional targets with those of known pluripotency-associated genes, including Nanog, Oct4, Sox2, Tcf3, Nac1, Dax1, Klf4, Smad1, Stat3, Esrrb, Zfp281, c-Myc, and Zfx. Hierarchical clustering of the above pluripotency genes revealed that the targets of Cnot3, Trim28, c-Myc, and Zfx formed a unique cluster (cluster II) (Fig. 4A, red box) that is distinct from the core transcriptional network formed by Nanog, Oct4, Sox2, Tcf3, Nac1, Dax1, and Klf4 (cluster I) (Fig. 5A, green box). In fact, Cnot3 and Trim28 share many common target genes with c-Myc and Zfx, but not with other self-renewal genes. In the common target promoter regions between Cnot3, Trim28, c-Myc, and Zfx, the binding sites localized in close proximity (Fig. 5B), suggesting that they may regulate target gene expression in a cooperative fashion. A total of 326 genes were found to be co-occupied by all four factors (P < 10−6, Monte Carlo simulation) (Supplemental Table S4). To examine the function of these genes in ES cells, we examined their expression during ES cell differentiation. We found that a large fraction of these genes were down-regulated over 14 d of EB formation (Fig. 5C,D), consistent with the notion that they may be important for self-renewal. Interestingly, GO analysis showed that targets of cluster I genes are enriched for genes involved in developmental processes while targets of cluster II genes are more enriched for transcription regulators involved in cell cycle and cell survival (Fig. 5E; Supplemental Table. S4). Based on these results we propose that specific regulation of cell cycle progression and survival is an integral component of ES cell self-renewal, and that Cnot3, Trim28, c-Myc, and Zfx control self-renewal by regulating these processes.
Figure 5.
The Cnot3, Trim28, Myc, and Zfx module in the self-renewal network. (A) Correlations of target gene occupancies between pluripotency genes. (B) Distances between Cnot3-, Trim28-, c-Myc-, and Zfx-binding sites in gene promoters occupied by any two of the four genes. Distance between Trim28- and Nanog-binding sites in their common targets was shown as a negative control. (C) Expression pattern of genes co-occupied by Cnot3, Trim28, c-Myc, and Zfx during EB differentiation. Genes were ordered based on their expression level in ES cells (0 h of EB differentiation). (D) Change in expression levels of genes occupied by all four factors (Cnot3, Trim28, Myc, and Zfx) between ES cells and 14-d EB. (Left panel) Heat map of log2 ratios between 14-d EB and ES for all genes detected on the array (All) and for genes co-occupied by all four factors (CTMZ). (Right panel) Average log2 ratios between 14-d EB and ES for all genes (All) and for genes co-occupied by four factors (CTMZ). (E) GO analysis (Ingenuity Pathway Analysis) of common target genes of Cnot3–Trim28–c-Myc–Zfx (left) and Nanog–Sox2–Oct4 (right). Only selected functional categories were shown. For complete lists, see Supplemental Table S4.
Discussion
ES cells have been used extensively as a model system to study early mammalian development and the molecular control of self-renewal and pluripotency (Keller 2005). They also bear promise for the development of stem cell based therapies for regenerative medicine (Murry and Keller 2008). In addition, further dissection of the components involved in self-renewal and pluripotency may also provide additional avenues for reprogramming of somatic cells into iPS cells (Loh et al. 2008; Mikkelsen et al. 2008). Here we present the first genome-scale functional genetic screen in mouse ES cells for pluripotency genes. Our approach differs from previous screens in ES cells in several ways. First, we performed an unbiased genome-scale RNAi screen to ensure that we would not bias toward genes expressed at higher levels in ES cells. Second, we used Oct4GiP cells and FACS assays to quantitatively measure the extent of self-renewal versus differentiation with high sensitivity. Third, using this rather than proliferation-based assays, we were able to identify genes that directly regulate self-renewal, thereby avoiding genes that simply interfere with general cellular physiology. Indeed, we rediscovered several known self-renewal genes and also identified many genes whose functions in self-renewal have not been established previously. However, as is true of all genome-wide screens, we also missed some self-renewal genes. The Dharmacon library does not contain siRNAs against all genes in the mouse genome (such as Nanog and Ronin) (Dejosez et al. 2008), and not all siRNAs in the library efficiently knocked down their target genes. In addition, by using Oct4GiP reporter assays, we were only able to identify genes that regulate ES-specific markers but not genes that only induce lineage markers (such as the Tip60–p400 complex) (Fazzio et al. 2008).
