Abstract
We compare typical qualitative protein identification data from two-dimensional (2D) polyacrylamide gel electrophoresis and reconstructed protein arrays, in the context of measuring protein expression by the Gram-negative periodontal pathogen Porphyromonas gingivalis. The arrays were assembled computationally from genome annotations and tandem mass spectrometry data from an off-line HPLC fractionation combined with 2D capillary HPLC analysis of whole proteome enzymatic digests. The 2D separation was carried out with a standard binary gradient HPLC system, modified only slightly with readily available components. Compared to 2D gels, the number of annotated open reading frames identified using the 3D HPLC approach was typically larger by at least a factor of 30. However, the newer technology is currently limited in its ability to reflect the many protein variants derived from posttranscriptional and posttranslational processing.
Introduction
Recently our laboratory has augmented existing 2D gel, HPLC and mass spectrometry capabilities with 2D capillary HPLC in the form that Washburn, Yates and coworkers have termed Multidimensional Protein Identification Technology (MudPIT).1,2 We were impressed by the coverage of the yeast proteome described in the publication of the MudPIT method.2 It was our desire to implement the method using existing binary gradient instrumentation with as few modifications as possible. Here we describe our initial experience with the MudPIT method as applied to a bacterium with a genome of 2.3 million base pairs (MBP) and 2,227 open reading frames (ORFs), according to the most recent annotations by The Institute for Genomic Research (TIGR, www.tigr.org). The organisation of the voluminous data sets produced in such an experiment into more manageable web-based reconstructed protein arrays is also described, in comparison with conventional 2D gel electrophoresis data. Porphyromonas gingivalis is of interest because of the role it plays in human periodontal disease, and certain systemic health problems that extend beyond the oral cavity.3 Measuring which genes are being turned on and off during interactions with the human host is an important strategy for understanding pathogenic mechanisms. The work described here was undertaken with the long-term goal of comparing protein expression by P. gingivalis during various stages of interaction with a target host cell system, gingival epithelial cells. An important part of examining changes in protein expression during the invasion process concerns defining the level of reproducibility of such whole proteome experiments. Before we could apply such an approach to hypothesis testing regarding biological questions, e.g. defining proteins that are differentially expressed during adhesion and invasion, the method needed to reach an acceptable level of reproducibility. In general, questions of reproducibility have not been sufficiently explored in the peer-reviewed literature. Here we describe our progress to date with respect to making such global measurements at the protein level for P. gingivalis.
For purposes of clarity, all references to open reading frames (ORFs) should be understood to mean an annotated, computationally predicted length of DNA that codes for a single polypeptide, with no implication that the protein has actually been expressed from cDNA or isolated from an organism. "Protein" refers to any one of a number of isoforms that may represent the mature gene product as it is actually isolated from a 2D polyacrylamide gel or other biochemical isolation scheme. Only in fortuitous cases does the situation exist in P. gingivalis where the Mr and sequence predicted from the annotated ORF for a single gene product are observed exactly as such at the expressed protein level. In other words, the rare situation where there is a single isoform with intact N and C-terminii, no covalent bonds to another subunit, no splicing, no co- or posttranslational modifications and no degradation either in the cell or through lab artifact.
Experimental
Chemicals
HPLC grade acetonitrile was from Burdick&Jackson (Muskegon, MI, USA); ammonium acetate, from Sigma (St. Louis, MO, USA); trifluoroacetic acid (TFA) from Fisher Scientific (Fair Lawn, NJ, USA); heptafluorobutyric acid (HFBA) and acetic acid from Aldrich (Milwaukee, WI, USA). High purity water was generated with a NANOpure UV system (Barnstead, Dubuque, IA, USA).
Sample preparation
Cell lysis and protein extraction
P. gingivalis strain 33277 was cultured under standard conditions.4 A pellet containing approximately 1×1010 cells was resuspended with 30 µl cold H2O in a 1.5 ml microcentrifuge tube, and was kept on ice; 30 µl DNAse/RNAse solution (1 mg/ml DNAse I, 500 µg/ml RNAse A, 50 mM MgCl2, 50 mM Tris-HCl at pH 7.0) was added into the cell suspension. Immediately, 240 µl of boiling lysis buffer was added to give a final solution with 1% CHAPS, 5 mM DTT, 50 mM Tris-HCl at pH 8.0. The cell suspension was kept in boiling water for 1.5 to 2.0 min, then vortexed vigorously, and centrifuged at 14k rpm for 1 to 2 min. The sample was frozen in liquid N2 and lyophilized to dry powder.
