Abstract
Coordination of cell proliferation and cell death is essential to attain proper organ size during development and for maintaining tissue homeostasis throughout postnatal life. In Drosophila, these two processes are orchestrated by the Hippo kinase cascade, a growth-suppressive pathway that ultimately antagonizes the transcriptional coactivator Yorkie (Yki). Here we demonstrate that a single phosphorylation site in Yki mediates the growth-suppressive output of the Hippo pathway. Hippo-mediated phosphorylation inactivates Yki by excluding it from the nucleus, whereas loss of Hippo signaling leads to nuclear accumulation and therefore increased Yki activity. We further delineate a mammalian Hippo signaling pathway that culminates in the phosphorylation of YAP, the mammalian homolog of Yki. Using a conditional YAP transgenic mouse model, we demonstrate that the mammalian Hippo pathway is a potent regulator of organ size, and that its dysregulation leads to tumorigenesis. These results uncover a universal size-control mechanism in metazoan.
INTRODUCTION
A longstanding question in biology is how the size of an organ is determined (Conlon and Raff, 1999). While environmental cues such as nutrient availability play an important role in regulating organ size, developing organs also possess intrinsic information about their final size. Accumulating evidence suggest that a “size checkpoint” operates at the level of the organ’s total mass, rather than the size or the number of the constituent cells. At present, the molecular nature of this organ size checkpoint remains a mystery.
Recent studies in the fruit fly Drosophila have implicated the Hippo signaling pathway as an intrinsic mechanism that restricts organ size in development (Edgar, 2006; Pan, 2007). This pathway is defined by a kinase cascade whereby the Ste20-like kinase Hippo (Hpo), facilitated by the WW-domain-containing adaptor protein Salvador (Sav), phosphorylates and activates the NDR family kinase Warts (Wts). Wts, in turn, phosphorylates and inactivates the transcriptional coactivator Yorkie (Yki), leading to transcriptional downregulation of target genes such as the cell-cycle regulator cyclin E, the cell death inhibitor diap1, and the microRNA bantam. Inactivation of the Hpo, Sav, or Wts tumor suppressors, or overexpression of the Yki oncoprotein, results in massive tissue overgrowth characterized by excessive cell proliferation and diminished apoptosis.
Several lines of evidence suggest that Yki represents the most critical effector of Wts in the Hippo growth-control pathway (Huang et al., 2005). First, Yki is phosphorylated by Wts in a Hpo-dependent manner. Second, Yki is required for normal diap1 transcription and tissue growth in Drosophila imaginal discs, and genetic analysis placed Yki downstream of hpo, sav, and wts. Most importantly, overexpression of Yki recapitulates the loss-of-function phenotypes of hpo, sav, and wts, such as increased diap1 and cycE transcription, increased cell proliferation, and diminished apoptosis, as well as tissue overgrowth. While these findings have established Yki as a critical nuclear effector of the Hippo pathway, the molecular mechanism by which Hippo signaling inactivates Yki function remains to be determined.
A longstanding issue in Hippo signaling concerns the composition and physiological role of this pathway in mammals. Despite the presence of mammalian homologs for all the known components of the Drosophila Hippo pathway (Mst1/2 for Hpo, WW45 for Sav, Lats1/2 for Wts, and YAP for Yki), previous studies in mammals have failed to unite these proteins in a physiologically relevant signaling cascade. Instead, these proteins have been associated with a wide range of biochemical and physiological roles. For example, Mst1/2 was reported to phosphorylate histone H2B (Cheung et al., 2003) and FOXO transcription factors (Lehtinen et al., 2006). Lats1/2 was reported to regulate mitosis and cytokinesis by interacting with cdc2 (Tao et al., 1999), zyxin (Hirota et al., 2000), and LIMK1 (Yang et al., 2004). YAP was reported to play diverse functions ranging from protein trafficking to transcription by binding to at least nine different targets (Sudol, 1994; Mohler et al., 1999; Yagi et al., 1999; Espanel and Sudol, 2001; Vassilev et al., 2001; Ferrigno et al., 2002; Komuro et al., 2003; Basu et al., 2003; Howell et al., 2004). Most critical to the establishment of a mammalian Hippo pathway is a physiologically relevant assay that measures pathway output, which unfortunately has not been available.
In this report, we first investigate the mechanism by which Hippo signaling antagonizes Yki in Drosophila. We show that Hippo signaling inactivates Yki by excluding it from the nucleus via phosphorylation of a critical residue (S168). We provide functional evidence that phosphorylation of this residue mediates the growth-suppressive output of the Hippo signaling pathway. Based on the functional conservation of this phosphorylation site in the mammalian YAP protein, we delineate a mammalian Hippo signaling pathway that links Mst1/2, WW45, and Lats1/2 to YAP phosphorylation. Finally, we explore the physiological function of the mammalian Hippo pathway using a conditional YAP transgenic mouse model. We demonstrate that the mammalian Hippo pathway is a potent regulator of organ size, and that its dysregulation leads to tumorigenesis. These results implicate the Hippo pathway as a universal regulator of tissue homeostasis in metazoan animals.
RESULTS
Hippo Signaling Causes Cytoplasmic Localization of Yki in Drosophila Cells
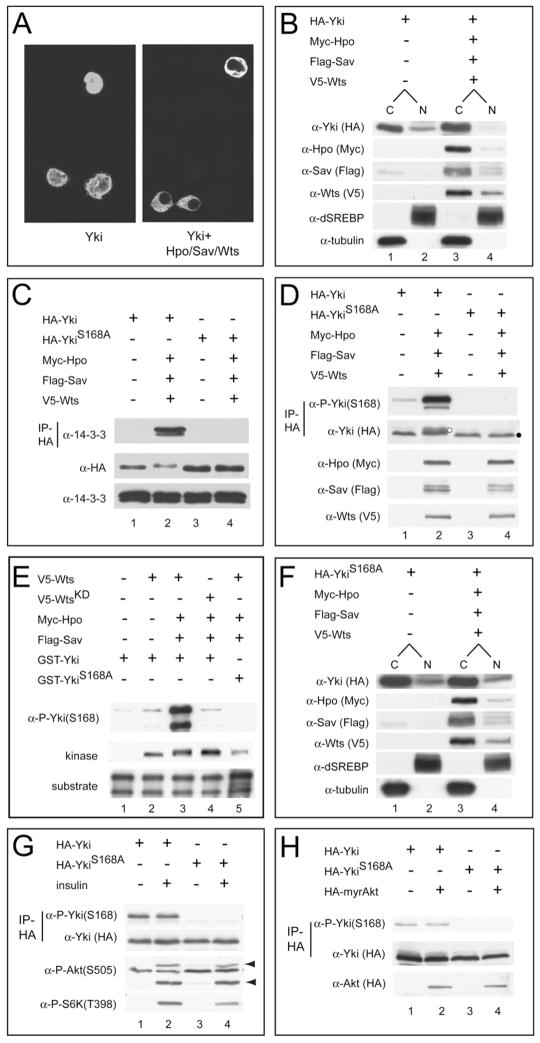
Protein phosphorylation is known to inactivate many transcriptional regulators by shuttling them to the cytoplasm, a process that often requires the binding of the phosphorylated proteins by the 14-3-3 proteins (Muslin and Xing, 2000). SCANSITE analysis (Obenauer et al., 2003) revealed a single optimal 14-3-3 binding motif—RxxS/T(phos)xP—at Yki S168 (RARS168SP), suggesting that Yki might be regulated via a similar mechanism. To test this possibility, we examined the subcellular localization of epitope-tagged Yki protein in Drosophila S2 cells. When expressed alone, Yki was present throughout the cytoplasm and the nucleus (Figure 1A). When coexpressed with Hpo, Sav, and Wts, a condition that induces Yki phosphorylation (Huang et al., 2005), Yki was mostly localized in the cytoplasm, with little signal detectable in the nucleus (Figure 1A). Cell fractionation revealed a similar cytoplasmic concentration of Yki under active Hippo signaling (Figure 1B). Consistent with 14-3-3 mediating the cytoplasmic shuttling of phosphorylated Yki, we found that Yki associates with 14-3-3 in the presence, but not in the absence, of activated Hippo signaling (Figure 1C). This binding also requires the predicted 14-3-3 binding motif because it was completely abolished when S168 was mutated to alanine (Figure 1C).
