Abstract
As a stress response, senescence is a dynamic process involving multiple effector mechanisms whose combination determines the phenotypic quality. Here we identify autophagy as a new effector mechanism of senescence. Autophagy is activated during senescence and its activation is correlated with negative feedback in the PI3K–mammalian target of rapamycin (mTOR) pathway. A subset of autophagy-related genes are up-regulated during senescence: Overexpression of one of those genes, ULK3, induces autophagy and senescence. Furthermore, inhibition of autophagy delays the senescence phenotype, including senescence-associated secretion. Our data suggest that autophagy, and its consequent protein turnover, mediate the acquisition of the senescence phenotype.
Keywords: Senescence, autophagy, oncogene
Cellular senescence is a state of stable cell cycle arrest with active metabolism. Similar to apoptosis, senescence can be a failsafe program against a variety of cellular insults. In contrast to apoptosis, however, in which the cytotoxic signals converge to a common mechanism, senescence is typically a delayed stress response involving multiple effector mechanisms. These effector mechanisms include epigenetic regulation (Narita et al. 2006; Adams 2007), the DNA damage response (Bartkova et al. 2006; Di Micco et al. 2006; Mallette et al. 2007), and the senescence-associated secretion phenotype (Kortlever et al. 2006; Acosta et al. 2008; Coppé et al. 2008; Kuilman et al. 2008; Wajapeyee et al. 2008). The relative contribution of these effectors varies depending on the trigger and cell type.
Oncogene-induced senescence (OIS) illustrates well the tumor-suppressive role of senescence (Collado and Serrano 2006). The initial phenotype of oncogene induction is a highly proliferative state, which mimics transformation. However, this mitotic burst is gradually replaced by senescence. Although it has been proposed that global and progressive epigenetic alterations play a crucial role in OIS (Narita et al. 2006), the precise mechanism by which cells can achieve such a dramatic change is still unclear.
Autophagy is a genetically regulated program responsible for the turnover of cellular proteins and damaged or superfluous organelles. This evolutionarily conserved process is characterized by the formation of double-membrane cytosolic vesicles, autophagosomes, which sequester cytoplasmic content and deliver it to lysosomes (Ohsumi 2001; Klionsky et al. 2007; Mizushima et al. 2008). Autophagy is often associated with acute metabolic changes and rapid protein replacement. For example, autophagy is required for preimplantation development, where maternal proteins are recycled by autophagy (Tsukamoto et al. 2008). Autophagy is also required for survival in the early neonatal starvation period (Kuma et al. 2004; Komatsu et al. 2005). In addition to these physiological conditions, cytotoxic stimuli can also activate autophagy, but its precise role as a stress response is unclear. Here we identify autophagy as a new effector mechanism of senescence, important for the rapid protein remodelling required to make the efficient transition from a proliferative to a senescent state.
Results and Discussion
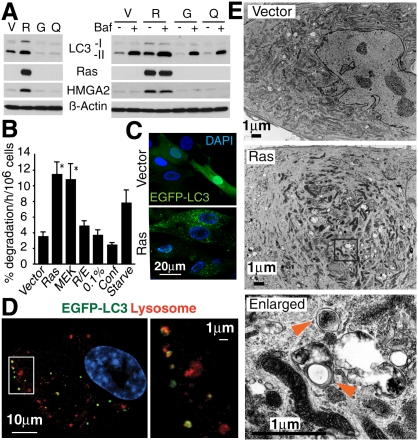
To determine whether autophagy is involved in senescence, we first measured the activity of autophagy during OIS in IMR90 human diploid fibroblasts (HDFs). MAP1LC3 (LC3) is a widely used marker of autophagy. The lipid-conjugated form of LC3 (LC3-II) localizes to the membranes of autophagosomes. It can be separated from the nonconjugated form (LC3-I) by immunoblotting. Senescence was induced by retroviral-mediated expression of oncogenic ras (H-rasV12) as described previously (Narita et al. 2006). Immunoblot analysis showed that LC3-II was up-regulated in Ras senescent cells compared with the control cells (Fig. 1A), indicating the accumulation of autophagosomes in Ras senescent cells. There was no increase of LC3-II in quiescent cells, indicating that the accumulation of autophagosomes is associated with senescence rather than cell cycle arrest (Fig. 1A). Similar results were obtained in other HDFs, including