Abstract
Translational regulation is heavily employed during developmental processes to control the timely accumulation of proteins independently of gene transcription. In particular, mRNA poly(A) tail metabolism in the cytoplasm is a key determinant for balancing an mRNA's translational output and its decay rate. Noncanonical poly(A) polymerases (PAPs), such as germline development defective-2 (GLD-2), can mediate poly(A) tail extension. Little is known about the regulation and functional complexity of cytoplasmic PAPs. Here we report the discovery of Caenorhabditis elegans GLD-4, a cytoplasmic PAP present in P granules that is orthologous to Trf4/5p from budding yeast. GLD-4 enzymatic activity is enhanced by its interaction with GLS-1, a protein associated with the RNA-binding protein GLD-3. GLD-4 is predominantly expressed in germ cells, and its activity is essential for early meiotic progression of male and female gametes in the absence of GLD-2. For commitment into female meiosis, both PAPs converge on at least one common target mRNA—i.e., gld-1 mRNA—and, as a consequence, counteract the repressive action of two PUF proteins and the putative deadenylase CCR-4. Together our findings suggest that two different cytoplasmic PAPs stabilize and translationally activate several meiotic mRNAs to provide a strong fail-safe mechanism for early meiotic progression.
Keywords: Translational control, poly(A) polymerases, PUF proteins, deadenylase, germ stem cell fate, meiotic progression
mRNA transcripts are the products of a stereotyped nuclear maturation process that is initiated at the site of RNA transcription and ends after its export from the nucleus into the cytoplasm. However, efficient protein synthesis is not an immediate consequence of mRNAs reaching the cytoplasm. In fact, a steadily increasing number of genes are found regulated post-transcriptionally, either before or at the level of translation. In particular, cytoplasmic events such as mRNA localization and storage precede bulk mRNA translation (Kindler et al. 2005; Keene 2007). mRNA fates are tightly controlled by their association with RNA-binding and RNA-modifying proteins that package mRNAs into higher-order ribonucleoprotein (RNP) complexes (Dreyfuss et al. 2002; Keene 2007).
The identity and function of cytoplasmic mRNA fate regulators stems to a large extent from their analysis during developmental processes (Kuersten and Goodwin 2003). One crucial mode of cytoplasmic regulation is centered on the 3′ end of an mRNA, controlling the dynamics of poly(A) tail metabolism. Its presence and length strongly influences the translational output and half-life of an mRNA (Richter 2007).
With each round of translation, the poly(A) tail is shortened; this eventually hampers translation and triggers RNA turnover (Jacobson and Peltz 1996). The conserved CCR4/NOT complex comprises deadenylase activity and was demonstrated in diverse organisms to trim the poly(A) tail of mRNAs (for review, see Parker and Sheth 2007). This complex consists of two 3′-to-5′ deadenylases—CCR4 and POP2/CAF1—and can be specifically targeted to mRNAs by their association with RNA-bound factors. Such evolutionarily conserved translational repressors include the RNA-binding proteins Smaug and Nanos in Drosophila (Semotok et al. 2005; Kadyrova et al. 2007), and members of the PUF (Pumilio/FBF) protein family in Saccharomyces cerevisae (Goldstrohm et al. 2006, 2007; Hook et al. 2007). Complete deadenylation triggers the 5′-to-3′ end mRNA degradation pathway. However, an RNA molecule with a short poly(A) tail can escape degradation and regain translational potential through cytoplasmic polyadenylation (Parker and Sheth 2007).
Our present molecular understanding of cytoplasmic polyadenylation is based largely on studies on Xenopus oocyte maturation (Richter 2007). During frog oogenesis, several maternal mRNAs are translationally dormant and are reactivated after meiosis resumes. In the case of cyclin B1 mRNA, an interplay between the activities of the noncanonical poly(A) polymerase (PAP) XGld2 and the poly(A)-specific ribonuclease (PARN) controls the dynamic length of the poly(A) tail (Kim and Richter 2006). PARN and XGld2 activities are influenced by the phosphorylation status of CPEB, which is bound to the cytoplasmic polyadenylation element (CPE) of a target mRNA. As PARN is the more active enzyme, the result is a net poly(A) tail shortening of the cyclin B1 mRNA. Upon progesterone stimulation, PARN dissociates from phosphorylated CPEB and XGld2 elongates the poly(A) tail of the CPEB-associated mRNA, thereby promoting its efficient translation and oocyte maturation (Kim and Richter 2006). Hence, meiotic progression is controlled by changing the character of CPEB from a translational repressor into a translational activator through phosphorylation. Recently, a connection between the ortholog of CPEB, Orb, and the germline development defective-2 (GLD-2)-type cytoplasmic PAP Wisp was established and found crucial for fly oogenesis (Benoit et al. 2008).
The development of the Caenorhabditis elegans hermaphrodite germline is another paradigm for studying translational control mechanisms. The underlying post-transcriptional events are best understood in two exemplary germ cell fate decisions: the mitosis/meiosis decision and the sperm/oocyte switch (Kimble and Crittenden 2007). Both decisions are controlled by a set of conserved RNA regulatory proteins that form a genetically redundant network in which the balance between translational activators and repressors enforces or restricts protein synthesis (Kimble and Crittenden 2007). For both decisions the target mRNAs are unique and illustrate the versatility of the molecular machinery. However, the molecular regulation of meiotic fate progression in worms and an involvement of poly(A) length control is poorly characterized.
At least two parallel genetic pathways are required in C. elegans for the entry into and commitment to meiosis (Kadyk and Kimble 1998). At the molecular level, two largely redundant PUF proteins, FBF-1 and FBF-2 (commonly referred to as FBF), promote adult stem cell mitosis by translationally repressing gld-1 and gld-3 mRNA, each encoding a member of the two dominating meiosis-promoting pathways (Crittenden et al. 2002; Eckmann et al. 2004). A commitment to the meiotic program is achieved by a combination of RNA-binding proteins that activate the meiotic cell fate and those that repress mitosis. In this context, a noncanonical cytoplasmic PAP complex was identified, consisting of the GLD-2/GLD-3 heterodimer, which enforces the switch into meiosis (Wang et al. 2002; Eckmann et al. 2004). GLD-3 is a Bicaudal-C type RNA-binding protein that stimulates GLD-2, the catalytic component of the PAP complex (Eckmann et al. 2002; Wang et al. 2002). Its only known target to date, gld-1 mRNA, encodes a conserved translational repressor protein that is proposed to enforce the meiotic switch by inactivating mitotic genes (Suh et al. 2006). Moreover, high GLD-1 accumulation is essential for meiotic commitment of oocytes; gld-1 mutant female germ cells fail to maintain meiosis and return from early pachytene to the mitotic cell cycle (Francis et al. 1995). However, GLD-1 levels are only partially controlled through the action of the GLD-2/GLD-3 PAP, and gld-2 mutant adults contain fully committed meiotic germ cells (Suh et al. 2006). Therefore, a more detailed molecular understanding of gld-1 mRNA regulation is pivotal to elucidate the regulation of meiotic progression.
In this study, we explore the complex regulation of gld-1 mRNA and thereby provide a molecular framework of likely cytoplasmic poly(A) tail dynamics in C. elegans meiosis. We identified a novel gene required for germline development, termed gld-4, that, like gld-2, is a member of the β-polymerase superfamily. GLD-4 polyadenylates and enhances the translation of reporter RNAs when tethered to an RNA target in Xenopus oocytes. However, this activity is further stimulated by the addition of GLS-1, which itself is an interactor of GLD-3. gld-4 activity is required for many aspects of germline development. Importantly, we find that gld-1 mRNA is a target of the GLD-4/GLS-1 complex and that gld-2 and gld-4 together are essential for high GLD-1 levels in the adult hermaphrodite germline. Further, we provide evidence that the conserved gene ccr-4, which encodes a putative deadenylase, may promote gld-1 mRNA degradation in part by being recruited through the translational repressor FBF. Our data on the regulation of gld-1 mRNA provide new insight into the molecular basis of how conserved RNA regulators ballance translational repression/degradation and activation to promote cell fates. Our finding of two different noncanonical cytoplasmic PAPs that may share a common targeting component—i.e., the RNA-binding protein GLD-3—is likely to be a general phenomenon.