Figure 1. Hippo Signaling Phosphorylates Yki S168 and Promotes Its Cytoplasmic Localization.
(A) S2 cells expressing HA-Yki (left) or HA-Yki plus Hpo, Sav, and Wts plasmids (right) were stained with α-HA antibody. Nuclear HA-Yki signal is detected in 100% of cells expressing HA-Yki alone, but in only 9% of cells coexpressing Hpo-Sav-Wts.
(B) Cells were transfected as in (A) and analyzed by subcellular fractionation. Note the decrease of nuclear Yki under active Hippo signaling (compare lanes 2 and 4). dSREBP (nuclear) and tubulin (cytosolic) were used as quality control for fractionation. C, cytoplasmic; N, nuclear.
(C) Cells were transfected as in (A), and α-HA immunoprecipitates were probed with α-14-3-3 (top gel). Cell lysates were also probed with the indicated antibodies (middle and bottom gels). Yki, but not YkiS168A, immunoprecipitated 14-3-3 under active Hippo signaling.
(D) Cells were transfected as in (A), and α-HA immunoprecipitates were probed with α-P-Yki(S168) and α-HA antibodies (top two gels). Cell lysates were also probed with the indicated antibodies (bottom three gels). Yki, but not YkiS168A, showed S168 phosphorylation, which was further increased under active Hippo signaling. Also note that Yki, but not YkiS168A, showed mobility shift under active Hippo signaling (compare lanes 2 and 4; indicated by white and black dots, respectively).
(E) Wts phosphorylates Yki at S168 in vitro. V5-tagged Wts (or kinase-dead WtsKD) was expressed alone or together with Hpo-Sav in S2 cells, immunoprecipitated, and incubated with GST-Yki (or GST-YkiS168A), and the reaction products were probed with α-P-Yki(S168). The input kinase and substrate are also shown (bottom two gels). Note that our GST-Yki preparation contains two Yki-related bands (Huang et al., 2005). Strong S168 phosphorylation was detected when Wts (lane 3), but not WtsKD (lane 4), was coexpressed with Hpo-Sav. No S168 phosphorylation was detected using GST-YkiS168A as a substrate (lane 5).
(F) Subcellular fractionation of YkiS168A mutant. The relative proportion of cytoplasmic and nuclear YkiS168A was not changed under active Hippo signaling.
(G) S2 cells expressing HA-Yki or HA-YkiS168A were treated with or without insulin. α-HA immunoprecipitates were probed with the indicated antibodies (top two gels). Cell lysates were also probed with the P-S6K or P-Akt antibodies (bottom two gels). Arrowheads mark the phospho-Akt signals. Insulin stimulates the phosphorylation of Akt and S6K, but not that of Yki (compare lanes 1 and 2).
(H) S2 cells expressing HA-Yki or HA-YkiS168A plus a constitutively active Akt mutant (myr-Akt) were analyzed as in (G). Similar P-Yki(S168) levels were seen in the presence or absence of myr-Akt (compare lanes 1 and 2).
Identification of Yki S168 as a Hippo-Responsive Phosphorylation Site
We noted that YAP contains a conserved 14-3-3 binding motif at S127 (RAHS127SP) that was reported to concentrate YAP in the cytoplasm upon Akt phosphorylation (Basu et al., 2003). Because both Akt and Wts belong to the AGC (protein kinases A, G, and C) group of protein kinases (Manning et al., 2002), we explored the possibility that Wts might phosphorylate the 14-3-3 binding motif of Yki. To facilitate this analysis, we generated a phosphospecific antibody against the Yki S168 motif. The phospho-Yki antibody detected a specific signal at the expected molecular weight of Yki (Figure 1D). The phospho-Yki signal was increased when Yki was coexpressed with Hpo, Sav, and Wts, and was abolished when S168 was mutated to alanine (Figure 1D). Similarly, in vitro kinase assay using immunoprecipitated Wts and bacterially purified GST-Yki fusion protein revealed a specific kinase activity toward Yki S168 when Wts was activated by Hpo and Sav (Figure 1E), further implicating S168 as a Wts-mediated phosphorylation site in response to active Hippo signaling.
We noted that in our cell culture assays, the S168A mutation not only eliminated the phospho-Yki signal as expected but also largely eliminated the mobility shift (Figure 1D) or the cytoplasmic shuttling (Figure 1F) of Yki induced by active Hippo signaling. These results suggest that S168 is a primary Hippo signaling-responsive phosphorylation site on Yki.
Yki Is Not Phosphorylated by Akt Signaling in Drosophila Cells
Our finding that Yki S168 is phosphorylated by Wts contrasts with a previous report showing that YAP is phosphorylated and inactivated by Akt at a conserved serine residue (S127) (Basu et al., 2003). We noted, however, that while the Yki S168 site (HSRARS168SP) matches perfectly to the optimal 14-3-3 binding motif (RxxS/T(phos)xP), it does not match the optimal Akt substrate sequence (RxRxxS/T) (Obata et al., 2000) in that histidine, rather than arginine, is present at the S-5 position. Indeed, SCANSITE analysis did not predict S168 as an Akt phosphorylation site. It is also difficult to explain how Akt could inactivate Yki, since both proteins are associated with growth-promoting activities in Drosophila (Verdu et al., 1999; Huang et al., 2005). Nevertheless, we tested whether Yki S168 might be phosphorylated by Akt, taking advantage of the phospho-Yki antibody we have generated. We induced Akt activation either by insulin treatment or expression of a constitutively active Akt mutant (Verdu et al., 1999). In contrast to the robust Hippo-induced Yki phosphorylation (Figures 1C and 1D), under neither condition of Akt activation could we observe any changes in the phospho-Yki signal or the mobility of Yki (Figures 1G and 1H). These results suggest that Yki, in particular its S168 residue, is a molecular target of Hippo rather than Akt signaling in Drosophila.
Yki S168 Phosphorylation Mediates the Growth-Suppressive Output of the Hippo Signaling Pathway In Vivo
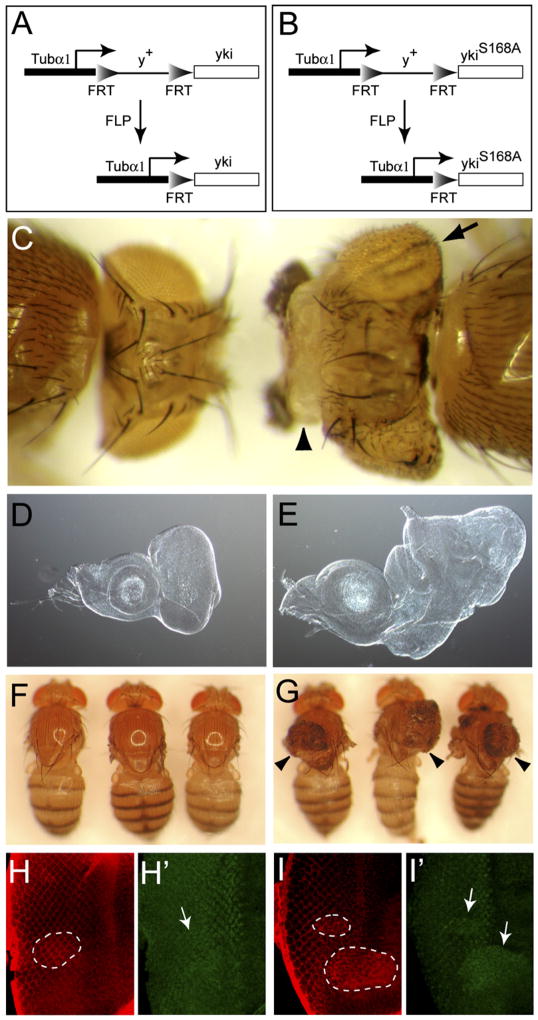
If phosphorylation of Yki at S168 mediates the growth-suppressing output of the Hippo pathway, we might expect a YkiS168A mutant to exhibit gain-of-function properties because it should no longer be inhibited by the endogenous Hippo signaling activity. We reasoned that the commonly used UAS/GAL4 system might not be adequate for comparing the activity of Yki and YkiS168A because overexpression of wild-type Yki is sufficient to drive massive overgrowth in Drosophila (Huang et al., 2005), potentially masking any intrinsic difference between these two forms. With this consideration, we turned to an expression system driven by the Tubulin α1 promoter. We have previously shown that a Tub-yki transgene completely rescues yki null animals (Huang et al., 2005). Indeed, fly strains in which the endogenous yki is replaced by Tub-yki have been continuously maintained as stable stocks in our laboratory. To compare the activity of Yki and YkiS168A, we initially constructed a Tub-ykiS168A trans-gene. In contrast to similarly constructed Tub-yki trans-gene, injection of Tub-ykiS168A into blastoderm embryos resulted in complete lethality. To circumvent this problem, we generated a transgene in which an FRT y+ FRT cassette (Basler and Struhl, 1994) (abbreviated as > y+ >) was placed between the Tub promoter and the yki (or ykiS168A) cDNA, thus blocking transcription downstream of the Tub promoter (Figures 2A and 2B). In transgenic flies carrying the Tub > y+ > yki (or Tub > y+ > ykiS168A) construct, the FRT cassette can be excised (or “flp-out”) by the FLP recombinase, thus allowing transcription downstream of the Tub promoter (Figures 2A and 2B).