BJ cells and embryonic skin fibroblasts (Supplemental Fig. S1). Since LC3-II is itself subject to autophagic protein degradation due to its localization to autophagosomes, LC3-II also accumulates when the process of autophagosome–lysosome fusion or downstream protein degradation is impaired (Klionsky et al. 2007). To exclude this possibility, we also assessed the accumulation of LC3-II in the presence of Bafilomycin A1, a specific inhibitor of the vacuolar type H(+)-ATPase, which inhibits lysosome acidification. As expected, rapid accumulation of LC3-II, but not LC3-I, was induced in growing and quiescent cells (Fig. 1A). In the same conditions, Ras senescent cells also exhibited further accumulation of LC3-II, but to a much higher level than that of the treated control cells. The same results were obtained using lysosomal protease inhibitors (data not shown). These data suggest that the autophagic flux is intact in these cells. To further confirm the activation of autophagy during OIS, we performed protein degradation assays. Senescence, induced either by Ras or its downstream effector MEK, showed a significant increase in the degradation of long-lived proteins, while quiescent conditions did not show any difference from growing cells (Fig. 1B). The adenoviral oncoprotein E1A, which blocks the senescence machinery, cancelled the increase in protein degradation and LC3-II accumulation caused by Ras, underscoring the specific association between OIS and autophagy (Fig. 1B; data not shown).
Figure 1.
Autophagic activity is up-regulated in OIS. (A) Immunoblot analysis of LC3 in IMR90 cells expressing H-rasV12 (R) or vector (V) at 7 d post-antibiotic selection (PS7), uninfected growing cells (G) and quiescent cells (Q) generated by 5 d confluency. (HMGA2) Marker of senescence; (β-actin) loading control; (Bafilomycin A1 [Baf] treatment) 100 mM for 3 h before extraction. (B) Long-lived protein degradation analysis in cells expressing Vector, H-rasV12 (Ras), MEKQ56P (MEK), or H-rasV12 and E1A (R/E) at PS9. Quiescent cells were generated either by 3 d 0.1% serum (0.1%) or 5 d confluency (Conf). Cells cultured in DMEM without amino acids or serum for 4 h (Starve) serve as a positive control. Values are mean ± SEM. (*) P < 0.05 relative to vector. (C) Confocal images of cells expressing EGFP-LC3B and either vector or H-rasV12. (D) Confocal images for EGFP-LC3B and LysoTracker in Ras senescent cells. Cells were labeled with 200 mM LysoTracker for 2 h. (E) Electron microscopy images of vector and Ras senescent cells. Arrowheads show representative autophagic vacuoles.
We also monitored LC3's subcellular localization pattern during senescence using EGFP-LC3-expressing cells. Growing cells exhibited a diffuse localization of EGFP-LC3, with occasional puncta in the cytoplasm indicating the basal activity of autophagy (Fig. 1C). In marked contrast, we found prominent EGFP-LC3 puncta in Ras senescent cells (Fig. 1C). The lysosomal compartment, probed using LysoTracker (a dye detecting low pH) was more prominent in OIS cells, indicating that the lysosomal arm of autophagy is also activated during OIS (data not shown). Smaller lysosomes, <1 μm, preferentially colocalized with the EGFP-LC3 puncta, indicating the formation of autolysosomes (Fig. 1D). These changes were confirmed by electron microscopy, which revealed that Ras-expressing cells exhibited the different stages of autophagic vacuole formation (Fig. 1E). Altogether, we concluded that OIS is accompanied by elevated autophagy activity.
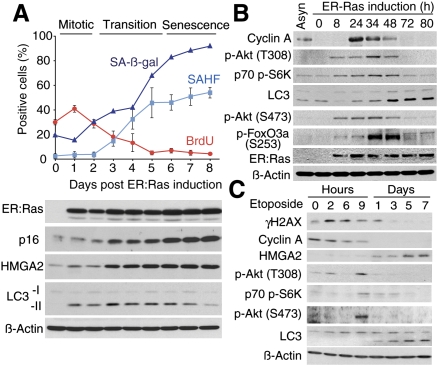
To determine the kinetics of autophagy regulation in OIS, we stably expressed a 4-hydroxytamoxifen (4-OHT)-inducible ER:Ras fusion protein in IMR90 cells. This protein is comprised of a mutant form of the estrogen receptor ligand-binding domain and H-RasV12. Typically, the initial “mitotic phase” in this system occurs around day 1 and the “senescence phase” after around days 5–6, with the “transition phase” in between (Fig. 2A). LC3-II was up-regulated, peaking during the transition phase. LC3-II levels gradually decreased in the later time points, likely due to continuously active autophagy (Fig. 2A).
