Abstract
The uaY gene encodes the transcriptional activator of purine catabolism genes in Aspergillus nidulans. uaY12 results in strongly defective growth on purines as nitrogen sources and in strongly diminished transcription of UaY-regulated genes. This mutation introduces an ATG codon 64 bp upstream of the uaY ATG, generating a 68-codon open reading frame (uORFA), overlapping with the uaY ORF. uaY12 revertants fall into three categories:
The majority eliminate the aberrant ATG. The growth and transcriptional phenotypes of these revertants are identical to those of the wild type.
Two revertants create a stop codon in frame with the uaY12 aberrant ATG, shortening the length of the uORFA, thus uORFA no longer overlaps the uaY ORF. The latter are partial suppressors of the uaY12 mutation, while chain termination suppressors, in turn, suppress this novel phenotype.
Two partial suppressors are unlinked to uaY. These two mutations result in a pleiotropic phenotype usually associated with ribosomal proteins.
We hypothesize that uORFA strongly diminishes translation of the uaY ORF and that revertants negate this effect by a number of different mechanisms. The first-AUG rule and the phenomena of translational inhibition and leaky scanning provide a coherent explanation of the results presented in this article.
THE fungus Aspergillus nidulans can use purines (adenine, hypoxanthine, xanthine, and uric acid) as sole nitrogen sources. The UaY transcription factor mediates uric acid induction of the majority of the purine-degradation pathway genes (Scazzocchio and Darlington 1968; Scazzocchio et al. 1982; Scazzocchio 1994). It is necessary for transcriptional induction of the three transporter genes uapA, uapC, and azgA (Gorfinkiel et al. 1993; Diallinas et al. 1995; Cecchetto et al. 2004) and of structural genes involved in purine degradation: nadA (adenine deaminase, Ribard et al. 2003), hxA (xanthine dehydrogenase, Glatigny and Scazzocchio 1995), hxB (MOCO sulfurase, necessary for the activity of the xanthine dehydrogenase, Amrani et al. 1999), xanA (xanthine α-ketoglutarate-dependent dioxygenase, Cultrone et al. 2007), and uaZ (urate oxidase, Oestreicher et al. 1993). These genes are also under the positive control of the GATA factor AreA, which is active in the absence of repressive nitrogen sources (such as ammonium and glutamine, Arst and Cove 1973; Kudla et al. 1990). With the exception of the nadA gene, where the two proteins have antagonistic effects (Oestreicher et al. 2008), AreA and UaY act in synergy and are both necessary for fully induced transcription (references as cited previously and Ravagnani et al. 1997).
The uaY gene encodes a 1060-amino-acid protein, containing a zinc binuclear cluster DNA-binding domain, which binds to DNA sequences of the form CCG-6X-CGG (Suárez et al. 1995). The uaY gene is transcribed at a very low level, and at variance with previous reports (Suárez et al. 1991b, 1995), it is mildly autoregulated, with an induction ratio of 1.4 (N. Oestreicher, unpublished results). Studies that have been summarized elsewhere have shown that the intracellular concentrations of UaY are strictly limiting for the expression of the cognate regulated genes (Scazzocchio, 1994). Thus growth phenotypes on different purines as nitrogen sources and mRNA levels of UaY-regulated genes give a sensitive, albeit qualitative estimation of the intracellular concentrations of UaY protein (Scazzocchio et al. 1982). Overexpression of uaY in the presence of its putative ligand, uric acid, is highly toxic to A. nidulans (N. Oestreicher, unpublished results), possibly due to squelching (Gill and Ptashne 1988).
The uaY gene has been extensively studied by classical genetics, with the isolation of null mutations and gain-of-function mutations. A fine structure map has being generated permitting to order linearly 21 uaY− alleles, of which 8 correspond to internal deletions (Scazzocchio et al. 1982; Suárez et al. 1991a). The identification of nine uaY− mutations by sequencing is in complete agreement with the fine structure map previously published (uaY2, 9, and 808: Suárez et al. 1995; uaY205: Oestreicher et al. 1997; uaY26, uaY27, 207, and 333: Suárez et al. 1991b and N. Oestreicher, unpublished results; and uaY109: N. Oestreicher, unpublished results).
Here, we identify the uaY12 allele, the most 5′ uaY− mutation isolated to date (Scazzocchio et al. 1982; Suárez et al. 1991b). Molecular characterization of this mutant and of 43 of its revertants strongly suggests that the uaY12 mutation is a mutation affecting the level of translation of the UaY protein. The phenotype of these mutants can be interpreted by the model of ribosome scanning and the first-AUG rule, together with the processes of reinitiation and leaky scanning. Kozak (2002 a,b) has proposed that leaky scanning has been recruited throughout evolution to limit intracellular concentrations of proteins, such as transcription factors, harmful to the cell when overproduced. The work presented here suggests that this mechanism operates for a thoroughly studied fungal transcription factor, UaY.
MATERIALS AND METHODS
Strains:
All the A. nidulans strains used and/or constructed in this work are listed in the supplemental material (see supplemental Table 1). Standard genetic markers are described in http://www.gla.ac.uk/Acad/IBLS/molgen/aspergillus/index.html. The null mutation uaY2 is a C-to-T transition at position +1173, which results in the introduction of amber codon at position 392 of the UaY protein (the wild-type protein being 1060 residues long, Suárez et al. 1995). uaY808 combines a deletion of 484 bp between positions +998 and +1482 with a 5-bp insertion (CTCGT); these modifications result in a frameshift generating a stop codon nine triplets downstream of the deletion terminus (Suárez et al. 1995). The leaky mutation uaY109 is a T-to-A transversion at position +334, which results in a Phe-to-Ile change at position 112 of the UaY protein (N. Oestreicher, unpublished results). uaY205 is a 16-bp deletion starting at position 2992, which results in premature translation termination and substitutes the C-terminal 63 amino acids by 13 new residues (Oestreicher et al. 1997).
The reversion experiment was carried out on strain NA9926; thus, all the revertants are, with the exception of 12r128, in the yA2 pyroA4 riboC3 background. The original 12r128 revertant is no longer available; thus, all the work has been carried out with strain NA0464 (yA2 pantoB100 pyroA4) obtained from the cross of 12r128 and NA0215.
Escherichia coli JM109 (Yanisch-Perron et al. 1985) was used for routine plasmid preparation.
Media, growth conditions, mutagenesis, and transformation techniques:
The media and growth conditions for A. nidulans were according to Scazzocchio et al. (1982). Mycelia used for DNA preparation were grown as described in Oestreicher and Scazzocchio (1993). The mycelia for RNA preparation were grown for 10 hr at 37° in supplemented minimal media with shaking at 150 rpm, in the presence of 5 mm urea as the sole nitrogen source (noninducing, nonrepressing conditions). Induction was carried out by addition of 0.1 mg/ml of acid uric for the last hour of growth. In the noninduced culture, urea was added again for the last hour.
uaY12 spores were mutagenized with nitroquinoline-1-oxide (Bal et al. 1977), resulting in <1% survival, followed by isolation of revertants grown on hypoxanthine as sole nitrogen source at both 37° and 25°, from where individual colonies were purified by single spore isolation.
Transformation of A. nidulans followed the method of Tilburn et al. (1983). Transformation of E. coli was carried out according to Hanahan (1983).