Figure 2. S168 Phosphorylation Mediates the Growth-Suppressive Output of Hippo Signaling In Vivo.
(A and B) A flp-out strategy to assay the activity of Yki (A) and YkiS168A (B) in vivo.
(C) Adult flies carrying Tub > y+ > yki (left) or Tub > y+ > ykiS168A (right) and an eye-specific FLP recombinase. The flies were photographed together to show their relative size. Note the enlarged and folded eyes (arrow) and excess head cuticle (arrowhead) in flies expressing Tub > ykiS168A (right).
(D and E) Eye discs carrying Tub > y+ > yki (D) or Tub > y+ > ykiS168A (E) and an eye-specific FLP recombinase. The images were taken under the same magnification.
(F and G) Adult flies carrying Tub > y+ > yki (F) or Tub > y+ > ykiS168A (G) and a heat-shock-inducible FLP source, and heat shocked at first instar larval stage. The wings had been removed for photographic purpose. Note the presence of thoracic overgrowth (arrowheads) in (G) only.
(H and H′) Eye disc containing Tub > y+ > yki flpout clones, stained for Yki (red) and Diap1 (green). The circled area in H indicates a flpout clone, which expressed a higher level of Yki. Diap1 levels were not affected in the clone (arrow in H′).
(I and I′) similar to H and H′ except that Tub > y+ > ykiS168A clones (outlined by circles in [I]) were analyzed. Note the increased levels of Diap1 in the clones (arrows in [I′]).
We first crossed the Tub > y+ > yki (or Tub > y+ > ykiS168A) flies with flies containing an eye-specific FLP source. Consistent with our previous report (Huang et al., 2005), expression of Tub > yki in the eye primordium did not result in any visible phenotype (Figure 2C), nor did it affect the viability of the animals (see Figure S1 in the Supplemental Data available with this article online) (four independent lines examined). In contrast, analogous expression of Tub > ykiS168A resulted in significant lethality (Figure S1), with rare survivors showing enlarged, folded eyes and excess head cuticle (Figure 2C), as well as massive overgrowth of the eye imaginal discs (compare Figures 2E and 2D) (five independent lines examined). These phenotypes are strikingly similar to those caused by selective removal of hpo, sav, or wts in the eye, suggesting that the S168A mutation leads to constitutive activation of Yki.
Next, we crossed the Tub > y+ > yki (or Tub > y+ > ykiS168A) flies with flies containing a heat-shock-inducible FLP source, which allows excision of the > y+ > cassette in random clones throughout the body. As expected, Tub > yki clones did not perturb normal epithelial structures (Figure 2F), nor did they affect the expression of Yki targets such as Diap1 (Figures 2H and 2H′). In contrast, Tub > ykiS168A clones resulted in dramatic overgrowth of epithelial structures such as notum, haltere, leg, and wing (Figure 2G and data not shown), as well as upregulation of Diap1 expression (Figures 2I and 2I′). These phenotypes are indistinguishable from those caused by loss of Hippo signaling in mosaic flies, providing further proof that phosphorylation of Yki at S168 mediates the growth-suppressive output of the Hippo pathway. The dissimilarity of these phenotypes to those caused by Akt inactivation (Verdu et al., 1999) also supports our cell culture assays showing that Yki is not a target of Akt signaling (Figures 1G and 1H).
Loss of Hippo Signaling Leads to Nuclear Accumulation of Yki In Vivo
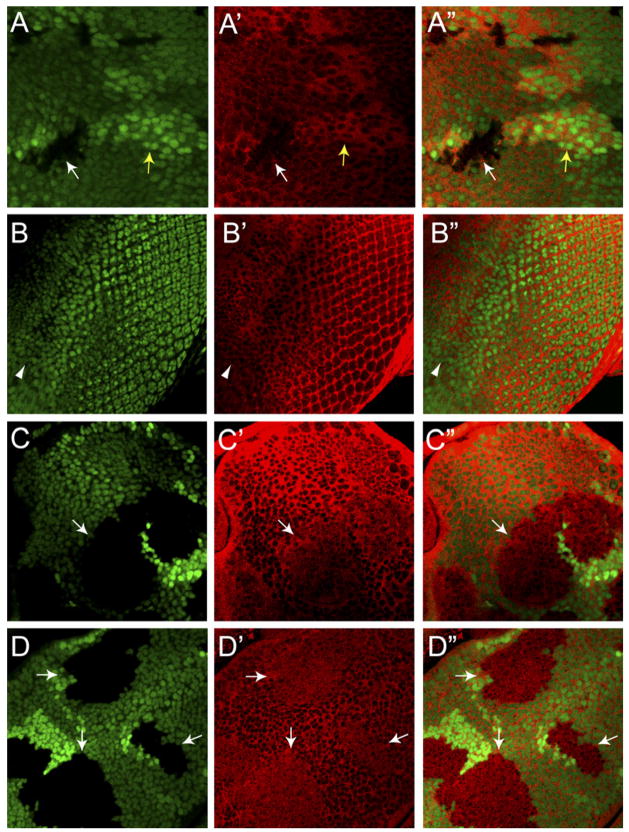
The results presented so far are consistent with a model wherein Hippo signaling leads to S168 phosphorylation and cytoplasmic localization of Yki, thereby compromising its function as a nuclear transcription regulator. To further corroborate this model, we examined the subcellular localization of endogenous Yki in imaginal discs using an antiserum against Yki. The specificity of the antibody was assessed in imaginal discs containing mutant clones of a deletion allele of yki in an otherwise heterozygous background. As shown in Figures 3A––3A″, there was a specific loss of staining in yki mutant clones. Moreover, a higher level of staining could be detected in the +/+ twin spot, demonstrating that this antibody could reliably detect a 2-fold difference in the yki gene dosage. Next, we examined the subcellular localization of Yki in third instar imaginal discs. In wild-type wing (Figures 3A––3A″) or eye imaginal discs (Figures 3B––3B″), Yki is mostly present in the cytoplasm with much less signal detectable in the nucleus, giving the appearance of a honeycomb-like pattern. In contrast, in mutant clones of wts (Figures 3C––3C″) or hpo (Figures 3D––3D″), Yki is distributed uniformly throughout the cell, showing comparable signals in both the nucleus and the cytoplasm. Thus, loss of Hippo signaling causes redistribution of Yki from the cytoplasm to the nucleus, further supporting our model that Hippo signaling promotes the cytoplasmic localization of Yki.
Figure 3. Inactivation of Hippo Signaling Leads to Nuclear Accumulation of Yki In Vivo.
In all panels, three images are shown, one of GFP (panels A, B, C, and D) (green), one of Yki (panels A′, B′, C′, and D′) (red), and one of superimposed GFP and Yki (panels A″, B″, C″, and D″). The GFP marker used here is concentrated in the nucleus.
(A–A″) Third instar wing disc containing ykiB5 clones and stained with α-Yki. −/− clones were marked by the absence of GFP (white arrow) while the +/+ twin spots were marked by 2×GFP signal (darker green, yellow arrow). Note the absence of Yki staining in the −/− clones and increased Yki staining in the twin spots, as well as the relative absence of Yki signal in the GFP-marked cell nuclei.