Figure 2.
Correlation between negative feedback on mTOR activity and autophagy induction during stress-induced senescence. (A) ER:Ras-expressing cells were given 100 nM 4-OHT and assessed for SA-β-gal, BrdU incorporation, senescence-associated heterochromatic foci (SAHF) formation, and expression of the indicated proteins at the various time points. Each value of SAHF and BrdU represents the mean ± SEM. For SA-β-gal a representative value is shown. p16 and HMGA2 serve as markers of senescence. (B) Cells were synchronized in G0 using low serum conditions (0.1%), then released by adding 4-OHT together with serum (10%). Cyclin A; cell cycle marker. (C) DNA damage was induced by a 48-h Etoposide (100 μM) treatment. γH2AX, cyclin A, and HMGA2 serve as markers of DNA damage, cell cycle, and senescence, respectively.
A key regulator of protein metabolism, the mammalian target of rapamycin (mTOR), is a serine/threonine protein kinase, which negatively regulates autophagy (Mathew et al. 2007). mTOR forms two distinct signaling complexes, mTOR complex 1 (mTORC1) and mTORC2 (Sabatini 2006). mTORC1 is a downstream effector of the Ras/PI3K pathway. Thus, Ras signaling should suppress autophagy. However, a recent study identified a negative feedback loop in the PI3K pathway that is critical for OIS (Courtois-Cox et al. 2006). To examine the activity of mTORC1 during Ras senescence, we assessed the phosphorylation status of p70 S6 Kinase (S6K), an mTORC1 substrate, in ER:Ras-IMR90 cells. In this experiment, to achieve maximum homogeneity in the cell population, we first synchronized the cells in G0 using low serum conditions and then added 4-OHT together with serum. As expected, the phosphorylated (p-) form of S6K was initially up-regulated in response to the mitotic signal, while it decreased during the transition phase (Fig. 2B). The timing of the down-regulation of mTORC1 activity correlated with the activation of autophagy and cell cycle exit assessed by cyclin A (Fig. 2B). The level of p-AKT on Thr308, an upstream regulator of mTORC1, showed a similar pattern to p-S6K (Fig. 2B). These data suggest that the delayed inactivation of the mTORC1 pathway is a mechanism for autophagy induction during OIS.
mTORC2 is an AKT S473 kinase, and AKT thus activated then phosphorylates FoxO3a and inhibits its transcriptional activity. Recent studies identified FoxO3a as a transcription factor responsible for a subset of autophagy-related (ATG) genes as well as other autophagy regulators (Mammucari et al. 2007; Zhao et al. 2007). We assessed mTORC2 kinase activity and found a transient increase in p-AKT (S473) and its target p-FoxO3a (S253). The kinetics of mTORC2 activity paralleled that of mTORC1, indicating that a negative feedback loop also exists in the mTORC2 pathway during Ras senescence (Fig. 2B).
We next tested whether autophagy activation and the concomitant negative feedback in the mTOR pathways occur during DNA damage-triggered senescence. Interestingly, etoposide or γ-irradiation treatment induced the transient activation of mTORC1 and TORC2 in the acute phase, even though these cells did not undergo the transient mitotic burst (Fig. 2C; data not shown). Subsequent down-regulation of the activity of mTORC1 and 2 coincided with LC3-II formation, cell cycle arrest, and senescence-associated HMGA2 up-regulation (Fig. 2C). The striking similarity in the kinetics of the mTORCs and autophagy activities during Ras- and DNA damage senescence suggests a mechanistic link between mTOR signaling and autophagy induction during senescence, although the possibility that an mTOR-independent mechanism could also induce autophagy is not excluded.