DNA manipulations:
Plasmid extraction from E. coli and other DNA manipulations were as described by Sambrook et al. (1989). DNA was prepared from A. nidulans as described by Specht et al. (1982). Southern blots were carried out as described in Oestreicher et al. (1993), using positive TM membranes (Qbiogene). DNA bands were purified from agarose gels using the Wizard PCR preps DNA purification system (Promega). Random Hexanucleotide Primer kit (Roche Applied Science) was used to label DNA molecules with [α-32P]dCTP to generate gene-specific probes. Routine PCR amplifications for genotype verification were carried out using Taq DNA polymerase of Promega and specific 17mer primers, and following these PCR conditions: 5 min at 95°, followed by 40 cycles with 1 min at 95°, 1 min at 58° and 2 min at 72°, and 1 final cycle of 10 min at 72°. Sequencing of PCR fragments was carried out with specific 17mer primers using the dideoxynucleotide chain termination procedure (Sanger et al. 1977) with the T7 Sequenase PCR product sequencing kit (Roche Applied Science).
Cloning and sequencing of uaY alleles, construction of the plasmid used in back-transformation experiments:
The uaY12 mutation:
The uaY12 mutation has been mapped by transformation in the 584-bp XhoI fragment, containing the wild-type uaY sequence from nucleotide −225 to +358 (numbering from the A of the ATG initiation codon). Amplification of a 1246-bp fragment (from position −540 to +708) was carried out on uaY12 genomic DNA with 17mer-specific primers and following the same conditions as above. After DNA purification and digestion by XhoI, the 584-bp fragment (−225 to +358) containing the uaY12 mutation, was cloned in the Bluescript KS(+) vector (Stratagene, plasmid named bAN754-12, see supplemental Table 2) and the whole insert of two independent clones was sequenced on double-stranded templates with M13 and M13 reverse primer using the T7 sequencing mixes (Roche Applied Science).
The revertant alleles:
For the 43 revertants, a 1286-bp fragment (from position −540 to +746) was amplified in the same conditions as explained above. The region from −225 to +40 was sequenced on the PCR product using a specific 19mer primer with its 5′ hybridizing at position −294 (see above).
Plasmids used in back-transformation experiments:
That the revertant alleles: uaY12r3, 12r17, 12r18, 12r113, 12r118, and 12r130 are responsible for the phenotype observed, was confirmed by partial gene replacement (Miller et al. 1985; Oestreicher et al. 1997) as described below. A 667-bp fragment (from position −294 to +373) was amplified [see above for PCR conditions, but using pfu polymerase (Promega) rather than Taq polymerase]. After purification, a 427-bp fragment (−225 to +203) corresponding to a XhoI–ClaI digest, was cloned in Bluescript KS(+), generating the bAN772-X plasmids (where X corresponds to the number of the uaY allele, see supplemental Table 2). Again, the whole inserts of these plasmids were sequenced (Fasteris, Switzerland), using the T3 primer, to ensure that no mutation was introduced by the PCR process. To expand the size of the insert on both sides of the XhoI–ClaI fragment, two successive steps were carried out: First, a 2836-bp ApaI–XhoI fragment (−2611 to −225) from plasmids bAN783 (Ribard et al. 2001) was inserted in all the six bAN772-X plasmids, creating the bAN755-X plasmids (see supplemental material). Second, a 1403-bp ClaI–EcoRI fragment (+203 to +1606) from plasmids bAN753 (Suárez et al. 1991b) was inserted in all the six bAN755-X plasmids, creating the final bAN756-X plasmids (see supplemental Table 2).
RNA manipulations:
Total RNA was isolated from A. nidulans as described by Lockington et al. (1985), and Northern blots were carried out as described in Oestreicher and Scazzocchio (1993) and modified as in Oestreicher et al. (2008). To monitor the RNA loading, an actin-specific probe was used (Fidel et al. 1988). The actin, hxA, uaZ, and uapA probes are described in Oestreicher et al. (2008). The uaY probe corresponds to the 3281-bp NcoI fragment from the bAN700 plasmid, which contains the genomic sequence of the uaY gene from the +1 position until the base pair before the stop codon. The intensities of radioactive signal were quantified with a 400A Phosphoimager (Amersham Biosciences). Data were analyzed with the ImageQuant software (Molecular Dynamics). All Northern blot experiments were carried out at least in triplicate.
The start of transcription was determined using the 5′-RACE method following the supplier's instructions (Boehringer), using RNAs extracted from wild-type mycelia grown in the presence of uric acid. The initial cDNA was obtained using the specific oligonucleotide 5′ GCTATTTCGCCGGCAGCTGG 3′ (5′ starting at the position +492). After purification and tailing with terminal transferase in the presence of dATP, the resulting product was amplified using a poly(dT) anchor primer (supplied by the kit) and the specific primer 5′ CGCCGGCAGCTGGTCCGTTTGCTGA 3′ (5′ starting at the position +487, this second specific primer is overlapping with the first one). The PCR conditions were: 2 min at 94°, followed by 10 cycles with 15 sec at 94°, 30 sec at 58°, and 40 sec at 72°, followed by 25 cycles with 15 sec at 94°, 30 sec at 58°, and 40 sec (+20 sec at each cycles) at 72°, and finally one round at 74° during 7 min. One-sixth of the PCR reaction was tested on an agarose gel, given a hardly visible 1-kb band. A second PCR was carried out on a 1-μl aliquot of a 1/10 dilution of the first PCR mixture, using the same conditions, with the exceptions of the annealing temperature (63° rather than 58°) and the two primers (with a new specific primer, overlapping with the second one: 5′ CGTTTGCTGAGCCCGCATATGCTTC 3′ and the anchor primer instead of the poly(dT) anchor primer). The resulting product was purified and digested by ClaI, which cuts inside the uaY sequence (at position +203) and in the anchor primer. The resulting products were cloned in the Bluescript KS(+) (Stratagene), giving nine clones (named bAN75′-1–9). These clones were sequenced using the M13 and reverse primers with the T7 sequencing mixes (Roche Applied Science).
RESULTS
Determination of the 5′ start point of the uaY mRNA:
The 5′ start point of the uaY mRNA had been estimated by primer extension (Suárez et al. 1995) to be at position −52/−46 from the ATG. This would result in an mRNA of ∼3.4 kb. The estimated size of the mRNA in Northern blots is 3.8–4.0 kb (not shown and Suárez et al. 1991b). As this work deals with the most 5′ mutation mapped in the uaY gene, we thought it important to address this apparent contradiction. Thus, we determined again the 5′ start point with 5′-RACE, a more up-to-date methodology (see materials and methods). At least seven start points have been identified at positions: −503 (two clones), −494 (two clones), −469, −457, −438, −431, and −284 (one clone each) (see supplemental Figure 1). These new start points are in agreement with the RNA size determined in Northern blots. This result has been confirmed using three PCR fragments as probes for Northern blots: one upstream of the published −52/−46 start point (region −540 to −220), one overlapping with the beginning of the ORF (region −294 to +16), and one internal to the ORF (+412 to +746). All the probes reveal the uaY messenger (data not shown).
The uaY12 mutation and its revertants:
We mapped uaY12 by transformation with linear fragments (see materials and methods) between nucleotides −225 to +358 of the uaY gene (numbering from the A of the ATG initiation codon). The corresponding sequence from a uaY12 strain showed a C-to-T transition at position −63 (Table 1). The sequence change is well within the transcribed sequences, but upstream of the UaY ATG.
TABLE 1.
