Abstract
The ocular surface is constantly exposed to a wide array of microorganisms. The ability of the cornea to recognize pathogens as foreign and eliminate them is critical to retain its transparency, hence preservation of sight. In the eye, as in other parts of the body, the early response against invading pathogens is provided by innate immunity. Corneal innate immune system uses a series of pattern recognition receptors to detect the presence of pathogens thus allowing for rapid host defense responses to invading microbes. A key component of such receptors is the “Toll-like receptors” (TLRs), which have come to occupy the center stage in innate immunity against invading pathogens. An increasing number of studies have shown that TLRs are expressed by a variety of tissues and cells of the eye and play an important role in ocular defense against microbial infection. Here in this review we summarize the current knowledge about TLR expression in human eye with main emphasis on the cornea, and discuss the future directions of the field.
Keywords: Toll-like receptors, Innate immunity, Adaptive immunity, Keratitis, Epithelial, Cornea, Eye, Human
1. INTRODUCTION
The unique structure of the human eye as well as exposure of the eye directly to the environment renders it vulnerable to a number of uncommon infectious diseases caused by bacteria, viruses, fungi and parasites [1, 2]. Host defenses directed against these microorganisms, once anatomical barriers are breached, are often insufficient to prevent infection which may lead to the loss of vision [3]. The cornea constitutes the outermost part of the eye and is in constant contact with potentially pathogenic microbes [4]. Under normal conditions, the cornea is highly resistant to microbial invasions. However, once the epithelial integrity is breached, pathogens may invade the cornea leading to microbial infections of the cornea, commonly termed infective keratitis, which is the leading cause of loss of vision in both developed and developing countries. In recent years the incidence of microbial keratitis has been significantly increased probably due to the increase in extended contact lens wear and LASIK surgery.
The immunity in vertebrates can be broadly classified as adaptive or innate immunity. Adaptive immunity is mediated by T and B lymphocytes that proliferate clonally in response to a specific pathogen or an antigen. The generation of adaptive immune responses (humoral and cellular) requires a number of days but is anamnestic through the generation of memory T and B lymphocytes. In contrast, the goals of the innate immune system are to provide protection in the first minutes to hours after an infectious challenge. Innate immunity was once thought to be a nonspecific response characterized by engulfment and digestion of microorganisms and foreign antigens by macrophages. Recently discovered mammalian innate receptors, called toll-like receptors (TLRs), provide considerable specificity for microbial pathogens and discrimination between pathogens and the host while providing an immediate response during pathogen invasion. In recent years the role of TLRs in the innate immune response has been the subject of intense investigation. Research on TLRs and their role in the innate immune response is focused on understanding how TLRs recognize pathogens and protect the host, and on determining the relationship between the innate immune response and the adaptive immune response.
2. COMPONENTS OF CORNEAL INNATE IMMUNITY
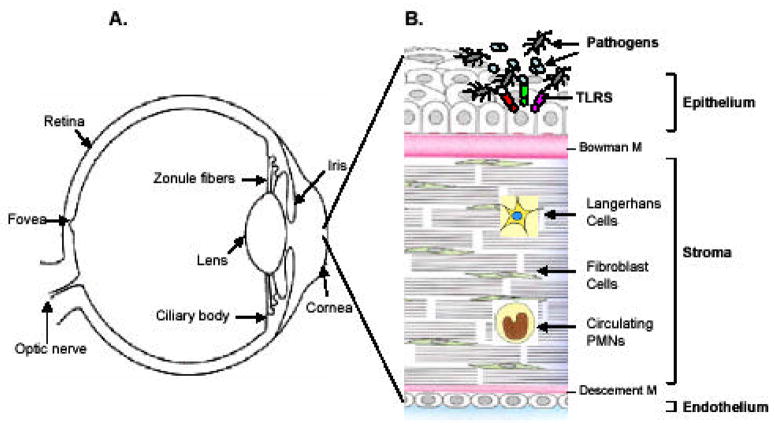
The cornea is a transparent, dome-shaped surface, measuring about 0.9 mm thick in the central and 1.1 mm in the peripheral region (Fig. 1A) [5]. The cornea serves two specialized functions: (1) provides a protective barrier between the external environment and the internal milieu and (2) constitutes the main refractive element of the visual system [6]. The barrier function of the cornea largely relies on the epithelial layer. The ongoing studies from our laboratory and others have documented that the outermost layer of the cornea, the epithelium, like other mucosal epithelial linings in the body, constitutes the first line of defense against microbial pathogens and possesses the ability to detect their presence [7–12]. The corneal innate immune system consists of multiple cell types. The first layer of defense is the corneal epithelium that lines the outermost surface of the cornea. Immediately beneath the layer of epithelial cells, is the stromal layer consisting of collageneous connective tissue produced by keratocytes (fibroblasts), which provides transparency, and the structural support to the cornea (Fig. 1B) followed by an inner most single layer of endothelial cells. In addition, the other two main components which provides corneal immunity are the Langerhans cells (dendritic cells), which modulate B and T lymphocyte activity in the cornea, and immunoglobulins (IgG and IgA), which are concentrated in the corneal stroma [13]. Upon injury or infection, the corneal epithelium releases chemotactic factors such as IL8 and CAP37 [14, 15], that initiate a local immune response to recruit polymorphonuclear cells (PMN), lymphocytes, and fibroblasts [16]. Thus, an efficient clearance of invading pathogens relies on the recognition of the pathogen by all cell types (Table 1), which may contribute to the innate immune response in the cornea.
Fig. (1). Components of corneal innate response.
(A) The cornea is a transparent, avascular, dome surface in front of the eye, in the cross section (B) it consists of an outer epithelial, middle stromal and inner endothelial layers. The epithelial cells being the outermost layer are in constant contact with microbes and their products. These cells recognize the pathogens possibly through various TLRs and secrete proinflammatory cytokines/chemokines such as IL-1β, IL-6, IL-8 and TNF-α. These cytokines either directly or indirectly recruit immune cells such as PMNs or dendritic cells (DCs) to the site of infection. The stromal fibroblast cells also express TLRs and might respond to the pathogen, when they reach the stromal layer after breaking epithelial integrity. Langerhans cells (LC) are abundantly present in peripheral cornea and less in the central cornea. These cells like DCs are powerful antigen presenting cells (APC) and present Ag to T and B cells, therefore they form the adaptive branch of immunity in the cornea. TLR expression patterns for various cell types are presented in Table 1.
Table 1.
TLR Expression in Various Cell Types Present in Human Eye
| PRRs | Corneal epithelial cells | Conjunctival epithelial cells | Stromal fibroblast cells† | Retinal pigment epithelial cells |
|---|---|---|---|---|
| TLR1 | + RNA[12, 76] | NA | + RNA | + RNA[55] |
| TLR2 | + RNA[76], protein *[12, 36] | + RNA, protein*[33, 34] | + RNA, protein* | + RNA[55], protein* |
| TLR3 | + RNA, protein*[76, 77] | NA | + RNA, protein* | + RNA, protein*[55] |
| TLR4 | + RNA[76], protein[51] | + RNA, protein[34] | + RNA, protein | + RNA[55], protein[56] |
| TLR5 | + RNA[76], protein*[11] | NA | + RNA, protein* | + RNA[55], |
| TLR6 | + RNA[12, 76] | NA | + RNA | + RNA[55] |
| TLR7 | + RNA[76], protein | NA | + RNA, protein | + RNA[55], protein |
| TLR8 | Not expressed[76] | NA | + RNA | Not expressed |
| TLR9 | + RNA[12, 76], protein | + RNA, protein[34] | + RNA, protein | + RNA[55], protein |
| TLR10 | + RNA[76] | NA | + RNA | + RNA |
indicates whether expression is positive at the RNA, protein level, or both
denotes that protein and RNA are expressed, and the receptor has been shown to be functional in response to the appropriate ligand.