(B–B″) Third instar eye disc. Note the relative absence of Yki signal in the GFP-marked cell nucleus. Arrowhead marks the MF.
(C–C″) Third instar eye discs containing wtsX1 clones. Note the relative absence of nuclear Yki in wild-type cells. In contrast, Yki is present throughout the cytoplasm and the nucleus in wtsX1 clones (arrow).
(D–D″) Third instar wing discs containing hpo42–47 clones. Note the relative absence of nuclear Yki in wild-type cells. In contrast, Yki is present throughout the cytoplasm and the nucleus in hpo42–47 clones (arrows).
Delineation of a Mammalian Hippo Signaling Pathway Leading to YAP Phosphorylation
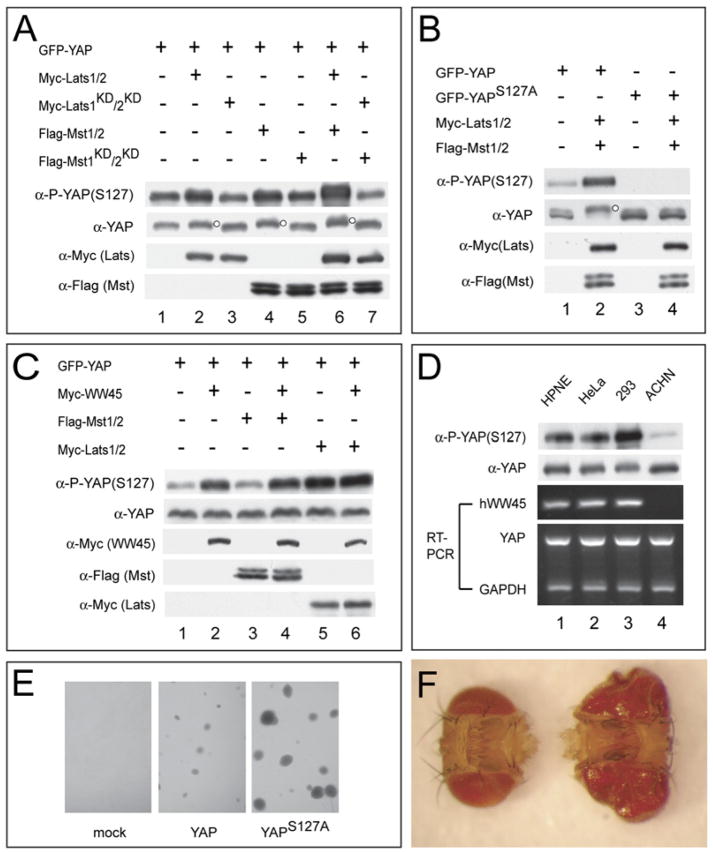
As discussed earlier, previous studies have failed to establish a mammalian Hippo pathway, largely due to the lack of a specific readout that measures pathway activity. The presence of a conserved motif at YAP S127 and Yki S168 suggests that YAP, and in particular the S127 site, might be a target of the purported mammalian Hippo pathway. We investigated this possibility in mammalian cells using a phosphospecific antibody against YAP S127. In HEK293 cells, expression of Lats1/2 or Mst1/2, but not the respective kinase-dead mutants, resulted in increased S127 phosphorylation as well as a mobility retardation of YAP on SDS-PAGE (Figure 4A). Coexpression of Lats1/2 and Mst1/2 resulted in synergistic S127 phosphorylation and greater mobility shift of YAP (Figure 4A), which was reversed by phosphatase treatment (Figure S2A). An S127A mutation not only eliminated the phospho-S127 signal but also largely reversed the mobility shift of YAP induced by Lats1/2 and Mst1/2 (Figure 4B), suggesting that S127 is a primary Hippo-responsive phosphorylation site on YAP. Similar results were seen in HPNE cells, an hTERT-immortalized nontransformed epithelial cell line derived from human pancreas (Lee et al., 2005) (Figure S2B). We further examined the subcellular localization of YAP in mammalian cells. Both YAP and YAPS127A can be detected in the nucleus as well as the cytoplasm, with YAPS127A showing a greater degree of nuclear signal (Figure S3). Importantly, coexpression of Lats1/2 and Mst1/2 resulted in relocalization of YAP, but not YAPS127A, to the cytoplasm (Figure S3). Thus, S127 phosphorylation promotes the cytoplasmic localization of YAP in a way similar to Yki phosphorylation in Drosophila.
Figure 4. Delineation of a Mammalian Hippo Signaling Pathway Leading to YAP S127 Phosphorylation.
(A) HEK293 cells were transfected with epitope-tagged constructs as indicated. Note the increased YAP(S127) phosphorylation and mobility retardation resulting from Lats1/2 (lane 2), Mst1/2 (lane 4), or both (lane 6), but not the respective kinase-dead forms or their combinations (lanes 3, 5, and 7). The slower-migrating species of YAP was indicated by a small dot to the right of the protein band.
(B) HEK293 cells expressing the indicated plasmids were analyzed. The slower-migrating species of YAP was indicated by a small dot to the right of the protein band.
(C) ACHN cells expressing the indicated plasmids were analyzed. Note the failure of Mst1/2 (lane 3), but not Lats1/2 (lane 5), to induce YAP(S127) phosphorylation, and the rescue of this defect by hWW45 (lane 4). Also note that hWW45 alone (lane 2) can induce YAP(S127) phosphorylation.
(D) Various cell lines were probed with α-P-YAP(S127) and α-YAP, and analyzed by RT-PCR for the expression of indicated genes. Note the absence of hWW45 expression and the much diminished levels of YAP(S127) phosphorylation in ACHN cells (lane 4).
(E) HPNE cells stably expressing YAP (middle), but not those expressing an empty vector (left), grew in soft agar to form colonies. HPNE cells stably expressing YAPS127A grew to significantly larger colonies (right). Similar results were obtained with multiple independently established cell lines.
(F) Adult heads from flies expressing YAP (left) or YAPS127A (right) in the eye. The genotypes are: GMRGal4/UASYAP (left) and GMRGal4/UASYAPS127A (right). The heads were photographed together to show their relative size.
The identification of YAP S127 as a Hippo-responsive phosphorylation site provides a powerful tool to interrogate additional components of the mammalian Hippo pathway. As an illustrative example, we investigated the relationship between hWW45, the human homolog of Sav, with respect to the mammalian Hippo pathway. We took advantage of the renal cancer cell line ACHN, which is known to have deleted hWW45 (Tapon et al., 2002). Compared to the other cell lines we have examined, such as HEK293, HeLa, and HPNE, the hWW45-deficient ACHN cells had greatly diminished levels of YAP S127 phosphorylation (Figure 4D). In contrast to HEK293 or HPNE cells, only Lats1/2, but not Mst1/2, promoted YAP S127 phosphorylation in ACHN cells (Figure 4C). This observation is consistent with a genetic placement of hWW45 upstream of Lats1/2 but downstream of Mst1/2. Interestingly, coexpression of hWW45 rescued the ability of Mst1/2 to promote YAP phosphorylation in ACHN cells (Figure 4C). In fact, hWW45 alone, which did not have any effect on YAP phosphorylation in the other cell lines we have tested (Figure S4A), substantially increased the basal level of YAP S127 phosphorylation in ACHN cells (Figure 4C). Taken together, these results provide the first evidence that functionally links hWW45 to the mammalian Hippo pathway and support the following order of signal propagation: Mst1/2 > hWW45 > Lats1/2 > YAP. Consistent with this conclusion, kinase-dead Lats1/2 efficiently suppressed Mst1/2-induced YAP phosphorylation (Figure S4B).
YAP S127 Phosphorylation Mediates the Growth-Suppressive Output of the Mammalian Hippo Pathway
If phosphorylation of YAP at S127 mediates the growth-suppressing output of the mammalian Hippo pathway, we would expect a YAPS127A mutant, like the YkiS168A mutant in Drosophila, to exhibit gain-of-function properties. Indeed, at comparable expression levels (Figure S5), HPNE cells that stably express YAPS127A grew to larger colonies in soft agar than cells expressing the wild-type YAP (Figure 4E), demonstrating an enhanced activity of YAPS127A to induce anchorage-independent growth.
