SUMMARY
An early step in the morphogenesis of the double-stranded DNA bacteriophage HK97 is the assembly of a precursor shell (prohead I) from 420 copies of a 384-residue subunit (gp5). Although formation of prohead I requires direct participation of gp5 residues 2–103 (Δ-domain), this domain is eliminated by viral protease prior to subsequent shell maturation and DNA packaging. The prohead I Δ-domain is thought to resemble a phage scaffolding protein, by virtue of its highly α-helical secondary structure and a tertiary fold that projects inward from the interior surface of the shell. Here, we employ factor analysis of temperature-dependent Raman spectra to characterize the thermostability of the Δ-domain secondary structure and to quantify the thermodynamic parameters of Δ-domain unfolding. The results are compared for the Δ-domain within the prohead I architecture (in situ) and for a recombinantly expressed 111-residue peptide (in vitro). We find that the α-helicity (~70%), median melting temperature (Tm = 58 °C), enthalpy (ΔHm = 50 ± 5 kcal·mol−1), entropy (ΔSm = 150 ± 10 cal·mol−1·K−1) and average cooperative melting unit (〈nc〉 ~3.5) of the in situ Δ-domain are altered in vitro, indicating specific interdomain interactions within prohead I. Thus, the in vitro Δ-domain, despite an enhanced helical secondary structure (~90% α-helix), exhibits diminished thermostability (Tm = 40 °C, ΔHm = 27 ± 2 kcal·mol−1, ΔSm = 86 ± 6 cal·mol−1·K−1) and noncooperative unfolding (〈nc〉 ~ 1) vis-à-vis the in situ Δ-domain. Temperature-dependent Raman markers of subunit side chains, particularly those of Phe and Trp residues, also confirm different local interactions for the in situ and in vitro Δ-domains. The present results clarify the key role of the gp5 Δ-domain in prohead I architecture by providing direct evidence of domain structure stabilization and interdomain interactions within the assembled shell.
Keywords: Prohead I, Δ-domain, thermostability, Raman spectroscopy, factor analysis
INTRODUCTION
The morphogenesis of icosahedral viruses typically involves the initial assembly of a precursor shell from a small number of virally encoded proteins, including the principal protein component of the icosahedral shell.1–6 The structural details of the precursor shell and its subsequent transformation into the architecture of the mature viral particle are rigorously controlled along the in vivo assembly pathway. Many structural details of precursor and mature shells have been determined from investigations of particles assembled in vitro using recombinantly expressed subunits of the requisite proteins.7–10 Efficient assembly usually involves, in addition to the constituent proteins of the shell, a virally encoded scaffolding protein to ensure the appropriate shell dimensions and assembly rate.11–16
The well-studied bacteriophage HK97 employs no separate scaffolding protein in the assembly process. Instead, following the co-translational removal of methionine at the N-terminus (Met 1), the major coat protein (gp5, residues 2–385) exploits its N-terminal domain (Δ-domain, residues 2–103) to assemble a precursor shell (prohead) in a manner similar to that expected of a phage scaffolding protein subunit.17,18 The initial precursor – prohead I – incorporates 420 copies of gp5 in a T = 7 surface lattice and encapsidates about 60 copies of a virally encoded protease (gp4).17 The next step in shell maturation involves proteolytic digestion of the Δ-domain and rearrangement of the lattice of cleaved coat subunits (gp5*, residues 104–385) to form the prohead II particle. Prohead II is competent to package the viral chromosome. During packaging it is transformed into the mature shell (head). Maturation is accompanied by rearrangement of gp5* subunit domains, intersubunit covalent crosslinking and shell expansion.17,19–20
A recent study of prohead I by electron cryomicroscopy (cryo-EM) and differential scanning calorimetry (DSC) shows that the Δ-domain projects radially inward from the inner surface of the shell and undergoes an unfolding transition at ~53 °C that is distinct from prohead I disassembly.19,21 Above the transition temperature the mean shell diameter is about 10% greater than that of prohead I, and the arrangement of subunits at the icosahedral six-fold axes resembles a previously identified expansion intermediate (EI-I).10 The enthalpy change (ΔH) for the subunit endothermic transition is approximately 14 kcal·mol−1. A high resolution structure of the Δ-domain in prohead I is not available; however, the predominantly α-helical structure revealed by Raman spectroscopy18 is consistent with structures determined for scaffolding proteins of bacteriophages P2222,23 and ϕ29.24
In the present work we report details of the organization, interactions and thermostability of the Δ-domain in two distinct environments, viz. as part of the gp5 subunit within the prohead I assembly (in situ), and free in solution as a recombinantly expressed 111-residue peptide (in vitro). For interpretation of the temperature-dependent Raman spectra we have introduced a novel factor analysis approach. The method allows the structural and thermodynamic information inherent in the data to be fitted to a biologically relevant molecular model. The present analysis demonstrates that (i) the structure of the Δ-domain is significantly thermostabilized within the prohead I assembly, (ii) this thermostabilization can be attributed to cooperative binding of several Δ-domains, and (iii) the stabilizing effect of cooperative Δ-domain interaction occurs at the expense of α-helical structure.
RESULTS AND INTERPRETATION
1. Ectopically Expressed Δ-Domain
(a) Hydrodynamic measurements
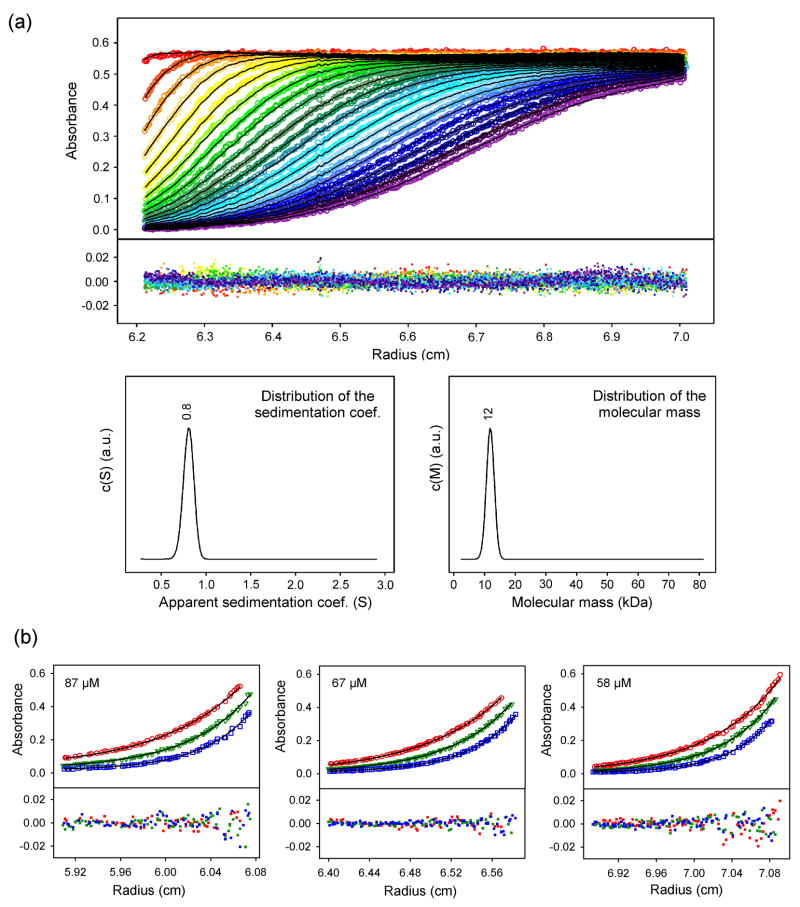
The oligomeric state of the purified 111-residue Δ-domain (see MATERIALS AND METHODS, below) was assessed by sedimentation velocity measurements (Figure 1a). Fitting of the monophasic sedimentation profiles to the Lamm equation indicates a single component of 0.8 S (s020,w = 1.3 S at standard conditions), corresponding to a protein with elongated shape (6:1 axial ratio)25 and molecular mass of 12 kDa (expected monomer mass = 12.5 kDa). The monomer state was confirmed by sedimentation equilibrium measurements on solutions at 58, 67 and 87 μM. Figure 1b shows the fit of the equilibrium profiles to a single-species model of 12 ± 1 kDa.