Expression pattern in stromal fibroblast cells is unpublished data (Kumar and Yu et al) NA; not analyzed
3. TOLL-LIKE RECEPTORs AND THEIR LIGANDs
Currently 13 TLRs have been identified; TLRs 1–9 are common to mouse and human, while TLR10 is only found in humans and TLRs 11–13 are unique to the mouse [17–19]. The TLRs have an extracellular domain containing leucine-rich repeats (LRRs) whereas the cytoplasmic domain shows a striking homology with that of the interleukin-1 receptor (IL-1R) and is referred to as the Toll/IL-1R (TIR) domain. Many but not all of these TLRs have been assigned a role in responding to particular ligand, so-called pathogen-associated molecular patterns (PAMPs). The PAMPs are conserved structural moieties of pathogens that are essential for their survival, hence make ideal targets for detection by the innate immune system [20]. This has three major advantages. First, PAMPs are produced only by microbes and not by host cells, enabling the innate immune system to distinguish between self and non-self. Second, as PAMPs are essential for microbial survival, mutations in or loss of patterns can be lethal, and therefore these patterns are not subject to high mutation rates. Third, PAMPs are invariant between microorganisms of a given class, which implies that only a limited number of germ line-encoded pattern recognition receptors are needed to detect the presence of a microbial infection [20]. Table 2 summarizes common PAMPs present on the pathogens frequently associated with infective keratitis and the TLRs that recognize them. The role of these individual TLRs in recognization of microbial pathogen associated with infective keratitis is described in the following sections.
Table 2.
Common Pathogens Causing Infective Keratitis and their PAMPs
| Pathogen | PAMP | PRR |
|---|---|---|
| Pseudumonas aeruginosa | Flagellin Lipoprotein LPS |
TLR5 TLR2 TLR4 |
| Staphylococcus aureus | Lipopeptide Peptidoglycan LTA |
TLR2 TLR2 TLR2 |
| HSV-1 | dsRNA ssRNA Glycoprotein |
TLR9 TLR7 TLR2 |
| Candida albicans | Zymosan Phospholipomannan |
TLR2 TLR2 |
3.1. TLRs Recognizing Microbial Cell Wall PAMPs
TLRs can be classified either on the basis of their localization (surface or intracellular) or by the nature of ligands recognized by them. Although there is always an overlap among the PAMPs recognized by various TLRs, still they can be broadly divided in two categories 1.) those recognizing the PAMPs mainly present on cell wall and 2.) those recognizing the PAMPs present on genetic material of the microorganisms (like DNA or RNA)
3.1.1. TLR2 is a Sensor of Diverse PAMPs
TLR2 mainly recognizes its ligands as heterodimers [21] with either TLR1 [22] or TLR6 [23]. Signaling does not appear to occur with TLR2 homodimers. The implication of the use of heterodimers of TLR2 with other TLRs for ligand recognition is significant, as it broadens the family of TLR2 dependent ligands. TLR1/TLR2 heterodimers recognize a variety of bacterial lipopeptides, including the 19 kDa mycobacterial lipoprotein [24], meningococcal lipoproteins [22], and the synthetic lipoprotein structure Pam3Cys [23]. TLR6/TLR2 heterodimers recognize mycoplasma lipoproteins (MALP) [23] and, potentially, peptidoglycan [25]. One major structural difference between these two groups of molecules (TLR1/2 specific versus TLR2/6 specific) is that most bacterial lipoproteins and Pam3Cys are triacylated while MALP and peptidoglycan are diacylated [26]. Recent reports demonstrated that triacylated lipoproteins are preferentially recognized by TLR1/TLR2 heterodimers [27].
There are a number of other ligands that appear to be dependent on TLR2 for induction of immune effects, but do not appear to need the presence of TLR1 or TLR6, implying heterodimerization with other non-TLR molecules. These include gram-positive cell walls [28], lipoteichoic acid (also be recognized by TLR4) [29], mycobacterial lipoarabinomannan [30], zymosan [21], heat shock protein 60 [31]. CD14, a major co-receptor required for LPS recognition and down-stream effects mediated by TLR4 [32] has also been shown to be required for signaling induced by TLR2 ligands. CD14 appears to bind a variety of different ligands, in addition to LPS, and can efficiently transfer these molecules to the appropriate TLRs. In the case of TLR2, the presence of CD14 enhances the efficiency of recognition by TLR2 to many specific ligands [32].
Recent studies have shown that human conjunctival [33, 34] and corneal epithelial cells abundantly express TLR2 [12, 35, 36] and TLR2 has been shown to play an active role in the chronic ocular inflammatory response to S. aureus in conjunctival epithelial cells [33]. We, on the other hand, have recently observed that human corneal epithelial cells respond to live S. aureus and its peptidoglycan (PGN) [12]. Contradicting reports have documented regarding PGN as TLR2 ligand, Travassos et al. [37] showed that peptidoglycan is not sensed by TLR2, whereas more recently Dziarski et al. reported TLR2-dependent recognition of S. aureus PGN [38]. We recently showed that corneal epithelial cells respond to Pam3Cys (a synthetic ligand for TLR2) in a TLR2-dependent manner [36], suggesting TLR2 is an innate receptor for S. aureus and functions as a Gram-positive bacterial sensor in the cornea. These data are consistent with a recent report showing that Pam3Cys stimulates PMN recruitment to the corneal stroma in a TLR2-dependent manner [10]. However, contradicting results have been reported regarding expression pattern of TLR2 in corneal epithelial cells. Ueta et al. showed that TLR2 is expressed intracellularly and that peptidoglycan fails to stimulate cytokine production above basal levels [35]. The reason for this discrepancy between the two laboratories in TLR2 cellular localization and function is not clear. Further studies, such as down-regulation of TLR2 in cells by siRNA silencing or dominant expression, are needed to clarify the role of TLR2 in the recognization of Gram-positive bacteria by human corneal epithelial cells.
3.1.2. TLR4: a Sensor for Gram-Negative Bacteria
One of the first mammalian toll-like receptors to be identified was TLR4 and its main role has been implicated in recognization of LPS which is the major component of Gram-negative bacterial cell wall [39, 40]. TLR4 is expressed in a variety of cell types, mostly in the cells of the immune system, including macrophages and dendritic cells (DCs). Recognition of LPS by TLR4 is complex and requires several co-receptors [41]. TLR4, MD2, and CD14 form a molecular complex that binds LPS and dramatically augments LPS responses [42, 43]. In DCs and macrophages, which enjoy the relatively sterile environment of the peripheral lymphoid tissues where they are situated, TLR4 is expressed at the cell surface and when the cells encounter LPS, transmits a signal rapidly for activation and initiation of immune responses, [44] [28] thereby allowing the cells to sense PAMPs readily when encountering them. Corneal epithelial cells, however, like some other epithelial cells, are in a unique position; they are in constant contact with microbes (pathogenic and commensal), and their products, [45] and yet to mount an inflammatory response to them on each encounter, would be detrimental to the host. Thus it would be beneficial for epithelia to be unresponsiveness to LPS. Several mechanisms have been reported for different epithelial cells to avoid unnecessary pro-inflammatory reactions to LPS exposure. These include expressing extremely low levels of TLR4 and no MD-2, a critical co-receptor of TLR4, in intestinal epithelial cells [46, 47] and intracellular localization of TLR4 in human pulmonary [48], intestinal [49, 50], and corneal epithelial cells [35].