Enzymatic digests
The powder obtained from cell lysis was redissolved in 300 µl of resolubilization solution (7M urea, 200 mM NH4HCO3, 20 mM CaCl2). The proteins were reduced with 5 mM DTT at 37°C for 30 min and then alkylated with 10 mM iodoacetamide at 30°C for 30 min in the dark; 5 µg of Lys-C (sequencing grade, Boehringer, Indianapolis, IN, USA) was added and the mixture was incubated at 37°C for 20 h. The mixture was diluted to give a solution of 2M urea, 100 mM NH4HCO3, 5mM CaCl2; 15 µg of trypsin (sequencing grade, Promega, Madison, WI, USA) was added and the mixture was incubated at 37°C overnight. The sample was centrifuged at 14k rpm and the supernatant was concentrated to 200 µl using a vacuum centrifuge (RC10-22, Jouan Inc. Winchester, VA, USA).
2D gel electrophoresis
The following procedure was based on previously published protocols.5,6 An aliquot of sample containing 60–120 µg protein was diluted with rehydration buffer (8M urea, 2% CHAPS, 10mM DTT) to 360 µl. The solution was vortexed vigorously, and centrifuged at 14k rpm for 2 min to remove insoluble material. The supernatant was loaded onto an 18 cm pH 3–10 immobilized pH gradient (IPG) strip (Amersham Pharmacia Biotech, Piscataway, NJ, USA) with bromophenol blue as color indicator. Isoelectric focusing (IEF) was run on a Multiphor II (Pharmacia) until the current was ~10 µA/strip. The IPG strip was incubated in equilibration solution (2% SDS, 6M urea, 30% glycerol, 0.05M Tris-HCl at pH 6.8) containing 2% DTT and 2.5% iodoacetamide sequentially to reduce and alkylate proteins. The equilibrated IPG strip was loaded on top of an 18cm × 18cm × 1mm, 10.5% SDS-PAGE gel. SDS-PAGE was run at constant current, 25 mA/gel, until the dye front reached the bottom of the gel. Gels were silver-stained7 and dried for further analysis. Spots on our reference state 2D gel maps (see Fig. 1) were analyzed using tandem mass spectrometry, as described previously.8 Peptide mass mapping based on MALDI-TOF MS data9 was also used in a few cases where the automated tandem MS approach failed to generate a good search result.
Fig. 1.
Silver-stained proteins from P. gingivalis 33277 separated by 2D polyacrylamide gel electrophoresis. Soluble proteins (160 µg) were separated in the first dimension using a nonlinear pH 3 – 10 IPG gel strip. The strip was then transferred to a 10.5% SDS-PAGE gel to perform the second dimension separation. Numbered spots illustrate examples of those proteins that have been identified using in situ trypsin digestion followed by mass spectrometry and database searching. Note the many variants that are often seen that map back to a single ORF.
Desalting
For the situation early in our work in which the off-line HPLC step was skipped and the whole cell digest was applied directly to the biphasic capillary column (see below), it was necessary to include a cleanup step. After enzymatic digestion, the solution was desalted using a ZipTipC-18(Millipore, Bedford, MA, USA), following the procedure from the manufacturer, with minor changes to the recommended volumes. Briefly, a ZipTip was rinsed with 50% acetonitrile and 0.1% TFA; 10 µl digest solution was mixed with 10 µl 1% TFA and loaded onto the ZipTip. After 3× 10 µl wash with 0.1% TFA, the peptides were eluted with 3× 10 µl wash with 50% acetonitrile in 0.1% TFA solution. The 30 µl eluent was reduced to 5 µl in the vacuum centrifuge; 15 µl 0.4% acetic acid in 2% acetonitrile solution was added to make the final volume about 20 µl.
3D HPLC combined with data dependent tandem mass spectrometry
Off-line HPLC fractionation
Approximately 100 µL of the supernatant from the digestion step described above was loaded onto a Poros R2 2.1 × 100 mm reversed-phase HPLC column (ABI, Foster City, CA, USA). The mobil phase was H2O and acetonitrile with 0.1% TFA. Peptides were eluted with increasing acetonitrile percentage (2% to 95% in 90 min) at 1.0 ml/min. Eluent was collected as 1 ml aliquots. The fractions were pooled into five combined fractions according to UV absorption at 214 nm. Each combined fraction was concentrated to 100 µl using the vacuum microcentrifuge. Acetic acid was added to a final concentration of 0.5% (v/v).