Sequence of the 43-uaY12 revertants
| Allele names | Sequence of the mutationb (uaY+: TCATTACGGGTCTTTCAGTT, uaY12: TCATTATGGGTCTTTCAGTT) | No. of revertants selected at
|
|||
|---|---|---|---|---|---|
| Nature of the reversiona | 25° | 37° | Total | ||
| uaY12r26 | T to A in position −63 | TCATTAAGGGTCTTTCAGTT | 1 | 0 | 1/43 |
| uaY12r11 | G to C in position −62 | TCATTATCGGTCTTTCAGTT | 3 | 3 | 6/43 |
| uaY12r17 | G to A in position −62 | TCATTATAGGTCTTTCAGTT | 13 | 3 | 16/43 |
| uaY12r49 | G to T in position −62 | TCATTATTGGTCTTTCAGTT | 7 | 2 | 9/43 |
| uaY12r43 | G to T in position −62 and −61 | TCATTATTTGTCTTTCAGTT | 1 | 0 | 1/43 |
| uaY12r44 | G to C in position −62 and −22 | TCATTATCGGTCTTTCAGTT | 1 | 0 | 1/43 |
| −26 CACCCGTCATTATATCCT | |||||
| uaY12r130 | G to T in position −62 and T to A in −66 | TCAATATTGGTCTTTCAGTT | 0 | 2 | 2/43 |
| uaY12r3 | Insertion of T between position −63 and −62 | TCATTATTGGGTCTTTCAGTT | 1 | 0 | 1/43 |
| See Figure 6 | |||||
| uaY12r18 | Deletion of 63 bp from position −91 to −29 | See Figure 6 | 1 | 0 | 1/43 |
| uaY12r113 | G to A in position −38 | See Figure 1 | 0 | 1 | 1/43 |
| uaY12r118 | G to A in position −42 | See Figure 1 | 0 | 2 | 2/43 |
| 12r122 (aas22) | Not in the uaY locus | ND | 0 | 1 | 1/43 |
| 12r128 (aas28) | Not in the uaY locus | ND | 0 | 1 | 1/43 |
The positions of these mutations are numbered from the A of the wild-type ATG initiation codon.
Modified bases in the revertants are in boldface type and in larger type, the underlined letter corresponding to the base mutated in the original uaY12 mutant. The given sequence begins at position −69 relative to the A of the uaY ATG, except indication.
To gain some insight into why this apparently anodyne mutation resulted in a phenotype only slightly more leaky (see below) than that shown by strains carrying null mutations (e.g., uaY2, uaY9, chain termination mutations; uaY207, a large insertion; uaY808, a deletion; Suárez et al. 1991b, 1995, for uaY2, see below), we isolated revertants by selection for growth on hypoxanthine as the sole nitrogen source at both 37° and 25° (see materials and methods). Forty-three revertants were isolated and analyzed. For each revertant, a −540 to +746 fragment of the uaY region was amplified and the −225 to +40 region was sequenced (see materials and methods). These revertants comprise 11 different sequence changes in the amplified region and two strains where the sequence obtained was identical to that of the uaY12 mutation. The nucleotide changes of the sequenced revertants are shown in Table 1. An exact reversion to the wild type (a T-to-C transition at position −63) was not represented in the sequence changes analyzed by us. One revertant, uaY12r26, shows a T-to-A transversion in nucleotide −63, where the original uaY12 mutation occurred. The majority of the revertants represented by uaY12r11, uaY12r17, and uaY12r49 show one of the three possible changes at nucleotide −62. The alleles uaY12r43, 12r44, and 12r130 carry a mutation at −62 together with a second one in its proximity. In uaY12r18 a deletion of a 63-bp region containing the uaY12 mutation (from position −91 to −29) has occurred while uaY12r3 carries an insertion of a T between position −63 and −62. Three revertants do not affect the nucleotides −63 and −62, but carry a transition of a G to A at nucleotide −42 (allele uaY12r118) or −38 (uaY12r113).
To confirm that the sequenced mutations were responsible for the reversion phenotype, a partial gene replacement of the uaY12 allele by the reverted alleles was carried out. A strain carrying the uaY12 mutation was transformed with a linear 4169-bp ApaI–EcoRI fragment containing the 427-bp XhoI-ClaI (interval −225 to +203) region carrying the uaY12r17, uaY12r3, uaY12r18, uaY12r113, uaY12r118, uaY12r130, or uaY+ sequences (see materials and methods). In every case, transformants able to grow on hypoxanthine as the sole nitrogen source were obtained. The original revertants are phenotypically heterogeneous (see below) and this heterogeneity is faithfully reproduced in the cognate transformants.
Finally two revertants (strains 12r122 and 12r128) still carry the uaY12 mutation with no additional mutation in the sequenced region. Each of these strains carry suppressors unlinked to the uaY locus. This was determined by crossing the revertant strains to a strain carrying the uaY12 allele together with three closely linked bracketing markers (cbxC34, carboxin resistance; oxpA5, oxypurinol resistance; and fpaD43, p-fluoro-phenylalanine resistance; order of markers: cbxC-0.5cM-oxpA-0.3cM-uaY-5cM-fpaD; Scazzocchio et al. 1982). This configuration allows one to distinguish the uaY12 allele of the revertant strain from the uaY12 allele of the marker strain. For both 12r122 and 12r128, the ability to grow on hypoxanthine segregated independently from the bracketing markers and by implication from the uaY12 allele. Analogous control crosses with strain carrying the 11 revertants discussed in previous paragraphs show a perfect cosegregation between the revertant phenotype and the bracketing markers in coupling with it.
No chromosomal rearrangement in the vicinity of the uaY locus was revealed by Southern blot analysis for any of the revertants (or for the uaY12 original mutant).
An interpretation of the nature of the uaY12 mutation and its revertants:
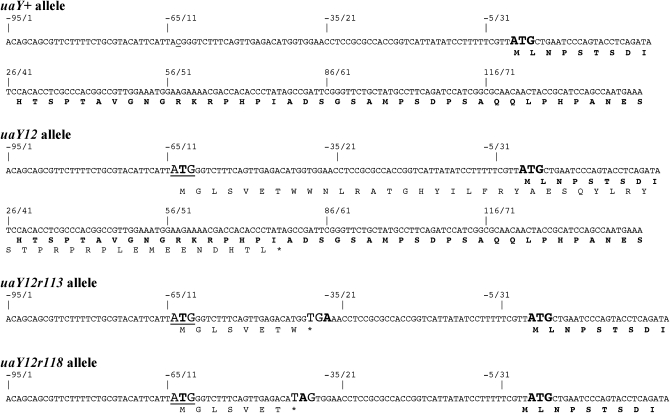
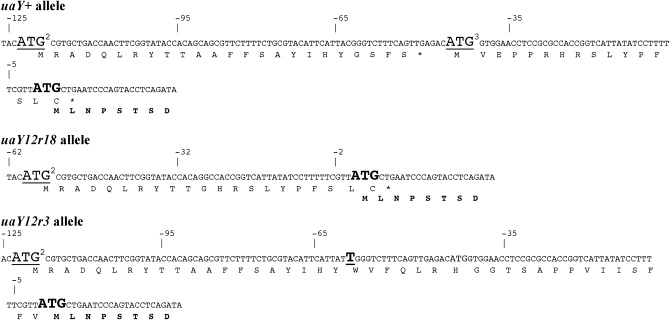
The uaY12 mutation changes the sequence (−70)TTCCATTACGGG into (−70)TTCCATTATGGG, which generates an ATG triplet 64 bp upstream of the uaY ATG codon. This new ATG is followed by a short open reading frame of 68 codons (Figure 1), which is overlapping and not in frame with the uaY open reading frame, this short mutant ORF ending at UaY codon 47. We shall call this ORF uORFA (for upstream ORF, aberrant). In revertant alleles occurring at position −63 and −62 the ATG created in the uaY12 mutation is lost; the same is a fortiori true for deletion uaY12r18. In both uaY12r113 and uaY12r118 the reversion introduces a stop codon (an opal codon TGA for uaY12r113 and an amber codon TAG for uaY12r118) that truncates to 8 and 7 residues, respectively, the open reading frame present in uaY12 (uORFA). These short ORFs do not overlap with the uaY ORF (Figure 1).