Among the self-renewal genes we identified, we selected Cnot3 and Trim28 for further studies. Cnot3 depletion resulted in differentiation primarily into the trophectoderm lineage, while Trim28 depletion resulted in differentiation into the primitive ectoderm lineage. Therefore, although they may have a common role in self-renewal (see below), they each prevent differentiation in a different direction. In fact, even though there is a strong overlap in their target genes, there are also unique targets for each factor, which may explain why they have different phenotypes in some respects.
The interconnectedness of the ES cell self-renewal network is illustrated by the fact that Cnot3 and Trim28 co-occupied many putative gene promoters with two other pluripotency genes, c-Myc and Zfx, and these four genes form a unique module in the self-renewal transcription network that is distinct from the module of Nanog–Sox2–Oct4. Target genes of all four factors (Cnot3, Trim28, c-Myc, and Zfx) are mostly down-regulated during ES cell differentiation, and are enriched for transcription regulators involved in cell cycle, cell death, and cancer, supporting the idea that ES cells specifically regulate these processes to maintain self-renewal. Furthermore, the enrichment for cancer genes also supports the idea that regulatory networks controlling self-renewal in stem cells may also be active in certain cancers (Ben-Porath et al. 2008). Therefore, our findings indicated additional layers of complexity in the self-renewal transcription network. The existence of these unique modules suggests that they may serve as hubs for signal integration, and may control self-renewal by translating signals from divergent pathways into the regulation of specific gene sets and functions.
Our screen identified many genes whose functions in self-renewal have not been reported previously. Further characterization of these genes for their roles in self-renewal, differentiation, and reprogramming will likely identify additional functional modules important for self-renewal, and may provide new insights into the mechanistic view of pluripotency. In addition, the functional genomic approach we employed can be adapted to study stem cell self-renewal and lineage commitment in other systems as well.
Materials and methods
Constructs
Mouse Cnot3 was PCRed with mCnotBioCHIP5 (5′-GTG CAATGCCCGGGGCGGACAAGCGCAAACTCCA AGG-3′) and mCnotBioCHIP3 5′-GTGCAATGCCCG GGCTGGAGGTCCCGGTCCTCCAGG-3′). Mouse Trim28 was PCRed with mTrimBioCHIP5 (5′-GTGC AATGCCCGGG GCGGCCTCGGCGGCAGCGAC-3′) and mTrimBioCHIP3 (5′-GTGCAATGCCCGGGGCC ATCACCAGGGCCAG-3′). The PCR products were digested with XmaI and cloned into the pEF1a-FlagBiotin plasmid (Kim et al. 2008) to generate Cnot3–pEF-FlagBiotin and Trim28–pEF-FlagBiotin plasmids.
siRNAs used in this study
Pou5f1: Invitrogen Stealth siRNAs: MSS237605, MSS237606; Dharmacon siGenome siRNAs: D-046256-02, D-046256-03; Nanog: Invitrogen Stealth siRNAs: MSS231180, MSS231181; Cnot3: Dharmacon siGenome siRNAs: D-052632-01, D-052632-02, D-052632-03, D-052632-04; Eny2: Dharmacon siGenome siRNAs: D-065225-01, D-065225-03; Fip1l1: Dharmacon siGenome siRNAs: D-063984-02, D-063984-03; Mga: Dharmacon siGenome siRNAs: D-045405-01, D-045405-02; Pcgf6: Dharmacon siGenome siRNAs: D-049359-01, D-049359-03; Pcid2: Dharmacon siGenome siRNAs: D-057926-01, D-057926-04; Smc1a: Dharmacon siGenome siRNAs: D-049483-01, D-049483-02; Trim28: Dharmacon siGenome siRNAs: D-040800-01, D-040800-02, D-040800-03, D-040800-04.