Results
GLD-4 is a putative PAP
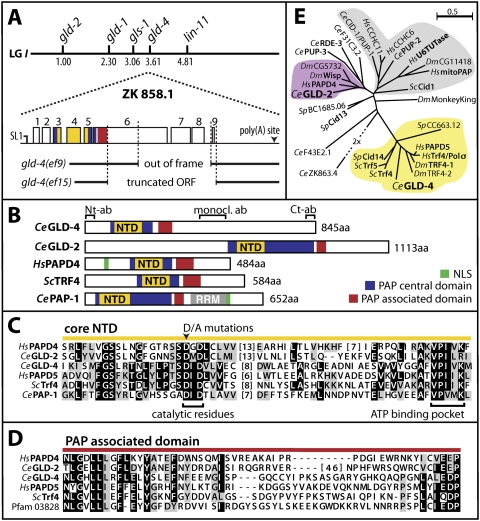
To identify additional proteins required for C. elegans germline development, we performed a yeast two-hybrid screen with GLS-1 (C36B1.8), a GLD-3-interacting protein, as bait. GLS-1 and GLD-3 regulate multiple germ cell fate decisions (A. Rybarska, M. Harterink, B. Jedamzik, A.P. Kupinski, M. Schmid, and C.R. Eckmann, in prep.). We analyzed ∼2 million transformants and isolated among the putative positive clones 19 cDNAs that correspond to the single predicted ORF ZK858.1, which we termed gld-4. Further cDNA analysis by RT–PCR of total RNA from mixed stage worms suggests that the gld-4 locus produces a single transcript of 3986 nucleotides (nt) in length that is SL1 trans-spliced (Fig. 1A). The gld-4 locus spans nine exons and eight introns encoding an 845-amino-acid-long protein (Fig. 1A,B).
Figure 1.
The gld-4 gene and its products. (A) The genetic position of gld-4 (ZK858.1) is predicted to map to position +3.60 on linkage group I. gld-4 mRNA is SL1 trans-spliced. Exons are shown as open boxes. gld-4 deletion alleles are indicated. (B) Predicted domain structure of several cytoplasmic PAPs and nuclear PAP-1. (NTD) nucleotidyltransferase domain; (RRM) RNA recognition motif; (NLS) nuclear localization signal; (Ab) Fragments for antibody production (N-terminal, C-terminal, monoclonal). (C) Amino acid sequence alignment of the core NTD. (Black boxes) Majority of identical amino acids; (gray boxes) majority of similar amino acids. Note that GLD-2 and GLD-4 are dissimilar to a large extent. (Ce) worm; (Hs) human; (Dm) fly; (Sc) budding yeast; (Sp) fission yeast. (D) Protein sequence alignment of the PAD; labels are as in C. (E) Unrooted phylogentic tree of all given nucleotidyl transferases based on a multiple sequence alignment of the Trf4/5 central domain (NTD and PAD). For details, see the Supplemental Material. Proteins with known biochemical activity are shown in bold. The gray branch contains mostly poly(U) polymerases.
GLD-4 contains two predicted functional domains: a nucleotidyltransferase domain (NTD), at amino acids 76–196, and a PAP-associated domain (PAD), covering amino acids 251–311 (Fig. 1B). NTDs transfer nucleotides onto 3′ hydroxyl groups of nucleic acids and are found in members of the DNA polymerase β super family (Holm and Sander 1995). PADs are paired with NTDs in proteins that possess polynucleotide transferase activity and are a hallmark of poly(A) and poly(U) polymerases (Martin and Keller 2007). Canonical PAPs are well conserved in evolution and interact with their RNA substrates through an RNA recognition motif (Fig. 1B). GLD-4 lacks any apparent RNA-binding domain, suggesting that it is more similar to the novel class of noncanonical RNA polymerases that includes PAPs such as Cid12 and GLD-2 (Wang et al. 2002; Win et al. 2006). In fact, the C-terminal half of GLD-4 is devoid of any predictable sequence motifs (Fig. 1B).
The overall domain conservation and organization of GLD-4 is most similar to budding yeast Trf4/5p and mammalian Pol σ/hPAPD5 proteins (Fig. 1B,E). GLD-4 and Trf4p share 50% and 52% similarity in the NTD and PAD, respectively (Fig. 1C,D). The conservation to the mammalian ortholog hPAPD5 is preserved to 50% similarity in the NTD and 52% similarity in the PAD. Furthermore, GLD-4 and its orthologs are similarly distant from the noncanonical PAP GLD-2 of C. elegans. The amino acid conservation in GLD-2's NTD and PAD in comparison with GLD-4 is 31% and 41%, respectively, and large gaps between the primary sequences exist (Fig. 1C,D). This evolutionary relationship places GLD-4 in the same branch of Trf4/5p-like molecules (Fig. 1E). Trf4/5p and Cid12 have been shown previously to possess polyadenylation activity (Motamedi et al. 2004; Haracska et al. 2005; Vanacova et al. 2005). All amino acids crucial for polymerase activity are conserved in GLD-4; the two aspartic acids essential for catalysis and the ATP-binding pocket within the core NTD are present (Fig. 1C). Therefore, GLD-4 shares key features with PAPs, suggesting its biochemical activity might be conserved. However, no significant conservation can be detected outside the functional domains of GLD-4 to its family members or GLD-2 (Fig. 1C).
We isolated two deletion mutations, gld-4(ef9) and gld-4(ef15), that affect the coding potential of the gld-4 locus (Fig. 1A). The gld-4(ef9) mutation removes half of exon 6 until the last intron 8 and is predicted to result in a C-terminal truncated form of GLD-4 (Fig. 1A). The gld-4(ef15) mutation eliminates more than half of the entire ORF, including the stop codon and a small part of the 3′ untranslated region (UTR) (Fig. 1A). Based on our phenotypic analysis of both alleles and their molecular defects, we rate gld-4(ef9) as a weaker allele than gld-4(ef15). Although gld-4(ef15) mRNA has the potential to code for the N-terminal domains of GLD-4, we were unable to detect the truncated protein by immunoblotting (see below). Hence, we consider gld-4(ef15) a strong loss-of-function allele and continued using it for our analysis to characterize GLD-4's biological functions.
GLD-4 is largely cytoplasmic and associates with P granules
As we expected gld-4 to be active in germ cells, we investigated GLD-4 protein distribution with the aid of several antibodies that were generated against three regions of recombinant GLD-4 (Fig. 1B). Two rabbit polyclonal sera, A161 and BABc, were raised against a C-terminal epitope and one rabbit serum, 449788, was raised against the N terminus. A mouse monoclonal antibody recognizing a central region of GLD-4 was also generated (Fig. 1B). All polyclonal sera were immunopurified and recognized consistently a single full-length (fl) protein of ∼125 kDa in wild-type extracts that was absent in both gld-4 mutants (Fig. 2A; Supplemental Fig. 1A). The observed size for GLD-4 deviates significantly from its theoretical weight of 94 kDa; however, the aberrant slower migration behavior of GLD-4 is consistently observed when expressed in various systems including bacteria (Supplemental Fig. 1B). Adult hermaphrodites and males expressed more GLD-4 than animals containing no germline (Fig. 2A). Although the N-terminal antiserum contained higher cross-reactivity to unspecific bands, we still detected a C-terminally truncated form of GLD-4 of the expected size difference in gld-4(ef9) but not in gld-4(ef15) mutants (Supplemental Fig. 1A). This is consistent with our genetic data of gld-4(ef15) being a strong loss-of-function mutation. We conclude that GLD-4 is predominantly expressed in germ cells and is absent in gld-4(ef15) animals.
Figure 2.
GLD-4 is enriched in the germline and associates with P granules. (A) GLD-4 (caret) is germline-enriched. Protein extracts of embryos (E), adult hermaphrodites of given genotypes probed with a rabbit GLD-4(Ct) antibody. (no GL) glp-1(q224ts) animals without germline; (Pos. cont.) Positive control of a LexA∷GLD-4 fusion produced in yeast. (B–F) Extruded germlines (B,C) and embryos (D–F) of wild-type (wt) and gld-4(ef15) adult hermaphrodites stained for chromatin (DAPI), P granules (anti-PGL-1), and GLD-4. (sp) Mature sperm at the proximal end (dashed line). Bar, 50 μm. Insets a–c in B represent a region in the mitotic germline as an optical section generated with an Apotome (Zeiss) microscope. (Inset a) DAPI. (Inset b) PGL-1. (Inset c) GLD-4 staining. (D) Two-cell embryo. (E) Approximately 100-cell embryo. (Arrowheads) Germ cell precursors Z2 and Z3. (F) Epitope-blocked anti-GLD-4(Ct) antibodies (F′′) fail to recognize P granules (F′) in a dividing two-cell wild-type embryo.