TLR4 and its co-receptor CD14 have been found to be expressed by a variety of ocular tissues (Table 1) and cells including corneal epithelial cells [51], corneal stromal fibroblasts [52], in human ciliary body, human iris endothelial cells (TLR4 only) [53], resident antigen presenting cells (APCs) in the normal human uvea [54] and retinal pigment epithelial cells [55, 56]. In a murine model of river blindness due to endosymbiotic Wolbachia bacteria, the induced inflammatory response in the cornea was dependent on expression of a functional TLR4 receptor on host cells [57]. A more recent study by Blais et al. showed that in the corneal epithelium, LPS binding protein (LBP) was mainly expressed by superficial and basal epithelial cells, whereas CD14, TLR4, and MD-2 expression were limited to the wing and basal epithelial cells. Moreover this study demonstrated that tear CD14 and LBP complemented the LPS receptor complex expressed by the corneal epithelia to trigger an immune response in the presence of LPS [58].
3.1.3. TLR5 Recognizes Flagellin
Flagellin is the major protein constituent of bacterial flagella, complex surface appendages that are involved in bacterial locomotion. More than 50 genes are known to be involved in the regulated expression and function of the flagellum, implying that motility and chemotaxis are critically important for bacterial survival [59, 60]. Flagellin possesses immunostimulatory properties [61]. Andersen et al. demonstrated that flagellated, but not non-flagellated bacteria activated TLR5, indicating that flagellin is a specific ligand for TLR5 [62, 63]. A stop codon polymorphism in the flagellin-binding domain of TLR5 is associated with susceptibility to legionnaires’ disease [64] and systemic lupus erythamatous [65], highlighting the importance of TLR5 in microbial recognition, particularly at a mucosal surface. Our laboratory and several others have shown that TLR5 is a major sensor of epithelial cells to detect Gram negative bacteria and to activate key signaling pathways leading to NF-κB and pro-inflammatory gene activation in tissues such as the cornea, the intestine, and airway/lung [66–68]. Furthermore, we showed that TLR5 is expressed on the cell surface of basal and wing corneal epithelial layers, but not superficial epithelial cells [11] as such, bacteria or bacterial products are separated from TLR5 by the apical layer. Thus, when barrier function is compromised, resulting in the exposure of the internal epithelial layers to pathogens, the innate response is initiated in corneal epithelial cells by pattern recognizing receptors such as TLR5. We believe this is a unique and key mechanism for epithelia to discriminate between pathogenic and non- pathogenic bacteria in vivo.
3.2. TLRs Recognizing Nucleotide PAMPs
Among the mammalian TLRs, four (3, 7, 8, and 9) recognize nucleic acids and are generally believed to be expressed on endosomal membranes, rather than the plasma membrane of cells; hence ligand-binding by the PRR motifs of these TLRs occurs in the lumen of the intracellular vesicles [69]. It has been suggested that nucleic acids from bacteria or viruses [70], multiplying within a cell, can be captured in membranous vesicles and brought to the TLRs in the endosomes. Alternatively, extracellular nucleic acids released from damaged tissues or cells, infected or uninfected, are endocytosed and presented to the internal TLRs [71].
3.2.1. TLR3 Induces Antiviral Response
TLR3 recognizes double-stranded (ds) RNA, which are found specifically in many viruses and thus are considered a viral PAMP. Previously, the interferon (IFN)-inducible RNA-dependent protein kinase R (PKR) was considered to be central in the interaction with dsRNA [72]. However, cells from PKR-deficient mice still respond to polyinosinic-polycytidylic acid poly(I:C), a synthetic dsRNA analog Alexopoulou et al. [73] showed that TLR3 mediates responses to poly(I:C), including activation of NF-κB and the production of type I IFNs. They further showed that TLR3 knockout mice were highly resistant to poly(I:C)-induced shock compared to wild-type mice [73]. These studies and several others suggest that TLR3 mediates cellular responses to poly(I:C), which has been extensively used to mimic viral infection. Recently Tabeta et al. [19] using Tlr3 −/− mice, documented that TLR3 contributes to murine CMV-induced type I IFN production, and to overall protection against the virus.
TLR3 localization varies according to the cell type, being expressed in the cell surface of fibroblasts and intracellularly in DCs [74, 75]. An intracellular location is consistent with a role in responding to viral nucleic acid. Interestingly, Ueta et al. recently reported that TLR3 is expressed on the surface of human corneal epithelial cells (HCECs) and its expression is amplified by poly(I:C) [76]. Consistent with what Ueta et al. reported, we also showed that stimulation of cultured human corneal epithelial cells with poly(I:C) elicited the elevated production of IL-6, IL-8 and IFN-beta.[76, 77] Furthermore, we documented that upon stimulation of TLR3, NF-κB was activated; IL-6, IL-8, and IP10 were secreted, and IFN-β, IP10, myxovirus resistance gene A, and 2, 5′-oligoadenylate synthetase mRNA were induced, indicating that activation of TLR leads to the induction of antiviral response in epithelial cells. Interestingly, our results suggest that [77] TLR 3 in HCEC is expressed intracellularly and poly(I:C) induced HCEC activation is sensitive to chloroquine, an endosomal acidification inhibitor that block poly(I:C) internalization and/or delivery to endosome where TLR3 is located, but not to TLR3 neutralizing antibody [78, 79]. To date, the role of TLR3 in mediating a host response to viral infection remains to be determined [80].
3.2.2. HSV-1 Infection Induced TLR7 Expression in HCECs
TLR7 and TLR8 are the most recent members of TLR family and their role has been implicated in responding to viral PAMPs [81]. Unlike other TLRs such as TLR3, 4 and 9, TLR7 expression is restricted to the interferon-producing plasmacytoid DC subset in humans and is induced in macrophages upon viral infection, suggesting the implication of TLR7 expression in antiviral responses. Originally, two imidazoquinoline compounds, imiquimod and resiquimod (or R-848) known to have potent anti-viral properties, were shown to activate murine macrophages through MyD88 and TLR7 [82]. Recently, Heil et al. [83] and Diebold et al. [84] demonstrated that TLR7 and TLR8 recognize the single-stranded RNAs (ssRNAs) found in many viruses, leading to IFN-α production in virus-infected macrophages and DCs. Furthermore, TLR7 recognizes the ssRNA viruses in vivo and mice deficient in TLR7 have reduced responses to in vivo infection with vesicular stomatitis virus (VSV) [85]. Diebold et al. [84] also showed that the production of large amounts of IFNα by pDCs in response to wild-type influenza virus required endosomal recognition of influenza genomic RNA and signaling via murine TLR7 and MyD88. Consistent with these studies, Lund et al. [86] has shown that TLR7 was required for pDC and B cell responses to another ssRNA virus, vesicular stomatitis virus (VSV). More recently Triantafilou et al. showed that human cardiac inflammatory responses triggered by Coxsackie B viruses are mainly TLR 8-dependent [87].
We investigated the response of cultured primary human corneal epithelial cells and HUCL cells to HSV-1 infection in vitro and demonstrated that HSV-1 infection activates the NF-κB, p38, and JNK signaling pathways in HCECs and induces expression and secretion of the proinflammatory cytokines IL-6, IL-8 and TNF-α. Moreover, the mRNAs levels of the type I IFNs (IFN-α, IFN-β) were also upregulated in HCECs in response to HSV-1 infection. HSV-1 infection also induced expression of TLR7 mRNA and protein in infected corneal epithelial cells [88]. Our results suggest that corneal epithelial cells possess the ability to recognize HSV-1 infection and TLR7 might be playing a role in corneal innate immune responses in herpetic keratitis.