Cell culture
ES cell lines used in this study (Oct4GiP, CCE, J1-BirA) were all cultured as described before on gelatin-coated plates in DMEM (Invitrogen) supplemented with 15% FBS, 10 μM 2-mercaptoethanol, 0.1 mM nonessential amino acids (Invitrogen), 1× EmbryoMax nucleosides (Millipore), 1000 U of ESGRO (Millipore) (Keller et al. 1993; Ying and Smith 2003; Kim et al. 2008). For RA-induced differentiation, ES cells were plated at 3 × 104 per square centimeter in six-well plates in ES cell medium, and differentiation was induced on the second day by changing medium to DMEM supplemented with 10% FBS and 1 μM all-trans-RA (Sigma). Cells were harvested 48 h after RA induction. For EB formation, ES cells were plated in DMEM supplemented with 10% FBS in Corning ultralow binding six-well plates at 5 × 105 per well. Medium was changed every other day, and cells were harvested on day 6.
ES cell transfections
For transfections in 96-well plates, ES cells were dissociated by trypsinization and plated at 10 × 103 per well in 100 μL of ES cell medium in gelatin-coated plates. For each well, 0.3–0.35 μL of Lipofectamine2000 was premixed with 10 μL of Opti-MEM (Invitrogen), and then mixed with 5 × 10−12 mol siRNAs. siRNA–lipids complexes were incubated at room temperature for 15–30 min and then added to the cells. For transfection in other vessels, the above protocol was scaled up or down accordingly.
Oct4GiP cell reporter assay
Oct4GiP cells were lifted and dissociated with 0.25% trypsin. Trypsin was then quenched with 10% FBS in DPBS (Invitrogen), and cells were resuspended into single-cell suspension by pipetting. GFP fluorescence of the cells was determined by FACS analysis on the LSRII FACS analyzer (BD Biosciences).
Alkaline phosphatase staining and SSEA-1 immunofluorescence
For Alkaline phosphatase staining, 20 × 103 ES cells were transfected with siRNAs at 100 nM in 96-well plates on day 1. Five percent to 20% of each well was replated into one well of the 12-well plates on day 2, and cultured in ES cell medium for another 5 d. On day 7, cells were fixed, permeablized, and stained for alkaline phosphatase activity with the Quantitative Alkaline Phosphatase ES Characterization Kit (Millipore).
For SSEA-1 immunofluorescence, 40 × 103 to 60 × 103 ES Oct4GiP cells were transfected with siRNAs at 50 nM in 24-well plates on day 1 and cultured in ES cell medium for 4 d. On day 4, cells were fixed, permeablized, and SSEA-1 immunofluorescence staining was carried out in the plates according to the manufacturer's suggested protocol, using SSEA-1 antibody (Abcam, Ab16285) and Alexa Fluor 488 goat anti-mouse (Invitrogen, A11001) secondary antibody (both 1:200 dilution). Pictures were taken with an inverted fluorescence microscope and the same exposure time was used to capture the SSEA-1 staining images in all the samples.
RNAi screen in Oct4GiP cells
To identify genes required for ES cell self-renewal, a high-throughput RNAi screen was performed with an arrayed siRNA library containing 16,683 siRNA pools targeting the vast majority of the mouse genome (Mouse siGENOME siRNA Library-Genome, G-015000-05, Thermo Fisher Scientific). Additional wells containing either buffer alone, a nontargeting control siRNA (siGENOME nontargeting siRNA #2, Dharmacon), and an siRNA pool directed against Polo like kinase one (PLK1, Dharmacon) were present on all plates transfected. Both primary and secondary screens were performed in duplicate.