Next, we compared the activity of YAP and YAPS127A in Drosophila. We have previously shown that overexpression of UAS-YAP by the eye-specific GMRGal4 driver does not cause any discernible overgrowth phenotype while partially suppressing the lethality of GMR-Gal4/UAS-Hpo animals (Huang et al., 2005), suggesting that YAP possesses similar, albeit weaker, activity than Yki when assayed in Drosophila. In contrast, we found that expression of UAS-YAPS127A by the GMR-Gal4 driver resulted in a dramatic increase in eye size (Figure 4F) similar to that observed in GMR-Gal4/UAS-Yki animals (six independent lines tested). Moreover, random clones overexpressing YAPS127A, but not YAP, resulted in overgrowth of epithelial structures such as the notum and the haltere (Figure S6), a phenotype that resembles clonal inactivation of hpo (or wts) or overexpression of Yki. Thus, like Yki S168 phosphorylation in Drosophila, YAP S127 phosphorylation mediates the growth-suppressive output of the mammalian Hippo pathway. These results further suggest that the Hippo pathway might play an analogous role in organ size control in mammals.
Reversible Control of Mammalian Organ Size by Conditional Activation/Inactivation of YAP
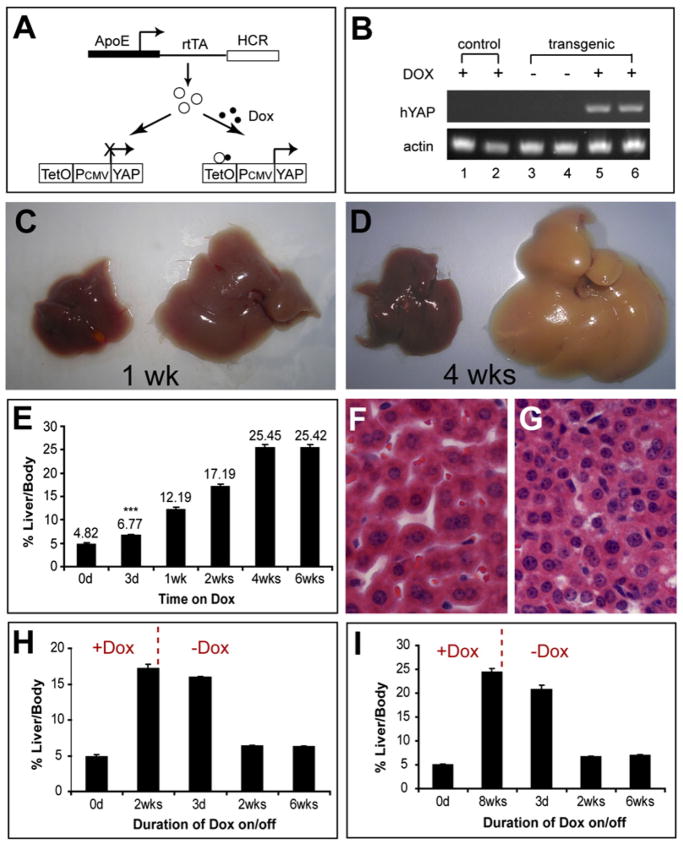
To directly test the role of Hippo signaling in mammalian organ size control, we asked whether conditional overexpression of YAP, the nuclear effector of the mammalian Hippo pathway, is sufficient to increase organ size in mammals. We chose mouse liver as our model, since liver size is known to be tightly regulated, constituting about5% of the body weight (Michalopoulos and DeFrances, 1997). We generated transgenic mice carrying two cointegrated DNA constructs, one with reverse tetracycline transactivator (rtTA) under the control of the liver-specific ApoE promoter, and the other with a human YAP cDNA (hYAP) driven by the minimal CMV promoter and a tetracycline (Tet)-response element (TRE) (Figure 5A). As expected, these transgenic mice, hereafter referred to as ApoE/rtTA-YAP, allowed conditional, Tet-dependent expression of YAP in the mouse liver (Figure 5B). A dose of 0.2 mg/ml doxycycline in drinking water was used throughout this study.
Figure 5. Reversible Control of Mammalian Organ Size by Conditional Activation/Inactivation of YAP.
(A) Schematic of the ApoE/rtTA-YAP mice.
(B) RT-PCR analysis of hYAP transgene expression in control and transgenic livers in the absence or presence of Dox for 1 week. Two mice were used for each genotype/condition.
(C and D) Livers from control (left) and ApoE/rtTA-YAP (right) mice kept on Dox for 1 (C) or 4 (D) weeks, starting at 3 weeks of age.
(E) The temporal course of YAP-induced hepatomegaly. YAP was induced as in (C) and (D), and the liver and body weight was measured at the indicated time. Values represent mean ± SD (n = 4; ***p < 0.001, t test).
(F and G) Hematoxylin and eosin (H&E) staining of control (F) and transgenic (G) livers after 4 weeks of Dox exposure. Both images were taken under the same magnification. Note the higher cell density in the transgenic liver.
(H and I) Reversal of liver overgrowth by Dox withdrawal. Three-week-old ApoE/rtTA-YAP mice were first kept on Dox for 2 (H) or 8 (I) weeks, followed by withdrawal of Dox from the drinking water. Values represent mean ± SD (n = 4).
To test the effect of YAP overexpression on liver size, control and ApoE/rtTA-YAP littermates were fed Dox-containing water starting at 3 weeks of age. As shown in Figures 5C–5E, YAP induction caused massive hepatomegaly. The increase in liver mass was detectable as early as 3 days after induction (Figure 5E) and reached 25% of total body weight after 4 weeks of Dox feeding (Figures 5D and 5E). Hepatocytes in the transgenic livers were smaller and more densely packed as compared to the control livers (Figures 5F and 5G), suggesting that the increase in liver mass was caused by an increase in cell number (hyperplasia) as opposed to increased cell size (hypertrophy). Consistent with this observation, transgenic Dox-exposed livers exhibited increased cell proliferation, as revealed by increased Ki67 expression (Figure 6A) and BrdU incorporation (Figures 6B and 6C).
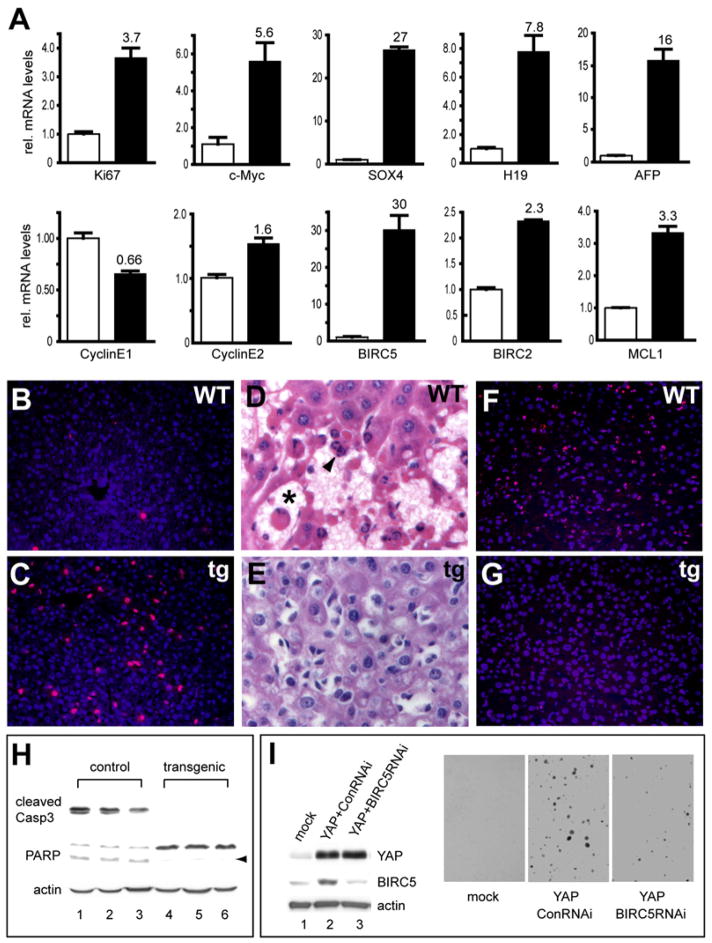
Figure 6. Coordinate Regulation of Cell Proliferation and Apoptosis by the Mammalian Hippo Pathway.