Figure 1.
(a) Sedimentation velocity of the in vitro Δ-domain. (Top) Absorbance at 280 nm as a function of radial position during Δ-domain (85 μM) sedimentation at 4 °C. Data were acquired in 90 s intervals. For clarity, the results for 15 min intervals are shown (colored circles). Sedimentation boundaries were fitted by the Lamm equation (black lines). Corresponding residuals are shown in the lower field. (Bottom) Fitted distributions of the apparent sedimentation coefficient (left) and molecular mass (right) reveal a single species of s* = 0.8 (corresponding to s020,w = 1.3) and 12 kDa, consistent with an elongated monomer. (b) Sedimentation equilibrium of the in vitro Δ-domain. Samples (87, 67 and 58 μM) at 4 °C were spun to equilibrium at 35,000 (red circles), 40,000 (green triangles) and 45,000 rpm (blue squares). The profiles were acquired at 280 nm and fitted with eq. 5 to determine the molecular mass of 12 ± 1 kDa.
(b) Raman spectroscopic measurements
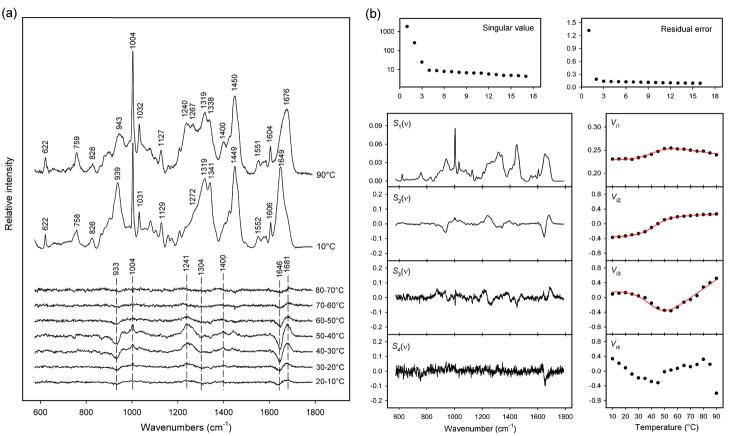
Raman spectra of the purified Δ-domain were acquired at intervals of 5 °C over the range 10–90 °C. In each spectrum the amplitudes were normalized to unit integrated intensity for the Raman amide I band (1610–1700 cm−1). Figure 2a compares spectra of the Δ-domain at 90 and 10 °C and shows the changes that occur for each 10 °C increment.
Figure 2.
(a) Temperature-dependence of the Raman spectrum of the in vitro Δ-domain. The spectra obtained at 10 and 90 °C and the difference spectra computed for each 10 °C interval between 10 and 80 °C are shown, as labeled. The major spectral changes, which correspond to Δ-domain unfolding, occur for 30–40 and 40–50 °C intervals. Raman intensities were normalized using the integrated intensity of the amide I band (see text). (b) Factor analysis of the temperature-dependent Raman spectra of the in vitro Δ-domain. Singular values (plotted on a logarithmic scale) and residual errors reveal that three factors are sufficient to describe all spectral changes associated with Δ-domain unfolding (factor dimension M = 3). The three factors were fitted to a two-state model using eqs. 3 and 4. The red lines show the precision of the fitted coefficients Vi1-Vi3.
The Raman signature of the Δ-domain at 10 °C exhibits several features diagnostic of a highly α-helical secondary structure, including amide I (1649 cm−1), amide III (1272–1300 cm−1) and skeletal mode (939 cm−1).18,26,27 Quantitative analysis of the secondary structure by decomposition of the amide I band28 indicates 90 ± 5% α-helix, 10 ± 1% β-strand and negligible irregular structure. A predominantly α-helical Δ-domain has also been proposed for the subunit of the prohead I shell.18 Similarly high percentages of α-helical secondary structure occur for scaffolding proteins of P22 and ϕ29.15,22,24
Thermal unfolding of the Δ-domain is not highly cooperative, although most occurs within the 30–50 °C interval (Figure 2a). Shifts in amide I (1649 → 1676 cm−1), amide III (1272–1300 → 1240–1267 cm−1) and the skeletal mode at 939 cm−1 signal the conversion of α-helix to β-strand, turns and irregular chain conformations.29,30 Changes in local environments of Δ-domain side chains with unfolding are also revealed in the data of Figure 2a. Thus, Raman markers of Phe (1004, 1032, 1606 cm−1), Asp and Glu (1400 cm−1) and other side chains (Table 1) are significantly affected by increasing temperature.
Table 1.