By immunocytochemistry, GLD-4 was found to be cytoplasmic in the hermaphrodite germline (Fig. 2B) and primordial germ cells throughout embryogenesis (Fig. 2D,E). Subcellularly, GLD-4 localizes also to cytoplasmic structures that are nuclear envelope-associated and the staining pattern is reminiscent of P granules, which are sites rich in RNA and RNA regulatory proteins (Kawasaki et al. 1998). Indeed, GLD-4 overlaps in colocalization experiments with PGL-1, a molecular marker for P granules (Fig. 2B [insets a–c], D–E′′). The staining pattern was observed with both C-terminal anti-GLD-4 antibodies and is reduced to background signals in the gld-4(ef15) allele (Fig. 2C), underscoring the specificity of our antibodies. The P granule association of GLD-4 was less apparent during pachytene, became obvious again in maturing oocytes, and was most prominently visible in developing embryos (Fig. 2D′′,E′′). In the late embryo, GLD-4 expression became limited to the germ cell lineage and its localization correlated perfectly with embryonic P granules (Fig. 2D′–E′′). We detected GLD-4 throughout embryonic development in the entire P-cell lineage, which gives rise to the two primordial germ cells Z2 and Z3 (Fig. 2E′′). Interestingly, we detected GLD-4 with GLS-1 coexpressed in the germline cytoplasm and colocalizing on germplasm granules (data not shown). As yet another specificity control, we performed immunocytochemistry with a peptide preblocked GLD-4 antibody, and no staining in the embryo was observed (Fig. 2F′′). Taken together, GLD-4 is a zygotic and maternally expressed protein that localizes predominantly to the cytoplasm of germ cells and associates with P granules. The continuous expression pattern of GLD-4 is consistent with a role during germline development and suggests putative cytoplasmic functions in mRNA control.
As yeast Trf4/5 proteins have been reported to reside in the nucleus (LaCava et al. 2005), we tested whether the cytoplasmic localization of GLD-4 is a unique feature of worms and expressed GLD-4 protein in HeLa tissue culture cells where it was found exclusively in the cell cytoplasm. hPAPD4 served as a nuclear-enriched protein control (Supplemental. Fig. 2). This observation suggests that GLD-4(fl) is, per se, a cytoplasmic protein.
gld-4 affects hermaphrodite germline development
To learn about biological functions of gld-4, we characterized our strongest allele of gld-4 in more detail. The majority of homozygote gld-4(ef15) hermaphrodites (∼74%, n = 86) are self-fertile with reduced brood sizes; the remaining gld-4(ef15) animals (∼26%, n = 86) were sterile. The sterility had two main reasons: an uncharacterized late oogenesis/fertilization defect (seven out of 86), and a failure of germ cells to adopt the oocyte cell fate (14 out of 86). In the latter case, only mature sperm cells were produced, suggesting either a cell fate switch defect or an early oogenesis defect. The process of spermatogenesis appears to be unaffected in hermaphrodites as mature sperm is produced and gld-4(ef15) males produce living cross-progeny with wild-type animals.
Although most adult gld-4(ef15) animals produced sperm and oocytes they were only partially fertile; gld-4(ef15) animals produced on average ∼65 progeny (n = 35), in contrast to ∼298 progeny of wild type (n = 20). Furthermore, due to high embryonic lethality, only ∼55% (n = 724) of the fertilized embryos eventually hatched (>99%, wild-type embryos). Some somatic defects were noted in hermaphrodites—for example, protruding vulvae—but their description is beyond the scope of this report. Given the low brood counts, fertile homozygote gld-4(ef15) hermaphrodites displayed abnormal germline development. Most strikingly, their germlines were on average ∼56% smaller than their wild-type counterparts (Fig. 2, cf. B and C); in the distal germline, ∼404 germ cells were present in a gld-4(ef15) (n = 10) gonadal arm compared with ∼954 germ cells in wild type (n = 10), suggesting a proliferation defect or premature entry into meiosis defect. Taken together, gld-4 is required for many aspects of germline development and embryogenesis. As some phenotypes are shared by the gls-1 mutant (see the Materials and Methods) a common molecular function is likely.
GLD-4 interacts specifically with GLS-1
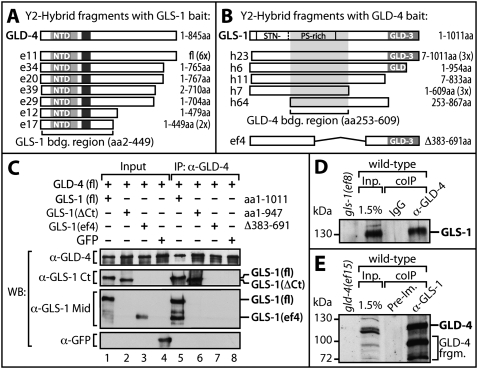
A specific interaction of GLS-1 and GLD-4 was originally discovered in yeast two-hybrid experiments (Fig. 3A,B). Using GLS-1 as a bait, we found that all gld-4 cDNAs identified encoded at least the N-terminal half of GLD-4, which covered all of its functional domains (Fig. 3A). Using GLD-4 as a bait, we screened 1.5 million transformants and recovered 16 gls-1 cDNAs (Fig. 3B). The region critical for GLD-4 interaction can be deduced from the various GLS-1 fragments isolated (Fig. 3B). Interestingly, this central region within GLS-1 is to a large extent deleted in the in-frame gls-1(ef4) mutant (Fig. 3B). All fragments recovered were tested for expression in the yeast (data not shown).
Figure 3.
GLD-4 specifically interacts with GLS-1. (A,B) Fragments recovered from a yeast two-hybrid screen with LexA-fused GLS-1(fl) (A) or GLD-4(fl) (B) as bait. The number of multiple isolates is shown in brackets. GLS-1 contains regions rich in serine (S)/threonine (T)/asparagine (N)/prolines (P) and has a GLD-3 interaction domain. Note that most of the GLD-4 interaction domain is missing in the in GLS-1(ef4) mutant form. (C) GLS-1 depends on the central region to bind GLD-4. Coexpression of GLS-1 variants with GLD-4 in insect cells with subsequent coimmunopurification (IP) using a monoclonal anti-GLD-4 antibody. GLS-1 variants are detected with antibodies specific for the C terminus (Ct) or middle portion (Mid) of GLS-1. Breakdown products of GLS-1(fl) are visible in lane 5. The GLD-3 interaction domain of GLS-1 is dispensable for GLD-4 binding. GFP as negative control. (D,E) An in vivo GLS-1/GLD-4 complex. GLS-1 and GLD-4 can be vice versa coimmunoprecipitated (coIP) from mixed stage worm extracts. Negative controls are given either as nonspecific IgG or preimmune serum (PI). (Inp) Input.
The interaction of GLD-4 and GLS-1 was recapitulated in insect cells using a baculovirus-mediated expression system (Fig. 3C). Complex formation was assessed by immunoprecipitation of GLD-4 and subsequent probing for GLS-1 derivatives with appropriate anti-GLS-1 antibodies. Coexpressed GFP protein served as a negative control. A specific interaction between full-length GLS-1 and GLD-4 was observed (Fig. 3C, lane 5), whereas GFP did not interact (Fig. 3C, lane 8). Additionally, derivatives of GLS-1 were probed for their interaction with GLD-4. GLS-1ΔCt, missing the C-terminal GLD-3 interaction region, is still capable of binding to GLD-4 (Fig. 3C, lane 6). However, a central region deleted GLS-1 protein, engineered after the gls-1(ef4) mutation was strongly compromised in associating with GLD-4 (Fig. 3C, lane 7).
To assess an in vivo interaction between GLD-4 and GLS-1 we used specific antibodies against each protein to copurify the respective proteins from worm extract (Fig. 3D,E). An immunoprecipitate using anti-GLD-4 antibodies, but not an equivalent amount of nonspecific anti-IgG, tested positive for endogenous GLS-1 (Fig. 3D). In the reciprocal experiment, we enriched GLD-4(fl) and several smaller pieces that are likely proteolytical N-terminal cleavage products of GLD-4 as they are absent in the mutant and preimmune control (Fig. 3E). In summary, we conclude that GLD-4 and GLS-1 are part of the same protein complex in vivo and that their interaction depends on specific domains. Furthermore, GLS-1 produced from the gls-1(ef4) allele is incapable of complex formation, which is consistent with its phenotypes being similar to gld-4 mutants.