Capillary HPLC system
A Magic 2002 HPLC (Michrom BioResources, Auburn, CA, USA) equipped with a 100 µl mixer cartridge and a variable ratio precolumn splitter10 was used for all separations. Bypassing the Michrom HPLC injector and UV detector, the mobile phase was delivered to a stainless steel tee (MT1XCS6, Valco Instruments, Houston, TX, USA) connected to both the precolumn splitter (Michrom) and the stock six-port injection valve (Cheminert™ C3-2006, Valco) mounted on the LCQ ion trap mass spectrometer. A biphasic capillary column2 (75 µm i.d., 34 cm long) packed in-house was connected to the LCQ injection valve and the ESI interface. The first packing (injection side) was a 4 cm section of 5 µm, 300 Å polysulfoethyl aspartamide (PSEA), the second was 11 cm of 5 µm, 200 Å Magic C18, (Michrom). When the flow rate was set at 160 µl/min at the HPLC, the flow rate at the outlet of the column was 300 nl/min at 2% mobile phase B. A 100 µm i.d. fused-silica capillary with a 1.0 µl volume was used as the injection loop for the stepwise salt gradients. Sample solution (2 µl) from a combined off-line fraction was loaded directly into the column pneumatically using helium. After loading, the column was reconnected to the injection valve and flushed with 2% mobile phase B at 300 nl/min for 4 to 5 min. This plumbing arrangement allowed us to make use of the helium bomb method10 of loading sample, while using pre-existing flow injection analysis (FIA) plumbing to admit the salt step gradients independently of the two solvent reservoirs on the HPLC.
Stepwise elution
The procedure was conducted manually. A reversed-phase elution was conducted first to detect peptides not strongly bound to the strong cation exchange (SCX) packing. The mobile phase A was 0.02% (v/v) HFBA in water, mobile phase B was 0.02% HFBA in acetonitrile. The linear gradients programmed were 2–25% B in 5 min, hold 5 min, 25–50% B in 60 min, hold 5 min, 50–2% B in 5 min and equilibrate for 15 min. The flow rate, as measured at the capillary column, was adjusted during the run such that it was reduced from 300 nL/min to 150 nL/min during the 25–50% B portion of the gradient. The SCX eluent was ammonium acetate (steps of 10, 25, 50, 100, 250 and 500 mM) in 0.02% HFBA + 2% acetonitrile. For each step, 1 µl was injected, followed by a 15 min wash with 2% B at 300 nl/min. Then, the ESI voltage was turned on and the reversed-phase gradient was started (Fig. 2). The column was cleaned with 4× 1 µl 500 mM ammonium acetate after each 250 mM elution from the SCX, packing, followed by 2 to 90% acetonitrile in 30 min with a 30 min hold. The column was then re-equilibrated for a minimum of 30 min with 2% mobile phase B, before injecting the next fraction from the off-line HPLC first dimension separation.
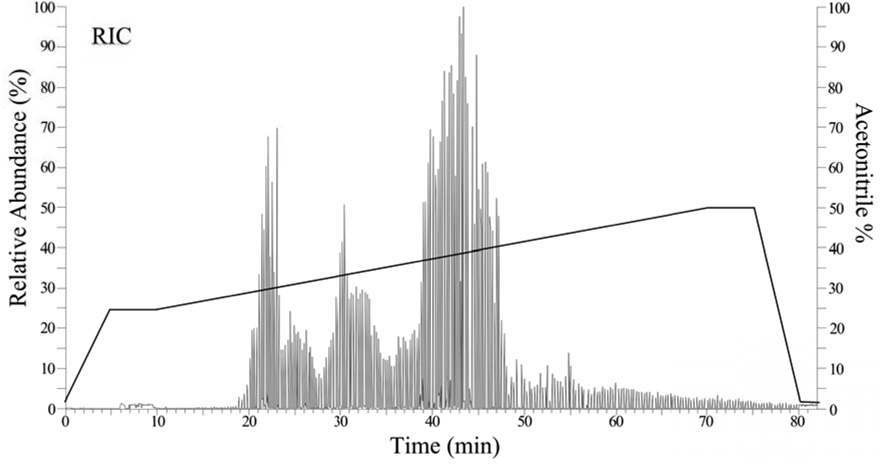
Fig. 2.
Representative automated collision-induced dissociation plot (Auto-CID) of reconstructed ion current versus time (min) from the 2D capillary HPLC-MS/MS analysis of whole cell digest. 2.0 µl of sample was loaded onto the biphasic column.1,2 Peptides were fractionated from the SCX packing onto C18. This plot was from fraction one out of seven. The solid line shows that portion of the acetonitrile gradient during which mass spectra are acquired, the percentage of acetonitrile scales with the relative intensity shown on the y-axis. Each dip in the trace indicates the instrument is switching from main beam (MS1 mode) to CID (MS2 mode).