| prohead I | prohead II | in situ Δ-domain | in vitro Δ-domain | assignments | ||||
|---|---|---|---|---|---|---|---|---|
| cm−1 | Irel | cm−1 | Irel | cm−1 | Irel | cm−1 | Irel | |
| 622 | 18 | 622 | 15 | 622 | 6 | 622 | 7 | F |
| 644 | 15 | 644 | 16 | Y | ||||
| 724 | 7 | 725 | 3 | 723 | 4 | 723b | 3 | M, Y |
| 749s | 6 | 750s | 7 | sc | ||||
| 759 | 30 | 759 | 25 | 759 | 9 | 758 | 9 | W |
| 803 | 2 | W | ||||||
| 826 | 19 | 826 | 19 | 825 | 2 | 826 | 5 | Y, F |
| 852 | 23 | 852 | 21 | Y | ||||
| 860s | 13 | 859s | 11 | 868 | 3 | 865 | 7 | |
| 879 | 15 | 878 | 4 | 883 | 8 | 881 | 10 | W |
| 900 | 24 | 900 | 11 | 900 | 11 | 902 | 14 | sc |
| 914 | 10 | 924s | 18 | 922s | 22 | sc | ||
| 935 | 49 | 935 | 22 | 937 | 26 | 939 | 31 | sc[ν(Cα-Cβ)] |
| 958 | 29 | 959 | 11 | 959s | 14 | 957s | 16 | sc |
| 978 | 30 | 979 | 21 | 976 | 8 | sc | ||
| 1004 | 142 | 1004 | 100 | 1004 | 53 | 1004 | 49 | F |
| 1013s | 33 | 1012s | 26 | 1016 | 9 | W | ||
| 1032 | 43 | 1033 | 30 | 1031 | 15 | 1031 | 17 | F |
| 1061 | 34 | 1060 | 32 | 1060 | 9 | sc[ν(C-C,C-O)] | ||
| 1082 | 36 | 1085 | 30 | 1080 | 10 | 1080 | 13 | K, sc[ν(C-C,C-N)] |
| 1103 | 32 | 1104 | 28 | 1103 | 6 | 1104 | 8 | sc[ν(C-C)] |
| 1127 | 41 | 1126 | 38 | 1129 | 9 | 1129 | 12 | W, sc[ν(C-C)] |
| 1158 | 17 | 1157 | 13 | 1156 | 5 | 1158 | 6 | sc[δ(CH3)] |
| 1173 | 17 | 1174 | 13 | 1172 | 4 | 1173 | 5 | Y, sc[δ(CH3)] |
| 1209 | 37 | 1209 | 29 | 1207 | 9 | 1208 | 8 | F, Y |
| 1235 | 6 | amide III (β-sheet) | ||||||
| 1248 | 58 | 1247 | 55 | amide III (turn,coil) | ||||
| 1271 | 55 | 1271 | 48 | 1285 | 15 | 1272s | 17 | amide III (α-helix) |
| 1294 | 42 | 1303 | 23 | amide III (α-helix) | ||||
| 1318 | 74 | 1317 | 51 | 1318 | 27 | 1319 | 32 | sc[δ(CH2)] |
| 1342 | 78 | 1341 | 60 | 1342 | 24 | 1341 | 30 | W, sc[δ(CH)] |
| 1403 | 37 | 1401 | 33 | 1407 | 11 | D, E [ν(CO2–)] | ||
| 1424 | 51 | 1424 | 41 | 1423 | 16 | 1425 | 17 | sc[δ(CH3),δ(CH2)] |
| 1450 | 114 | 1451 | 97 | 1450 | 33 | 1449 | 38 | sc[δ(CH2)] |
| 1458s | 101 | 1459s | 92 | W | ||||
| 1553 | 22 | 1553 | 21 | 1551 | 5 | 1552 | 5 | W |
| 1578 | 4 | 1578 | 4 | 1575b | 5 | F, W | ||
| 1586 | 9 | 1585 | 7 | 1585 | 4 | 1586 | 5 | F, W |
| 1606 | 31 | 1605 | 24 | 1606 | 11 | 1606 | 12 | F |
| 1620 | 27 | 1619 | 26 | Y, W | ||||
| 1655s | 91 | 1652 | 32 | 1649 | 37 | amide I (α-helix) | ||
| 1667 | (100) | 1668 | 90 | 1664s | 25 | amide I (β-strand) | ||
Data were collected at 10 °C. Wavenumbers are accurate to ± 1 cm−1 and intensities (Irel) are relative to an arbitrary assignment of 100 for the 1667 cm−1 band of prohead I. Bands, including broad (b) and shoulder (s) features, are assigned to specific amino acid side chains, as indicated by standard one-letter symbols; sc indicates assignments to unspecified side chains. Where specific bond stretching (ν) and deformation (δ) modes are known, they are indicated by chemical symbols in parentheses.
(c) Thermodynamics of Δ-domain unfolding
Qualitative and quantitative assessment of Δ-domain unfolding has been carried out by factor analysis of the Raman signature (600–1750 cm−1) using the SVD algorithm. The results, inclusive of the first four principal factors (eq. 1, j = 1, 2, 3, 4), are shown in Figure 2b. The first three factors (j = 1, 2, 3), which account for 90% of the variance in the series of eq. 1, describe all of the spectral changes from Δ-domain unfolding. The remaining 10% of the variance is attributable to random noise in the experimental data. Thus, the factor dimension is three (eq. 1, M = 3).
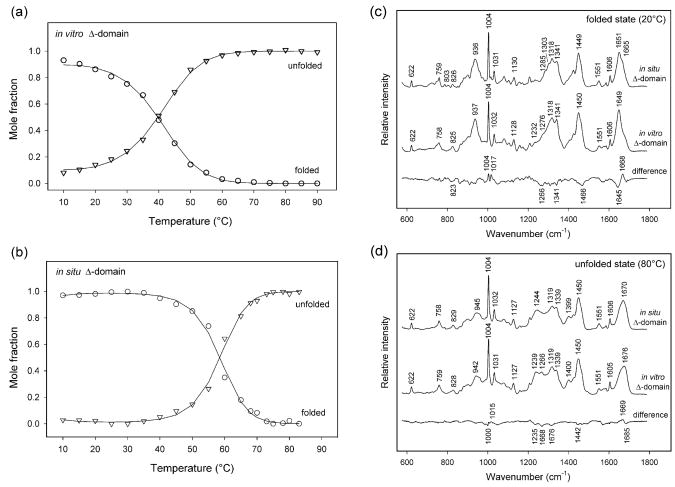
The three relevant factors were fitted to different models. The best fit of the data was achieved assuming a simple two-state equilibrium (NΔ ⇆ UΔ) between natively folded (NΔ) and unfolded (UΔ) states and a temperature-dependent Raman spectrum for the unfolded state. The fitted parameters ΔHm, ΔSm and ΔCp are listed in Table 2 and the derived Raman spectra of the NΔ and UΔ states using this model are shown in Figures 3c and 3d. The temperature dependence of the Raman spectrum of the UΔ state is very feeble, representing no more than 0.5% of the difference between the NΔ and UΔ states. The precision of the fit was also tested by comparison of the mole fractions of NΔ and UΔ as a function of temperature (Figure 3a), as determined either from the fitted thermodynamic parameters (eqs. 3 and 4) or from decomposition of the experimental spectrum into the spectra of species NΔ and UΔ.
Table 2.
Thermodynamic parameters for thermal unfolding of the gp5 Δ-domain in prohead I (in situ) and the ectopically expressed 111-residue Δ-domain (in vitro)a
| Δ-domain | ΔHm kcal·mol−1 | ΔSm cal·mol−1·K−1 | ΔCp kcal·mol−1·K−1 | ΔG310K kcal·mol−1 | Tm °C |
|---|---|---|---|---|---|
| in vitro | 27 ± 2 | 86 ± 6 | 0.9 ± 0.1 | 0.2 ± 0.1 | 40 ± 1 |
| in situ | 50 ± 5 | 150 ± 10 | 1.5 ± 0.2 | 2.2 ± 0.2 | 58 ± 1 |
ΔHm and ΔSm refer to the median melting temperature, Tm, ΔCp is considered temperature-independent, and ΔG310K refers to the HK97 physiological temperature (37 °C).