GLD-4 and GLS-1 together form a functional PAP
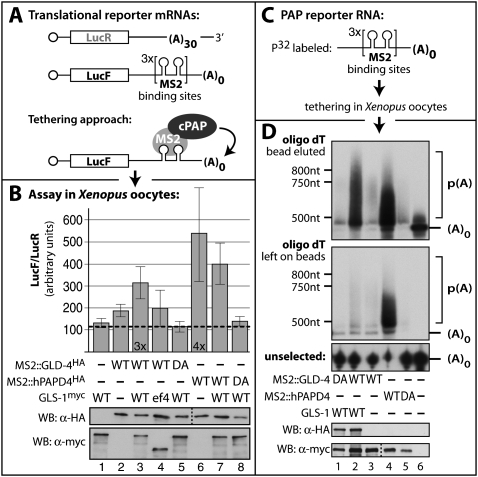
Given its sequence similarity to other nucleotidyl transferases, we investigated the biochemical function of GLD-4 and a possible role for its interaction partner, GLS-1 (Fig. 4). To test for a likely PAP activity, we probed for a translational stimulation of an mRNA reporter gene that encodes firefly luciferase (LucF). To this end, we made use of an RNA-tethering assay to circumvent the need for an intrinsic RNA-binding moiety, exploiting the high affinity and selective interaction between MS2 RNA-binding protein and its cognate target RNA-binding site (MS2bs) (Fig. 4A). We tested translational efficiency in Xenopus oocytes, which have been successfully employed to describe several poly(A) and poly(U) polymerases from various organisms (Kwak et al. 2004; Kwak and Wickens 2007). Our test proteins included a wild-type and a presumed active site DA mutant form of GLD-4 and hPAPD4, a known PAP (Kwak et al. 2004). The DA mutant contains an alanine substitution of one of the catalytic aspartic acids within the NTD (Fig. 1C). Two capped reporter mRNAs were transcribed in vitro: a LucF reporter mRNA containing six MS2bs but no poly(A) tail in its 3′UTR, and a renilla luciferase (LucR) reporter mRNA containing no MS2bs but a poly(A) tail of 30 nt length in its 3′UTR. The LucR reporter mRNA serves as an internal control for background translation activity (Fig. 4A). The assay was performed by sequentially injecting the mRNAs encoding test proteins and the RNA translational reporters into Xenopus oocytes. The luciferase activities of several independent experiments were measured and compared.
Figure 4.
GLD-4 is a PAP and requires GLS-1 for biochemical activity. (A) Translational simulation tested by a tethering assay in Xenopus oocytes. Schematic diagram of mRNAs encoding LucF and LucR. The LucR reporter mRNA contains a poly(A) tail of 30 adenosines (A). The LucF reporter contains MS2 RNA-binding sites in its 3′UTR and no poly(A) tail. MS2-fused PAPs are specifically recruited to the LucF mRNA reporter. (B) Measurements of relative luciferase activities from oocyte extracts. The expression levels of all test proteins are detected with Tag-specific antibodies. The translation of the LucF reporters is stimulated by wild-type (WT) hPAPD4 alone and by GLD-4 mostly the presence of wild-type GLS-1. Catalytic inactive mutants (DA) of both PAPs have no effect on translational enhancement. Data represented as ±SEM. (C) Tethering assay to asses the length of the poly(A) tail with a smaller MS2 RNA substrate. (D) GLD-4 and GLS-1 are required for efficient poly(A) tail elongation. Total RNA was extracted and poly(A) RNA selected on oligo dT beads. A water eluate is given on the top and the remaining strongly bead-bound RNAs were denatured and heat-eluted (middle). RNA polyadenylation is visible as an upward smear in polyacylamide gels. (Bottom) The unselected RNA in the extract supernatant is given as an integrity control.
In our dual luciferase assay, the stimulatory activity of wild-type hPAPD4 on LucF production was observed to about fourfold on average, when compared with the catalytic compromised form of hPAPD4 (Fig. 4B). Also, full-length tethered GLD-2 protein enhanced translation of the LucF reporter (data not shown). Wild-type GLD-4 was able to confer a modest ∼1.7-fold increase in LucF activity (Fig. 4B). This was independent of the GLD-4 concentration or the location of the MS2 fusion tag (data not shown). In order to test a possible positive influence of the known protein interactor GLS-1 onto GLD-4 activity, we coproduced in Xenopus oocytes GLS-1 by preinjection of a GLS-1-coding mRNA. The presence of wild-type GLS-1, but not GLS-1(ef4), increased the level of firefly luciferase activity by approximately threefold when coexpressed with wild-type, but not DA mutant, GLD-4 (Fig. 4B). This effect is GLD-4-specific, as coexpression of GLS-1 and hPAPD4 did not positively stimulate the measured relative luciferase activities compared with hPAPD4 alone (Fig. 4B). Little degradation of both luciferase mRNAs was observed during the course of the experiment and affected both mRNAs equally (data not shown). We conclude that the presence of full-length GLS-1 enhances the activity of tethered GLD-4 to stimulate the translational output of a luciferase reporter mRNA.
To visualize the effects of GLD-4 on the reporter mRNA more directly, we again exploited the RNA-tethering assay. However, we minimized the target RNA size to essentially the MS2-binding sites to facilitate better analysis by denaturing polyacrylamid gel electrophoresis (Fig. 4C). Radiolabeled target RNA was injected into Xenopus oocytes after the production of individual test proteins, and poly(A)-containing RNA was retrieved from total RNA preparations via oligo dT affinity purification. Wild-type, but not DA mutant, hPAPD4 displayed strong poly(A) activity, which is visible by a significantly slower migrating fraction of oligo dT-captured MS2 RNA (Fig. 4D, lane 4,5). Consistent with conferring a weak translational activity, we found that GLD-4 alone displayed little poly(A) activity by itself (Fig. 4D, lane 3). However, upon coexpression of GLS-1, wild-type GLD-4 became active in elongating the reporter RNA by ∼400 nt (Fig. 4D, lane 2). No activity was observed when the catalytic DA mutant was expressed in the presence of GLS-1 (Fig. 4D, lane 1). Taken together, GLD-4 promotes the addition of a poly(A) tail on an artificial substrate RNA that is otherwise lacking such a tail. Its PAP activity depends on an intact NTD domain and is enhanced by GLS-1, which serves as a stimulatory cofactor. The regulation of GLD-4 PAP activity by GLS-1 distinguishes GLD-4 from GLD-2, which is a more active PAP when tethered to mRNAs (Kwak et al. 2004; data not shown).
GLD-4 and GLD-2 are essential for meiotic progression
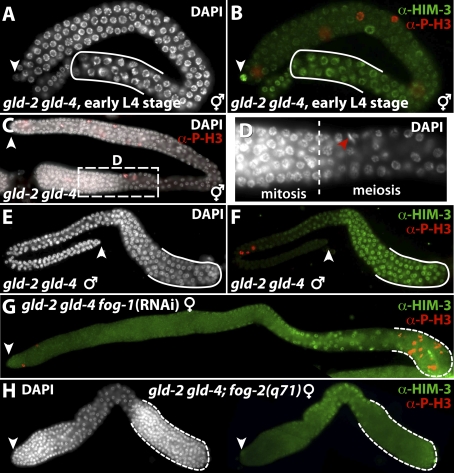
Germ cells of gld-2 single-mutant animals initiate meiosis and progress through pachytene of meiosis I; however, depending on their sexual fate, they arrest in later stages of meiosis (Kadyk and Kimble 1998). In contrast, most gld-4 single mutant germ cells complete meiosis upon initiation (this study). To examine the relationship between gld-2 and gld-4 we created animals lacking both PAP activities (Fig. 5). As expected, adult gld-2 gld-4 double-mutant animals of either sex were fully sterile, and displayed either an earlier meiotic arrest than gld-2 single mutants or a failure to commit to meiotic progression and a return to mitosis.
Figure 5.
GLD-4 and GLD-2 are essential for meiotic progression. (A–H) Extruded germlines stained for chromatin morphology (DAPI), active mitosis (anti-Phosho Histone3, P-H3), and the meiotic chromosome-localizing protein HIM-3, a lateral component of the synaptonemal complex (Dernburg et al. 1998; Zetka et al. 1999). (White arrowhead) Distal end. (A,B) L4 hermaphrodite gld-2 gld-4 mutant germlines possess meiotic cells at their proximal end that accumulate HIM-3 slowly on the developing chromosomal axis (solid line). The onset and progression of meiosis appears to be delayed compared with wild type. (C,D) Adult gld-2 gld-4 hermaphrodite germlines fail to commit to meiosis and return to mitosis (dashed line). (Red arrowhead) Anti-PH3-positive prometaphase nuclei. (E,F) Adult gld-2 gld-4 double-mutant hermaphrodite germlines are most proximally arrested in pachytene (solid line). (G,H) Feminized adult germlines of a fog-1(RNAi)-treated gld-2 gld-4 mutant and gld-2 gld-4; unc-51 fog-2 mutant hermaphrodite (shown in H). Proximal proliferating cells are present (dashed line). Note that a larger germline, concomitant with the presence of more proliferating cells in the distal part, is present in G.