Ion trap mass spectrometry
An LCQ ion trap mass spectrometer (Thermo Finnigan, San Jose, CA, USA) equipped with an ESI interface built in-house was used for mass analysis. The ESI microsprayer was made from a 0.15 mm bore stainless steel union connector (MU1XCS6, Valco) and a 10 µm i.d. "no coating" PicoTip (FS360-20-10-N, New Objective, Inc. Woburn, MA, USA). The ESI voltage was 1.8 kV; heated capillary temperature 165 °C; scan range 400–2000 m/z units. A full scan (about 1.5 s) contained 3 micro scans with a maximum injection time of 200 ms. A 3 segment automated and data dependent acquisition program was used: 0–10 min, main beam (MS1) scan; 10–80 min, data dependent collision-induced dissociation (CID) scan (MS2) and 80–90 min, main beam scan (see Fig. 2). The default parameters under Xcalibur 1.2 were used for CID, with the following exceptions: activation amplitude 35%, isolation width 3.0 m/z units. Eight MS2 scans for the four most intense ions from each pre-scan were taken. Dynamic exclusion was activated during all acquisitions. This MudPIT2 procedure differs from that employed by the Yates laboratory in that we do not apply power to the electrospray ionization (ESI) source when ammonium acetate solution is being pumped through the capillary column.
Post-run data reduction using SEQUEST, DTASelect and D2g
Our use of tandem mass spectrometry coupled with SEQUEST database searching11,12 and ORF databases prepared from the Porphyromonas gingivalis genome have been described.8 Since our initial publication an annotated ORF database has become available from TIGR in a preliminary form. In order to compare the large number of SEQUEST output files, many thousands for each whole proteome extract, the program DTASelect was used.13 This software replaced previous summary programs and allows the rapid preparation of reports from large numbers of SEQUEST searches, limited in any practical sense only by computer memory size and data storage capacity (see more detail at http://fields.scripps.edu/DTASelect/). The issues surrounding assigning cutoff values for the SEQUEST search parameters have been discussed at length previously with respect to the P. gingivalis genome and ORF database.8 Briefly, two peptides with an Xcorr value11,12 greater than 0.9 for singly charged precursors, 1.4 for doubly charged, and 2.2 for triply charged precursors, and a Delta CN value > 0.08 were required to be retained in the data set. Hits with peptides shorter than four residues were rejected.
For each complete analysis of the P. gingivalis proteome, the DTASelect output file was used to generate 2D plots of the reconstructed proteins based on whole protein molecular mass and isoelectric point or hydrophobicity index. The plots are created over the web using a short Java program, D2g, created in-house. Each point on the graph represents a URL link on our local servers to the appropriate summary for the protein in the DTASelect output file. Collectively, the points on the graph make up a reconstructed protein array that describes each experiment in terms of annotated ORFs rather than peptides. Multiple experiments can be overlayed graphically in different colors, as well as the entire ORF database, allowing the rapid visual comparison of expressed ORFs under different growth conditions and regulation states, analogous to nucleic acid microarray technology. Each DTASelect entry accessed by clicking on the graph contains the degree of sequence coverage, the sequences of the peptides isolated from the particular ORF, the location in the ORF database for the protein, its theoretical neutral Mr, isoelectric point and SEQUEST command line parameters. We have also included a URL link from the graphing program output directly to the ORF database for each point, a concept that can be readily expanded in the future to include URLs for DNA or RNA microarray data or a remote link to the annotated gene at TIGR or another set of annotations prepared at the Los Alamos National Laboratory (see Table 1).
Table 1.
Examples of ORFs identified from 2D gel spots (Fig. 1) using SEQUEST searches against the TIGRa and Los Alamosb databases for P. gingivalis strain W83
| Spots | ORFTIGR | ORFLANL | MWgelc | MWcalcd | Protein Description from LANL annotationsc |
|---|---|---|---|---|---|
| 1 | PG2132 | PG1865 | 70–120 | 41 | major fimbrillin A |
| 2 | PG0178 | PG0163 | 88, 42 | 50d | 67 kDa fimbrillin/PGA67/minor fimbrillin A |
| 3 | PG2024 | PG1768 | 50 | 186 | arginine-specific cysteine proteinase; gingipain R1 |
| 4 | PG1844 | PG1605 | 42 | 188 | porphypain polyprotein; lys-X proteinase/hemagglutinin |
| 5 | PG1551 | PG1357 | 22 | 15.5 | tonB-dependent receptor HmuY |
| 6 | PG1823 | PG1592 | 22 | 24 | probable integral outer membrane protein P20 |
| 7 | PG2102 | PG1838 | 20 | 61 | LPS-modified surface protein P59 |
| 8 | PG2124 | PG1857 | 18 | 36 | glyceraldehyde 3-phosphate dehydrogenase |
| 9 | PG1858 | PG1621 | 17 | 19 | flavodoxin A |
Los Alamos National Lab: http://www.stdgen.lanl.gov/oragen/bacteria/pgin/
Molecular weight estimated from spot location on the gel in kDa (see Fig. 1).
Molecular weight in kDa from Los Alamos database.