3.3.3. TLR9: a Sensor for Bacterial and Viral CpG DNA
TLR9 recognizes DNA containing unmethylated CpG motifs common to both bacterial and viral genomes [89]. The CpG motifs in bacterial DNA are unmethylated and occur more frequently [90]. Mammalian DNA, on the other hand, has a low frequency of CpG dinucleotides, and these are mostly methylated; therefore, mammalian DNA does not have immunostimulatory activity [89]. CpG-DNA induces a strong T-helper-1 (Th1) inflammatory response. Moreover, TLR9 shows a restricted cellular and sub-cellular pattern of expression. In contrast to other TLR agonists, CpG-DNA is superior in activation of DCs and induction of co-stimulatory molecules (e.g., CD80, CD86) and cytokines such as interleukin (IL)-12 and IL-18. This qualifies CpG-DNA as a Th1-promoting adjuvant. During infection, recognition of CpG-DNA of intracellular pathogens skews and fine-tunes the ongoing immune response and induces a long-lasting Th1 milieu. In the eye of B6 mice, the end result of such a Th1 mediated response to P. aeruginosa infection is devastating and results in corneal perforation, while a T helper 2 (Th2) predominant response, as in BALB/c mice, results in healing [91, 92]. An important role for TLR9 in P. aeruginosa keratitis has been shown in B6 mice and the efficacy of silencing TLR9 using siRNA technology to modulate disease correlated with reduced pro-inflammatory cytokine production, but increased bacterial load in the cornea [92].
Unmethylated CpG dinucleotide motifs are also found in abundance in some viral genomes, such as herpes simplex virus (HSV). Recently, both HSV-1 and HSV-2 have been shown to activate pDCs to produce type I IFNs through TLR9 [93, 94]. In addition, purified HSV-2 DNA was capable of inducing IFNα from pDCs [94]. In Tlr9 −/− mice injected with HSV-2, no IFNα was detected, although mice lacking either TLR9 or MyD88 were capable of controlling HSV-1 replication after local infection, suggesting that TLR9- and MyD88-independent pathways in cells other than pDCs can effectively compensate for defective responses to HSV-1 [93, 94]. Recognition of HSV-2 by pDCs did not require virus replication and was through an endocytic pathway that was inhibited by chloroquine or bafilomycin A. This is consistent with the fact that TLR9 is located in, and signals from intracellular endosomal compartments. Recognition of MCMV by DC occurs through TLR9 causing cytokine secretion and viral clearance by natural killer (NK) cells [93]. Induction of IFN-α in DCs and other cells by HSV-1 is mediated by both TLR9 dependent and independent pathways [95].
4. TLR SIGNALING PATHWAYS
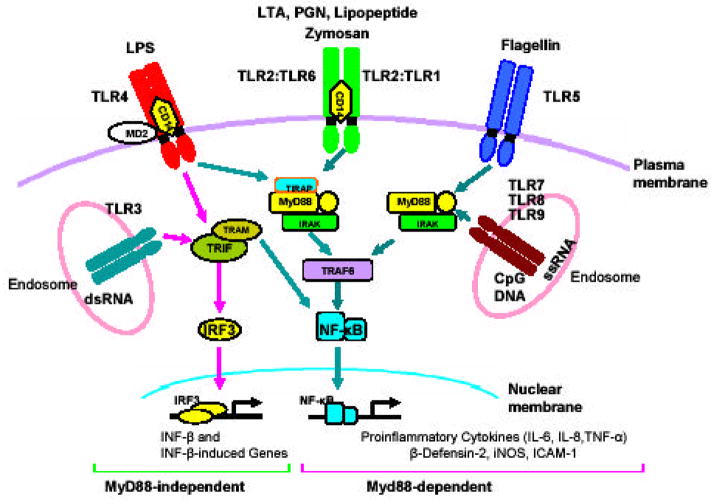
In recent years, several studies have been conducted to determine how the activation of different TLRs upon ligation with their specific PAMPs lead to different patterns of gene expression [96, 97]. Initially, all TLRs were thought to signal only through the common adaptor molecule myeloid differentiation primary-response protein 88 (MyD88). Recently, three new adaptor proteins have been identified which include: TIR-domain-containing adaptor protein (TIRAP), also known as MyD88-adaptor-like protein (MAL); TIR-domain-containing adaptor inducing interferon b or TRIF, also known as TIR-domain-containing adaptor molecule 1 (TICAM-1); and TRIF-related adaptor molecule (TRAM), also known as TIR-domain-containing adaptor molecule 2 (TICAM-2) [98]. With the identification of these new adaptor molecules, the TLR down-streaming signaling pathways can be divided into either MyD88-dependent or MyD88-independent pathways. Fig. (2) summarizes the down-stream signaling pathways, and their role in mediating innate response in corneal epithelial cells. In case of MyD88-dependent signaling, TIRAP (MAL) is essential for signaling through TLR2 and TLR4 and leads to activation of nuclear factor kappaB (NF-kB) [99, 100]. On the other hand, TRIF functions in the MyD88-independent pathway downstream of TLR3 and TLR4 to induce IFN-β TLR4 [101] and TRAM is involved in the activation of the MyD88-independent/TRIF-dependent signaling pathway through TLR4 and leads to the expression of IFN-inducible genes [102].
Fig. (2). TLR signaling pathways in corneal epithelium.
TLRs such as TLR2, 4, and 5 at the cell surface of HCEC recognize PAMPs such as lipoproteins, LPS, and flagellin, respectively. Similarly nucleic acids released from damaged infected tissues or cells or captured from multiplying bacteria or viruses are recognized by TLRs such (TLR3, 7, 8, and 9) expressed in endosomes. The recognition of PAMPs by specific TLRs leads to the activation of cascade of intracellular signaling pathways in a MyD88 dependent or MyD88-independent manner. The MyD88-dependent pathway (all TLRs except TLR3) utilizes MyD88 which transduces signal through IRAK and TRAF6, leading to the activation of NF-κB pathways and production of proinflammatory cytokines and anti-microbial peptides. On the other hand, The MyD88-independent pathways, uses TRIF as an adaptor and transduces signal through TBK1 and IRF3/7, leading to the expression of IFNs and the IFN-induced genes. TLR2 and TLR4 also utilize non-TLR receptors such as CD14 and MD2 for initial binding of their ligands.
TLRs utilize some non TLR receptors such as CD14 (for TLR2 and TLR4) and MD2 (for TLR4) for proper ligation of their PAMPs. Following ligand binding, TLRs dimerize and undergo the conformational changes required for the recruitment of downstream signaling molecules. In case of MyD88-dependent pathways TLRs recruit the adapter molecule MyD88 through homotypic interactions with a TIR domain present in C-terminus of MyD88 [96, 103]. MyD88, in turn, recruits IL-1R-associated kinase (IRAK) and IRAK-4 to the TLR complex via death-domain interaction. The binding of MyD88 to IRAK4 and IRAK results in their phosphorylation and activation. Phosphorylated IRAK then dissociates from MyD88, and becomes associated with tumor necrosis factor receptor-associated factor 6 (TRAF6) [104]. TGF-β-activated protein kinase (TAK-1), constitutively associated with TAK-1-binding proteins, TAB-1 and TAB-2, then associates with the TRAF6-IRAK complex, whereby TRAF6 in a ubiquitination-dependent manner triggers the phosphorylation and activation of TAK1 [104]. Activated TAK1 phosphorylates the inhibitor of the NF-kB (IkB)-kinase (IKK) complex, which consists of IKKa, IKKb and NF-kB essential modulator (NEMO)/IKKg, and thereby induces activation of the NF-kB-dependent transcription. In addition, TAK1 activates mitogen-activated protein (MAP) and stress-associated protein (SAP) kinases, such as extracellular signal-related kinase (ERK), p38 and Jun N-terminal kinase (JNK). Activation of NF-kB leads to the production of proinflammatory molecules such as TNF-α, IL-6, IL-8, IL-18, MIP-1.