(A) Quantitative real-time RT-PCR analysis of selected genes from control (white columns) and ApoE/rtTA-YAP (black columns) livers after 2 weeks of Dox exposure. Gene expression was measured in triplicate and expressed as mean ± SEM. The subtle changes of Cyclin E1 and Cyclin E2 are insignificant (p = 0.1, t test).
(B and C) YAP overexpression drives cell proliferation. Control (B) and ApoE/rtTA-YAP (C) livers after 1 week of Dox exposure were analyzed for BrdU incorporation (red), counter-stained with DAPI (blue).
(D–H) YAP overexpression confers potent resistance to apoptosis. Control and ApoE/rtTA-YAP littermates were kept on Dox for 1 week, injected with Jo-2, and analyzed 3 hr postinjection by H&E staining (D and E), TUNEL (F and G), and western blotting for cell death markers (H). Note the widespread hemorrhage (asterisk) and apoptotic nuclei (arrowhead) in the control (D), but not the transgenic livers (E). Also note the extensive TUNEL staining in the control (F), but not the transgenic livers (G). In (H), cleavage of caspase-3 (Casp3) and PARP was detected in the control, but not the transgenic, livers (arrowhead marks the cleaved PARP product). Three animals were analyzed for each genotype.
(I) RNAi knockdown of BIRC5/survivin reduces the transforming activity of YAP. Left: western blots showing increased BIRC5/survivin expression in YAP-HPNE cells compared to mock transfected cells, and the knockdown of BIRC5/survivin expression in YAP-HPNE cells stably expressing BIRC5/survivin shRNA. Right: soft agar assay showing anchorage-independent growth of YAP-HPNE cells, a feature that is not observed in the mock-transfected HPNE cells. Note that YAP-HPNE cells stably expressing BIRC5/survivin shRNA formed reduced numbers of colonies.
The ApoE/rtTA-YAP transgenic mice also allowed us to investigate whether the YAP-induced increase in organ size was dependent on continuous YAP expression. We induced YAP expression for 2 or 8 weeks, at which point the livers reached approximately 17% or 25% of body weight, respectively. We then withdrew Dox from the diet. Remarkably, in both cases, the enlarged liver returned to near normal size 2 weeks after Dox withdrawal (Figures 5H and 5I), a process that was accompanied by apoptosis (Figure S7). The reversible control of liver size by YAP further supports our hypothesis that the Hippo pathway regulates organ size in mammals.
Coordinate Regulation of Cell Proliferation and Apoptosis by the Mammalian Hippo Pathway
To investigate the mechanism by which Hippo signaling regulates mammalian organ size, we used microarrays to identify YAP-induced genes in murine livers (Table S1). Selected genes from the microarray analysis were validated by real-time quantitative PCR analysis (Figure 6A). YAP induced the transcription of many genes that are normally associated with hepatocyte proliferation, such as Ki67, c-myc, SOX4, H19, and AFP (Figure 6A), and caused a dramatic increase in BrdU incorporation (Figures 6B and 6C). In contrast to Drosophila, our microarray analysis did not identify Cyclin E1 or Cyclin E2 as transcriptional targets of YAP. Real-time PCR analysis showed that Cyclin E1 and Cyclin E2 transcript levels were not significantly changed by YAP induction (Figure 6A). Thus, cyclin E is unlikely to be a transcriptional target of YAP, at least in the context of murine liver. We note that even in Drosophila, the functional relevance of cycE transcription in Hippo signaling remains to be determined—unlike other targets such diap1, which are regulated cell autonomously by the Hippo pathway in all imaginal tissues, cycE transcription is regulated by the Hippo pathway in a tissue (eye imaginal discs only)- and region (close to the morphogenetic furrow)-specific manner (Huang et al., 2005).
YAP also induced the expression of several negative regulators of apoptosis, such as the IAP family members BIRC5/survivin and BIRC2/cIAP1, and the BCL2 family gene MCL1 (Figure 6A). Of note is the massive induction (30-fold) of BIRC5/survivin, a gene that has been implicated as a cell-death inhibitor as well as a mitotic regulator (Altieri, 2003). In support of the functional relevance of BIRC5/survivin induction by YAP, RNAi knockdown of BIRC5/survivin significantly reduced the ability of YAP to induce anchorage-independent growth of HPNE cells (Figure 6I). Another IAP family member induced by YAP, BIRC2/cIAP1, is physically linked to YAP in mouse (chromosome 9qA1) and human (chromosome 11q22), and both genes are coamplified in certain tumors (Zender et al., 2006; Overholtzer et al., 2006). The induction of BIRC2/cIAP1 expression by YAP suggests a feed-forward mechanism by which the YAP-BIRC2/cIAP1 amplicon mediates the antiapoptotic function.
The induction of antiapoptotic genes suggests that like its Drosophila counterpart, YAP might confer resistance to apoptosis. To investigate this possibility, we employed the well-characterized paradigm of Jo-2-induced hepatocellular apoptosis (Ogasawara et al., 1993). The Fas-agonist antibody Jo-2 was administered to control and transgenic mice that had been kept on Dox for 1 week, and the livers were analyzed 3 hr after Jo-2 injection. Control livers showed disorganization of the hepatic architecture, hemorrhage into sinusoidal spaces, and extensive apoptotic cells characterized by condensed and fragmented chromatin (Figure 6D). In contrast, transgenic livers were markedly less hemorrhagic, displaying intact lobular hepatic architecture without necrotic foci or apoptosis (Figure 6E). Consistent with liver histology, transgenic livers showed greatly diminished TUNEL staining (Figures 6F and 6G). Furthermore, cleavage of caspase-3 or poly-(ADP)-ribose-polymerase (PARP), both hallmarks of apoptosis, was absent from the transgenic livers (Figure 6H). Thus, YAP activation confers potent resistance to apoptosis, further implicating the antiapoptotic function as an evolutionarily conserved output of the Hippo pathway from Drosophila to mammals.
Dysregulation of the Mammalian Hippo Pathway Leads to Tumorigenesis
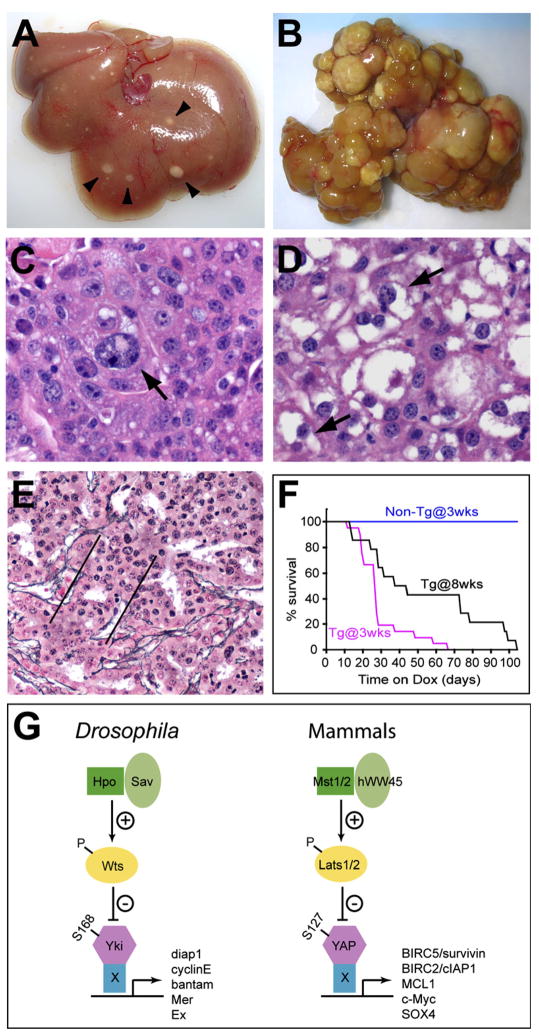
Increased liver size caused by short-term YAP induction is characterized by uniform expansion of liver mass with no overt evidence of discrete nodules. However, when the ApoE/rtTA-YAP mice were exposed to Dox for over 8 weeks, they developed numerous discrete nodules throughout the livers (Figure 7A). More widespread nodule development was observed if YAP induction was initiated at birth, by giving Dox water to the nursing mothers (Figure 7B). These nodules were comprised of discrete, yet expansive areas of proliferative hepatocytes that compressed the surrounding nontumorous liver and displayed many characteristics of hepatocellular carcinoma (HCC). Cytologically, the tumors displayed the characteristics of mature hepatocytes with abundant cytoplasm, moderate nuclear to cytoplasmic ratio, and a round nucleus with distinct nucleolus. Regions within the tumor showed areas of cellular pleiomorphism manifested as increase in cell size (Figure 7C) or loss of cytoplasmic staining (Figure 7D) and tumor necrosis (data not shown). Finally, the tumors showed a thickening of hepatocyte plates and expansion of the reticulin network in a trabecular pattern (Figure 7E), which is strikingly similar to what is seen in the trabecular type of human HCC. The transgenic mice eventually succumbed to liver tumors, with a mean survival of 7 weeks (from birth) for mice in which YAP was induced starting at 3 weeks of age, and a mean survival of 14 weeks (from birth) in which YAP was induced starting at 8 weeks of age (Figure 7F). Taken together, these results reveal a direct link between dysregulation of the Hippo size-control pathway and tumorigenesis.