A single complete data set for P. gingivalis, under one experimentally defined state (see Fig. 3), requires about 1.0 Gbytes of disk space. Each data set consisted of *.raw (raw data) files from the LCQ, *.dta files (preprocessed CID mass spectra in a form suitable for SEQUEST), *.out files (SEQUEST search output) and DTASelect summary files. Each of the six combined off-line fractions was divided into seven cuts from the SCX packing, yielding 42 capillary reversed-phase gradient runs of the type shown in Fig. 2.
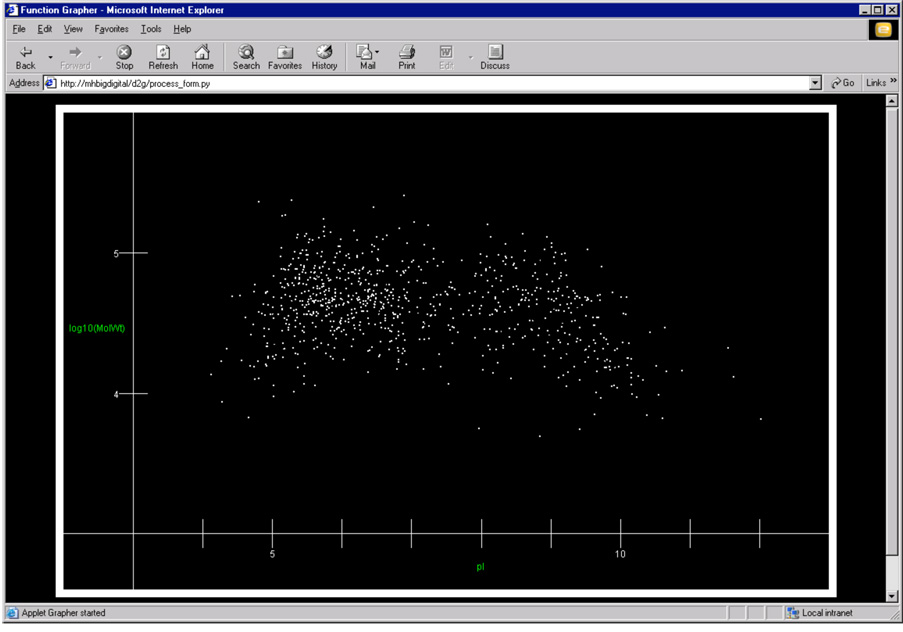
Fig. 3.
A single layer reconstructed protein array display of 967 out of approximately 2000 possible proteins that can potentially be expressed by P. gingivalis. The display shows one complete data set for a single growth condition. The plot is generated in any web browser by a separate Java program, D2g, that reads output from DTASelect.13 In the screen image above, the x-axis is pI (calculated) and the y-axis is log Mr, also calculated from the putative ORF. Each point on the plot also serves as a web-based link to the full entry in our P. gingivalis ORF database, the DTASelect summary for the particular protein represented by the dot, the mass spectral data and a BLAST link back to the original genome database. The hot links embedded in the plot make a convenient user interface to the many types of data used in our studies.
Results and discussion
A representative 2D gel map for P. gingivalis, acquired under our standard growth conditions4 is shown in Fig. 1. A CRT screen image of the web-based user interface to the reconstructed protein array for P. gingivalis grown under the same conditions is shown in Fig. 3. The total number of ORFs identified on our 2D gels to date is less than 30, although the number of silver-stained gel spots that we have mapped back to the genome is many times that number. This is due to the many isoforms derived from a relatively small number of ORFs that make up a typical data set. Table 1 summarizes results for nine groups of spots isolated from the gel shown in Fig. 1. Because the 2D gel maps are based on electrophoretic migration velocity for proteins in their mature forms, truncation variants, co- and posttranslational modifications will be reflected in the gel map. However, the gel map probably reflects a relatively small percentage of the entire P. gingivalis proteome, that portion that is well behaved under standard conditions used for isoelectric focusing and SDS-PAGE, respectively.14 "Well behaved" in this context means avoiding extremes of pI, hydrophobicity and molecular weight. However, for those proteins that are amenable, it can reasonably be assumed that many, if not all, of the changes made to the protein primary structure during biosynthesis and maturation will be seen in the gel map.