The MyD88-independent signaling as in case of TLR3 or TLR4 leads to the phosphorylation of the transcription factor IFN regulatory factor 3 (IRF3). Phosphorylated IRF3 translocates to the nucleus and activates the production of type I IFN-β, which subsequently induces the expression of IRF7 and further production of IFN-α and IFN-β. A more detailed description of the molecular mechanisms that mediate TLR signaling can be found in the reviews by Akira &Takeda, and Beutler [96, 97].
We, along with others, have recently shown that corneal epithelial cells constitutively or inducibly express several functional TLRs in vitro and in vivo, including TLR2 (ligands e.g.: lipopeptide) [10, 36], TLR3 (dsRNA) [76, 77], TLR4 (LPS) [51, 58]), TLR5 (flagellin) [11], TLR7 (ssRNA, HSV-1) [88] and TLR9 (CpG DNA) [10, 92]. These studies have documented the production of proinflammatory cytokines/chemokines such as IL-6, IL-8, and TNF-α, via stimulation by specific TLR ligands as indicated, suggesting that TLRs play a critical role in providing innate immunity in the cornea.
5. ROLE OF TLRs IN SECRETION OF ANTI-MICROBIAL PEPTIDES (DEFENSINS)IN THE CORNEA
Defensins are small cationic peptides containing sulfide bonds that exert their effect by damaging the bacterial cell membrane [105]. Besides demonstrating broad anti-microbial properties, defensins have chemotactic properties as well [106]. While α-defensins are expressed in neutrophils and the Paneth cells of the intestine, β-defensins are produced by various epithelial cells such as those in the skin, respiratory tract, and gastrointestinal tract [107, 108]. Human β-defensin-1 is constitutively expressed while β-defensin-2 and -3 are induced by bacterial infection LPS TNF-α and IL-1.
In addition to recognizing pathogens and producing proinflammatory cytokines and chemokines, the corneal epithelium is also known to function in the innate immune response through the secretion of antimicrobial peptides [109–116]. Although most β-defensins are typically constitutively expressed, human β-defensin-2 (hBD-2) is remarkable in that it is inducible in a variety of epithelial cell types including those of the cornea [115, 116]. These studies suggest that TLRs might be implicated in the expression of β-defensins in the cornea.
Recently we demonstrated that TLR2-dependent pathways can stimulate β-defensin-2 expression by corneal epithelial cells [36]. These data suggest that, like other epithelial cells, HCECs respond to PAMPs by secreting antimicrobial peptides. Thus, secretion of various anti-microbial peptides by corneal epithelial cells is likely to be regulated by TLR-mediated recognition of PAMPs. Since many TLRs including TLR2, 4, and 5 share the same signaling pathways leading to NF-κB and MAPK activation, it is likely other pathogens/TLRs may also induce hBD2 expression in HCECs. Indeed, induced hBD2 expression was also reported in HCECs in response to P. aeruginosa infection [117]. McDermott et al. showed that the expression of hBD2 in HCECs, unlikely hBD1 and hBD3, is stimulated by proinflammatory cytokines such as IL-1beta, acting through mitogen-activated protein (MAP) kinase and nuclear factor (NF)-kappaB pathways [116]. Thus, NF-κB is required for hBD2 expression and other TLR-mediated signaling pathways contributing to pathogen-induced hBD2 expression in HCECs.
6. NEGATIVE REGULATORS OF TLR SIGNALING PATHWAYS
The inflammatory cytokines produced as a result of TLR signaling initiate innate response to rid the cornea of invaders. However, if the production of proinflammatory cytokines is left unchecked, excessive cytokines can lead to severe inflammatory disease and scar formation in the cornea. Little is known about how TLR pathways are negatively regulated. Molecules suggested to be negative regulators of TLR signaling pathways include MyD88 short [118], SIGIRR (single immunoglobulin IL-1R-related molecule)[119], Tollip (suppressing IRAK) [120], and ST2 which sequestrates the adaptors MyD88 and Mal [121]. Tollip, a Toll/IL-1R (TIR) domain-containing inhibitory protein that is bound to IRAK [120] has shown to be abundantly expressed in intestinal epithelial cells, which are poorly responsive to LPS [122]. Furthermore, Tollip expression increases in LPS- or LTA-treated intestinal epithelial cells and is associated with hyporesponsiveness to these PAMPs [47, 123].
Recently, another TIR-containing inhibitory molecule called SIGIRR or Tir8 has been identified and its expression has also been documented in the intestinal epithelial cells [119]. Animals that are deficient in TIR8/SIGIRR are more susceptible to colitis induced by dextran sodium sulfate (DSS) [124, 125]. Nothing is known about the functions of SIGIRR in bacterial keratitis, although preliminary evidence (Xi, H. et al., 2006 ARVO abstract 1906) suggest that if the levels of SIGIRR are reduced by antibody neutralization, P. aeruginosa induced keratitis is worsened in normally resistant BALB/c mice and corneal perforation is significantly enhanced. The trend of these preliminary studies suggest that expression and activity of inhibitors of TLR signaling might be an important mechanism to limit corneal inflammation and that reduced expression of these inhibitors might contribute to severe corneal damage.
7. ROLE OF TLRs IN THE REGULATION OF THE ADAPTIVE IMMUNE RESPONSE
Although innate immunity is efficient at either preventing infection or greatly reducing the pathogen load, complete elimination of invading microbial agents or control of an infection is achieved only when adaptive immunity is induced. However, because of the need for expansion of antigen-specific T- and B-lymphocyte populations with clonally distributed receptors, an efficient adaptive response is induced in several days after a primary infection. Innate immunity and adaptive immunity are not simply sequential and complementary mechanisms of resistance to microorganism they regulate each other, through cellular contacts and soluble mediators [126].
The recognition of PAMPs through TLRs initiates an inflammatory response characterized by the recruitment of cells to the sites of infection to augment the killing of invading pathogens and to prevent their spread [127]. Acute inflammatory cellular infiltrate consists of innate immune cells such as monocytes, neutrophils, basophils, eosinophils and NK cells. Both PMNs and NK cells are critical effector cells that protect the host by killing pathogenic microbes and infected cells, respectively. To provide maximal surveillance for infectious agents, in addition to the inflammation-induced cell recruitment, most tissues of the body, particularly at the mucosal surfaces that represent portals of pathogen entry, are interlaced with resident innate leukocytes such as DCs, macrophages and mast cells. The generation of adaptive immunity begins with DCs capturing microbial antigens in the peripheral tissues. Subsequently, DCs migrate to the draining lymph nodes to present the processed peptides to naive T lymphocytes in the context of MHC molecules. The stimulation of immature DCs with TLR ligands results in CCR6 down regulation and CCR7 upregulation [128], which enhances the ability of DCs to migrate from the peripheral tissues to the draining lymph node [129, 130]. In transit, DCs also undergo a maturation program that endows the cells with the ability to stimulate naive T lymphocytes. It is only after encountering microbial pathogens that DCs begin the process of maturation [131, 132]. Increasing number of studies have documented expression of TLRs on DCs and their role in the activation of adaptive immunity [132]. Studies of DC subsets isolated from humans and mice have shown that TLRs have distinct expression patterns. In murine species, all splenic DC subsets express TLRs 1, 2, 4, 6, 8, and 9, [132] but some DC subsets (plasmacytoid DC) lack TLR3. A given DC population will respond only to the pathogens for which they have appropriate TLRs.