In the primary screen, Oct4GiP cells were transfected with smart pools of siRNAs in 96- or 384-well plates at 50 nM final concentration and then cultured in ES cell medium with daily medium change. Cells were dissociated by trypsinization 4 d after transfection and FACS analyzed. Percent of differentiation was defined as the percentage of GFP-negative cells. Wells with a percent of differentiation greater than two standard deviations above the plate mean in both duplicates were considered primary hits. All four individual siRNAs against the primary hits were cherry-picked and arrayed in 96- or 384-well plates for the secondary screen. In the secondary screen, Oct4GiP cells were transfected with individual siRNAs against the primary hits at 50 nM, cultured for 4 d, and FACS-analyzed similarly as in the primary screen. siRNAs in wells with a percent of differentiations greater than two standard deviations above the mean of the buffer-alone and negative control wells in both duplicates were considered final hits.
qPCRs
Total RNAs were prepared from cells with the RNAeasy-plus kit (Qiagen), and cDNAs were generated from 1–2 μg of total RNA with SuperscriptIII reverse-transcriptase and oligo-dT primers (Invitrogen) according to the manufacturers’ instructions. qPCRs were performed with the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) on Applied Biosystems 7500 Fast Real-Time PCR System. Triplicate or quadruplicate PCR reactions were carried out for each sample. See Supplemental Table S5 for primers used.
Western blot
Cells were harvested and lysed in SDS-PAGE sample buffer at 2 × 107 cells per milliliter. Equal amount of lysates were loaded in each lane. Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane (Millipore), and probed with the indicated antibodies. Primary antibodies used in this study were as follows Oct4 (Abcam AB19857), Nanog (Millipore, AB9220), Streptavidin-peroxidase polymer (Sigma, S2438-250UG), Flag-HRP (Sigma, A8295), Cnot3 (Novus, H00004849-M01), Trim28 (Abcam, Ab10484), Smad1-5-8-phospho-specific (Cell Signaling, 9512), Smad1 (Cell Signaling, 9511), Stat3-S727-phospho-specific (Cell Signaling, 9134), Stat3–Y705-phospho-specific (Cell Signaling, 9145), Stat3 (Cell Signaling, 9132), p42/p44-phospho-specific (Cell Signaling, 9101), p42/p44 (Cell Signaling, 9102), Ran (BD, 610340).
Biotin-ChIP
To expression biotin-tagged Cnot3 or Trim28 in the J1-BirA cells, Cnot3–pEF-FlagBiotin or Trim28–pEF-FlagBiotin plasmids were transfected into the J1-BirA cells with Lipofectamine2000. Transfected cells were selected with puromycin (1.5 μg/mL) from 48 h after transfection for 4 d, and then replated to form colonies. Several clones were picked, and expression of Flag-Biotin-tagged Cnot3 or Trim28 was verified by Western blot with HRP-conjugated Flag antibody and HRP-conjugated streptavidin, as well as qPCRs. Clones Cnot3–pEF-FlagBiotin-9 and Trim28–pEF-FlagBiotin-8 were used for the Biotin-ChIP experiments. The parental J1-BirA cells were used as the negative control.
Biotin-ChIP was performed similarly as described previously (Kim et al. 2008). Briefly, cells from two 15-cm plates (∼1 × 108) were used for ChIP in each replicate, and triplicates were performed for each sample: J1-BirA (as control), Cnot3–pEF-FlagBiotin-9, and Trim28–pEF-FlagBiotin-8. Cells were crosslinked, washed, and collected. Cell pellets were resuspended in SDS-ChIP buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl at pH 8.1, 150 mM NaCl, protease inhibitors), and sonicated to fragment genomic DNA (average size 0.5–1 kb). Samples were centrifuged at 12,000 rpm for 10 min at 4°C to remove insoluble materials, and then precleared with protein-A beads for 1 h at 4°C. They were incubated with streptavidin beads (Dynabeads MyOne Streptavidin T1) overnight at 4°C, and the beads were successively washed with buffer I (2% SDS), buffer II (0.1% Deoxycholate, 1% Triton X-100, 1 mM EDTA, 50 mM HEPES at pH 7.5, 500 mM NaCl), buffer III (250 mM LiCl, 0.5% NP-40, 0.5% Deoxycholate, 1 mM EDTA, 10 mM Tris-Cl at pH 8.1), and TE buffer (10 mM Tris-Cl at pH 7.5, 1 mM EDTA). At the end, SDS elution buffer was added and incubated with the beads overnight at 65°C to elute DNAs and reverse cross-link protein–DNA complexes. Elutions were treated with RNase A and Proteinase K and extracted with phenol:chloroform, and DNAs were ethanol precipitated and resuspended in water.