Figure 7. Dysregulation of the Mammalian Hippo Pathway Leads to Tumorigenesis In Vivo.
(A) Liver from an ApoE/rtTA-YAP mouse raised on Dox for 8 weeks, starting at 3 weeks after birth. Note the presence of discrete nodules scattered throughout the liver (arrowheads).
(B) Liver from an ApoE/rtTA-YAP mouse raised on Dox for 3 months, starting at birth. Note the widespread development of HCC throughout the liver.
(C–E) Histolopathologic examination of murine liver nodules reveals characteristics of hepatocellular carcinoma. Mice were fed Dox-water as in (A). (C) shows cellular pleiomorphism of YAP-induced HCC, with a large cell (arrow) surrounded by smaller cells. (D) shows loss of cytoplasmic staining (arrows), or the so called “clear cell change.” (E) shows expanded hepatic plates. A reticulin stain highlights the edges of the hepatic plates (indicated by parallel lines), which are wider in HCC than the typical 1 to 2 cells in a nonneoplastic liver.
(F) Survival curves of control (Non-Tg) and ApoE/rtTA-YAP (Tg) littermates raised on Dox, starting at 3 or 8 weeks of age as indicated.
(G) Conserved Hippo kinase cascade in Drosophila and mammals. The corresponding proteins in Drosophila and mammals are indicated by matching colors and shapes. The critical phosphorylation site on Yki and YAP is also indicated. “X” denotes unknown DNA-binding protein(s) that partner with Yki or YAP to regulate target gene transcription.
Consistent with a direct link between YAP misregulation and tumorigenesis, we found that YAP is overexpressed in a wide spectrum of human cancer cell lines and primary tumors (Figure S8). Interestingly, we observed two distinct patterns of YAP distribution in tumors, one with cytoplasmic labeling in conjunction with prominent nuclear accumulation, and the second with uniform cytoplasmic and nuclear staining without nuclear accumulation (Figure S8C). The proportion of cases conforming to these two patterns varied with tumor types. We will discuss the implication of these staining patterns later.
DISCUSSION
The Hippo Signaling Pathway in Drosophila and Mammals
The Hippo pathway has recently emerged as a central mechanism that restricts organ size in Drosophila (Edgar, 2006; Pan, 2007). This pathway impinges on the transcriptional coactivator Yki to regulate the transcription of target genes involved in cell growth, proliferation, and survival. Given its pivotal position in the Hippo pathway, understanding the molecular and cellular mechanism of Yki regulation should provide important insights into this size-control pathway. In this study, we provide several lines of evidence demonstrating that Hippo signaling antagonizes Yki function by changing its subcellular localization. Hippo signaling promotes Yki cytoplasmic localization in cultured Drosophila cells, and accordingly, loss of Hippo signaling promotes nuclear accumulation of Yki in imaginal discs. This is further supported by the ability of phosphorylated Yki (but not unphosphorylated Yki) to bind to 14-3-3 proteins, which are known to promote the cytoplasmic shuttling of other transcription factors in a phosphorylation-dependent manner (Muslin and Xing, 2000). Importantly, we identify S168 as a primary Hippo-responsive phosphorylation site on Yki both in vitro and in vivo: the S168A mutation not only abrogates Hippo-induced Yki phosphorylation and cytoplasmic shuttling in S2 cells but, more significantly, causes constitutive Yki activation in developing tissues. These results demonstrate that S168 mediates the growth-suppressive output of the Hippo signaling pathway.
The conservation of this phosphorylation site in mammalian YAP provides us the first opportunity to functionally link Mst1/2, WW45, and Lats1/2 in a single kinase cascade that culminates in YAP S127 phosphorylation (Figure 7G). We further demonstrate that the mammalian Hippo signaling pathway antagonizes YAP function by promoting its cytoplasmic localization in a S127 phosphorylation-dependent manner. The identification of S168/S127 as Wts/Lats-mediated phosphorylation site in Yki/YAP is rather unexpected given that previous studies have implicated this residue as an Akt phosphorylation site (Basu et al., 2003). Our observation that both YkiS168A and YAPS127A result in a loss-of-Wts rather than a loss-of-Akt phenotype in Drosophila strongly suggests that this site is regulated by the Hippo pathway rather than Akt under normal physiological conditions.
The identification of a single phosphorylation site as the functional output of the Hippo pathway, and the constitutive active Yki/YAP mutants described in this study, will greatly facilitate future investigation of this important size-control pathway in multiple species. For example, the constitutive active Yki/YAP mutants can be conveniently used to modulate the Hippo pathway in animal models and in genetic epistasis studies to characterize new components of the pathway; the phospho-Yki/YAP antibodies should provide a sensitive assay to link a specific protein to the Hippo pathway. These tools are especially important for the mammalian system, where a functional readout of the Hippo pathway has so far been unavailable. Indeed, we were able to place hWW45 in the mammalian Hippo pathway using phospho-YAP as a convenient readout (Figure 4C).
Control of Mammalian Organ Size by the Hippo Signaling Pathway
Despite the conservation of many Hippo pathway components between flies to mammals, previous studies have not revealed a direct role for this pathway in mammalian organ size control. Several recent studies have focused on their involvement in tumorigenesis. For example, YAP was recently shown to transform immortalized mammary epithelial cells in vitro (Overholtzer et al., 2006) and to accelerate tumorigenesis in conjunction with p53 loss and c-myc overexpression (Zender et al., 2006). While suggestive of an involvement of the Hippo pathway in mammalian tumorigenesis, these observations alone do not necessarily prove a direct requirement for the Hippo pathway in the control of organ size, since perturbations of many cellular processes in addition to growth control can contribute to tumorigenesis (Hanahan and Weinberg, 2000). It is worth noting that knockout mice have been generated for several components of the mammalian Hippo pathway. However, these mice are either viable, lacking any overt overgrowth characteristic of the respective Drosophila mutants (e.g., Lats1) (St John et al., 1999), or embryonic lethal, thus preventing a critical assessment of their involvement in organ size regulation (e.g., Lats2 and YAP) (McPherson et al., 2004; Morin-Kensicki et al., 2006).
The identification of YAP as the nuclear effector of the mammalian Hippo pathway provides a powerful tool to manipulate this pathway in mammals, in much the same way that Yki overexpression recapitulates loss of Hippo signaling in Drosophila (Huang et al., 2005). By manipulating YAP activity in a conditional and tissue-specific manner, we demonstrate here that modulating Hippo pathway activity is sufficient to cause a rapid and reversible change of organ size (up to 500%), therefore offering the first direct evidence implicating the Hippo pathway in mammalian organ size control. We further demonstrate that, like its Drosophila counterpart, the mammalian Hippo pathway coordinately regulates both cell proliferation and apoptosis. The dual function of YAP in promoting cell proliferation and suppressing apoptosis distinguishes it from a conventional oncogene such as c-myc, whose mitogenic activity is coupled with a proapoptotic activity (Pelengaris et al., 2002). We suggest that this dual activity in promoting cell proliferation and suppressing apoptosis underlies the rapid and uniform expansion of liver mass in the ApoE/rtTA-YAP mice. The ability of YAP to induce organomegaly in postnatal mice is consistent with the notion that the Hippo pathway not only controls organ size during development as demonstrated in Drosophila but also regulates tissue homeostasis in postnatal life.