The cornea also has resident Langerhans cells (Fig. 1) which behave like DCs. The Langerhans cells which participate in Ag presentation to the B and T cells (both CD4 and CD8) constitute the adaptive mucosal immune system in the cornea. These cells are concentrated in the epithelium of the peripheral cornea and conjunctiva but sparse in the central cornea [16]. Like macrophages, they possess receptors for immunoglobulins, complement, and antigen. The Langerhans cells recognize, phagocytize, and process certain antigens for presentation via the epithelial surface and stroma [13]. Langerhans cells also stimulate helper T and B cells that collaborate with other lymphocytes (killer, suppressor T cells) to enlist a strong cellular immune response [3]. During inflammation Langerhans cells migrate toward the center of the cornea and may participate in the secretion or release of inflammatory mediator substances [133]. Expression of TLRs in Langerhans cells of the cornea has not been evaluated so far. Studies are warranted to explore the link between TLR expression and adaptive immune response in the cornea.
8. CONCLUSION AND FUTURE PERSPECTIVES
In the past few decades, most immunologic research has been focused on adaptive immune responses. Interest in innate immunity followed the discovery of Toll receptors in the fruit fly, Drosophila, and their counterparts in humans, TLRs, which have provided exciting insights into host-pathogen interactions. Research into the molecular bases of infective keratitis and treatments for these diseases has turned in an exciting new direction. Until very recently, most of the evidence favoring a link between innate immunity and keratitis has been indirect, but now we have data from experimental animal models [10, 92] that directly implicate the role TLR signaling pathways play in infective keratitis. Before we can begin to develop effective therapies, it will be necessary to expand our understanding of what are the relevant ligands, signaling pathways, and cell types involved? Where can we effectively intervene? How to manipulate the TLR system to minimize hyper-inflammation during minor infection, yet is highly sensitive to detect potentially dangerous pathogens that would be advantageous for the cornea. Answering these questions will provide a better understanding of these basic molecular bases of infective keratitis which can potentially be translated in many exciting ways, including using small molecules to inhibit TLR signaling or employing selective TLR ligands as adjuvant to generate tolerance.
Acknowledgments
The preparation of this manuscript and the part of the studies reviewed have been supported by the NIH grants RO1-EY14080, EY10869 (FSY), Fight for sight (AK) and by an unrestricted grant from the Research to Prevent Blindness to the Department of Ophthalmology, Wayne state university.
References
- 1.Hazlett LD. Prog Retin Eye Res. 2004;23:1–30. doi: 10.1016/j.preteyeres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Klotz SA, Penn CC, Negvesky GJ, Butrus SI. Clin Microbiol Rev. 2000;13:662–685. doi: 10.1128/cmr.13.4.662-685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazlett LD. DNA Cell Biol. 2002;21:383–390. doi: 10.1089/10445490260099665. [DOI] [PubMed] [Google Scholar]
- 4.Ladage PM. Eye Contact Lens. 2004;30:194–197. doi: 10.1097/01.icl.0000140224.70483.2f. discussion 205–196. [DOI] [PubMed] [Google Scholar]
- 5.Kurpakus-Wheater M, Kernacki KA, Hazlett LD. Prog Histochem Cytochem. 2001;36:185–259. [PubMed] [Google Scholar]
- 6.Suzuki K, Saito J, Yanai R, Yamada N, Chikama T, Seki K, Nishida T. Prog Retin Eye Res. 2003;22:113–133. doi: 10.1016/s1350-9462(02)00042-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Wu XY, Yu FS. Curr Eye Res. 2005;30:527–534. doi: 10.1080/02713680590968150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaidi T, Mowrey-McKee M, Pier GB. Invest Ophthalmol Vis Sci. 2004;45:4066–4074. doi: 10.1167/iovs.04-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue ML, Willcox MD, Lloyd A, Wakefield D, Thakur A. Clin Exp Ophthalmol. 2001;29:171–174. doi: 10.1046/j.1442-9071.2001.00410.x. [DOI] [PubMed] [Google Scholar]
- 10.Johnson AC, Heinzel FP, Diaconu E, Sun Y, Hise AG, Golenbock D, Lass JH, Pearlman E. Invest Ophthalmol Vis Sci. 2005;46:589–595. doi: 10.1167/iovs.04-1077. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Xu K, Ambati B, Yu FS. Invest Ophthalmol Vis Sci. 2003;44:4247–4254. doi: 10.1167/iovs.03-0219. [DOI] [PubMed] [Google Scholar]
- 12.Kumar A, Zhang J, Yu FS. Invest Ophthalmol Vis Sci. 2004;45:3513–3522. doi: 10.1167/iovs.04-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sack RA, Nunes I, Beaton A, Morris C. Biosci Rep. 2001;21(4):463–80. doi: 10.1023/a:1017943826684. [DOI] [PubMed] [Google Scholar]
- 14.Ruan X, Chodosh J, Callegan MC, Booth MC, Lee TD, Kumar P, Gilmore MS, Pereira HA. Invest Ophthalmol Vis Sci. 2002;43:1414–1421. [PubMed] [Google Scholar]
- 15.Pereira HA, Ruan X, Gonzalez ML, Tsyshevskaya-Hoover I, Chodosh J. Invest Ophthalmol Vis Sci. 2004;45:4284–4292. doi: 10.1167/iovs.03-1052. [DOI] [PubMed] [Google Scholar]
- 16.Nassif KF. Infections of the eye. New York, N.Y.: Little, Brown and Company; 1996. [Google Scholar]
- 17.Takeda K, Kaisho T, Akira S. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 18.Zhang D, Zhang G, Hayden MS, Greenblatt MB, Bussey C, Flavell RA, Ghosh S. Science. 2004;303:1522–1526. doi: 10.1126/science.1094351. [DOI] [PubMed] [Google Scholar]
- 19.Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, Shamel L, Sovath S, Goode J, Alexopoulou L, Flavell RA, Beutler B. Proc Natl Acad Sci USA. 2004;101:3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medzhitov R. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 21.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. PNAS. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyllie DH, Kiss-Toth E, Visintin A, Smith SC, Boussouf S, Segal DM, Duff GW, Dower SK. Journal Of Immunology (Baltimore, Md: 1950) 2000;165:7125–7132. doi: 10.4049/jimmunol.165.12.7125. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Journal Of Immunology (Baltimore, Md: 1950) 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 25.Imler JL, Hoffmann JA. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- 26.Akira S. Curr Opin Immunol. 2003;15:5–11. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 27.Akira S. Biochem Soc Transact. 2000;28:551–556. doi: 10.1042/bst0280551. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 29.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 30.Means TK, Lien E, Yoshimura A, Wang S, Golenbock DT, Fenton MJ. J Immunol. 1999;163:6748–6755. [PubMed] [Google Scholar]