Figure 1.—
Sequence of uaY12 and of some of its revertants. The DNA and translated peptide of the relevant sequence of the uaY gene are shown for uaY+ (wild type), uaY12, and for two intragenic suppressors of the uaY12 mutation. Numbers indicate the nucleotide position from the A of the uaY ATG. The wild-type ATG and the wild-type NH2 terminal sequence of the UaY protein are shown in boldface type, with the ATG in a larger type. The C underlined in position −63 of the wild-type sequence is the nucleotide mutated in the uaY12 allele. The uaY12 aberrant ATG is in a larger type and underlined, the mutated base shown in boldface type. The stop codons introduced by the reversion mutations found in 12r113 and 12r118 are in larger type with the mutated base in boldface type.
These data lead to the following hypothesis: The appearance of a new ORF in uaY12 would diminish the translation initiation efficiency at the physiological uaY ATG, while all the suppressors would restore completely or partially initiation at the physiological ATG.
Growth and transcriptional phenotypes of uaY12 and its intragenic revertants:
uaY12:
The strains carrying the uaY12 mutation do not use purines (adenine, hypoxanthine, xanthine, and uric acid) as sole nitrogen sources, but their growth on uric acid is intermediate between the residual growth of null uaY mutants (e.g., bearing uaY2 and uaY9 alleles) and the weak growth of leaky uaY mutants (uaY109 and uaY110) (Scazzocchio et al. 1982) (Figure 2A). This utilization of uric acid suggests that the mutant strains must express to some degree the uaZ gene (encoding urate oxidase) and at least one of the uric acid transporters. Both hxA (encoding xanthine dehydrogenase) and uaZ are still weakly inducible in uaY12 strains (Figures 3–5). This strongly reduced level of induction is consistent with a mutation that by affecting translation diminishes substantially the intracellular level of the UaY protein, without affecting its functionality.
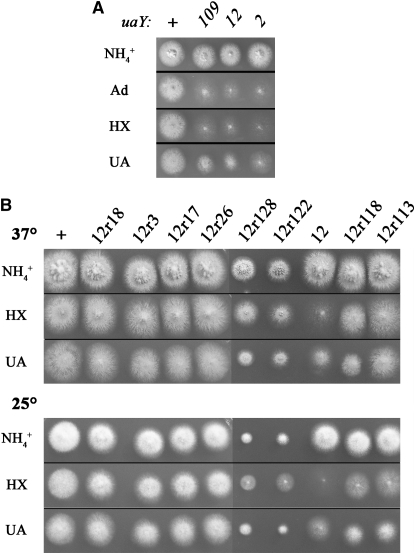
Figure 2.—
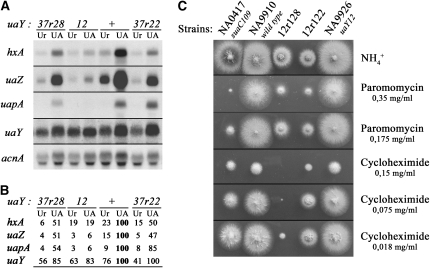
Growth tests of the uaY12 mutant and of some of its revertants. (A) Comparison of the growth on adenine, hypoxanthine, and uric acid of uaY12 and some other uaY− mutants. uaY2 corresponds to a null uaY allele and uaY109 to a leaky allele (see supplemental material for full genotypes, and materials and methods for the sequence of the mutations). These strains were plated on supplemented minimal media containing 5 mm of ammonium tartrate (NH4+) or 0.1 mg/ml of adenine (Ad), hypoxanthine (HX), or uric acid (UA) as sole nitrogen sources. Plates were incubated for 2 days at 37°. At the top, the uaY alleles carried by each strain are indicated. A. nidulans coniospores are able to germinate on media without any utilizable nitrogen source, leading to low hyphal density and sparse sporulation, as illustrated by the residual growth of the null uaY2 mutant. (B) Growth test of uaY12 revertants. Conditions as in A, except that the plates at 25° were incubated for 4 days and the test on adenine was not included. “ +” indicates the wild-type strain (NA9910) and “12” the uaY12 strain used in the reversion experiment (NA9926). All the strains are in the same genetic background (yA2 riboC3 pyroA4) except the 12r128 strain (yA2 pantoB100 pyroA4).
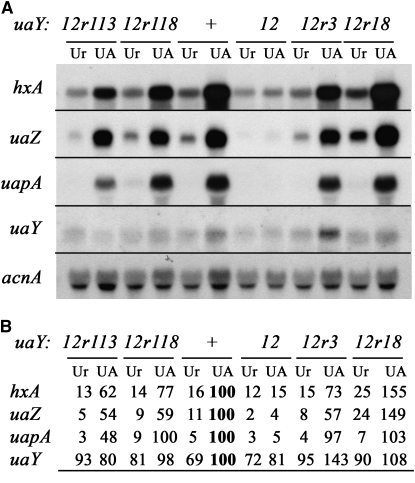
Figure 3.—
Expression of the hxA, uaZ, uapA, and uaY genes in the uaY12 revertants 12r113, 12r118, 12r3, and 12r18. (A) Northern blot analysis of hxA, uaZ, uapA, and uaY gene expression. The relevant uaY allele of each strain is shown at the top. See materials and methods for growth conditions and probe description. Mycelia were grown on urea as a nitrogen source for a total of 10 hr at 37°, after 9 hr growth uric acid (UA, induced) or urea (Ur, noninduced) was added at the concentrations indicated in materials and methods. (B) Quantification of the hxA, uaZ, uapA, and uaY transcripts. The Phosphoimager readings were corrected using the corresponding acnA transcript signal intensity as a measure of RNA loading. All values are expressed in relation to the wild-type mycelia induced by uric acid and correspond to the average values of three independent Northern blots. The maximal SD values found are 15% for hxA, 17% for uaZ, 14% for uapA, and 17% for uaY. The reference values (corresponding to the arbitrary value of 100) are shown in boldface type.
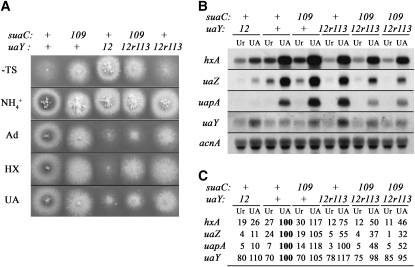
Figure 4.—
Effect of the suaC109 mutation on the uaY12r113 allele. (A) Growth tests. The relevant genotypes for the suaC and uaY genes are given at the top. All the strains, except the uaY12 strain, are in an sB43 background, a mutation in the sulfate transporter, which can be supplemented by thiosulfate. The sB43 allele is suppressed by the suaC109 mutation (Roberts et al. 1979). The different strains were inoculated on supplemented minimal media containing 5 mm of ammonium tartrate (NH4+), or 0.1 mg/ml of adenine (Ad), or hypoxanthine (HX), or uric acid (UA) as sole nitrogen sources. All the plates except the one in the first panel (noted −TS) are supplemented with thiosulfate. The plates were incubated for 2 days at 37°. (B and C) Expression of the hxA, uaZ, uapA, and uaY genes in the 12r113 revertant in a suaC+ or suaC109 genetic background. The relevant genotypes for the suaC and uaY genes are given at the top. Experimental conditions and symbols are as in Figure 3. The values shown in C correspond to the average values of three independent Northern blots. The maximal SD values found are 11% for hxA, 15% for uaZ, 14% for uapA, and 18% for uaY. Note: the suaC109 uaY+ strain is in an oxpA5 genetic background. In the presence of this partial loss of function of the adB gene (encoding adenylosuccinate synthetase), the expression of the UaY-dependent genes is slightly enhanced (Ribard et al. 2001).