Figure 3.
(a) Plots of the mole fractions of folded and unfolded states of the in vitro Δ-domain as a function of temperature. The fractions were determined from the fitted thermodynamic parameters using eqs. 3 and 4 (lines) and by spectral decomposition of the temperature-dependent Raman spectra of the in vitro Δ-domain into the spectra determined for folded and unfolded states of the in vitro Δ-domain (circles and triangles, respectively). (b) Plots of the mole fractions of folded and unfolded states of the in situ Δ-domain as a function of temperature. The fractions were determined from the fitted thermodynamic parameters using eqs. 3 and 4 (lines) and by spectral decomposition of the temperature-dependent Raman spectra of the in situ Δ-domain into the spectra determined for folded and unfolded states of the in situ Δ-domain (circles and triangles, respectively). (c) Comparison of the Raman spectra of the folded Δ-domain in situ (top) and in vitro (middle). The difference spectrum (bottom) reveals changes in secondary structure and local environments of Trp and Phe side chains of the in situ Δ-domain (see text). (d) Comparison of the Raman spectra of the unfolded Δ-domain in situ (top) and in vitro (middle). The difference spectrum (bottom) shows that changes in secondary structure and Trp and Phe local environments are smaller than those occurring for the folded Δ-domain (cf. bottom trace of panel (c)).
2. Δ-Domain of the Prohead Subunit
(a) Raman spectroscopic measurements
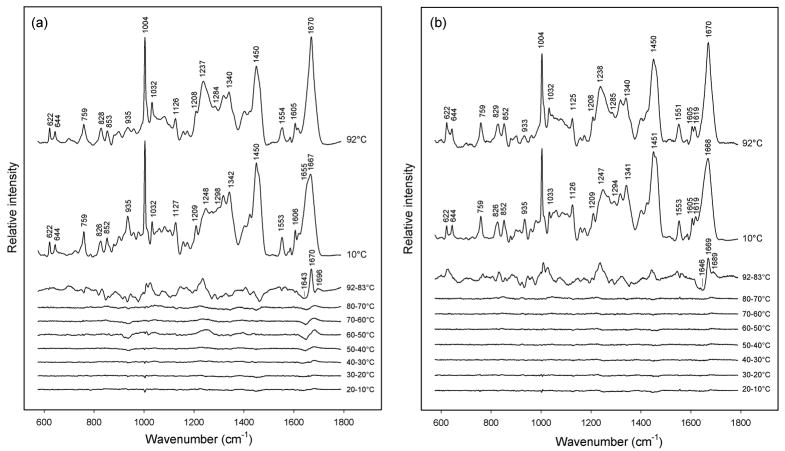
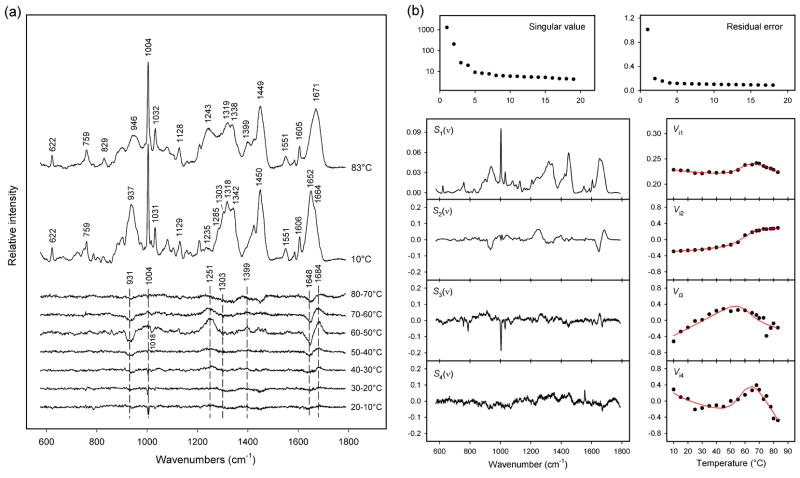
Temperature-dependent Raman spectroscopy of purified prohead I and prohead II was carried out using the protocol described in MATERIALS AND METHODS, below. Figure 4 compares Raman signatures of prohead I and prohead II at 10 and 92 °C, as well as corresponding difference spectra computed for each 10 °C increase in temperature between 10 and 92 °C. At 10 °C both proheads exhibit the Raman amide I and amide III markers expected for the subunit α/β fold.7 Nevertheless, the greater prominence of the α-helix markers (viz. shoulders at 1655 and 1298 cm−1) in prohead I indicate more α-helix than in prohead II. We attribute the greater α-helicity of prohead I to the subunit Δ-domain, consistent with previous results.18
Figure 4.
(a) Temperature-dependence of the Raman spectrum of prohead I. The spectra obtained at 10 and 92 °C and the difference spectra computed for the indicated intervals between 10 and 92 °C are shown, as labeled. The largest spectral change, which corresponds to prohead I disassembly, occurs above 83 °C. The other major spectral transition, which occurs primarily between 50 and 60 °C, corresponds to Δ-domain unfolding within the prohead I assembly. Raman intensities were normalized to the integrated intensity of the amide I band. (The intensity of the tyrosine marker at 644 cm−1 is also invariant to temperature.) (b) Temperature-dependence of the Raman spectrum of prohead II, showing a single transition above 83 °C, which corresponds to prohead II disassembly.
More detailed analysis of Figure 4a reveals two distinct stages of subunit unfolding in prohead I. The first transition, which occurs between 50 and 60 °C, can be attributed to unfolding of the subunit Δ-domain.19 The Raman markers affected with increasing temperature reflect the loss of α-helical secondary structure (troughs at 936, 1303, 1343 and 1648 cm−1) and a concomitant increase in β-strand and/or turns (peaks at 1250 and 1684 cm−1). On the other hand, relatively little change is observed between 50 and 60 ° C for Raman markers assigned to subunit side chains (Table 1). The second prohead I transition, which occurs between 83 and 92 °C, reflects global subunit unfolding coupled with disassembly of the prohead I particle.19 The corresponding difference profile between 83 and 92 °C exhibits strong peaks and troughs throughout the spectral interval 700–1700 cm−1. Accordingly, both mainchain conformation (secondary structure) and side chain environments (tertiary structure) of the subunits are altered above 83 °C. The amide I band prevailing at 92 °C (1670 cm−1) is very strong and sharp, suggesting a preponderance of β-strand structure in the disassembled subunits. This presumably reflects aggregation of the denatured subunits.
Figure 4b shows that the Raman signature of the prohead II subunit is virtually unchanged between 10 and 80 °C. This is consistent with the attribution of the spectral changes observed only for prohead I between 50 and 60 °C to Δ-domain unfolding (Figure 4a). Between 83 and 92 °C, on the other hand, the prohead II spectrum exhibits the same large-scale spectral changes observed for prohead I. The prohead II spectral difference between 83 and 92 °C is therefore attributed to subunit unfolding and shell disassembly. As noted above, the affected Raman markers indicate conversion of α-helix to β-strand and extensive changes in side-chain local environments. The similar high-temperature difference profiles (83 → 92 °C) shown in panels (a) and (b) of Figure 4 indicate that the unfolding transition of residues 104–385 (gp5*) is independent of the Δ-domain.