- 31.Vabulas RM, Ahmad-Nejad P, da Costa C, Miethke T, Kirschning CJ, Hacker H, Wagner H. J Biol Chem. 2001;276:31332–31339. doi: 10.1074/jbc.M103217200. [DOI] [PubMed] [Google Scholar]
- 32.Wetzler LM. Vaccine. 2003;21(Supplement 2):S55–S60. doi: 10.1016/s0264-410x(03)00201-9. [DOI] [PubMed] [Google Scholar]
- 33.Cook EB, Stahl JL, Esnault S, Barney NP, Graziano FM. Ann Allergy Asthma Immunol. 2005;94:486–497. doi: 10.1016/S1081-1206(10)61120-9. [DOI] [PubMed] [Google Scholar]
- 34.Bonini S, Micera A, Iovieno A, Lambiase A, Bonini S. Ophthalmology. 2005;112:1528.e1521–1528.e1528. doi: 10.1016/j.ophtha.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Ueta M, Nochi T, Jang MH, Park EJ, Igarashi O, Hino A, Kawasaki S, Shikina T, Hiroi T, Kinoshita S, et al. J Immunol. 2004;173:3337–3347. doi: 10.4049/jimmunol.173.5.3337. [DOI] [PubMed] [Google Scholar]
- 36.Kumar A, Zhang J, Yu FSX. Microbes Infect. 2006;8(2):380–389. doi: 10.1016/j.micinf.2005.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travassos LH, Girardin SE, Philpott DJ, Blanot D, Nahori MA, Werts C, Boneca IG. EMBO Rep. 2004;5:1000–1006. doi: 10.1038/sj.embor.7400248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dziarski R, Gupta D. Infect Immun. 2005;73:5212–5216. doi: 10.1128/IAI.73.8.5212-5216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 40.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 41.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. J Exp Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 43.Wright SD. J Exp Med. 1999;189:605–609. doi: 10.1084/jem.189.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akashi S, Shimazu R, Ogata H, Nagai Y, Takeda K, Kimoto M, Miyake K. J Immunol. 2000;164:3471–3475. doi: 10.4049/jimmunol.164.7.3471. [DOI] [PubMed] [Google Scholar]
- 45.Williams JM, Fini ME, Cousins SW, JS R. In: Cornea. Krachmer JH, Mannis MJ, Holland EJ, editors. St. Louis: Mosby; 1997. pp. 128–162. [Google Scholar]
- 46.Abreu MT, Vora P, Faure E, Thomas LS, Arnold ET, Arditi M. J Immunol. 2001;167:1609–1616. doi: 10.4049/jimmunol.167.3.1609. [DOI] [PubMed] [Google Scholar]
- 47.Abreu MT, Fukata M, Arditi M. J Immunol. 2005;174:4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 48.Guillot L, Medjane S, Le-Barillec K, Balloy V, Danel C, Chignard M, Si-Tahar M. J Biol Chem. 2004;279:2712–2718. doi: 10.1074/jbc.M305790200. [DOI] [PubMed] [Google Scholar]
- 49.Hornef MW, Frisan T, Vandewalle A, Normark S, Richter-Dahlfors A. J Exp Med. 2002;195:559–570. doi: 10.1084/jem.20011788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hornef MW, Normark BH, Vandewalle A, Normark S. J Exp Med. 2003;198:1225–1235. doi: 10.1084/jem.20022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song PI, Abraham TA, Park Y, Zivony AS, Harten B, Edelhauser HF, Ward SL, Armstrong CA, Ansel JC. Invest Ophthalmol Vis Sci. 2001;42:2867–2877. [PubMed] [Google Scholar]
- 52.Kumagai N, Fukuda K, Fujitsu Y, Lu Y, Chikamoto N, Nishida T. Invest Ophthalmol Vis Sci. 2005;46:114–120. doi: 10.1167/iovs.04-0922. [DOI] [PubMed] [Google Scholar]
- 53.Brito BE, Zamora DO, Bonnah RA, Pan Y, Planck SR, Rosenbaum JT. Exp Eye Res. 2004;79:203–208. doi: 10.1016/j.exer.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 54.Chang JH, McCluskey P, Wakefield D. Invest Ophthalmol Vis Sci. 2004;45:1871–1878. doi: 10.1167/iovs.03-1113. [DOI] [PubMed] [Google Scholar]
- 55.Kumar MV, Nagineni CN, Chin MS, Hooks JJ, Detrick B. J Neuroimmunol. 2004;153:7–15. doi: 10.1016/j.jneuroim.2004.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kindzelskii AL, Elner VM, Elner SG, Yang D, Hughes BA, Petty HR. J Gen Physiol. 2004;124:139–149. doi: 10.1085/jgp.200409062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saint Andre A, Blackwell NM, Hall LR, Hoerauf A, Brattig NW, Volkmann L, Taylor MJ, Ford L, Hise AG, Lass JH, Diaconu E, Pearlman E. Science. 2002;295:1892–1895. doi: 10.1126/science.1068732. [DOI] [PubMed] [Google Scholar]
- 58.Blais DR, Vascotto SG, Griffith M, Altosaar I. Invest Ophthalmol Vis Sci. 2005;46:4235–4244. doi: 10.1167/iovs.05-0543. [DOI] [PubMed] [Google Scholar]
- 59.Macnab RM. Ann Rev Gene. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 60.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 61.Eaves-Pyles T, Murthy K, Liaudet L, Virag L, Ross G, Soriano FG, Szabo C, Salzman AL. J Immunol. 2001;166:1248–1260. doi: 10.4049/jimmunol.166.2.1248. [DOI] [PubMed] [Google Scholar]
- 62.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Proc Natl Acad Sci USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 64.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, Skerrett SJ, Beutler B, Schroeder L, Nachman A, et al. J Exp Med. 2003;198:1563–1572. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hawn TR, Wu H, Grossman JM, Hahn BH, Tsao BP, Aderem A. Proc Natl Acad Sci USA. 2005;102:10593–10597. doi: 10.1073/pnas.0501165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bambou JC, Giraud A, Menard S, Begue B, Rakotobe S, Heyman M, Taddei F, Cerf-Bensussan N, Gaboriau-Routhiau V. J Biol Chem. 2004;279:42984–42992. doi: 10.1074/jbc.M405410200. [DOI] [PubMed] [Google Scholar]
- 67.Tallant T, Deb A, Kar N, Lupica J, de Veer MJ, DiDonato JA. BMC Microbiol. 2004;4:33. doi: 10.1186/1471-2180-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 69.Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, Si-Tahar M. J Biol Chem. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 70.Bowie AG, Haga IR. Mol Immunol. 2005;42:859–867. doi: 10.1016/j.molimm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 71.Sen GC, Sarkar SN. Cytokine Growth Factor Rev. 2005;16:1–14. doi: 10.1016/j.cytogfr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 72.JACOBS BL, LANGLAND JO. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 73.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 74.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, Seya T. J Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 75.Matsumoto M, Funami K, Oshiumi H, Seya T. Microbiol Immunol. 2004;48:147–154. doi: 10.1111/j.1348-0421.2004.tb03500.x. [DOI] [PubMed] [Google Scholar]
- 76.Ueta M, Hamuro J, Kiyono H, Kinoshita S. Biochem Biophys Res Commun. 2005;331:285–294. doi: 10.1016/j.bbrc.2005.02.196. [DOI] [PubMed] [Google Scholar]
- 77.Kumar A, Zhang J, Yu FSX. Immunology. 2006;177:11–21. [Google Scholar]
- 78.Matsumoto M, Kikkawa S, Kohase M, Miyake K, Seya T. Biochem Biophys Res Commun. 2002;293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 79.Schaefer TM, Fahey JV, Wright JA, Wira CR. J Immunol. 2005;174:992–1002. doi: 10.4049/jimmunol.174.2.992. [DOI] [PubMed] [Google Scholar]