Figure 5.—
Extragenic suppression of the uaY12 mutation and suppressor-dependent drug sensitivity. (A and B) Expression of the hxA, uaZ, uapA, and uaY genes in the 12r122 and 12r128 strains. Experimental conditions and symbols are as in Figure 3. The values of B correspond to the average values of three independent Northern blots. The maximal SD values found are 8% for hxA, 14% for uaZ, 14% for uapA, and 11% for uaY. (C) Growth tests in the presence of paromomycin and cycloheximide. The strain numbers and relevant genotypes are shown above each panel. The different strains were inoculated on supplemented minimal media containing 5 mm of ammonium tartrate (NH4+) with or without the relevant inhibitor. The final concentration of paromomycin or cycloheximide in the plate is shown on the right. The plates were incubated for 2 days at 37°.
Revertants carrying a modification at position −63/−62:
As expected, all strains carrying reversion mutations at positions −62 and −63, whether or not they carry an additional mutation, show a wild-type growth phenotype on all purines tested at both 25° and 37°. This class of revertants is represented by the 12r26 and 12r17 strains in Figure 2B. Similarly wild-type growth on purines as a nitrogen source was seen for the insertion uaY12r3 and the deletion uaY12r18. We have checked the mRNA steady-state levels of three genes strictly controlled by UaY, hxA, uaZ, and uapA (encoding the specific urate-xanthine transporter), and the uaY gene itself. Quantitative estimation of the steady-state uaY levels suggests that at variance to what was reported before the uaY gene itself, it is slightly (∼1.4-fold) induced by uric acid and that this induction is UaY dependent. In 12r26, 12r49, 12r17, 12r11, 12r43, 12r44, and 12r130 strains, uaY, hxA, uaZ, and uapA mRNA levels are fully restored to wild-type levels (not shown). In the 12r3 strain, the expression of hxA and uaZ is lower than the wild-type level, uapA is expressed at wild-type levels, while the induced level for the uaY gene is slightly higher than in the wild type (Figure 3).
In the 12r18 revertant, the expression levels of the three structural genes tested are actually higher than in the wild-type strain. This result has been confirmed by three independent biological repetitions (Figure 3).
Revertants 12r113 and 12r118:
The uaY12r113 and uaY12r118 revertants conserve the uaY12 mutation, but they each carry a chain termination codon that truncates the uORFA (see above). Strains carrying these two intragenic suppressors grow less than the wild type on adenine, hypoxanthine, xanthine, and especially uric acid as sole nitrogen sources (Figure 2B). This limited growth on purines is more apparent at 25° than 37°. Thus, the uaY12 mutation is only partially suppressed by these two intragenic suppressors, at variance with the revertants that modify the −63/−62 positions (see above). Northern blot analysis carried out on the 12r118 and 12r113 strains (Figure 3) shows that the hxA and uaZ messengers' induced steady states are lower than those obtained in the wild type. The Northern blot shown in Figure 3 has been carried out with strains grown at 37°. When mRNAs steady states are determined for mycelia grown at 25°, the reduction of the induced mRNA levels of UaY-dependent genes in 12r118 and 12r113 strains is even more noticeable with a downfall of near 25% compared to their induced levels at 37° (data not shown). Thus both growth tests and mRNA steady-state levels indicate that uaY12r118 and uaY12r113 act as partial intragenic cryosensitive suppressors of uaY12.
Antisuppression: effect of chain termination suppression on the uaY12r113 allele:
If the chain termination codons present in uaY12r118 and uaY12r113 act by truncating the uORFA present in strains carrying the uaY12 allele, the presence in the same strain of a chain termination suppressor should act as an antisuppressor for uaY12, i.e., should restore to some or to the full extent the uaY12 phenotype.
The translational suppressors described in A. nidulans map in four loci (Roberts et al. 1979; Martinelli and Roberts 1983; Martinelli et al. 1984; see Martinelli 1994 for a review). The suaB and suaD loci are thought to specify a tRNA, this has been confirmed for suaB, which codes a gln-tRNA (Espeso et al. 2005). The suaA and suaC genes are thought to be ribosomal mutants affecting translational fidelity (Martinelli 1984; Bratt and Martinelli 1988). Several of the mutations suppressed by alleles of these genes have been shown to be chain termination mutations (areA600 amber: Kudla et al. 1990; areA1601 opal: Stankovich et al. 1993; alcR125 amber: M. Mathieu, personal communication).
Four suppressor mutations, representing four different genes (suaA101, suaB111, suaC109, and suaD103) were crossed with a strain carrying the uaY12r113 allele. The crosses were homozygous for sB43, which is suppressed by every one of the suppressor alleles mentioned above (Roberts et al. 1979), thus allowing us to score the segregation of these suppressors in the progeny. sB43 is a chain termination mutation in the sulfate transporter sB gene of A. nidulans, resulting in a thiosulfate or organic sulfate requirement (Roberts et al. 1979). The uaY+ locus in the chain termination suppressor carrying strain was flanked by closely linked markers, cbxC34, oxpA5 on the centromere proximal side, and fpaD43 on the centromere distal side of the uaY gene (Scazzocchio et al. 1982, see above) permitting the identification of the uaY+ or uaY12r113 alleles by the segregation of the external makers. Partial suppression of the uaY12r113 revertant phenotype was obtained with the suaC109 mutation, the other suppressor mutations affording no visible effect. Strains carrying uaY12r113 suaC109 show an intermediate growth between that of the uaY12 mutant and that of the uaY12r113 revertant, this reduced growth is more marked on adenine than on uric acid, and nearly not visible on hypoxanthine (Figure 4A). This partial suppression is confirmed by Northern blot analysis of the relevant transcripts (Figure 4, B and C). The steady-state mRNA levels in the uaY12r113 suaC109 strain are intermediate between the messenger levels seen in the uaY12 mutant and the one of the uaY12r113 revertant, the hxA gene being less affected than the uapA and uaZ genes. The agreement between the growth phenotypes of these strains and the mRNA levels of UaY regulated genes illustrates the exquisite sensitivity of this system to assess, albeit qualitatively, the intracellular activity of this transcription factor.
Characterization of the uaY12 strains carrying extragenic suppressors (12r122 and 12r128):
Growth tests showed that 12r122 and 12r128 strains are partially suppressed for growth on purines as sole nitrogen sources (Figure 2B), but also that these strains have restricted growth on every other media tested (see growth on ammonium in Figure 2B), this restricted phenotype being more severe at 25° than at 37°. Northern blot analysis (Figure 5, A and B) show that induced transcription of hxA, uaZ, and uapA is partially restored in both 12r122 and 12r128 strains.
Conventional genetic crosses show that the suppressor mutations carried by 12r122 and 12r128 strains are allele specific for uaY12. The suppressor carried by 12r122 does not suppress the uaY2 (amber, Suárez et al. 1995), uaY205 (frameshift, Oestreicher et al. 1997), and uaY808 (deletion, Suárez et al. 1991b, 1995) mutations. The suppressor carried by 12r128 does not suppress uaY808 (other alleles not tested).
The cold sensitivity and restricted growth shown by 12r122 and 12r128 stains is shared with known ribosomal translational suppressors (Bratt and Martinelli 1988; Martinelli 1994). We thus tested the above strains for increased sensitivity to translational inhibitors such as cycloheximide and the aminoglucoside paromomycin. Sensitivity to these inhibitors is a component of the ribosomal mutants pleiotropic phenotype (Martinelli 1984, 1994). Figure 5C shows that both strains show sensitivity to paromomycin intermediate between that of the wild type and the suaC109 translational suppressor mutant. 12r122 is as sensitive to cycloheximide as suaC109, while 12r128 is strikingly hypersensitive to this antibiotic (Figure 5C). Additionally, the mutation carried by 12r128 results in complete sterility in homozygous crosses, while that present in 12r122 results in drastically diminished fertility. Crosses with strains carrying uaY-flanking markers, as described above, confirm for both 12r122 and 12r128 strains that suppression of uaY12, restricted growth, cryosensitivity, and hypersensitivity to cycloheximide and paromomycin cosegregate (not shown).