First, we analyzed meiotic progression in the last larval stage of gld-2 gld-4 mutant hermaphrodites (n = 13). We observed that proximal germ cells had initiated meiosis but, unlike their single-mutant counterparts, had not yet progressed to pachytene I. Instead, their nuclei appeared to be only in leptotene and zygotene of prophase I (Fig. 5A,B). Although early pachytene nuclei were present in all young hermaphrodite gld-2 gld-4 adults, germ cells did not progress further in their meiotic program. Instead, germ cell nuclei either returned to mitosis or arrested in late pachytene I (68% or 32%, respectively; n = 150) (Fig. 5C,D). In contrast, gld-2 gld-4 mutant male germlines arrested exclusively in pachytene I (100%, n = 50) (Fig. 5E,F).
The apparent sex-specific difference of the meiotic progression defects argues that hermaphroditic gld-2 gld-4 germlines may exhibit a male fate when arresting in pachytene. In support of this hypothesis, we find that all feminized germlines of hermaphrodites display a return to mitosis phenotype (Fig. 5G,H). Taken together, our results suggested that the two PAPs, GLD-4 and GLD-2, are essential for early meiotic progression of male and female germ cells. Furthermore, female germ cells depend more strongly on both activities for meiotic commitment. The occurrence of pachytene I-arrested germ cells in hermaphrodites can be interpreted as a consequence of a sexual fate change. Alternatively, these germ cells might have escaped the first meiotic requirement for both PAP activities and arrested in a second even more PAP-sensitive step of meiosis. Interestingly, a return to the mitosis phenotype was also observed in the gls-1(ef4) gld-2 double mutant (M. Harterink and C.R. Eckmann, unpubl.).
GLD-4 and GLD-2 promote GLD-1 accumulation in the hermaphrodite
Strikingly, the gld-2 gld-4 adult hermaphrodite germline fate organization appears similar to the gld-1-null mutant phenotype; female, but not male, meiosis requires gld-1 activity for meiotic commitment during early pachytene (Francis et al. 1995). Given these phenotypic similarities, we speculated that GLD-4 and GLD-2 may act in parallel to regulate gld-1 next to other genes required for meiosis. Indeed, GLD-2 was found previously to influence the poly(A) status of gld-1 mRNA. In gld-2(0) mutants, GLD-1 protein accumulation is reduced in the L4 larvae but returns to wild-type levels in the adult animal (Suh et al. 2006). This prompted us to examine GLD-1 protein levels in various gld-4 genetic backgrounds.
We prepared extruded germlines from staged adult hermaphrodites under identical conditions and compared overall GLD-1 accumulation by immunocytochemistry (Fig. 6). As expected, we found GLD-1 expression in the gld-2 and gld-4 adult single mutants comparable with wild type (Fig. 6B,D,F). The double mutant of gld-2 gld-4, however, expressed very little to no GLD-1, suggesting a direct translational activation role of both PAPs (Fig. 6H).
Figure 6.
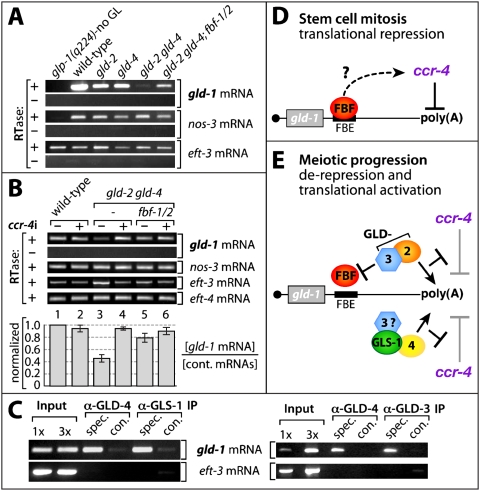
Efficient GLD-1 accumulation is dependent on gld-4 and gld-2 PAPs. (A–L) Extruded distal germlines stained with DAPI (left column) and for GLD-1 (right column). Note that double mutants of the same gene class are shortened and separated by a dash; e.g., fbf-1/2 is a double mutant of fbf-1 and fbf-2. ccr-4i indicates a ccr-4 knockdown by feeding RNAi. (M) Quantification of the immunofluorescence signal from the above germlines. Measured GLD-1 levels from unprocessed images were normalized to wild-type germlines. Data are represented as ±SEM. Genotype labels are as in C–L.
One potential complication might have been that the translational repressor FBF continuously prevented GLD-1 accumulation in the gld-2 gld-4 double mutant (Crittenden et al. 2002). Therefore, we constructed the gld-2 gld-4; fbf-1 fbf-2 quadruple mutant (Fig. 6J). Surprisingly, GLD-1 levels in the quadruple mutant remained very low; only a small increase compared with the gld-2 gld-4 double mutant was visible. This is consistent with the view that GLD-2 and GLD-4 provide a positive and rather direct effect on gld-1 mRNA translational activation.
Yeast and fly FBF-like proteins were found to act as translational repressors by causing mRNA turnover. They recruit the CCR4/NOT deadenylase complex, which leads ultimately to 5′ end-mediated RNA decay (Goldstrohm et al. 2006, 2007; Hook et al. 2007; Kadyrova et al. 2007). Hence, we speculated that FBF-mediated GLD-1 expression might work in a similar fashion. However, to untangle the two roles of a poly(A) tail, mRNA stability and translatability, we addressed the impact of CCR-4 reduction in the absence of both GLD-type PAPs and knocked down CCR-4 activity by feeding RNAi. No rescue of GLD-1 levels was observed in the gld-2 gld-4 double mutant (data not shown) or the gld-2 gld-4; fbf-1 fbf-2 quadruple mutant (Fig. 6L). These results further support an active role of GLD-4 and GLD-2 as a gld-1 translational activator and not just a role for inhibiting translational repression. This is consistent with our observation that tethered GLD-4 can extend the poly(A) tail of an mRNA.
GLD-4 and GLD-2 together prevent CCR-4-mediated gld-1 mRNA decay
Although GLD-4 and GLD-2 act as translational activators, we did observe an effect on mRNA decay. To investigate the cause of the low GLD-1 levels in the gld-2 gld-4 double mutant, we measured gld-1 mRNA levels by semiquantitative RT–PCR (Fig. 7A,B). We normalized mRNA amounts to ubiquitously expressed genes eft-3 and eft-4 and the germ cell-specific gene nos-3 (Kraemer et al. 1999; Maciejowski et al. 2005). Wild-type germ cells express high levels of gld-1 mRNA (Fig. 7A,B). gld-2 and gld-4 single-mutant animals produce less but robust levels of gld-1 mRNA, consistent with a high GLD-1 expression. On the contrary, gld-1 mRNA expression is strongly reduced in gld-2 gld-4 double mutants, consistent with their low GLD-1 protein levels. Upon FBF elimination in gld-2 gld-4 germ cells, we observed a reproducible increase of gld-1 mRNA levels, suggesting that PUF activity enhances gld-1 mRNA turnover in the absence of both PAPs (Fig. 7A,B). In situ hybridization experiments revealed consistently reduced amounts of gld-1 mRNA in all gld-2 gld-4 germlines (100%, n = 66 of one set) (Supplemental Fig. 3B,D). In contrast, gld-1 mRNA is expressed well in the most distal part of wild-type germlines and accumulated in meiotic germ cells (88%, n = 72 of one set) (Supplemental Fig. 3A,C).