In contrast, the reconstructed protein array presentation (Fig. 3) of the MudPIT type of experiment (see Fig. 2) is referencing limited and necessarily incomplete protein sequence data back to a genomic database, using the database of inferred sequences to generate the 2D plot (see Fig 3). The plot represents expressed genes, not proteins in their complete covalent form. The plot is "real" in that for a point to be included experimental evidence that the protein is being expressed must be mined from the DTASelect summary. The plot shown in Fig. 3 is "virtual" in that the molecular weights and pI's are all calculated rather than observed and do not reflect deviations or additions to the theoretical amino acid sequence inferred from the gene. The primary advantage of this type of experiment, relative to conventional 2D gels, is increased coverage of the proteome. The total number of *.dta files, or non-redundant searchable CIDs, was about 60,000. This would be the approximate equivalent of 30 peptides per protein for each annotated ORF in the genome. Coverage in the data set shown in Fig. 3 varied from two unique peptides per annotated ORF to as many as 40 for larger proteins. The most recent information to date (see www.stdgen.lanl.gov/oragen/bacteria/pgin/properties.html) suggests that the total number of proteins expressed under any given set of conditions may not exceed about 1300. For purposes of comparison, a plot of Mr versus pI is given for all 2,227 predicted ORFs in our database, see Fig. 5. The actual number of proteins expressed by strain 33277 under the growth conditions used here is unknown. If the estimate of 1300 is correct, then we are at present recovering peptides representing about 75% of the gene products being expressed, far more, by at least 30x, then we can identify from 2D gels (see Table 2). In the absence of sophisticated automated peak parking technology for our ion trap mass spectrometer,15 we found it necessary to include an off-line reversed-phase fractionation step. This step served to reduce the sample loading for each 2D capillary HPLC step such that the mass spectrometer could acquire many more CIDs than if we used the biphasic capillary column alone. Our experiments that involved applying the digested proteins directly to the capillary column after a desalting step did not yield adequate coverage. Without the off-line step, it was only possible to map peptides back to about 100 ORFs, in the absence of peak parking.
Fig. 5.
Our entire ORF database plotted as a function of log Mr and pI, as in the observed ORFs shown in Fig. 3. Note the similarity in the two distributions.
Table 2.
Comparison of duplicate runs of P. gingivalis whole cell digest.
| Totala | Commonb | Uniquec | |
|---|---|---|---|
| 1st run | 957 | 683 (60%) d | 274 (24%) |
| 2nd run | 865 | 683 (60%) | 182 (16%) |
| 1st + 2nd | 1139 |
Number of proteins identified using the parameter set described in the Experimental section.
Number of proteins identified in both runs
Number of proteins identified only in one run
Percentages are calculated using the combined total number, 1139.
The data shown in Fig. 3 can also be plotted by calculated hydrophobicity, as shown in Fig. 4. The hydrophobicity plot is generally less useful as a working display of the data set, due to the dense clustering about certain values. However, it does provide some indication that our recoveries are not being biased against more hydrophobic proteins. The scatter shown in Fig. 4, based on real data, is quite similar in distribution to that shown by a plot of the database itself, see Fig. 6. Consistent with prior observations using the yeast model,2 the analytical scheme seems to be equally efficient across the range of hydrophobicities. We expected that certain proteins with a very high percentage of their amino acids contained in domains completely within the membrane would be missed. The number of such proteins in P. gingivalis appears to be quite small, based on our initial results, but a more thorough comparison of our data and the predicted membrane domains coded in the genome needs be done before any definitive statement can be made. The methods employed here are well known to have difficulties generating proteolytic fragments from such domains. Thus far we have avoided using chemical cleavage reagents that will work with organic solvent systems, e.g. chloroform/methanol, that will solubilize such hydrophobic proteins.
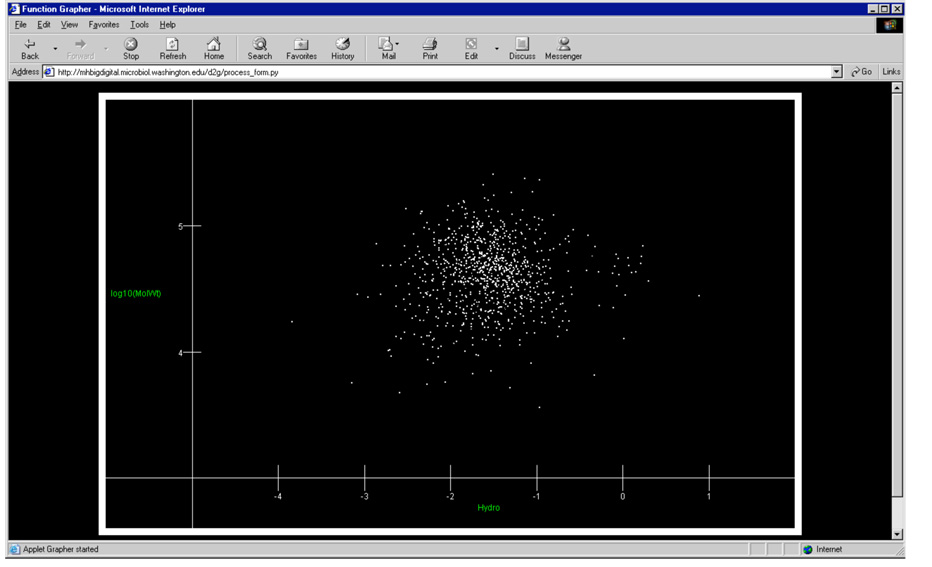
Fig. 4.