Initially isolated as a yes-associated protein (Sudol, 1994), YAP has since been reported to bind to a large number of proteins in cultured mammalian cells, including EBP50 (Mohler et al., 1999), Smad7 (Ferrigno et al., 2002), ErbB4 (Komuro et al., 2003), p53BP-2 (Espanel and Sudol, 2001), p73 (Basu et al., 2003), and hnRNAP U (Howell et al., 2004), as well as Runt (Yagi et al., 1999) and TEAD (Vassilev et al., 2001) transcription factors. However, it has been difficult to ascertain whether any of these binding partners mediate YAP function in vivo. The anti-apoptotic activity we have observed in the transgenic mouse liver is clearly distinct from the reported ability of YAP to potentiate p73-mediated apoptosis in response to DNA damage in cultured mammalian cells (Basu et al., 2003). Given that p73-deficient mice are viable (Yang et al., 2000) while YAP-deficient mice die at embryonic day 8.5 (Morin-Kensicki et al., 2006), p73 is unlikely to be a critical partner for YAP in mouse development.
Dysregulation of Size-Control Mechanism and Tumorigenesis
Studies from both insects and mammals support the existence of an intrinsic size checkpoint that monitors organ size at the tissue, rather than the cellular, level (Conlon and Raff, 1999). For example, while constitutive activity of the myc oncogene drives the growth of individual Drosophila cells, it has little effect on the size of imaginal disc compartments (Johnston et al., 1999). Therefore, increased cell growth or cell proliferation does not automatically lead to a corresponding increase in tissue size, unless the size checkpoint is simultaneously perturbed. It follows that such intrinsic size-control mechanism must be overridden to permit the sustained overgrowth of tumors. Our finding that YAP overexpression leads to immediate organomegaly followed by tumor formation provides direct support for this hypothesis. The widespread upregulation of YAP in diverse tumor types further suggests that the Hippo pathway represents a common mutational target that allows cancer cells to evade the intrinsic size-control mechanisms that normally maintain tissue homeostasis in animals.
Our observation of two distinct patterns of YAP distribution in tumor cells (Figure S8C)—with or without nuclear accumulation—implicates two possible mechanisms by which Hippo signaling may be dysregulated in cancer cells. Based on the mechanism of Yki/YAP inactivation by Hippo signaling as revealed by the current study, we suggest that the former pattern could result from inactivation of tumor suppressors upstream of YAP, mutation of the S127 phosphorylation site, or perturbation of the nuclear-cytoplasmic shuttling machinery, whereas the latter pattern could be caused by YAP overabundance, either via gene amplification, increased transcription, or protein stabilization. We further speculate that these mechanisms may also be employed in normal physiological contexts to regulate the activity of the Hippo pathway in flies and mammals. Thus, besides phosphorylation, mechanisms that regulate Yki/YAP transcription or stability are likely relevant to the modulation of Hippo signaling activity in vivo.
EXPERIMENTAL PROCEDURES
Drosophila Genetics
The Tub > y+ > yki transgene was constructed by cloning a FRT y+ FRT cassette (Basler and Struhl, 1994) into Tub-yki (Huang et al., 2005). The Tub > y+ > ykiS168A transgene was constructed similarly. Flies of the following genotype were used for analysis: y w; Tub > y+ > yki (or ykiS168A)/ey-FLP (for eye-specific flp-out), or y w hsp-flp; Tub > y+ > yki (or ykiS168A) (for random clones in all imaginal discs; first instar larvae subjected to 10 min heat shock at 38°C).
For comparison of YAP and YAPS127A, flies of the genotype y w hsp-flp; Act > CD2 > Gal4; UAS-YAP (or YAPS127A) were subjected to 6 min of 38°C heat shock at first instar larval stage.
Phosphospecific Antibodies
Phosphospecific antibodies against Yki S168 were produced by immunizing rabbits with the HHSRAR(pS)SPA phosphopeptide. Antibodies reactive with the nonphosphopeptide were removed by adsorption to a nonphosphopeptide affinity column. Antibodies that flowed through this column were next passed over a column of immobilized phosphopeptide; after washing, antibodies were eluted at low pH and dialyzed. Phosphospecific antibodies against YAP S127 were obtained from Cell Signaling Technology.
Transgenic Mice
To generate the Tet-On inducible YAP transgene, a human YAP cDNA (Huang et al., 2005) was cloned into the pTRE2 vector (BD Biosciences). A modified tetracycline transactivator, rtTAs-M2 (Lamartina et al., 2002), was inserted into the liver-specific expression cassette pLIV 11 (Fan et al., 1994). Both transgenes were excised from the respective vectors and coinjected into fertilized (C57Bl/6 × SJL) F2 hybrid mouse eggs. Three founders were identified, of which two showed Dox responsiveness. Unless otherwise indicated, 3-week-old transgenic and nontransgenic littermates were fed 0.2 mg/ml doxycycline (Sigma) in drinking water supplemented with 2.5% sucrose. For all the experimental conditions, at least four mice were used to calculate average liver/body ratio and standard deviation.
Supplementary Material
Acknowledgments
We would like to thank Elizabeth Chen and Keith Wharton for critical reading of the manuscript. We apologize to colleagues whose work was not cited in this paper due to publisher’s space constraints. This work was supported by grants from the National Institutes of Health to D.P. (EY015708), A.M. (R01CA113669 and R21DK072532), and R.A.A. (DK067187). D.P. is a Scholar of the Leukemia & Lymphoma Society.
Footnotes
Supplemental Data
Supplemental Data include eight figures, one table, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/130/6/1120/DC1/.
References
- Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Espanel X, Sudol M. Yes-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J Biol Chem. 2001;276:14514–14523. doi: 10.1074/jbc.M008568200. [DOI] [PubMed] [Google Scholar]
- Fan J, Wang J, Bensadoun A, Lauer SJ, Dang Q, Mahley RW, Taylor JM. Overexpression of hepatic lipase in transgenic rabbits leads to a marked reduction of plasma high density lipoproteins and intermediate density lipoproteins. Proc Natl Acad Sci USA. 1994;91:8724–8728. doi: 10.1073/pnas.91.18.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L’Hoste S, Camonis J, Atfi A, Mauviel A. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149:1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Borchers C, Milgram SL. Heterogeneous nuclear ribonuclear protein U associates with YAP and regulates its co-activation of Bax transcription. J Biol Chem. 2004;279:26300–26306. doi: 10.1074/jbc.M401070200. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Lamartina S, Roscilli G, Rinaudo CD, Sporeno E, Silvi L, Hillen W, Bujard H, Cortese R, Ciliberto G, Toniatti C. Stringent control of gene expression in vivo by using novel doxycycline-dependent trans-activators. Hum Gene Ther. 2002;13:199–210. doi: 10.1089/10430340252769734. [DOI] [PubMed] [Google Scholar]
- Lee KM, Yasuda H, Hollingsworth MA, Ouellette MM. Notch 2-positive progenitors with the intrinsic ability to give rise to pancreatic ductal cells. Lab Invest. 2005;85:1003–1012. doi: 10.1038/labinvest.3700298. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- McPherson JP, Tamblyn L, Elia A, Migon E, Shehabeldin A, Matysiak-Zablocki E, Lemmers B, Salmena L, Hakem A, Fish J, et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Kreda SM, Boucher RC, Sudol M, Stutts MJ, Milgram SL. Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J Cell Biol. 1999;147:879–890. doi: 10.1083/jcb.147.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O’Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, Itoh N, Suda T, Nagata S. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- Tao W, Zhang S, Turenchalk GS, Stewart RA, St John MA, Chen W, Xu T. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Yang X, Yu K, Hao Y, Li DM, Stewart R, Insogna KL, Xu T. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol. 2004;6:609–617. doi: 10.1038/ncb1140. [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.