ChIP samples were amplified by ligation-mediated PCR, fragmented, and biotin labeled with the Affymetrix GeneChip WT Double-Stranded DNA Terminal Labeling Kit. Three biological replicates of hybridization were performed on Affymetrix GeneChip Mouse promoter 1.0R arrays at Microarray Core Facility, Dana-Farber Cancer Institute. MAT (model-based analysis of tiling array) was applied to predict the target loci with a P-value ≤1 × 10−6. Genomic regions between 8 kb upstream of and 2 kb downstream from TSS from March 2006 released (mm8) mouse genome annotation were used, and only one transcript was used for genes with multiple transcripts with same TSS presented in RefSeq (total 19,253 genes).
To validate target gene promoters identified in the ChIP experiments, we tested 20 Cnot3 target loci and Trim28 target loci. Primer pairs for quantitative ChIP–PCR were designed using ±150-base-pair (bp) genomic sequence information specific to the predicted target loci to generate 100-bp to 125-bp amplified products. qPCRs were performed with Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) on input genomic DNA (as negative control), Cnot3 Biotin-ChIP DNA, and Trim28 Biotin-ChIP DNA. Triplicate or quadruplicate PCR reactions were carried out for each sample. For each locus, fold enrichment was calculated by comparing relative quantity of PCR product from the ChIP samples to that from the total input genomic DNA, using Gfi1b locus (nontarget for either Cnot3 or Trim28) for normalizations.
Trim28 ChIP
Trim28 ChIP was carried out similarly as described above in J1 ES cells. Trim28 antibody (Abcam, ab10483) and protein A sepharose (Invitrogen, 101041) was used for the IP. A control ChIP was also performed using protein A sepharose only (no Trim28 antibody in the IP). qPCR verifications of 10 of the predicted Trim28 loci were carried out similarly as described above on control and Trim28 ChIP DNA. Triplicate or quadruplicate PCR reactions were carried out for each sample, and fold enrichment was calculated similarly as described above using Gfi1b locus for normalizations.
GO analysis
GO analysis was performed with the Panther Classification System (Thomas et al. 2003) or the Ingenuity Pathway Analysis software (http://www.ingenuity.com).
Bioinformatics analysis
Microarray expression data used in this study were obtained from previous studies: RA differentiation (Ivanova et al. 2006), EB differentiation (StemBase experiment 201) (Porter et al. 2007), and GNF SymAtlas gene expression in mouse tissues (Su et al. 2002). The MAS5 normalized intensity values with call signs of the RA and EB differentiation data, and the gcRMA-analyzed expression values with call signs for the GNF SymAtlas data were imported into the R program for further analysis. For EB differentiation, replicates were collapsed to the average expression value for each probe.
For Figure 1C, probes corresponding to the 148 genes identified in the screen were extracted from both the RA and EB differentiation data. A single probe with the largest expression range across all differentiation time points was selected for genes with multiple probes. One-hundred-twenty-eight out of the 148 genes were found to be represented on the Affymetrix array. Changes in expression between ES and day 4 RA differentiation or ES and day 4 EB differentiation were calculated from the intensity values for each gene, and genes with log2(day 4/ES) < −0.5 were classified as down-regulated. Heat maps of the down-regulated genes were plotted with the R program.