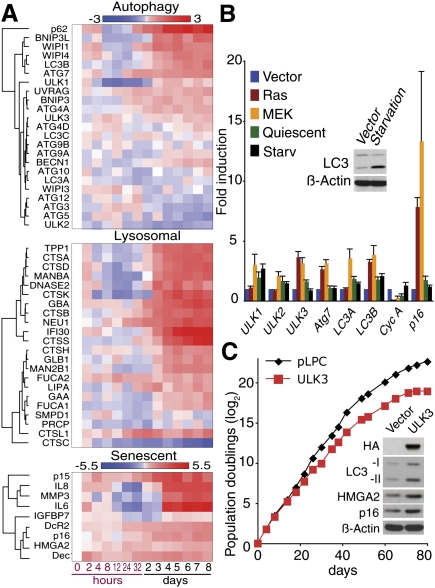
To further examine the involvement of the mTORC2–FoxO3a axis, we assessed the dynamic transcriptional profile of genes involved in autophagy and senescence in microarray experiments. A subset of ATG genes and other autophagy regulators were initially diminished during the mitotic phase, where FoxO3a was highly phosphorylated (Figs. 2B, 3A). As the level of phosphorylation of FoxO3a dropped (Fig. 2B), the suppression of these genes was removed and hence they were up-regulated during the transition to the senescence phase. This indicates a dynamic correlation between FoxO3a activity and the expression of these genes (Fig. 3A), which include direct targets of FoxO3a: LC3B (a human homolog of Atg8), Bnip3, and Bnip3L (although Bnip3 and Bnip3L also have other functions) (Mammucari et al. 2007; Zhao et al. 2007). Notably, LC3B mRNA showed a steady up-regulation after the mitotic phase (Fig. 3A; Supplemental Fig. S2). Together with the pattern of LC3-II levels, which rather decreased at the later time points (Fig. 2A), these data further suggest highly active autophagy during OIS. Consistently, using ONCOMINE (http://www.oncomine.org), a Web-based data-mining platform, we found that some autophagy regulators, including LC3 and Bnip3L, were up-regulated in human nevi and prostatic intraepithelial neoplasia, models of in vivo OIS (Supplemental Fig. S3; Chen et al. 2005; Michaloglou et al. 2005; Majumder et al. 2008).
Figure 3.
Regulation of autophagy and senescence genes after Ras induction. (A) Transcriptional profiles of autophagy, lysosomal and senescent genes during OIS. Total RNA was extracted from ER:Ras cells at the indicated time points after 4-OHT addition and assayed on Illumina microarrays. The data are the weighted mean of three independent experiments. Log fold changes compared with time 0 are shown. (B) Expression of the indicated genes was assessed by qPCR in cells expressing H-rasV12 (Ras), MEKQ56P (MEK) or Vector at PS8. (Quiescent) Five days confluent. Amino acid-starved cells (Starv) were generated as in Figure 1B. Cyclin A confirms cell cycle arrest. (p16) Senescence marker. Values are mean ± SEM. (C) Growth curves of IMR90 cells expressing vector or HA-ULK3. Data are representative of three independent experiments. Inset shows immunoblot analysis at day 18.
Consistent with the prominent lysosomal compartments in Ras senescent cells, many lysosomal genes showed a similar expression pattern to the up-regulated ATG genes (Fig. 3A). One such gene was GLB1, whose product is believed to be responsible for senescence-associated β-galactosidase (SA-β-gal) activity (Lee et al. 2006). These data suggest a coordinated regulation of a subset of ATG and lysosomal genes during OIS.
Senescence-associated secreted factors, such as IL6, IL8, IGFBP7, and MMP3, also showed similar expression kinetics, while other senescence-associated genes showed a rather steady increase over the time course (Fig. 3A). These secreted factors were highly abundant, and their concomitant expression with genes involved in autophagy is further indicative of rapid protein turnover during OIS.
Quantitative PCR (qPCR) confirmed a distinct ATG gene expression pattern in Ras-, MEK-, and etoposide-induced senescent cells, but not in quiescent cells (Fig. 3B; Supplemental Fig. S4). The ATG gene expression profile of the senescent cells was unique, since amino acid starvation, which mimics a physiological trigger of autophagy, did not induce a similar expression pattern of ATG genes (Fig. 3B).