A hydrophobicity plot of the same reconstructed protein array as shown in Fig. 3. The hydropobicity calculation was based on the scale published by Engelman, Steitz and Goldman22 and only takes into account primary structure, that is the relative hydrophobicity of the amino acid monomers averaged over the entire ORF. In the plot above the x-axis is hydrophobicity (calculated) and the y-axis is log Mr. More hydrophobic proteins have positive values that graph to the right side of the image.
Fig. 6.
Our entire ORF database plotted by hydrophobicity, similarly to the data plotted in Fig. 4. Note the similarity of the distribution of the observed ORFs in Fig. 4 and the purely theoretical construct shown here.
The system can fail to return a match when proteins or variants of known proteins unique to our P. gingivalis strain are encountered. For example, the major and minor fimbria (Table 1) are shown as the groups of proteins at positions 1 and 2 on the gel map (Fig. 1). These highly expressed proteins from two separate genes (fimA, mfa1) are clearly distinguished on the 2D gel. Early in our work, hits were returned for the minor fimbria (mfa1) only, which is known to be similar in W83 and 33277. Interestingly, with the exception of the major fimbria gene (fimA) that is known to differ between W83, used to generate the genome sequence, and 33277, the strain used here, we encountered little difficulty mapping most of our proteins to the ORF databases derived from the W83 genome. Once a variant gene or a sequence error at the DNA level has been identified, we augment our ORF database accordingly. With a proper fimA sequence for 33277 now included in our database, the gel data and the reconstructed array data now agree. When the non-gel based protein methods become more common, we expect they will contribute substantially to the overall annotation process required for the genomic databases to mature.
The method suffers from poor reproducibility at the level of individual peptide CIDs. However, the inherent redundancy of the experiment, with many proteolytic fragments available per protein, limits these problems at the level of identifying expressed ORFs. Duty cycle limitations of the ion trap, combined with the large number of parent ions present (see Fig. 2), limit the number of CID spectra that can be acquired per unit time, relative to the ideal situation in which every parent ion above a predetermined S/N threshold yields a CID. Thus, the run-to-run repeatability with respect to which parent ions are chosen for fragmentation at any given retention time is relatively poor. However, coverage of the P. gingivalis proteome was consistent when duplicate experiments were compared, see Table 2. The most relevant observation made to date from the large amount of data summarized in Table 2 is that the ORF identifications are reproducible to within about 60% of the total data set. Those ORFs that were unique to only one of the duplicates tended to share two common features, low abundance and (or) predicted molecular masses smaller than 7 kDa. As discussed above, there was no obvious discrimination based on either isoelectric point (Fig. 3) or hydrophobicity (Fig. 4) when our data set was compared with the distributions observed for the entire genome (Fig. 5 and Fig 6). The distribution of observed ORFs for the duplicate data set is shown in Fig. 7. The similarity in appearance observed when comparing Figure 3 and Figure 7 is born out by a closer inspection of the data summarized in Table 2.
Fig. 7.
The duplicate data set of 865 observed ORFs (see Fig. 3 and Table 2), plotted by log Mr and pI.
What is intriguing, and also frustrating at present, is the awareness that much of the information desired regarding multiple forms, posttranslational modifications, etc. is at least in theory inherently present in the raw tandem MS data. If one makes the approximations that each peptide generated in a proteolytic digest of an organism is recovered and searchable back to an ORF, and that all remaining peptides represent some kind of modification or deviation from the inferred sequence, and that the remaining peptides can be sequenced, one could then propose to computationally reassemble this vast puzzle into a map that more accurately reflects the real nature of the proteins in their mature, expressed forms. Such a scheme remains highly speculative because the assumptions mentioned above are violated in practice. Getting enough sequence back to map to an ORF is now relatively routine. However, getting complete sequence coverage with the quantitative recoveries required to deduce variant forms of the same ORF or to distinguish reliably among ORFs with a high degree of sequence similarity is beyond the scope of existing separation, tandem MS or bioinformatic technology.
The reconstructed array experiment in its present form contains information gained at the protein level for what the genes are doing, what is being turned on and turned off, in a way that bypasses certain of the issues surrounding the relationship of mRNA arrays to actual protein expression,16, 17 and thus may have practical value. At present, we have not encoded relative signal intensity into graphical presentation of the array (Fig. 3 and Fig 4), but such information is retained in the data set and could be coded into a multiple pixel and (or) multicolor presentation of each protein. How much the use of isotopic labels for quantitation, either through chemical methods18 or metabolic labeling,19 will improve the data in terms of ability to answer biological questions is still an issue, especially given the onerous expense of commercial reagent kits being marketed for quantitative proteomics. Our thinking at present is that application of isotope dilution mass spectrometry in the context of host-pathogen interactions is best left for subsets of proteins that have already been identified qualitatively in sufficient detail that their relevance to pathogenic mechanisms has been established.