For Figure 1D, probes corresponding to the 148 genes identified in the screen were extracted from the GNF SymAtlas data. Only probes with present calls in more than four tissues were selected, and a single probe with the largest expression range across all differentiation time points was used for genes with multiple probes. One-hundred-three probes passed the above filters. For each probe, their expression values were converted to Log2 values and then mean subtracted across all tissues. Of the 61 tissues in GNF SymAtlas, 10 tissues were classified as embryonic tissues: oocyte, fertilized egg, blastocysts, embryo day 6.5, embryo day 7.5, embryo day 8.5, embryo day 9.5, embryo day 10.5, ovary, and testis. Three out of the above 10 were further classified as preimplantation tissues: oocyte, fertilized egg, and blastocysts. Sixteen tissues were classified as nervous tissues: amygdale, cerebellum, cerebral cortex, dorsal root ganglia, dorsal striatum, frontal cortex, hippocampus, hypothalamus, medial olfactory epithelium, olfactory bulb, preoptic, retina, spinal cord lower, spinal cord upper, substantia nigra, and trigeminal. The rest 35 tissues were classified as “others.” The expression values for each probe in embryonic tissues and preimplantation tissues were tested against expression values in other tissues with single-sided Mann-Whitney test to obtain P-values under the alternative hypothesis that the overall expression in embryonic tissues or preimplantation tissues is greater than in other tissues. Expression difference between the nervous tissues and other tissues was calculated similarly. We define embryonic enrichment as a significantly higher expression level in either embryonic tissues or preimplantation tissues compared with other tissues (P < 0.05), and nervous enrichment as a significantly higher expression level in nervous tissues compared with other tissues (P < 0.05). Genes were organized according to their expression pattern: enriched in embryonic tissues or preimplantation tissues (43), enriched in nervous tissues but not the above two (16), and the rest (46).
For Figure 4C, sequences from the top 50 Cnot3- or Trim28-binding sites (−150 bp to +150 bp surrounding the peak positions) were submitted to Weeder Web (Pavesi et al. 2004) to identify consensus motifs bound by Cnot3 and Trim28.
For Figure 4D, lists of H3K4me3 and H3K27me3 methylated gene promoters were downloaded from a previous study (Kim et al. 2008). Methylation status of the Cnot3 and Trim28 target gene promoters were deduced from the above lists. Fold enrichment = number of methylated promoters/number of total promoters in all genes, Cnot3 target genes, Trim28 target genes, or Cnot3 and Trim28 common target genes.
For Figure 5A, correlations of target gene occupancies between pluripotency factors were calculated as follows. Target gene lists for Cnot3 and Trim28 were from this study. Target gene list for Tcf3 was obtained from Cole et al. (2008). Target gene lists for Nanog, Sox2, Oct4, Nac1, Dax1, Klf4, Zfp281, and c-Myc were obtained from Kim et al. (2008). Chromosome loci coordinates for Esrrb, Smad1, Stat3, and Zfx were obtained from (Chen et al. 2008) and translated into gene lists by mapping them to genomic regions between 8 kb upstream of and 2 kb downstream from TSS from March 2006 released (mm8) mouse genome annotation. Only one transcript was used for genes with multiple transcripts with same TSS presented in RefSeq, adding up to a total 19,253 genes in the mouse genome. A target gene list matrix was then assembled using the 19,253 genes as rows, and the target gene lists of the 15 pluripotency factors as columns. In each column, the target gene list was converted to a list of 1s and 0s, with 1s in target genes of that pluripotency factor and 0s in nontarget genes. Overall similarities between the 15 pluripotency factors were determined by performing hierarchical clustering on the above matrix using Euclidian distance as metric, and the correlation coefficients between every two factors were calculated using the above matrix as well.
For Figure 5C, MAS5 analyzed EB differentiation data was obtained from StemBase experiment 201 (Porter et al. 2007). Only the 18,367 probes, corresponding to 11,662 genes, with present calls in more than three of the 33 samples (11 time points, three replicates for each time point) were selected. The expression values of these probes were converted to Log2 values and then cross time points mean subtracted for each probe. Target genes of Cnot3, Trim29, c-Myc, and Zfx were obtained as described above, and 326 genes were found to be co-occupied by all four. Probes against these 326 genes were extracted from the filtered EB differentiation data, resulting in a total of 481 probes, corresponding to 264 genes. Out of these probes, 420 of them, corresponding to 241 genes, showed a significant change in expression level during the entire course of differentiation (defined by maximum Log2 value − minimum Log2 value > 0.5), and were used to generate the heat map. These probes were sorted based on their expressions in ES cells (time 0).