ATG1, a serine/threonine protein kinase, is an upstream regulator of autophagy under mTORC1 regulation, and overexpression of the Drosophila homolog of Atg1 is sufficient to induce autophagy in fed animals (Scott et al. 2007). There are potentially five human homologs of Atg1, among which UNC-51-like kinase (ULK) 1 and ULK3 are consistently up-regulated in senescent cells, and ULK3 in particular was commonly up-regulated in all senescent cells tested (Fig. 3A,B; Supplemental Fig. S4). Thus, we assessed the impact of ULK3 overexpression on autophagy and senescence. Since ULK3 has not been implicated in autophagy as yet, we first tested whether ULK3 localizes to the elongating isolation membrane (the precursor of the autophagosome), to which the ATG12-5 conjugate localizes. Similar to results previously shown for ULK1/2 (Hara et al. 2008), ULK3 showed a punctate pattern upon autophagy activation by amino acid and serum depletion (Supplemental Fig. S5). Some of the ULK3 puncta colocalized to endogenous ATG12, indicating that ULK3 is involved in autophagy. Next, we found that stable overexpression of ULK3 was sufficient to induce autophagy as well as premature senescence, suggesting a causative role for active autophagy in senescence (Fig. 3C). Altogether, our data suggest that the mTORC1–ATG1 and mTORC2–FoxO3a pathways are coordinately involved in the induction of autophagy during senescence.
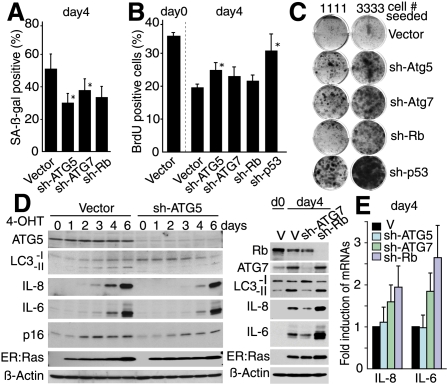
To assess the requirement for autophagy in senescence, we generated shRNAs targeting ATG5 and ATG7, essential genes for autophagy (Kuma et al. 2004; Komatsu et al. 2005). Knockdown of ATG5 or ATG7 attenuated the activation of autophagy by Ras (see Fig. 4D). ATG gene knockdown caused SA-β-gal activity to be delayed (Fig. 4A; Supplemental Fig. S6A). SA-β-gal activity is the most widely used marker of senescence, although its functional relevance in senescence is unclear. Our data suggest that SA-β-gal activity, and perhaps GLB1 up-regulation (Fig. 3A), might reflect the activity of autophagy, at least in part. Next, we assessed the functional requirement for autophagy in senescence. Although IMR90 cells expressing sh-ATG5 or sh-ATG7 showed only a modest impact on BrdU incorporation and chromatin phenotype during the transition phase of Ras senescence (Fig. 4B; Supplemental Fig. S6B), colony formation analysis in ER:Ras-expressing BJ cells, in which bypass is easier than in IMR90 cells, showed a substantial increase in senescence bypass by depletion of ATG5 or ATG7, to levels higher than those seen with Rb depletion (Fig. 4C). Our preliminary data showed that knockdown of ATG5/7 in established ras senescent BJ cells failed to reverse the senescence arrest (data not shown). These data suggest that autophagy contributes to, at least, the establishment of senescence arrest.
Figure 4.
Autophagy contributes to the transition to the OIS phenotype. (A) SA-β-gal activity and (B) BrdU incorporation in ER:Ras IMR90 expressing the indicated shRNAs at day 4 after 4-OHT treatment. *P < 0.05 relative to vector. (C) The indicated ER:Ras populations of BJ cells were assessed for colony formation in the presence of 4-OHT. (D,E) Autophagy deficiency delays cytokine production as assessed by immunoblotting (D) and qPCR (E) of ER:Ras IMR90 cells. Values are mean ± SEM of at least three experiments (A,B,E).
The dramatic phenotypic change during the transition phase is accompanied by increased secretion of several key cytokines that play a prominent role in OIS, including IL6 and IL8 (Acosta et al. 2008; Kuilman et al. 2008). We reasoned that rapid protein turnover, which involves autophagy coupled with active protein synthesis, might facilitate this process through protein remodeling. Indeed, senescent cells are typically enlarged, and we confirmed that the protein quantity per cell was higher in Ras senescent cells (Supplemental Fig. S7A). In addition, we observed an increased amino acid uptake in Ras senescent cells (Supplemental Fig. S7B), indicating that protein synthesis is also active during senescence. Strikingly, ATG5 or ATG7 depletion delayed IL6 and IL8 production (Fig. 4D). This effect was not observed by knockdown of Rb, suggesting that the inhibition of IL6/8 production is not a consequence of delayed senescence. The mRNA levels of IL6/8 were often even higher in sh-ATG5/7-expressing cells, indicating that the repression of IL6/8 is post-transcriptional (Fig. 4E). Of note, IL6/8 were further up-regulated both in protein and mRNA levels by sh-Rb, which did not perturb autophagy activity (Fig. 4D,E). These results highlight the functional relevance of autophagy in senescence establishment.