The suppressor mutations carried by both 12r122 and 12r128 are recessive for compact growth, the cryosensitive phenotype, and suppression of uaY12 in diploids heterozygous for the suppressor and homozygous for the uaY12 mutation. The two suppressor mutations fully complement in a homozygous uaY12 diploid, i.e., no suppression of the uaY12 phenotype can be observed. However, no complementation is seen for the restricted growth and cold-sensitive phenotype. It proved impossible to cross strains carrying these mutations to each other, and given the contradictory results of the complementation tests we cannot establish whether they are mutant at the same or at two different loci. We thus called them aas22 and aas28 (for aberrant ATG suppressor), without a locus designation.
DISCUSSION
We propose that the phenotype of uaY12 and its revertants can be accounted for by the current models of translational initiation. The proposed mechanism involves ribosome scanning and the first-AUG rule (Kozak 2002a, 2005 for review), where the eukaryotic small (40s) ribosomal subunits enter at the 5′ end of the messenger and migrate along the mRNA until they reach the first AUG, where they initiate translation. Initiation at the first, upstream AUG, (uAUG) would inhibit initiation of translation on a downstream AUG (first-AUG rule). Translation inhibition of the major downstream ORF has been demonstrated in numerous cases of naturally occurring uORFs (see for review Kozak 2002a). An overlap between the uORF and the major ORF results in a strong inhibition effect, because this situation impairs drastically ribosomal reinitiation of the major ORF (Kozak 2002a and references therein). There are a number of examples, where a newly generated mutant AUG in the 5′-UTR followed by an uORF overlapping the major ORF generates a mutant phenotype (see for review Kozak 2002b).
The uaY12 mutation is a C-to-T transition at position −63, creating an out-of-frame AUG codon, which is followed by a 68-codon ORF (uORFA), ending within the UaY ORF. While we had not measured directly intracellular UaY concentrations, the negative impact of uORFA on the expression of UaY-regulated genes is wholly consistent with a strong diminution of UaY translation as predicted by the model outlined above.
Leaky scanning (see for review Kozak 2002a) seems to occur in the context of the uaY12 mutation. Indeed, the leaky growth on uric acid and the slight inducibility of the UaY-dependent genes in this mutant indicate that the uaY12 mutation results in a low, albeit not nil, intracellular concentration of the UaY protein. This implies that some 40S ribosomal subunits could initiate translation at the downstream physiological AUG. Leaky scanning has been described in A. nidulans at the areA locus where it was proposed that the context GXX AUG (C/U)CX (where X represents any base and AUG is the initiation codon) prevents leaky scanning (Arst and Sheerins 1996). The mutant uaY12 ATG, which apparently allows leaky scanning, is in a sequence context (A UU AUG GGU), which does not fall in the “strong” initiation codon category of Arst and Sheerins (1996), (see Table 2), while it agrees with the consensus context proposed by Balance (1986) and Kozak (1986, 1997).
TABLE 2.
AUG context
| Positions
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Consensusa | −4 | −3 | −2 | −1 | AUG | +4 | +5 | +6 |
| Balance (1986) | C | A | C/A | A/C | AUG | G/U | C | |
| Hamilton et al. (1987) | A/C | A | A | A/C | AUG | U | C | U/C |
| Kozak (1986, 1997) | C | A/G | C | C | AUG | G | ||
| Arst and Sheerins (1996) | G | X | X | AUG | C/U | C | ||
| Positions
|
||||||||
| Flanking sequencesb | −4 | −3 | −2 | −1 | AUG | +4 | +5 | +6 |
| AUG of uaY12 uORFA | C | A | U | U | AUG | G | G | U |
| AUG of uaY ORF | C | G | U | U | AUG | C | U | G |
| uAUG 3 in −44 of uaY | A | G | A | C | AUG | G | U | G |
| uAUG 2 in −122 of uaY | C | U | A | C | AUG | C | G | U |
| uAUG 1 in −338 of uaY | A | G | U | C | AUG | C | G | C |
Consensus sequences flanking the AUG start codon according to different references in the literature.
Sequences flanking the AUG codon of the uaY ORF and of the new uaY12 aberrant AUG (uORFA), together with the three natural AUGs, which are found in the wild-type uaY 5′-UTR at positions −338, −122, and −44.
Reversion of the uaY12 mutation has been obtained by 4-nitroquinoline-1-oxide (NQO) mutagenesis. This mutagen is supposed to affect specifically GC base pairs producing both transitions and transversions (Prakash et al. 1974). Nearly all the reversion mutations obtained affect GC base pairs (18 transitions G to A, 12 transversions G to T, and 8 transversions G to C). Nevertheless, four mutations do not follow this pattern: two affect TA base pairs (transversions T to A), one is 1-bp insertion (a T), and the fourth one is a deletion of 63 bp. The fact that NQO attacks preferentially GC base pairs, may explain why no true reversion was obtained, because this would imply the transition TA to CG, a mutation not described for this mutagen (Prakash et al. 1974).
The elimination of the ATG of uORFA occurs in 38 revertants. All these strains show a wild-type phenotype. In two other revertants (12r18 and 12r3) this uATG is lost, but by a mutation other than a point substitution, they will be discussed separately.
The two intragenic suppressors create a stop codon in frame with the uaY12 aberrant uATG, which shortens the length of uORFA. In these revertants, 12r113 and 12r118, the uORF is 8 and 7 codons long, respectively, rather than the 68 codons found for the uaY12 mutation. Thus, the new uORFAs, at variance with the uaY12 uORFA, do not overlap the UaY ORF, terminating 37 and 40 bp upstream of the UaY ATG, respectively. Suppression afforded by both 12r113 and 12r118 is partial, suggesting defective initiation of the UaY ORF. The fact that a known chain termination suppressor (suaC109) acts as an antisuppressor of uaY12r113 is completely consistent with the proposal that premature chain termination of uORFA, which would occur in this revertant, is responsible for its suppressor phenotype.
An analogous mutation occurs in hereditary thrombocythaemia, due to overproduction of the thrombopoietin (TPO) cytokine, which regulates megakaryopoiesis and platelet production. The human TPO cytokine is normally present at very low levels in the serum because of its low level of translation due to the presence in its 5′-UTR of seven uORFs and essentially to the last uAUG (no. 7), which overlaps with the TPO ORF (Ghilardi et al. 1998). In hereditary thrombocythaemia, the increase production of TPO is due to a mutation introducing a stop codon within the upstream uORF no. 7 with the consequence that uORF 7 terminates 31 nucleotides before the TPO ATG and thus no longer overlaps with it (Ghilardi et al. 1999).
The efficiency of ribosomal reinitiation relies on several constraints such as the size of the uORF and the distance between the two ORFs (Kozak 1987; Luukkonen et al. 1995). Luukkonen et al. (1995) have shown that an intercistronic distance shorter than 37 nucleotides acts negatively on the reinitiation frequency at a downstream AUG. The suppression of the uaY12 mutation in the 12r113 revertant is less marked than in the 12r118 strain, the only difference between these two strains being the distance between the stop suppressor and the UaY AUG. We have no obvious explanations of the fact that suppression in both strains is less efficient at 25° than at 37°. It is possible that this is only a trivial consequence of physiological differences of requirements for purine utilization between these two temperatures, but it may also reveal more interesting differences in the efficiency of the translational machinery (in the reading of the physiological AUG) or in the transcriptional machinery (a lower requirement for UaY at 37° in UaY-dependent promoters).
Revertants 12r122 and 12r128 carry suppressor mutations segregating independently from the uaY locus. The pleiotropic nature of these suppressors, including their hypersensitivity to translational inhibitors, is consistent with their mapping in genes coding for proteins of the translational machinery, perhaps ribosomal proteins or translation initiation factors, and further supports our hypothesis that translation of uORFA is responsible for the uaY12 phenotype.