For Figure 5D, heat maps for the ΔLog2 values between day 14 and day 0 for “all genes” (11,662 genes in the filtered EB differentiation data, see above) and the Cnot3, Trim29, c-Myc, and Zfx common target genes “CTMZ” (264 genes, see above) were plotted with the R program. The average ΔLog2 values for “all genes” and “CTMZ” were calculated and the two-tailed Student t-test was used to determine the P-value.
For Figure 5E, 245 genes whose promoters were co-occupied by Nanog, Sox2, and Oct4 were obtained from Kim et al. (2008). Three-hundred-twendy-six genes whose promoters were co-occupied by Cnot3, Trim28, c-Myc, and Zfx were identified as described above. GO analysis was done with Ingenuity pathway analysis software.
Acknowledgments
We thank Dr. Austin G. Smith for the Oct4GiP cells, Rene Maehr for scientific advice, and the Gene Expression Microarray Core at the DFCI for sample processing. G.H. is a fellow of the Helen Hay Whitney Foundation. J.K., Q.X., and Y.L. are Howard Hughes Medical Institute Research Associates. S.J.E. and S.H.O. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1769609.
Supplemental material is available at http://www.genesdev.org.
References
- Ben-Porath I., Thomson M.W., Carey V.J., Ge R., Bell G.W., Regev A., Weinberg R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T., Stracey C., Chambers I., Nichols J., Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- Cammas F., Mark M., Dolle P., Dierich A., Chambon P., Losson R. Mice lacking the transcriptional corepressor TIF1β are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- Cammas F., Herzog M., Lerouge T., Chambon P., Losson R. Association of the transcriptional corepressor TIF1β with heterochromatin protein 1 (HP1): An essential role for progression through differentiation. Genes & Dev. 2004;18:2147–2160. doi: 10.1101/gad.302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cole M.F., Johnstone S.E., Newman J.J., Kagey M.H., Young R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes & Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A., Timmers H.T. The eukaryotic Ccr4-not complex: A regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- Dejosez M., Krumenacker J.S., Zitur L.J., Passeri M., Chu L.F., Songyang Z., Thomson J.A., Zwaka T.P. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio T.G., Huff J.T., Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes & Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Keller G., Kennedy M., Papayannopoulou T., Wiles M.V. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S.Y., Qin J., Wong J., Cooney A.J., Liu D., et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Ng J.H., Ng H.H. Molecular framework underlying pluripotency. Cell Cycle. 2008;7:885–891. doi: 10.4161/cc.7.7.5636. [DOI] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin L., Puisieux A. C. elegans homologue of the Caf1 gene, which encodes a subunit of the CCR4–NOT complex, is essential for embryonic and larval development and for meiotic progression. Gene. 2005;358:73–81. doi: 10.1016/j.gene.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G., Mereghetti P., Mauri G., Pesole G. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32 (Web Server issue):W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter C.J., Palidwor G.A., Sandie R., Krzyzanowski P.M., Muro E.M., Perez-Iratxeta C., Andrade-Navarro M.A. StemBase: A resource for the analysis of stem cell gene expression data. Methods Mol. Biol. 2007;407:137–148. doi: 10.1007/978-1-59745-536-7_11. [DOI] [PubMed] [Google Scholar]
- Ryan R.F., Schultz D.C., Ayyanathan K., Singh P.B., Friedman J.R., Fredericks W.J., Rauscher F.J., 3rd KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: A potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wolf D., Goff S.P. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Smith A.G. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Nichols J., Evans E.P., Smith A.G. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Piao Y., Zhong J., Matoba R., Carter M.G., Wang Y., Goldberg I., Ko M.S. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr. Patterns. 2006;6:213–224. doi: 10.1016/j.modgep.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaessinger S., Busseau I., Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- Zhang J.Z., Gao W., Yang H.B., Zhang B., Zhu Z.Y., Xue Y.F. Screening for genes essential for mouse embryonic stem cell self-renewal using a subtractive RNA interference library. Stem Cells. 2006;24:2661–2668. doi: 10.1634/stemcells.2006-0017. [DOI] [PubMed] [Google Scholar]