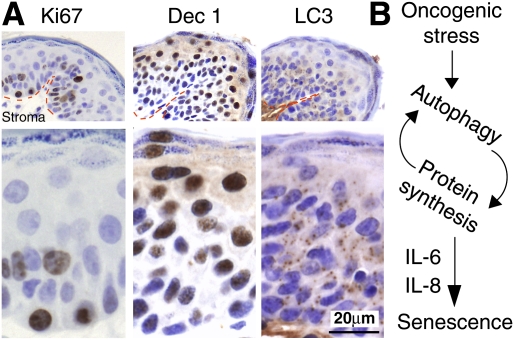
To test the relevance of autophagy during OIS in vivo, we assessed chemically induced murine papillomas, which commonly harbor a constitutively active mutation of H-ras, for Ki67, LC3, and a senescence marker Dec1 (Collado and Serrano 2006; Acosta et al. 2008). Consistent with previous reports, these papillomas contained proliferating (Ki67-positive) cells in the layer closest to the tumor stroma (Fig. 5A). The suprabasal cells, which were Ki67-negative and well separated from the proliferative area, were largely positive for Dec1, showing a predominant nuclear pattern with some cytoplasmic staining (Fig. 5A; Giatromanolaki et al. 2003). Accumulation of LC3 puncta was observed in the Dec1-positive regions of the papillomas and was often most pronounced in the transition areas from proliferative to nonproliferative regions (Fig. 5A). These data support our in vitro evidence pointing to the critical role of autophagy in the mitotic/transition phase of senescence establishment.
Figure 5.
Immunohistochemical analysis of Ki67, Dec1, and LC3 in the same DMBA/TPA-induced mouse papillomas. (A) A representative example is shown from a total of seven papillomas from seven different animals. (Red dotted lines) Basement membrane. (B) Model of the relationship between autophagy and OIS.
Here we show that autophagy is activated upon the acute induction of senescence and contributes to the establishment of senescence. Autophagy has been implicated in tumorigenesis, but its precise role is ambiguous. Autophagy contributes to tumor survival under low nutrient conditions or during chemotherapy (Degenhardt et al. 2006; Amaravadi et al. 2007). On the other hand, the tumor-suppressive role of autophagy has been shown in mouse models, where Beclin-1 (Atg6 homolog) heterozygous and Atg4C-deficient mice are tumor prone (Qu et al. 2003; Yue et al. 2003; Karantza-Wadsworth et al. 2007; Marino et al. 2007). It is conceivable that autophagy has different roles in the different stages, or contexts, of tumorigenesis. Our data focusing on senescence establishment, an early window during tumor development, provides new insight into the complex roles of autophagy in tumorigenesis.
Evidence for the functionality of senescent cells is emerging. For instance, many functional molecules are produced upon the induction of senescence, and these factors often reinforce the senescence phenotype, while potentially also affecting neighboring cells and the microenvironment in vivo. The requirement of autophagy for the efficient production of these factors indicates that autophagy can modulate the functional activity of senescent cells. Thus, our data suggest that autophagy not only facilitates the process but also controls the quality of the phenotype. The molecular mechanism behind this and how autophagy and protein synthesis are both activated is still unclear, but it is tempting to speculate that autophagy promotes rapid protein turnover and facilitates the translation of proteins highly necessary for OIS establishment (Fig. 5B). Thus, autophagy might be a complementary system to epigenetic regulation to achieve the specific biochemical alterations observed during the acute induction of senescence.
Materials and methods
Vectors and cell culture
Vector details are in the Supplemental Material. shRNA target sequences are in Supplemental Table S1. IMR90 and BJ cells (ATCC) were cultured in DMEM supplemented with 10% FBS. Cells were maintained in physiological 5% O2, except for protein degradation assays, microarray experiments, and population doubling analysis.
Protein degradation assays
Protein degradation assays were performed as described (Young et al. 2006).