Figure 7.
gld-1 mRNA levels and efficient translation require gld-4 and gld-2. (A) GLD-4/2 polymerases prevent FBF mediated gld-1 mRNA decay. Semiquantitative gene specific RT–PCR analysis of total RNA (labels are as in Fig. 6). cDNA production in the presence (+) or absence (−) of reverse transcriptase (RTase). glp-1(q224) animals lack a germline. (B) gld-1 mRNA decay depends on ccr-4. Semiquantitative RT–PCR analysis of control and ccr-4(RNAi)-treated animals. A representative set of experiments is given. The quantification of gld-1 mRNA levels is normalized to three control mRNAs. Data are represented as ±SEM. (C) gld-1 mRNA is an in vivo target of the GLD-4/GLS-1 complex. RT–PCR analysis of RNA molecules that coimmunoprecipitated with anti-GLD-4, anti-GLS-1, and anti-GLD-3 antibodies from wild-type extracts. (D,E) A model (based on genetic and biochemical evidence) of factors regulating gld-1 mRNA translation: negatively (barred lines) and positively (arrows). (D) The translational repressor FBF associates with gld-1 mRNA via the cis-regulatory sequence element FBE and may promote gld-1 mRNA translational repression by recruiting the putative CCR-4/NOT deadenylase complex. (E) GLD-3/GLD-2 and GLD-4/GLS-1 PAPs synergize to stabilize and translationally activate gld-1 mRNA for sufficient GLD-1 production to promote female meiotic progression. Further meiotic progression past pachytene I of male and female gametes may involve other mRNA targets in a similar manner. Both PAPs are likely to be targeted to the mRNA by GLD-3. The Bicaudal C-type RNA-binding protein GLD-3 may directly inhibit FBF (see the text).
Next, we carefully examined the effects of ccr-4 RNAi on gld-1 mRNA levels (Fig. 7B). Upon ccr-4 knockdown, gld-1 mRNA levels were restored to wild-type levels in gld-2 gld-4 mutants. Similarly, ccr-4 reduction in the gld-2 gld-4; fbf-1 fbf-2 quadruple mutant rescued gld-1 mRNA levels (Fig. 7B). This set of experiments suggests that gld-1 mRNA levels are highly responsive to a balance between the redundant activities of the PAPs GLD-4 and GLD-2, FBF, and the putative deadenylase CCR-4. We conclude that gld-1 mRNA degradation is enhanced by ccr-4 in the absence of both PAPs.
A prediction of this finding is a physical association of gld-1 mRNA with the GLD-4/GLS-1 PAP complex, as observed for the GLD-2/GLD-3 PAP complex. Therefore, we used anti-GLD-4 antibodies to coimmunopurify mRNA molecules specifically bound to the GLD-4 complex. Repeatedly, gld-1 mRNA, but not eft-3 mRNA, was detected in the immunoprecipitates from whole-worm extracts (Fig. 7C). Specific antibodies directed against GLS-1 and GLD-3 also enriched for gld-1 mRNA in the immunoprecipitate (Fig. 7C). We conclude that gld-1 mRNA is a direct target of the GLD-4/GLS-1 complex in vivo.
Discussion
In this study, we report the discovery of the novel P granular PAP GLD-4. GLD-4 is a noncanonical PAP found in a complex with GLS-1, a GLD-3-interacting protein. GLS-1 stimulates the enzymatic function of GLD-4 and both proteins affect several aspects of germline development. Strikingly, we find that two PAPs, GLD-4 and GLD-2, are together essential for early meiotic progression in either sex and are required for GLD-1 protein accumulation in the hermaphrodite germline. Consistent with this, we identified gld-1 mRNA as a target of the GLS-1/GLD-4 PAP complex. Furthermore, we find that the activity of both PAPs prevents gld-1 mRNA degradation mediated by two PUF proteins and CCR-4, a putative deadenylase. The regulation of a single mRNA by two redundantly operating PAPs provides a fail-safe mechanism that protects the mRNA from degradation and at the same time stimulates translation. These findings explain the observed genetic redundancies that have hampered a more thorough molecular understanding of underlying meiotic commitment in C. elegans.
GLD-4 is a new cytoplasmic PAP most similar to Trf4/5p
NTD-containing proteins are a large and diverse protein family that, when combined with a PAD, resemble the highly conserved canonical PAP (Holm and Sander 1995). Nuclear PAP is the prototype of all known polynucleotide polymerases and contains intrinsic RNA affinity, provided by an RNA-recognition-like motif (Martin et al. 2000). However, other nuclear noncanonical PAPs were identified, of which Trf4p and Trf5p from budding yeast represent the best-known examples (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006; Houseley and Tollervey 2006; Stevenson and Norbury 2006; Song and Kiledjian 2007; Wang et al. 2008). Trf4/5p associates with Air1/2p and Mtr4p to form the TRAMP complex and mediates nuclear poly(A) tail-stimulated RNA degradation (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). GLD-4 appears to be the closest worm ortholog of the conserved Trf4/5p family; its NTD and PAD regions are most similar to Trf4/5p and no other gene in the worm genome shows higher similarity. Evolutionary distance analysis of all NTD/PAD-containing proteins in the worm and their orthologous proteins from various organisms suggests a clear positioning of GLD-4 as the sole C. elegans representative of the Trf4/5p branch, which otherwise contains two highly similar protein pairs in investigated species (Fig. 1E). Nevertheless, the overall sequence conservation of GLD-4 to Trf4/5 proteins outside the catalytic region is extremely low and apparent functional differences exist. Trf4/5p functions in RNA surveillance to promote the degradation of aberrantly produced RNAs and RNA processing intermediates by the addition of a short poly(A) tail (LaCava et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006). In fission yeast, Cid12, an ortholog of Trf4/5p and GLD-4, is implicated in eliminating a wide variety of nuclear RNA targets (Wang et al. 2008). On the contrary, GLD-4 is predominantly cytoplasmic in C. elegans germ cells and when expressed in human tissue culture cells. We find gld-4 activity important for stabilizing gld-1 mRNA and putatively directly promoting efficient GLD-1 synthesis. In Xenopus oocytes, we observed that tethered GLD-4 possesses enzymatic activity similar to PAPs when coexpressed with GLS-1; a reporter mRNA is translationally up-regulated and a target RNA acquires a roughly 300- to 450-nt long poly(A) tail. We argue that GLD-4 and Trf4/5p contain similar enzymatic PAP functions, but their enzymatic processivity and biological functions differ, most likely due to the association with distinct cofactors in worms and yeast, respectively. The discovery of a stimulatory protein like GLS-1 might explain why several NTD-containing proteins were found to be biochemically inactive in similar tethering assays (Kwak et al. 2004; Kwak and Wickens 2007).
GLD-4 is a regulatory PAP and P granule component
The first described noncanonical cytoplasmic PAP similar to GLD-4 is GLD-2 (Wang et al. 2002). GLD-2 forms a heterodimer with the Bicaudal-C ortholog GLD-3 and regulates meiotic entry (Wang et al. 2002; Eckmann et al. 2004). GLD-2 relies on GLD-3 interaction for its polyadenylation activity and target specificity in vitro (Wang et al. 2002). However, a tethered GLD-2 fragment, covering the central NTD/PAD region, is able to adenylate a target mRNA without the presence of GLD-3 in Xenopus oocytes, suggesting an intrinsic enzymatic activity of GLD-2 (Kwak et al. 2004). These aspects of regulatory enzymatic behavior are similar to but distinct from GLD-4. GLD-4 resides in a complex with GLS-1, a novel cytoplasmic protein that regulates multiple cell fate decisions in the germline and is essential for oogenesis (A. Rybarska, M. Harterink, B. Jedamzik, A.P. Kupinski, M. Schmid, and C.R. Eckmann, in prep.). GLS-1 contains no predicted RNA-binding domains; however, it is a known interactor and regulator of the multi-KH domain-containing protein GLD-3 (A. Rybarska, M. Harterink, B. Jedamzik, A.P. Kupinski, M. Schmid, and C.R. Eckmann,, in prep.). Interestingly, GLD-3 uses a region partially overlapping with its first KH domain to bind to GLD-2, whereas GLD-3 binds to GLS-1 using a different region (Eckmann et al. 2004; A. Rybarska, M. Harterink, B. Jedamzik, A.P. Kupinski, M. Schmid, and C.R. Eckmann, in prep.), suggesting that GLD-3 might serve for both cytoplasmic PAPs as a link to their mRNA targets as GLD-3 associates with gld-1 mRNA in vivo (this study; Suh et al. 2006). However, we find GLD-4 PAP activity to be dependent on GLS-1; tethered GLD-4 is a poor PAP by itself but is stimulated by the coexpression of GLS-1. Strikingly, the GLS-1/GLD-4 complex requires an interaction surface on GLD-4 that overlaps entirely with its enzymatic domains, suggesting that GLS-1 might induce a structural rearrangement of GLD-4 crucial for catalytic activity. Remarkably, GLD-2's binding region to GLD-3 also involves the functional core region, including NTD and PAD, of GLD-2 (Wang et al. 2002). These features of GLD-2 and GLD-4 suggest a mechanistic principle of all GLD-type PAPs, providing a safety mechanism to prevent arbitrary translational activation of bulk mRNAs. Both GLD-type catalytic moieties are intrinsically inactive as monomers in solution and they carry no high RNA affinity by themselves. To gain access to a distinct subset of target mRNAs they rely on RNA-binding proteins and, in the case of GLD-4, even on an adaptor molecule; i.e., GLS-1. These features allow for a high degree of regulation and support their broad roles during development.