(b) Structural interpretation
The Raman signature and structure of the prohead I Δ-domain in the 10–83 °C range is well approximated by a subtraction of the prohead II spectrum from that of prohead I.18,19 Because tyrosine is absent from residues 2–103, the Tyr phenoxyl marker at 642 cm−1 is a reliable internal intensity standard for the spectral subtractions (Figure 5a).31
Figure 5.
(a) Temperature-dependence of the Raman spectrum of the in situ Δ-domain. The spectra obtained at 10 and 83 °C and the difference spectra computed for each 10 °C interval between 10 and 80 °C are shown, as labeled. The major spectral changes, which correspond to Δ-domain unfolding, occur principally in the 50–60 °C interval. Raman intensities were normalized using the integrated intensity of the amide I band. (b) Factor analysis of the temperature-dependent Raman spectra of the in situ Δ-domain. Singular values (plotted on a logarithmic scale) and residual errors reveal a factor dimension M = 4. The four factors were fitted to a two-state model using eqs. 3 and 4. The red lines show the precision of the fitted coefficients Vi1-Vi4.
Quantitative analysis28 of the in situ Δ-domain secondary structure at 10 °C indicates 69 ± 6% α-helix, 14 ± 2% β-strand and 17 ± 3% irregular structure, in agreement with previous estimates.18 The spectral changes that occur with Δ-domain unfolding consist of shifts in amide I (1648 → 1684 cm−1) and amide III (1303 → 1251 cm−1) markers, pronounced intensity reductions in other bands of the peptide backbone (937 and 1342 cm−1), and changes in the local environments of multiple side chains (Table 1).30–33 These occur principally between 50 and 60 °C (Figure 5a) and signal the loss of α-helix and altered tertiary structure. Of particular interest is the trough at 1018 cm−1, which is assignable to Trp.18,31,34 The parent band is observed only in the spectrum of the folded Δ-domain of prohead I. Its absence from spectra of both the unfolded Δ-domain of prohead I and the ectopically expressed Δ-domain (Figure 2a) suggests an interaction specific to W89 of the assembled prohead I subunit. Figure 5a also shows that Raman spectral changes below 40 °C are small, indicating a thermostable subunit fold at physiological temperature.
(c) Thermodynamics of Δ-domain unfolding
Quantitative analysis of Δ-domain unfolding in prohead I has been carried out by factor analysis of the prohead I and prohead II difference spectra (600–1750 cm−1, Figure 5a) using the SVD algorithm. Figure 5b shows the first four principal factors [(eq. 1, j = 1, 2, 3, 4)], which represent 88% variance in the spectral series. Within the observed noise level these completely describe all spectral changes due to Δ-domain unfolding and indicate a factor dimension of four (eq. 1, M = 4).
The four relevant factors were fitted assuming a two-state unfolding model (NΔ and UΔ) and linear temperature-dependence of the corresponding Raman spectra. The fitted thermodynamic parameters (ΔHm, ΔSm, ΔCp) are summarized in Table 2. The derived Raman spectra of the NΔ and UΔ states using this model are shown in Figures 3c and 3d. The temperature dependencies of the UΔ and NΔ states are very weak (<0.5% of the amplitude of the difference Raman spectrum). The mole fraction distribution of NΔ and UΔ as a function of temperature (Figure 3b) clearly reflects cooperative unfolding between 45 and 70 °C with substantially increased stability of the Δ-domain in prohead I. The transition midpoint (Tm) is 58 ± 1 °C. The mole fractions determined from the fitted thermodynamic parameters (eqs. 3 and 4) correlate well with those derived from decomposition of the experimental spectra into NΔ and UΔ, in support of the two-state model.
DISCUSSION
In the dsDNA bacteriophages P22 and ϕ29 shell assembly proceeds with the help of a scaffolding protein,11–14 which adopts an elongated shape in solution and may exploit α-helical coiled-coils to facilitate function.15,22,24,35,36 The scaffolding function in HK97 shell assembly may be fulfilled by the Δ-domain of the prohead I subunit.18,37 Like scaffolding subunits of P22 and ϕ29, the HK97 Δ-domain is rich in α-helix and may exploit coiled-coils to project from the inner shell surface toward the prohead I interior.18,19,21,37 Here we have characterized the α-helical structure of the HK97 Δ-domain and quantified its thermodynamic stability in the prohead I environment (in situ) and in solution (in vitro).
1. Oligomerization of the Δ-Domain upon Prohead I Assembly
Our sedimentation experiments show that the in vitro Δ-domain does not spontaneously self-assemble, but remains ~100% monomeric up to 1 mg/mL concentration. Therefore, gp5 self-assembly into capsomers and shells at similar concentrations cannot be attributed simply to interactions between subunit Δ-domains. Evidently, residues 104–385 are involved. The present results suggest that the Δ-domain serves to lock or stabilize the assembled subunit in its proper orientation within a capsomer or prohead I shell rather than to initiate assembly.
In addition to demonstrating a stable monomeric state for the in vitro Δ-domain, the sedimentation results establish that the molecule is a prolate ellipsoid (6:1 axial ratio) in solution. Similar nonspherical shapes have been established for the scaffolding proteins of bacteriophages λ and P22 at comparable experimental conditions.35,38 Raman spectra show further that both the secondary and tertiary structures of the in vitro Δ-domain are distinct from those of the in situ Δ-domain. This presumably reflects interactions in prohead I between neighboring Δ-domains and/or between Δ-domains and domains of residues 104–385.
2. Structure and Stability of the In Vitro Δ-domain in Solution
The secondary structure of the in vitro Δ-domain at 10 °C is shown by Raman spectroscopy to be primarily α-helical (90%), containing only short segments of β-strands and/or turns. The heat-induced transition observed between 10 and 60 °C (Figures 2a and 3a) involves the loss of mainchain α-helicity and modest changes in side-chain local environments. The in vitro Δ-domain is not fully folded at the HK97 physiological temperature (37 °C).
Thermodynamic parameters governing the unfolding of the in vitro Δ-domain have been determined by van’t Hoff analysis using a two-state model with constant heat capacity change (Table 2). The enthalpy (ΔHm) and entropy (ΔSm) changes are diagnostic of a noncooperative transition, typical of a protein that lacks a robust folding core.22,29 The small positive change in heat capacity (ΔCp) implies that hydrophobic residues become only slightly more solvent exposed with unfolding. This is consistent with the small intensity increase of the phenylalanyl Raman marker (1004 cm−1) and the minor perturbations to aliphatic side chain Raman markers (Table 1) with unfolding. A similar Raman unfolding signature has been observed for the P22 scaffolding protein, which also lacks a robust folding core.22,29 We conclude that the in vitro Δ-domain structure is similar to those of P22 and ϕ29 scaffolding proteins with regard to hydrodynamic shape, α-helicity and unfolding pathway.