Cell proliferation, SA-β-Gal assays and electron microscopy (EM)
BrdU incorporation, SA-β-Gal, population doubling, and colony formation were assessed as described (Narita et al. 2006). EM was performed as reported (Shimizu et al. 2004).
Gene expression
Immunoblotting analysis was carried out as described (Narita et al. 2006). Antibody details are in the Supplemental Material. qPCR was performed as described (Narita et al. 2006). Primer sets are in Supplemental Table S2. For microarray experiments, cRNA was hybridized to Human-6 v2 Expression BeadChips (Illumina) as per the manufacturer's instructions. Three biological replicates were performed for each experimental condition. The bead-level data were preprocessed and analyzed using the R beadarray package (Dunning et al. 2007).
Immunohistochemistry (IHC)
Papillomas were generated and analyzed as described (Owens and Watt 2001). IHC was performed using standard methods. Antigen unmasking was performed by heat-mediated retrieval using either pH6 citrate buffer (Dec1 and Ki67) or pH9 Tris-EDTA buffer (LC3).
Acknowledgments
We thank the University of Cambridge, Cancer Research UK, Hutchison Whampoa, Ltd., and EMBL for support. We thank A. Harris, D. Beach, and J. Gil for reagents; D. Tuveson and his laboratory for critical reading of the manuscript; D. Rubinsztein and M. Komatsu for helpful discussion; L. Blackburn for editing; and V. Failla and J. Skepper for technical support. We thank our Institute's core facilities (Genomics, Histopathology and Microscopy).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.519709.
Supplemental material is available at http://www.genesdev.org.
References
- Acosta J.C., O'Loghlen A., Banito A., Guijarro M.V., Augert A., Raguz S., Fumagalli M., Da Costa M., Brown C., Popov N., et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Adams P.D. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene. 2007;397:84–93. doi: 10.1016/j.gene.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R.K., Yu D., Lum J.J., Bui T., Christophorou M.A., Evan G.I., Thomas-Tikhonenko A., Thompson C.B. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M., Serrano M. The power and the promise of oncogene-induced senescence markers. Nat. Rev. Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- Coppé J.P., Patil C.K., Rodier F., Sun Y., Munoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:e301. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S., Genther Williams S.M., Reczek E.E., Johnson B.W., McGillicuddy L.T., Johannessen C.M., Hollstein P.E., MacCollin M., Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre M., Nuciforo P.G., Bensimon A., et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dunning M.J., Smith M.L., Ritchie M.E., Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- Giatromanolaki A., Koukourakis M.I., Sivridis E., Turley H., Wykoff C.C., Gatter K.C., Harris A.L. DEC1 (STRA13) protein expression relates to hypoxia- inducible factor 1-α and carbonic anhydrase-9 overexpression in non-small cell lung cancer. J. Pathol. 2003;200:222–228. doi: 10.1002/path.1330. [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes & Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Cuervo A.M., Seglen P.O. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlever R.M., Higgins P.J., Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Vredeveld L.C., Douma S., van Doorn R., Desmet C.J., Aarden L.A., Mooi W.J., Peeper D.S. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lee B.Y., Han J.A., Im J.S., Morrone A., Johung K., Goodwin E.C., Kleijer W.J., DiMaio D., Hwang E.S. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- Majumder P.K., Grisanzio C., O'Connell F., Barry M., Brito J.M., Xu Q., Guney I., Berger R., Herman P., Bikoff R., et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14:146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette F.A., Gaumont-Leclerc M.F., Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes & Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Marino G., Salvador-Montoliu N., Fueyo A., Knecht E., Mizushima N., Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J. Biol. Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- Mathew R., Karantza-Wadsworth V., White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M., Narita M., Krizhanovsky V., Nunez S., Chicas A., Hearn S.A., Myers M.P., Lowe S.W. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Owens D.M., Watt F.M. Influence of β1 integrins on epidermal squamous cell carcinoma formation in a transgenic mouse model: α3β1, but not α2β1, suppresses malignant conversion. Cancer Res. 2001;61:5248–5254. [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D.M. mTOR and cancer: Insights into a complex relationship. Nat. Rev. Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- Scott R.C., Juhasz G., Neufeld T.P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Wajapeyee N., Serra R.W., Zhu X., Mahalingam M., Green M.R. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.R., Chan E.Y., Hu X.W., Kochl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]