A second common feature of GLD-type PAPs in the germline is their association with cytoplasmic RNP structures, termed P granules. P granules are large RNP complexes specific to germ cells, implicated in RNA regulation and germ cell identity. Both subunits of the GLS-1/GLD-4 complex are found on P granules. We observed GLD-4 association with P granules throughout the adult germline and the entire embryonic germ cell progenitor lineage. GLD-2 associates with P granules most prevalently during early embryonic germ cell development and to a lesser extent in the post-embryonic germline (Wang et al. 2002). Interestingly, P granules harbor several RNA-binding proteins, some of which are conserved and might serve as RNA-targeting molecules for GLD-type PAPs such as the Bicaudal-C ortholog GLD-3 (Eckmann et al. 2002; Strome 2005). Together with their different single-mutant phenotypes, these temporal and spatial differences imply unique regulatory aspects for both PAPs. Yet their common subcellular location highlights P granules as a hub for translational RNA activation and regulation of disparate PAP activities.
Meiotic progression is regulated by two PAP complexes converging on a single target mRNA
In order to produce gametes, germ cells initiate and execute a complex program known as meiosis, which is coupled to a sex-specific differentiation process. The goal of meiosis is to produce haploid cells; however, developmental differences exist between individual stages of female and male meiosis. An essential factor for oogenic meiosis in C. elegans hermaphrodites is gld-1; its protein expression parallels early meiotic progression in oocytes (Francis et al. 1995; Jones et al. 1996). However, gld-1 mRNA is expressed ubiquitously throughout the entire germline and its translational control events were only partly understood (Jones et al. 1996; Crittenden et al. 2002; Suh et al. 2006).
The discovery of the GLS-1/GLD-4 PAP complex as a gld-1 mRNA regulator allows for a more detailed model of how GLD-1 accumulation might be controlled in the adult hermaphrodite germline. Our extended model of gld-1 mRNA translational regulation during mitosis and meiosis is presented in Figure 7, D and E. gld-1 mRNA is translationally repressed in the mitotic region by FBF, which is essential for adult mitosis (Crittenden et al. 2002; Lamont et al. 2004). We find that translational repression may involve the putative deadenylase CCR-4. In analogy to other PUF proteins, we suggest that FBF might lead to deadenylation of gld-1 mRNA and, ultimately, mRNA turnover by recruiting the putative worm CCR-4/NOT complex (Fig. 7D). Indeed, gld-1 mRNA instability, mediated by fbf and ccr-4, is clearly visible in the absence of both GLD-type PAPs; an elimination of all four factors stabilizes gld-1 mRNA.
Oogenic meiotic commitment especially requires gld-1 translational activation mediated by two redundantly acting PAP complexes. Upon entry into meiosis and/or during early meiosis, translational repression by FBF and CCR-4 is alleviated by the activity of two PAP complexes; i.e., GLD-2/GLD-3 and GLS-1/GLD-4 (Fig. 7E). In the GLD-type PAP double mutants, GLD-1 accumulation is dramatically impaired even in the absence of both fbf and ccr-4 activities. Concomitantly, female germ cells fail to progress into meiosis and return to mitosis instead. The simplest interpretation of these findings is that all involved factors act by controlling the length of the gld-1 mRNA poly(A) tail. Consistent with this view are our measurements of gld-1 mRNA and GLD-1 protein levels in various genetic backgrounds. These indicate a stronger reduction in GLD-1 protein accumulation than the difference in mRNA abundance would suggest (cf. Figs. 6M and 7B). Therefore, we propose that in mitotic germ cells the poly(A) tail of gld-1 mRNA is kept short by FBF and CCR-4, and in meiotic cells the gld-1 mRNA poly(A) tail is elongated and maintained after the sequential or combinatorial action of two PAPs (Fig. 7E). nos-3, an ortholog of Drosophila Nanos, was genetically implicated in the promotion of GLD-1 accumulation in parallel to gld-2, yet how NOS-3 influences gld-1 gene expression remains unclear (Hansen et al. 2004). We hypothesize that a synergism with gld-4 may be possible.
gld-1 translational activation might be developmentally regulated. In comparison with wild type, GLD-1 accumulation is less abundant in the last larval stage than in adults of gld-2 mutants (Suh et al. 2006). We find that most gld-4 mutant meiotic L4 germlines express GLD-1 levels comparable with wild type (M. Schmid and C.R. Eckmann, unpubl.). Therefore, prior to adulthood the GLS-1/GLD-4 PAP seems either not fully active or not important for gld-1 regulation (Fig. 7E). Alternatively, the added poly(A) tail of the GLD-4 PAP complex may not provide robust translational activity of gld-1 mRNA; hence, GLD-1 accumulates more slowly over time in meiotic cells of gld-2 mutants.
Interestingly, the long form of GLD-3 negatively influences FBF's RNA-binding activity by binding to FBF (Eckmann et al. 2002). Hence, GLD-3 was proposed to be an integral part of a translational switch mechanism (Suh et al. 2006). As GLD-3 interacts with GLD-2 and GLS-1 directly, it seems likely that GLD-3 might serve as a targeting unit for both PAP proteins (Fig. 7E). In this scenario, GLS-1 serves as a molecular bridge between GLD-4 and GLD-3, next to its role as enhancer of GLD-4 catalytic activity. This idea is supported by the overlapping protein expression of all three proteins in the cytoplasm and their localization to P granules (this study; A. Rybarska, M. Harterink, B. Jedamzik, A.P. Kupinski, M. Schmid, and C.R. Eckmann, in prep.). It is also consistent with the fact that gld-3(RNAi) knockdowns result in tumorogenic germlines that fail to progress through meiosis, suggesting that GLD-3 does have a more central role in promoting the meiotic cell fate (Belfiore et al. 2004). Whether both PAP complexes form on a single GLD-3 molecule, and thus on the same mRNA, is unclear, and is not a necessity for their communal function. Although our current and previous work support a crucial role of GLD-3 as a master regulator of the meiotic cell fate, it seems likely that other RNA-binding factors may target the GLS-1/GLD-4 PAP activity to regulate cell fate decisions. Additionally, it is clear that these redundant PAP complexes also have a larger regulatory repertoire than the gld-1 mRNA. For example, male germ cells require both PAPs for progressing past pachytene of meiosis I, although gld-1 activity is not required for male meiosis. Therefore, other shared mRNA targets are likely.
Our proposed model of gld-1 mRNA translational control differs from the mechanism observed in frog oocytes for cyclin B1 mRNA but shows similarities to other developmentally regulated mRNAs. Several PUF proteins were implicated recently in cytoplasmic poly(A) tail metabolism by recruiting the CCR4/NOT deadenylation machinery, either directly as in yeast or indirectly through Nanos as in Drosophila, and induce RNA turnover (Goldstrohm et al. 2007; Kadyrova et al. 2007). Whether and how FBF recruits CCR-4 to the mRNA in C. elegans is not known.
The use of targeting proteins to regulate the role of PAP activities in cell fate and patterning decisions is likely to be a general, broadly used mechanism in animal development. A conceptually analogous situation was recently reported underlying Drosophila oogenesis (Benoit et al. 2008). A sequential requirement of two PAPs, a canonical nuclear PAP isoform (Hiiragi) and a GLD-2-type cytoplasmic PAP (Wisp), was found to orchestrate oogenic meiotic progression. Intriguingly, both PAPs are found in one complex and bind the Orb protein, a CPEB ortholog. Wisp PAP is active by itself when tethered to mRNAs; however, the influence of Orb on PAP activity remains unexplored. Nevertheless, sharing a common RNA-binding protein, Orb, as a potential targeting device has obvious similarities to GLS-1/GLD4 and GLD-2 PAP activities sharing GLD-3. Our work supports the emerging theme that there are many levels at which PAP activity can be regulated and restricted to specific targets to carry out developmental decisions. For example, the existence of two functionally redundant PAPs provides an explanation of why a knockout mouse of mGLD-2 showed no apparent meiotic progression defect (Nakanishi et al. 2007). Both PAP activities are likely to have a surprisingly large and diverse set of social interactions with many targeting partners and mRNA targets. The changing dynamics of GLS-1/GLD-4 and GLD-2 PAP activities are a founding exemplar of what promises to be a general mechanism for developmental regulation.