- 80.Schroder M, Bowie AG. Trends Immunol. 2005;26:462–468. doi: 10.1016/j.it.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 81.Crozat K, Beutler B. Proc Natl Acad Sci U S A. 2004;101:6835–6836. doi: 10.1073/pnas.0401347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira, et al. Nature Immunology. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 83.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 84.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 85.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. PNAS. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Triantafilou K, Orthopoulos G, Vakakis E, Ahmed MA, Golenbock DT, Lepper PM, Triantafilou M. Cell Microbiol. 2005;7:1117–1126. doi: 10.1111/j.1462-5822.2005.00537.x. [DOI] [PubMed] [Google Scholar]
- 88.Li H, Zhang J, Kumar A, Zheng M, Atherton SS, Yu FSX. Immunology. 2006;117:167–176. doi: 10.1111/j.1365-2567.2005.02275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 90.Cardon LR, Burge C, Clayton DA, Karlin S. Proc Natl Acad Sci USA. 1994;91:3799–3803. doi: 10.1073/pnas.91.9.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kwon B, Hazlett LD. J Immunol. 1997;159:6283–6290. [PubMed] [Google Scholar]
- 92.Huang X, Barrett RP, McClellan SA, Hazlett LD. Invest Ophthalmol Vis Sci. 2005;46:4209–4216. doi: 10.1167/iovs.05-0185. [DOI] [PubMed] [Google Scholar]
- 93.Krug A, French AR, Barchet W, Fischer JA, Dzionek A, Pingel JT, Orihuela MM, Akira S, Yokoyama WM, Colonna M. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 94.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mattsson E, Persson T, Andersson P, Rollof J, Egesten A. Clin Diagn Lab Immunol. 2003;10:485–488. doi: 10.1128/CDLI.10.3.485-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Akira S, Takeda K, Kaisho T. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 97.Beutler B. Mol Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 98.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 99.Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, Takeuchi O, Kobayashi M, Fujita T, Takeda K, Akira S. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 100.Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 101.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 102.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 103.Burns K, Martinon F, Esslinger C, Pahl H, Schneider P, Bodmer JL, Di Marco F, French L, Tschopp J. J Biol Chem. 1998;273:12203–12209. doi: 10.1074/jbc.273.20.12203. [DOI] [PubMed] [Google Scholar]
- 104.Song HY, Regnier CH, Kirschning CJ, Goeddel DV, Rothe M. PNAS. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ganz T. C R Biol. 2004;327:539–549. doi: 10.1016/j.crvi.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 106.Ganz T. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 107.O’Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, Kagnoff MF. J Immunol. 1999;163:6718–6724. [PubMed] [Google Scholar]
- 108.Witthoft T, Pilz CS, Fellermann K, Nitschke M, Stange EF, Ludwig D. Dig Dis Sci. 2005;50:1252–1259. doi: 10.1007/s10620-005-2768-5. [DOI] [PubMed] [Google Scholar]
- 109.Ikeda A, Sakimoto T, Shoji J, Sawa M. Jpn J Ophthalmol. 2005;49:73–78. doi: 10.1007/s10384-004-0163-y. [DOI] [PubMed] [Google Scholar]
- 110.McIntosh RS, Cade JE, Al-Abed M, Shanmuganathan V, Gupta R, Bhan A, Tighe PJ, Dua HS. Invest Ophthalmol Vis Sci. 2005;46:1379–1385. doi: 10.1167/iovs.04-0607. [DOI] [PubMed] [Google Scholar]
- 111.Huang LC, Jean D, McDermott AM. Eye Contact Lens. 2005;31:34–38. doi: 10.1097/01.ICL.0000146320.64438.8C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou L, Huang LQ, Beuerman RW, Grigg ME, Li SF, Chew FT, Ang L, Stern ME, Tan D. J Proteome Res. 2004;3:410–416. doi: 10.1021/pr034065n. [DOI] [PubMed] [Google Scholar]
- 113.Haynes RJ, McElveen JE, Dua HS, Tighe PJ, Liversidge J. Invest Ophthalmol Vis Sci. 2000;41:3026–3031. [PubMed] [Google Scholar]
- 114.Haynes RJ, Tighe PJ, Dua HS. Br J Ophthalmol. 1999;83:737–741. doi: 10.1136/bjo.83.6.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McDermott AM, Redfern RL, Zhang B. Curr Eye Res. 2001;22:64–67. doi: 10.1076/ceyr.22.1.64.6978. [DOI] [PubMed] [Google Scholar]
- 116.McDermott AM, Redfern RL, Zhang B, Pei Y, Huang L, Proske RJ. Invest Ophthalmol Vis Sci. 2003;44:1859–1865. doi: 10.1167/iovs.02-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mc NN, Van R, Tuchin OS, Fleiszig SM. Exp Eye Res. 1999;69:483–490. doi: 10.1006/exer.1999.0722. [DOI] [PubMed] [Google Scholar]
- 118.Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. J Exp Med. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wald D, Qin J, Zhao Z, Qian Y, Naramura M, Tian L, Towne J, Sims JE, Stark GR, Li X. Nat Immunol. 2003;4:920–927. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 120.Burns K, Clatworthy J, Martin L, Martinon F, Plumpton C, Maschera B, Lewis A, Ray K, Tschopp J, Volpe F. Nat Cell Biol. 2000;2:346–351. doi: 10.1038/35014038. [DOI] [PubMed] [Google Scholar]
- 121.Liew FY, Liu H, Xu D. Immunol Lett. 2005;96:27–31. doi: 10.1016/j.imlet.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 122.Abreu MT, Arnold ET, Thomas LS, Gonsky R, Zhou Y, Hu B, Arditi M. J Biol Chem. 2002;277:20431–20437. doi: 10.1074/jbc.M110333200. [DOI] [PubMed] [Google Scholar]
- 123.Otte JM, Cario E, Podolsky DK. Gastroenterology. 2004;126:1054–1070. doi: 10.1053/j.gastro.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 124.Garlanda C, Riva F, Polentarutti N, Buracchi C, Sironi M, De Bortoli M, Muzio M, Bergottini R, Scanziani E, Vecchi A, Hirsch E, Mantovani A. PNAS. 2004;101:3522–3526. doi: 10.1073/pnas.0308680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mantovani A, Locati M, Polentarutti N, Vecchi A, Garlanda C. J Leukoc Biol. 2004;75:738–742. doi: 10.1189/jlb.1003473. [DOI] [PubMed] [Google Scholar]
- 126.Shimosato T, Kitazawa H, Katoh S, Tohno M, Iliev ID, Nagasawa C, Kimura T, Kawai Y, Saito T. Biochem Biophys Res Commun. 2005;326:782–787. doi: 10.1016/j.bbrc.2004.11.119. [DOI] [PubMed] [Google Scholar]
- 127.Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 128.Dieu MC, Vanbervliet B, Vicari A, Bridon JM, Oldham E, Ait-Yahia S, Briere F, Zlotnik A, Lebecque S, Caux C. J Exp Med. 1998;188:373–386. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, Lipp M. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 130.Gunn MD, Kyuwa S, Tam C, Kakiuchi T, Matsuzawa A, Williams LT, Nakano H. J Exp Med. 1999;189:451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Banchereau J, Steinman RM. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 132.Iwasaki A, Medzhitov R. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 133.Hazlett LD, McClellan SA, Rudner XL, Barrett RP. Invest Ophthalmol Vis Sci. 2002;43:189–197. [PubMed] [Google Scholar]