In order to get a more useful and complete picture of what the proteins are doing, the type of data represented by Figure 3 and Figure 4 needs to be combined with mass measurements of the intact proteins in a systematic fashion for the entire protein complement of the cell. Even though there is not necessarily an obvious relationship between the Mr calculated for an inferred sequence and the Mr measured experimentally (see Table 1), the intact protein data will help clarify the issue of how many isoforms are present, which of several closely related ORFs are being expressed, etc. It will provide at least some evidence for the structural basis for deviations from the inferred sequence. The future may very well involve a proteome wide "top down" type of experiment in which the whole protein is mass analyzed and then fragmented to generate a sequence tag adequate to match the intact molecular mass data with a gene. Recent work with fragmentation of intact single proteins in McLafferty's lab20,21 and others certainly suggests that one day this will be technically feasible as a high throughput experiment.
Acknowledgements
We thank David L. Tabb and John R. Yates III for DTASelect and Contrast, and Kobi Alfandari for D2g. Additional computer support was provided by M. Taylor Monacelli. Funding was provided by the NIH through NIDCR DE11111, with additional support provided by Michrom BioResources and Immunex Corporation. We thank TIGR for prepublication licensing of the PG genome and preliminary annotations. Annotations of the PG genome were also provided by Los Alamos National Laboratory.
References
- 1.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III Nature Biotech. 1999;17:676. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 2.Washburn MP, Wolters D, Yates JR., III Nature Biotech. 2001;19:242. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 3.Lamont RJ, Jenkinson HF. Microbiol. Mol. Biol. Rev. 1998;62:1244. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park Y, Lamont RJ. Infect. Immun. 1998;66:4777. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabilloud T, Adessi C, Giraudel A, Lunardi J. Electrophoresis. 1997;18:307. doi: 10.1002/elps.1150180303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi SY, Li Y, Szyroki A, Giles IG, Moir A, Connor CDO. Molec. Micro. 1995;17:523. doi: 10.1111/j.1365-2958.1995.mmi_17030523.x. [DOI] [PubMed] [Google Scholar]
- 7.Shevchenko A, Wilm M, Vorm O, Mann M. Anal. Chem. 1996;68:850. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Laidig KE, Park Y, Park K, Yates JR, III, Lamont RJ, Hackett M. Analyst. 2001;26:52. doi: 10.1039/b008012h. [DOI] [PubMed] [Google Scholar]
- 9.Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C. Proc. Natl. Acad. Sci. USA. 1993;90:5011. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt DF, Alexander JE, McCormack AL, Martino PA, Michel H, Shabanowitz J, Sherman N, Moseley MA, Jorgenson JW, Deterding LJ, Tomer KB. In: Techniques in Protein Chemistry II. Villafranca JJ, editor. New York: Academic Press; 1991. p. 441. [Google Scholar]
- 11.Eng JK, McCormack AL, Yates JR., III J. Am. Soc. Mass Spectrom. 1994;5:976. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 12.Yates JR, III, Eng JK, McCormack AL, Schietz D. Anal. Chem. 1995;67:1426. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 13.Tabb DL, McDonald WH, Yates JR., III J. Proteome Res. 2002;1:21. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aebersold R, Rist B, Gygi SP. Ann. N.Y. Acad. Sci. 2000;919:33. doi: 10.1111/j.1749-6632.2000.tb06865.x. [DOI] [PubMed] [Google Scholar]
- 15.Hunt DF. J. Proteome Res. 2002;1:15. doi: 10.1021/pr020300a. [DOI] [PubMed] [Google Scholar]
- 16.Pandey A, Mann M. Nature. 2000;405:837. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 17.Lockhart DJ, Winzeler EA. Nature. 2000;405:827. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 18.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat. Biotech. 2000;17:994. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Desiderio DM. Mass Spectrom. Rev. 1996;15:213. doi: 10.1002/(SICI)1098-2787(1996)15:4<213::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 20.Ge Y, Lawhorn BG, El Naggar M, Strauss E, Park JH, Begley TP, McLafferty FW. Jour. Amer. Chem. Soc. 2002;124:672. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 21.Sze SK, Ge Y, Oh H, McLafferty FW. Proc. Natl. Acad. Sci. USA. 2002;99:1774. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman DM, Steitz TA, Goldman A. Annu. Rev. Biophys. & Biophys. Chem. 1986;15:321. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]