Materials and methods
Strains and RNAi experiments
gld-4(ef9) and gld-4(ef15) were isolated in an EMS-induced PCR-based deletion screen (Kraemer et al. 1999), outcrossed against wild type (Bristol, N2) 10 times, and balanced with hT2g (I;III). Strains used in this study were LGI, gld-2(q497), gls-1(ef4); LGII, fbf-1(ok91), fbf-2(q704); LGIII, glp-1(q224); LGV, unc-51(e1189), and fog-2(q71). The gls-1(ef4) mutation is a hypomorph, deleting in-frame amino acids 384–692. Although the strain can be kept in homozygosis, oogenesis and embryogenesis defects were observed; spermatogenesis appears unaffected (see also A. Rybarska, M. Harterink, B. Jedamzik, A.P. Kupinski, M. Schmid, and C.R. Eckmann, in prep.). The generation of multiple mutant combinations and RNAi knockdown experiments are described in the Supplemental Material. All adult animals were staged as mid-L4 larvae and analyzed 24 h later.
GLD-4 and GLS-1 antibodies
The polyclonal rabbit antibodies 449788, BABc, and A161 were raised against GST fusions of an N-terminal (amino acids 1–68) and a C-terminal GLD-4 fragment (amino acids 784–845), respectively. The antibodies were affinity-purified using the same GLD-4 peptide fused to maltose-binding protein and coupled to a Hitrap-NHS column (GE Healthcare) according to the manufacturer's instructions. A monoclonal antibody, mo402A42-3, was raised against a large portion of GLD-4 (amino acids 421–845) fused to GST. The epitope recognized is between amino acids 420 and 525 (see the Supplemental Material). An affinity-purified rabbit polyclonal GLS-1 antibody was made against amino acids 249–576 (mid-antibody: rbC5C0) and was used for Figure 3, C and D. A mouse monoclonal antibody against amino acids 989–1011 (Ct-antibody: mo184C16) was used for Figure 3C.
Immunocytochemistry and GLD-1 quantification
Extruded germlines were prepared as described in Crittenden and Kimble (1999). Antibodies against GLD-1(1:400), HIM-3(1:200), and PGL-1(1:20000) were used as described previously (Jones et al. 1996; Kawasaki et al. 1998; Zetka et al. 1999). For secondary antibodies and embryo staining, see the Supplemental Material.
Germlines for each tested phenotype were prepared and stained together in the same well for GLD-1 quantifications. Unique pronounced phenotypic defects were used to identify individual genotypes and only equally counterstained germlines were used. Pictures were taken with identical settings on a Zeiss Imager Z1 epifluorescent upright microscope equipped with an AxioCam MRm CCD camera and were processed with AxioVision software version 1.4 (Zeiss). To measure anti-GLD-1 intensities, a focal plane was chosen where the syncytium was at its maximum width. The pixel intensities of germline stainings were measured in Photoshop C2 (Adobe). Three identical square boxes covering approximately two germ cell nuclei in width (100 × 100 pixels) were placed over the rachis of the distal arm along the distal-to-proximal axis at positions where the GLD-1 signal was the highest in wild type; i.e., just before the transition zone, in the transition zone, and in early pachytene. Equivalent regions were chosen in mutant germlines. The intensity values of three measurements were taken and averaged over three germlines for each genotype examined.
Biochemistry and GLD-4 PAP-tethering assays
Worm extract preparations, protein–protein, and protein–RNA coimmunopurifications; in vitro protein interaction assays; as well as the Xenopus tethering assay are described in detail in the Supplemental Material.
The GenBank ID of gld-4 is FJ610054, and the GenBank ID of gls-1 is FJ610055.
Acknowledgments
We thank members of the Eckmann laboratory, Karla Neugebauer, Dave Rudel, and Eli Knust for comments on the manuscript; the MPI-CBG protein expression facility and antibody facility for help in generating GLD-4 antibodies; the MPI-CBG animal care facility for maintaining a Xenopus laevis colony; Bianca Habermann for bioinformatic assistance; Britta Jedamzik for sharing her expertise in coimmunoprecipitation experiments; Joanne Stamford for technical help with the deletion library; Agata Rybarska and Ryuji Minasaki for characterizing several GLD-4 antisera; and Tim Schedl, Susan Strome, and Monique Zetka for antisera. C.E. is an MPI-CBG investigator, and this work was financially supported through the Max Planck Society.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.494009.
Supplemental material is available at http://www.genesdev.org.
References
- Belfiore M., Pugnale P., Saudan Z., Puoti A. Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development. 2004;131:2935–2945. doi: 10.1242/dev.01154. [DOI] [PubMed] [Google Scholar]
- Benoit P., Papin C., Kwak J.E., Wickens M., Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1979. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S.L., Kimble J. Confocal methods for Caenorhabditis elegans. Methods Mol. Biol. 1999;122:141–151. doi: 10.1385/1-59259-722-x:141. [DOI] [PubMed] [Google Scholar]
- Crittenden S.L., Bernstein D.S., Bachorik J.L., Thompson B.E., Gallegos M., Petcherski A.G., Moulder G., Barstead R., Wickens M., Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald K., Moulder G., Barstead R., Dresser M., Villeneuve A.M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Eckmann C.R., Kraemer B., Wickens M., Kimble J. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell. 2002;3:697–710. doi: 10.1016/s1534-5807(02)00322-2. [DOI] [PubMed] [Google Scholar]
- Eckmann C.R., Crittenden S.L., Suh N., Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D.E., Henras A.K., Chanfreau G.F. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Barton M.K., Kimble J., Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Seay D.J., Hook B.A., Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- Haracska L., Johnson R.E., Prakash L., Prakash S. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol. Cell. Biol. 2005;25:10183–10189. doi: 10.1128/MCB.25.22.10183-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- Hook B.A., Goldstrohm A.C., Seay D.J., Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 2007;282:15430–15438. doi: 10.1074/jbc.M611253200. [DOI] [PubMed] [Google Scholar]
- Houseley J., Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A., Peltz S.W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Jones A.R., Francis R., Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Kadyk L.C., Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kadyrova L.Y., Habara Y., Lee T.H., Wharton R.P. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.H., Kirchner J., Kaminker J., Wood W.B., Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Keene J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Richter J.D. Opposing polymerase–deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kimble J., Crittenden S.L. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in C. elegans. Annu. Rev. Cell Dev. Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Kindler S., Wang H., Richter D., Tiedge H. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B., Crittenden S., Gallegos M., Moulder G., Barstead R., Kimble J., Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Goodwin E.B. The power of the 3′ UTR: Translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Kwak J.E., Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.E., Wang L., Ballantyne S., Kimble J., Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lamont L.B., Crittenden S.L., Bernstein D., Wickens M., Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Maciejowski J., Ahn J.H., Cipriani P.G., Killian D.J., Chaudhary A.L., Lee J.I., Voutev R., Johnsen R.C., Baillie D.L., Gunsalus K.C., et al. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics. 2005;169:1997–2011. doi: 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Keller W., Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Nakanishi T., Kumagai S., Kimura M., Watanabe H., Sakurai T., Kashiwabara S., Baba T. Disruption of mouse poly(A) polymerase mGLD-2 does not alter polyadenylation status in oocytes and somatic cells. Biochem. Biophys. Res. Commun. 2007;364:14–19. doi: 10.1016/j.bbrc.2007.09.096. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Richter J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Semotok J.L., Cooperstock R.L., Pinder B.D., Vari H.K., Lipshitz H.D., Smibert C.A. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Song M.G., Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson A.L., Norbury C.J. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Strome S. Specification of the germ line. The C. elegans Research Community, editor. WormBook. 2005 doi: 10.1895/wormbook.1.9.1. WormBook. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Suh N., Jedamzik B., Eckmann C.R., Wickens M., Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Wang S.W., Stevenson A.L., Kearsey S.E., Watt S., Bahler J. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol. Cell. Biol. 2008;28:656–665. doi: 10.1128/MCB.01531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win T.Z., Stevenson A.L., Wang S.W. Fission yeast Cid12 has dual functions in chromosome segregation and checkpoint control. Mol. Cell. Biol. 2006;26:4435–4447. doi: 10.1128/MCB.02205-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Zetka M.C., Kawasaki I., Strome S., Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]