3. Structure and Stability of the Δ-Domain in Prohead I
Despite the high α-helicities of both in situ and in vitro Δ-domains, the former exhibits more β-strand than the latter. This is true for the respective folded (20 °C) and unfolded (80 °C) states, as is evident from the Raman amide I difference peak near 1668 cm−1 in Figures 3c and 3d. At 20 °C, the in situ Δ-domain includes about 8% more of its mainchain (~8 residues) in stable β-strand structure due to interactions specific to the prohead I assembly. The stabilizing interactions within prohead I also perturb Raman bands of various side chains in the 800–1100 cm−1 spectral interval (Table 1). Raman markers of Phe (825 cm−1) and Trp (803 and 1018 cm−1), for example, exhibit distinct band shapes, intensities and peak positions for the in situ and in vitro environments (Figure 3c). These distinctions are diminished in the respective unfolded Δ-domains (Figure 3d). We note that the majority of the Δ-domain Phe residues (F77, F96, F101) as well as the unique Trp residue (W89) occur near the C-terminus, where interactions with the gp5* arm (residues 104–132) are presumably facilitated.
An important conclusion of this study is that the thermostability of the Δ-domain is greatly enhanced by the prohead I environment (ΔTm = 18 °C). At the physiological temperature of the prohead I particle (37 °C), the in situ Δ-domain is virtually completely folded (Figure 3b), whereas the in vitro Δ-domain is less than 60% folded (Figure 3a).
Although the Δ-domain contains less α-helix in the in situ environment, the thermodynamic parameters of unfolding are clearly greater (Table 2). The significantly more robust thermodynamic parameters associated with unfolding of the in situ Δ-domain are attributable to a cooperative effect, i.e. the simultaneous unfolding of multiple Δ-domains within prohead I.39 Conway et al.21 observed a thermal transition at 53 °C in prohead I, using differential scanning calorimetry. We propose that this is the same thermal transition that we have monitored between 50 and 60 °C using the Raman signature. Conway et al.21 employed cryo-EM to show that the principal structural change associated with the 53 °C DSC transition is disordering of most of the Δ-domains, and this supports our assignment of the Raman spectral changes in the 50–60 °C transition primarily to denaturation of the Δ-domains. Comparison of the van’t Hoff enthalpy change derived from the Raman spectra (ΔHm = 50 ± 5 kcal·mol−1) with the calorimetric value reported previously (ΔHcal = 14 kcal·mol−1),19 indicates an average cooperative unit 〈nc〉 = ΔHm/ΔHcal of 3.6 ± 0.4. This implies the association of Δ-domains of neighboring prohead I subunits into trimers and higher-order oligomers, which is consistent with the proposal of Conway et al.21 that such Δ-domains form trimeric and pentameric clusters on the inner shell surface.
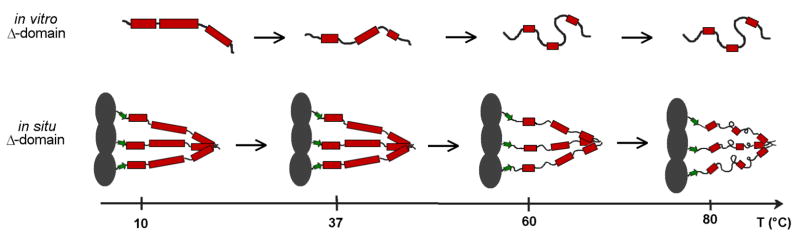
A plausible scheme for the evolution of Δ-domain structure as a function of temperature is shown in Figure 6. In situ (lower pathway), the cooperative and largely trimeric melting unit exhibits little structural change between 10 and 37 °C. Between 37 and 60 °C, about half of the Δ-domain α-helicity is lost in a cooperative manner in favor of turns and irregular conformations. Δ-domain unfolding within the prohead I assembly also impacts shell architecture.21 Above 60 °C, the remaining α-helical structure is further diminished, also cooperatively. In vitro (upper pathway), the monomeric and more richly α-helical Δ-domain, melts noncooperatively and the process is essentially complete at 60 °C.
Figure 6.
A comparison of thermally induced perturbations to Δ-domain secondary structure in vitro (top) and in situ (bottom). The models incorporate results of the Raman amide I band analyses and the sizes of the cooperative units (〈nc〉) determined from sedimentation and thermodynamic data (see text). A key feature is that at physiological temperature (37 °C) the structural organization of the in situ Δ-domain is greater than that of the in vitro Δ-domain. In particular, each chain of the in situ trimer retains full α-helicity, whereas the in vitro monomer is partially unfolded.
The present study demonstrates (i) that the Δ-domain of the HK97 prohead I subunit exhibits a highly organized submolecular architecture at physiological temperature, (ii) that the precise details of the Δ-domain chain conformation (secondary structure) as well as the local environments of domain side chains (tertiary structure) are determined by interactions involving neighboring domains of prohead I subunits, (iii) that such interdomain interactions within prohead I confer a robust thermodynamic stability to the otherwise marginally stable Δ-domain structure in vitro, and (iv) that the average cooperative unit of in situ Δ-domain unfolding is greater than three. We have also shown that the free (in vitro) Δ-domain is monomeric and cannot self-assemble at physiological temperature. This distinguishes the HK97 Δ-domain from the dimeric scaffolding proteins of other dsDNA viruses, such as P22 and ϕ29.14,15 The failure of the in vitro Δ-domain to oligomerize may account for its inability to function productively in the assembly of HK97 shells.
MATERIALS AND METHODS
1. Sample Preparations
A 111-residue peptide comprising the Δ-domain (2–103) with a C-terminal tag (SLEH6) was expressed using the One Shot® BL21(DE3)pLysS E. coli cell system (Invitrogen, Carlsbad, CA). The His6 tag was engineered at the C-terminus in order to minimize possible interference with the putative oligomerization sequence of the native gp5 N-terminus.21 Additionally, in the native subunit the C-terminal residue of the Δ-domain (Lys103) is connected to residues 104–385 (gp5*) via an unstructured segment (N-arm, residues 104–132).7 The expressed protein was separated from the cell lysate via Ni-affinity chromatography (HisTrap® HP Column, GE Healthcare Bio-Sciences Corp., Piscataway, NJ) and purified by anion-exchange chromatography (HiTrap® Q column, GE Healthcare BioSciences Corp.). The purified Δ-domain was exchanged into 25 mM NaCl, 20 mM Tris, pH 7.5 solution (buffer A), before concentration to ~20 mg/mL in a Centricon® device (Millipore, Bedford, MA) for Raman spectroscopic analysis.