The determination of the uaY mRNA start points (see results) shows a 284- to 503-long 5′-UTR, unusually long for A. nidulans. Analysis of the uaY 5′-UTR reveals the presence of three upstream naturally occurring AUG in position −338, −122, and −44, respectively (to be called, respectively, uAUG1, uAUG2, and uAUG3; Table 2, Figure 6, and see supplemental Figure 1), followed by short ORFs of 16, 24, and 16 codons, respectively, the last one overlapping with the UaY ORF on four nucleotides (to be called, respectively, uORF1, uORF2, and uORF3). The uORF1 might not be present in all the uaY messengers, as the shorter mRNA begins at position −284. It can be proposed that these ORFs result in physiological inhibition of UaY translation, which is consistent with the strictly limiting concentrations of UaY present in the cell as discussed in Scazzocchio et al. (1982) and Scazzocchio (1994). It may be relevant that overexpression of UaY, in the presence of its presumed ligand, uric acid, is lethal for the organism (N. Oestreicher, unpublished results).
Figure 6.—
Sequence of the uaY12r18 and uaY12r3 alleles. The DNA and translated peptide sequence of the relevant portion of the uaY gene are shown for the uaY12r18, uaY12r3, and uaY+ alleles. Numbers indicate nucleotide position (+1 corresponding to the A of the uaY ATG). The uaY ATG and the wild-type NH2 terminal sequence of the UaY protein are given in boldface type, the ATG being in a larger type. The ATGs of uATG2 and uATG3, respectively, at −122 and −44 (see text) are underlined and in a larger type. uATG3 is deleted in uaY12r18. The inserted T between positions −63 and −62 in the uaY12r3 allele is in boldface type, underlined, and in a larger type.
There are numerous examples where upstream ORFs are part of the physiological mechanism, which limits translation (Kozak 2002a,b). Two of our revertant alleles (uaY12r18 and uaY12r3) suggest that this process may actually occur in the uaY wild-type gene. The uaY12r18 mutation deletes nucleotides −91 to −29, eliminating the uaY12 uORFA but also uORF3, thus bringing the uaY AUG closer to a new ORF, which contains 16 of the 24 codons of uORF2 (starting at −122 in uaY+ but at −58 in uaY12r18). This new ORF of 21 codons (to be called uORFN), overlaps by four nucleotides with the uaY ORF (Figure 6). In this revertant, the induced transcription of the UaY-dependent genes is actually higher than in the wild type (Figure 3), without any increase in the uaY mRNA itself. We can speculate that this phenotype reflects a more efficient translation of the UaY protein. This, in turn, may reflect that the AUG of uORFN (which is obviously identical to that of uORF2, Figure 6B, Table 2, and see supplemental Figure 1) is in a less favorable context than the AUG of uORF3, this allowing more efficient initiation of translation by leaky scanning of the UaY AUG. In revertant uaY12r3, a T insertion eliminates the uORFA present in the uaY12 allele, but also places uORF2 in the same reading frame as the UaY ORF, potentially lengthening the UaY ORF by 41 residues. In this revertant the transcription of UaY-dependent genes (hxA and uaZ) is lower than in the wild type (Figure 3). This may be due to less favorable context for the AUG of uORF2 compared with the physiological UaY AUG (Table 2) and/or to less efficient UaY function due to the N-terminal additional sequences.
The data presented above illustrate the use of an A. nidulans transcriptional regulatory system whose exquisite sensitivity to the intracellular concentrations of the cognate transcription factor, results in mutant phenotypes that can be most simply interpreted making use of the first-AUG rule and the occurrence of leaky scanning. Moreover, the isolation of two extragenic suppressors of uaY12 phenotype could be useful to identify factors involved in the initiation of translation in this organism.
Acknowledgments
We thank H. M. Sealy-Lewis for kindly providing some A. nidulans strains. N.O. thanks C. Velot to have allowed this work to be completed in his laboratory. This work was supported by the Centre National de la Recherche Scientifique, the Université Paris-Sud, and the Institut Universitaire de France.
References
- Amrani, L., G. Cecchetto, C. Scazzocchio and A. Glatigny, 1999. The hxB gene, necessary for post-translational activation of purine hydroxylases in Aspergillus nidulans, is independently controlled by the purine utilization and the nicotinate utilization transcriptional activating systems. Mol. Microbiol. 31 1065–1073. [DOI] [PubMed] [Google Scholar]
- Arst, H. N. Jr., and D. J. Cove, 1973. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126 111–141. [DOI] [PubMed] [Google Scholar]
- Arst, H. N. Jr., and A. Sheerins, 1996. Translational initiation competence, ‘leaky scanning’ and translational reinitiation in areA mRNA of Aspergillus nidulans. Mol. Microbiol. 19 1019–1024. [DOI] [PubMed] [Google Scholar]
- Bal, J., E. M. Kajtaniak and N. I. Pieniazek, 1977. 4-nitroquinoline-1-oxide, a good mutagen for Aspergillus nidulans. Mutat. Res. 56 153–156. [Google Scholar]
- Balance, D. J., 1986. Sequences important for gene expression in filamentous fungi. Yeast 2 229–236. [DOI] [PubMed] [Google Scholar]
- Bratt, R., and S. D. Martinelli, 1988. Every ribosomal suppressor mutation in Aspergillus nidulans has a unique and highly pleiotropic phenotype. Curr. Genet. 14 29–36. [DOI] [PubMed] [Google Scholar]
- Cecchetto, G., S. Amillis, G. Diallinas, C. Scazzocchio and C. Drevet, 2004. The AzgA purine transporter of Aspergillus nidulans. Characterization of a protein belonging to a new phylogenetic cluster. J. Biol. Chem. 279 3132–3141. [DOI] [PubMed] [Google Scholar]
- Cultrone, A., Y. Reyes Dominguez, C. Drevet, C. Scazzocchio and R. Fernández-Martin, 2007. The tightly regulated promoter of the xanA gene of Aspergillus nidulans is included in a helitron. Mol. Microbiol. 63 1577–1587. [DOI] [PubMed] [Google Scholar]
- Diallinas, G., L. Gorfinkiel, H. N. Jr Arst, G. Cecchetto and C. Scazzocchio, 1995. Genetic and molecular characterisation of a gene encoding a wide-specificity purine permease of Aspergillus nidulans reveals a novel family of transporters conserved in procaryotes and in eucaryotes. J. Biol. Chem. 270 8610–8622. [DOI] [PubMed] [Google Scholar]
- Espeso, E. A., L. Cobeno and H. N. Arst, Jr, 2005. Discrepancies between recombination frequencies and physical distances in Aspergillus nidulans: implications for gene identification. Genetics 171 835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidel, S., J. H. Doonan and N. R. Morris, 1988. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a γ-actin. Gene 70 283–293. [DOI] [PubMed] [Google Scholar]
- Ghilardi, N., A. Wiestner and R. C. Skoda, 1998. Thrombopoietin production is inhibited by a translational mechanism. Blood 92 4023–4030. [PubMed] [Google Scholar]
- Ghilardi, N., A. Wiestner, M. Kikuchi, A. Ohsaka and R. C. Skoda, 1999. Hereditary thrombocythaemia in a Japanese family is caused by a novel point mutation in the thrombopoietin gene. Br. J. Haematol. 107 310–316. [DOI] [PubMed] [Google Scholar]
- Gill, G., and M. Ptashne, 1988. Negative effect of the transcriptional activator GAL4. Nature 333 721–724. [DOI] [PubMed] [Google Scholar]
- Glatigny, A., and C. Scazzocchio, 1995. Cloning and molecular characterization of hxA, the gene coding for the xanthine dehydrogenase (purine hydroxylase I) of Aspergillus nidulans. J. Biol. Chem. 270 3534–3550. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel, L., G. Diallinas and C. Scazzocchio, 1993. Sequence and regulation of the uapA gene, encoding a uric acid-xanthine permease in the fungus Aspergillus nidulans. J. Biol. Chem. 268 23376–23381. [PubMed] [Google Scholar]
- Hamilton, R., C. Watanabe and H. A. De Boer, 1987. Compilation and comparison of the sequence context around the AUG start codons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 15 3571–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D., 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166 557–580. [DOI] [PubMed] [Google Scholar]
- Kozak, M., 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak, M., 1987. Effects of intercistronic length on the efficiency of reinitiation by eukaryotic ribosomes. Mol. Cell Biol. 7 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M., 1997. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in position +5 and +6. EMBO J. 16 2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M., 2002. a Pushing the limits of the scanning mechanism for initiation of translation. Gene 299 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M., 2002. b Emerging links between initiation of translation and human disease. Mamm. Genome 13 401–410. [DOI] [PubMed] [Google Scholar]
- Kozak, M., 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361 13–37. [DOI] [PubMed] [Google Scholar]
- Kudla, B., M. X. Caddick, T. Langdon, N. Martinez-Rossi, C. F. Bennett et al., 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockington, R. A., H. M. Sealy-Lewis, C. Scazzocchio and R. W. Davies, 1985. Cloning and characterization of the ethanol utilization regulon in Aspergillus nidulans. Gene 33 137–149. [DOI] [PubMed] [Google Scholar]
- Luukkonen, B. G. M., W. Tan and S. Schwartz, 1995. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 69 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli, S. D., 1984. Interactions of ribosomal antibiotics and informational suppressors of Aspergillus nidulans. J. Gen. Microbiol. 130 575–582. [DOI] [PubMed] [Google Scholar]
- Martinelli, S. D., 1994. Translational suppression, pp. 733–762 in Aspergillus nidulans: 50 Years On, edited by S. D. Martinelli and J. R. Kinghorn, Elsevier Scientific, Amsterdam.
- Martinelli, S. D., and T. J. Roberts, 1983. Diagnosis of nonsense mutations in Aspergillus nidulans. Biosci. Rep. 3 11–17. [DOI] [PubMed] [Google Scholar]
- Martinelli, S. D., T. J. Roberts, H. M. Sealy-Lewis and C. Scazzocchio, 1984. Evidence for a nonsense mutation in the niaD locus of Aspergillus nidulans. Genet. Res. 43 241–248. [DOI] [PubMed] [Google Scholar]
- Miller, B. L., K. Y. Miller and W. E. Timberlake, 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell Biol. 5 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreicher, N., and C. Scazzocchio, 1993. Sequence, regulation and mutational analysis of the gene encoding urate oxidase in Aspergillus nidulans. J. Biol. Chem. 268 23382–23389. [PubMed] [Google Scholar]
- Oestreicher, N., H. M. Sealy-Lewis and C. Scazzocchio, 1993. Characterization, cloning and integrative properties of the gene encoding for the urate oxidase in Aspergillus nidulans. Gene 132 185–192. [DOI] [PubMed] [Google Scholar]
- Oestreicher, N., C. Scazzocchio and T. Suárez, 1997. Mutations in a dispensable region of the UaY transcription factor of Aspergillus nidulans differentially affect the expression of structural genes. Mol. Microbiol. 24 1189–1199. [DOI] [PubMed] [Google Scholar]
- Oestreicher, N., C. Ribard and C. Scazzocchio, 2008. The nadA gene of Aspergillus nidulans, encoding adenine deaminase, is subject to a unique regulatory pattern. Fungal Genet. Biol. 45 760–775. [DOI] [PubMed] [Google Scholar]
- Prakash, L., J. W. Stewart and F. Sherman, 1974. Specific induction of transitions and transversions of G-C base pairs by 4-nitroquinoline-1-oxide in iso-1-cytochrome c mutants of yeast. J. Mol. Biol. 85 51–65. [DOI] [PubMed] [Google Scholar]
- Ravagnani, A., L. Gorfinkiel, T. Langdon, E. Adjadj, S. Demais et al., 1997. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 16 3974–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribard, C., C. Scazzocchio and N. Oestreicher, 2001. The oxpA5 mutation of Aspergillus nidulans is an allele of adB, the gene encoding adenylosuccinate synthetase. Mol. Genet. Genomics 266 701–710. [DOI] [PubMed] [Google Scholar]
- Ribard, C., M. Rochet, B. Labedan, B. Daignan-Fornier, P. Alzari et al., 2003. Sub-families of α/β barrel enzymes: a new adenine deaminase family. J. Mol. Biol. 334 1117–1131. [DOI] [PubMed] [Google Scholar]
- Roberts, T., S. Martinelli and C. Scazzocchio, 1979. Allele specific, gene unspecific suppressors in Aspergillus nidulans. Mol. Gen. Genet. 177 57–64. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanger, F., S. Nicklens and A. R. Coulson, 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 70 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio, C., 1994. The purine degradation pathway: genetics, biochemistry and regulation, pp. 221–257 in Aspergillus nidulans: 50 Years On, edited by S. D. Martinelli and J. R. Kinghorn. Elsevier Scientific, Amsterdam. [PubMed]
- Scazzocchio, C., and A. J. Darlington, 1968. The induction and repression of the enzymes of purine breakdown in Aspergillus nidulans. Biochim. Biophys. Acta 166 557–568. [DOI] [PubMed] [Google Scholar]
- Scazzocchio, C., N. Sdrin and G. Ong, 1982. Positive regulation in a eukaryote, a study of the uaY gene of Aspergillus nidulans: I. Characterization of alleles, dominance and complementation studies, and a fine structure map of the uaY–oxpA cluster. Genetics 100 185–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht, C. A., C. C. DiRusso, C. P. Novotny and R. C. Ullrich, 1982. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal. Biochem. 119 158–163. [DOI] [PubMed] [Google Scholar]
- Stankovich, M., A. Platt, M. X. Caddix, T. Langdon, P. M. Schaffer et al., 1993. C-terminal truncation of the transcriptional activator encoded by areA in Aspergillus nidulans results in both loss-of-function and gain-of-function phenotypes. Mol. Microbiol. 7 81–87. [DOI] [PubMed] [Google Scholar]
- Suárez, T., N. Oestreicher, J. Kelly, G. Ong, T. Sankarsingh et al., 1991. a The uaY positive control gene of Aspergillus nidulans: Fine structure, isolation of constitutive mutants and reversion patterns. Mol. Gen. Genet. 230 359–368. [DOI] [PubMed] [Google Scholar]
- Suárez, T., N. Oestreicher, M. A. Penalva and C. Scazzocchio, 1991. b Molecular cloning of the uaY regulatory gene of Aspergillus nidulans reveals a favoured region for DNA insertions. Mol. Gen. Genet. 230 369–375. [DOI] [PubMed] [Google Scholar]
- Suárez, T., M. V. de Queiroz, N. Oestreicher and C. Scazzocchio, 1995. The sequence and binding specificity of UaY, the specific regulator of the purine utilization pathway in Aspergillus nidulans, suggest an evolutionary relationship with the PPR1 protein of Saccharomyces cerevisiæ. EMBO J. 14 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilburn, J., C. Scazzocchio, G. G. Taylor, J. H. Zabicky-Zissman, R. A. Lockington et al., 1983. Transformation by integration in Aspergillus nidulans. Gene 26 205–221. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron, C., J. Vieira and J. Messing, 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33 103–119. [DOI] [PubMed] [Google Scholar]