Prohead I and prohead II particles were prepared from HK97 shell (gp5) and protease (gp4) subunits expressed using plasmids pT7-5Hd2.9 and pT7-5Hd2.9(fdBstBI).37 Subunits were purified by a combination of differential centrifugation, polyethylene glycol precipitation and velocity sedimentation protocols, as previously described.37 For Raman spectroscopy samples were concentrated by centrifugation and resuspension to ~20 mg/mL in 25 mM Tris, 100 mM KCl, 1 mM β-mercaptoethanol, pH 7.5 (buffer B).
2. Raman Spectroscopy
Solutions of the 111-residue Δ-domain, prohead I particle and prohead II particle, each at 20 mg/mL, were sealed in glass capillary cells (KIMAX 34502) for Raman spectroscopy. Solutions were thermostated to a precision of ±0.5 °C in the sample chamber of the Raman spectrometer throughout the spectral data collection protocols. Raman spectra were excited using ~100 mW of 532 nm radiation from a NdY:VO4 diode laser (model Verdi 5 W, Coherent, Santa Clara, CA) and collected on a single-grating spectrograph, as previously described.40. Spectra were collected in the temperature interval 10 < t < 92 °C, at increments of 2 to 5 °C depending upon the rate of change of the spectrum with temperature. Data collection protocols were repeated three times on independently prepared specimens of each sample.
3. Factor Analysis of Raman Spectra
The structural information inherent in the temperature-dependent Raman spectra collected in this study, was assessed qualitatively and quantitatively by use of singular value decomposition (SVD).41 Using the SVD algorithm, a series of N Raman spectra denoted as Yi(ν) (i = 1, …, N) where ν is the number of data points in each spectrum is transformed into a set of N orthonormal components or factors, given in order of decreasing statistical weight Wj, as Sj(ν) (j = 1, …, N). The relative contribution of the j-th factor to the i-th Raman spectrum is expressed in terms of the unitary matrix Vij by eq. 1.
| (1) |
The series of N Raman spectra Yi(ν) can be approximated within experimental error by a subset of factors Sj (j = 1, 2, …, M, where M < N). The value of M also represents the number of independent spectral species in the original series and constitutes the dimension of the factor analysis.41 Each Raman spectrum is also a linear combination of the M species, which represent discrete states of the system in accordance with eq. 2,
| (2) |
where Zn(ν) are the spectra of the M individual states normalized to unit concentration. The coefficient γin represents the molar concentration of state n in Raman spectrum i. In the case of protein melting, the set M is expected to include the native and unfolded states of the protein. The results of SVD can be fitted to the concentrations of the individual states γin via a rotation matrix ρnj, which relates the factors Sj(ν) and the spectra Zn(ν).42 In our analysis, we assumed a weak linear dependence of Zn(ν) on temperature to allow for protein chain flexibility or protein-solvent interactions that may occur without significant changes in the average secondary or tertiary structures.39,43
In the present application the concentrations γin were derived from a simple two-state model of Δ-domain unfolding using the van’t Hoff (eq. 3) and Gibbs-Helmholtz relations. (eq. 4) at each temperature (Ti).
| (3) |
| (4) |
R is the molar gas constant and Tm is the temperature at which half of the molecules in the system are unfolded. Ki and ΔGi indicate the equilibrium constant and the Gibbs free energy, respectively, at temperature Ti. The heat capacity change was considered temperature independent in this approximation.
4. Sedimentation Velocity
The Δ-domain and a corresponding reference (buffer A) were spun at 60,000 rpm in a Beckman XL-A ultracentrifuge using the An60-Ti rotor. The sedimentation process was scanned in 90 s intervals for 7 h. To avoid thermal convection, the sample-loaded rotor was incubated for 1 h at 4 °C prior to sedimentation and thermostated at 4 °C until the end of the run. The experimental data were acquired at 280 nm and analyzed using SEDFIT software (http://www.analyticalultracentrifugation.com/default.htm).44 Input parameters such as the partial specific volume, buffer density and viscosity were determined from the Δ-domain sequence using Sednterp software (http://www.jphilo.mailway.com/download.htm).
5. Sedimentation Equilibrium
Solutions of the Δ-domain at three concentrations (58, 67 and 87 μM) and corresponding reference solutions were loaded into the sedimentation equilibrium cell of the Beckman XL-A rotor (An60-Ti). Data were collected at 280 nm for speeds of 35,000, 40,000 and 45,000 rpm. Equilibrium was reached after 18 h and confirmed by comparison of subsequent scans acquired in three 3 h intervals. Only the last scan at a given speed was used for data analysis (HeteroAnalysis software, http://www.biotech.uconn.edu/auf/).45 A single species model was used to fit the sedimentation equilibrium profiles for each sample at all speeds, in accordance with eq. 5.
| (5) |
A(r) is the total absorbance at radius r, A0 is the absorbance at the reference radius r0, M is the molar mass, ν̄ is the partial specific volume, ρ is the buffer density and ω is the angular velocity.
Acknowledgments
This research was supported by grants GM50776 (GJT) and GM47795 (RWH) from the National Institutes of Health.
ABBREVIATIONS
- cryo-EM
electron cryomicroscopy
- ds
double-stranded
- DSC
differential scanning calorimetry
- HDX
hydrogen-deuterium exchange
- IPTG
isopropyl-β-D-thiogalactopyranoside
- PMSF
phenylmethylsulfonyl fluoride
- SVD
singular value decomposition
- Tris
tris[hydroxymethyl]aminomethane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simon LD. Infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope: T4 head morphogenesis. Proc Natl Acad Sci U S A. 1972;69:907–911. doi: 10.1073/pnas.69.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 3.Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD. Intermediates in the assembly pathway of the double-stranded RNA virus ϕ6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King J, Chiu W. The Procapsid to Capsid Transition in Double-stranded DNA Bacteriophages. In: Chiu W, Burnett RM, Garcea R, editors. Structural Biology of Viruses. Oxford University Press; New York: 1997. pp. 288–311. [Google Scholar]
- 5.Turner BG, Summers MF. Structural biology of HIV. J Mol Biol. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 6.Canady MA, Tihova M, Hanzlik TN, Johnson JE, Yeager M. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis ω virus (NωV) J Mol Biol. 2000;299:573–584. doi: 10.1006/jmbi.2000.3723. [DOI] [PubMed] [Google Scholar]
- 7.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 8.Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- 9.Gan L, Speir JA, Conway JF, Lander G, Cheng N, Firek BA, Hendrix RW, Duda RL, Liljas L, Johnson JE. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure. 2006;14:1655–1665. doi: 10.1016/j.str.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Wikoff WR, Conway JF, Tang J, Lee KK, Gan L, Cheng N, Duda RL, Hendrix RW, Steven AC, Johnson JE. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J Struct Biol. 2006;153:300–306. doi: 10.1016/j.jsb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 11.King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- 12.D’Halluin JC, Martin GR, Torpier G, Boulanger PA. Adenovirus type 2 assembly analyzed by reversible cross-linking of labile intermediates. J Virol. 1978;26:357–363. doi: 10.1128/jvi.26.2.357-363.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman G, Bachenheimer SL. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 14.Thuman-Commike PA, Greene B, Malinski JA, Burbea M, McGough A, Chiu W, Prevelige PE., Jr Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys J. 1999;76:3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu CY, Morais MC, Battisti AJ, Rossmann MG, Prevelige PE., Jr Molecular dissection of ϕ29 scaffolding protein function in an in vitro assembly system. J Mol Biol. 2007;366:1161–1173. doi: 10.1016/j.jmb.2006.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fane BA, Prevelige PE., Jr Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 17.Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- 18.Benevides JM, Bondre P, Duda RL, Hendrix RW, Thomas GJ., Jr Domain structures and roles in bacteriophage HK97 capsid assembly and maturation. Biochemistry. 2004;43:5428–5436. doi: 10.1021/bi0302494. [DOI] [PubMed] [Google Scholar]
- 19.Ross PD, Conway JF, Cheng N, Dierkes L, Firek BA, Hendrix RW, Steven AC, Duda RL. A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J Mol Biol. 2006;364:512–525. doi: 10.1016/j.jmb.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross PD, Cheng N, Conway JF, Firek BA, Hendrix RW, Duda RL, Steven AC. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. EMBO J. 2005;24:1352–1363. doi: 10.1038/sj.emboj.7600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conway JF, Cheng N, Ross PD, Hendrix RW, Duda RL, Steven AC. A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J Struct Biol. 2007;158:224–232. doi: 10.1016/j.jsb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuma R, Prevelige PE, Jr, Thomas GJ., Jr Structural transitions in the scaffolding and coat proteins of P22 virus during assembly and disassembly. Biochemistry. 1996;35:4619–4627. doi: 10.1021/bi952793l. [DOI] [PubMed] [Google Scholar]
- 23.Tuma R, Parker MH, Weigele P, Sampson L, Sun Y, Krishna NR, Casjens S, Thomas GJ, Prevelige PE., Jr A helical coat protein recognition domain of the bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:81–94. doi: 10.1006/jmbi.1998.1916. [DOI] [PubMed] [Google Scholar]
- 24.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage ϕ29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- 25.Teller DC. Accessible area, packing volumes and interaction surfaces of globular proteins. Nature. 1976;260:729–731. doi: 10.1038/260729a0. [DOI] [PubMed] [Google Scholar]
- 26.Overman SA, Thomas GJ., Jr Amide modes of the α-helix: Raman spectroscopy of filamentous virus fd containing peptide 13C and 2H labels in coat protein subunits. Biochemistry. 1998;37:5654–5665. doi: 10.1021/bi972339c. [DOI] [PubMed] [Google Scholar]
- 27.Benevides JM, Overman SA, Thomas GJ., Jr . Raman spectroscopy of proteins. In: Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT, editors. Current Protocols in Protein Science. John Wiley & Sons; New York: 2005. pp. 17.8.1–17.8.35. [DOI] [PubMed] [Google Scholar]
- 28.Berjot M, Marx J, Alix AJP. Determination of the secondary structure of proteins from the Raman amide I band: The reference intensity profiles method. J Raman Spectrosc. 1987;18:289–300. [Google Scholar]
- 29.Tuma R, Thomas GJ., Jr Mechanisms of virus assembly probed by Raman spectroscopy: The icosahedral bacteriophage P22. Biophys Chem. 1997;68:17–31. doi: 10.1016/s0301-4622(97)00001-x. [DOI] [PubMed] [Google Scholar]
- 30.Overman SA, Thomas GJ., Jr Raman markers of nonaromatic side chains in an α-helix assembly: Ala, Asp, Glu, Gly, Ile, Leu, Lys, Ser, and Val residues of phage fd subunits. Biochemistry. 1999;38:4018–4027. doi: 10.1021/bi982901e. [DOI] [PubMed] [Google Scholar]
- 31.Overman SA, Thomas GJ., Jr Raman spectroscopy of the filamentous virus Ff (fd, fl, M13): structural interpretation for coat protein aromatics. Biochemistry. 1995;34:5440–5451. doi: 10.1021/bi00016a015. [DOI] [PubMed] [Google Scholar]
- 32.Thomas GJ, Jr, Prescott B, Day LA. Structure similarity, difference and variability in the filamentous viruses fd, If1, IKe, Pf1 and Xf. Investigation by laser Raman spectroscopy. J Mol Biol. 1983;165:321–356. doi: 10.1016/s0022-2836(83)80260-5. [DOI] [PubMed] [Google Scholar]
- 33.Grygon CA, Perno JR, Fodor SP, Spiro TG. Ultraviolet resonance Raman spectroscopy as a probe of protein structure in the fd virus. Biotechniques. 1988;6:50–55. [PubMed] [Google Scholar]
- 34.Overman SA, Bondre P, Maiti NC, Thomas GJ., Jr Structural characterization of the filamentous bacteriophage PH75 from Thermus thermophilus by Raman and UV-resonance Raman spectroscopy. Biochemistry. 2005;44:3091–3100. doi: 10.1021/bi048163d. [DOI] [PubMed] [Google Scholar]
- 35.Parker MH, Stafford WF, III, Prevelige PE., Jr Bacteriophage P22 scaffolding protein forms oligomers in solution. J Mol Biol. 1997;268:655–665. doi: 10.1006/jmbi.1997.0995. [DOI] [PubMed] [Google Scholar]
- 36.Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J Mol Biol. 2005;348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995;247:636–647. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- 38.Ziegelhoffer T, Yau P, Chandrasekhar GN, Kochan J, Georgopoulos C, Murialdo H. The purification and properties of the scaffolding protein of bacteriophage λ. J Biol Chem. 1992;267:455–461. [PubMed] [Google Scholar]
- 39.Cooper A. Thermodynamics of Protein Folding and Stability. In: Allen G, editor. Protein: A Comprehensive Treatise. JAI Press Inc; 1999. pp. 217–270. [Google Scholar]
- 40.Movileanu L, Benevides JM, Thomas GJ. Temperature dependence of the Raman spectrum of DNA. I Raman signatures of premelting and melting transitions of poly(dA-dT)·poly(dA-dT) J Raman Spectrosc. 1999;30:637–649. doi: 10.1002/bip.10022. [DOI] [PubMed] [Google Scholar]
- 41.Malinowski ER. Factor Analysis in Chemistry. John Wiley & Sons, Inc; USA, New York: 2002. [Google Scholar]
- 42.Hanus J, Chmelova K, Stepanek J, Turpin PY, Bok J, Rosenberg I, Tocik Z. Raman Spectroscopic Study of Triplex-like Complexes of Polyuridylic Acid with the Isopolar, Non-isosteric Phosphonate Analogues of Diadenosine Monophosphate. J Raman Spectrosc. 1999;30:667–676. [Google Scholar]
- 43.Hanus J, Nemecek D, Stepanek J, Turpin PY. Structural compatibility of novel nucleotide modifications with shortened linkages designed for antigene/antisense therapy. J Raman Spectrosc. 2004;35:418–425. [Google Scholar]
- 44.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole JL. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]