Abstract
Lentiviruses, including HIV-1, have transmembrane envelope (Env) glycoproteins with cytoplasmic tails that are quite long compared with those of other retroviruses. However, mainly because of the lack of biochemical studies performed in cell types that are targets for HIV-1 infection, no clear consensus exists regarding the function of the long lentiviral Env cytoplasmic tail in virus replication. In this report, we characterize the biological and biochemical properties of an HIV-1 mutant lacking the gp41 cytoplasmic tail. We find that the gp41 cytoplasmic tail is necessary for the efficient establishment of a productive, spreading infection in the majority of T cell lines tested, peripheral blood mononuclear cells, and monocyte-derived macrophages. Biochemical studies using a high-level, transient HIV-1 expression system based on pseudotyping with the vesicular stomatitis virus glycoprotein demonstrate that in HeLa and MT-4 cells, mutant Env incorporation into virions is reduced only 3-fold relative to wild type. In contrast, gp120 levels in virions produced from a number of other T cell lines and primary macrophages are reduced more than 10-fold by the gp41 truncation. The Env incorporation defect imposed by the cytoplasmic tail truncation is not the result of increased shedding of gp120 from virions or reduced cell-surface Env expression. These results demonstrate that in the majority of T cell lines, and in primary cell types that serve as natural targets for HIV-1 infection in vivo, the gp41 cytoplasmic tail is essential for efficient Env incorporation into virions.
The HIV-1 envelope (Env) glycoprotein is synthesized as a precursor, gp160, that is proteolytically cleaved by a cellular protease during transport to the cell surface (1, 2). Cleavage of gp160 produces the two components of the mature Env glycoprotein complex: the surface Env glycoprotein gp120 and the transmembrane Env glycoprotein gp41. After its arrival at the plasma membrane, the gp120/gp41 complex is incorporated into nascent, budding virus particles by a mechanism that remains poorly understood.
Lentiviruses, including HIV-1, encode transmembrane glycoproteins with cytoplasmic tails (CTs) that are quite long compared with those of other retroviruses. For example, the CT of the HIV-1 and HIV-2 transmembrane glycoproteins are generally around 150 residues in length; in contrast, those of the avian and murine oncoretroviruses are typically 20–30 aa long. Although the role of the long lentiviral Env CT has been the focus of a number of studies, no clear consensus has emerged regarding the function of this unique domain. Some studies observed that deletions of more than 19 (3) or 43 (4) residues from the gp41 C terminus blocked virus infectivity; this phenotype was attributed in part to reduced Env incorporation into virions. Another study (5) observed that the gp41 CT played no major role in Env incorporation, but was required for virus infectivity. Wilk et al. (6) reported that a mutant lacking essentially all of the gp41 CT efficiently established a spreading infection in the MT-4 T cell line. It is important to note that the role of the gp41 CT in Env incorporation generally has been assessed in transfected COS, CV-1, or HeLa cells, rather than in cells that are targets for productive, spreading HIV-1 infections. Although a variety of biochemical (7, 8) and genetic (9–11) data support the existence of an interaction between HIV-1 matrix (MA) and the gp41 CT, understanding the requirements for Env incorporation into HIV-1 virions is complicated by the finding that a variety of heterologous retroviral (12–14) and nonretroviral (15, 16) glycoproteins, as well as a number of host cell surface proteins (17), are incorporated into budding HIV-1 particles. In addition to potential roles in Env incorporation and virus infectivity, the gp41 CT contains determinants that promote rapid Env internalization (18–20), direct basolateral virus release in polarized epithelial cells (21), induce pore formation in membranes (22, 23), and interact with calmodulin (24).
We previously observed that the majority of CT truncation mutations did not block Env incorporation in transfected HeLa cells (9, 10). Relatively small truncations caused substantial defects in virus infectivity in the single-cycle multinuclear activation of a galactosidase indicator (MAGI) assay, whereas larger truncations that removed the majority of the CT had no measurable effect on infectivity in this single-cycle assay. Preliminary results suggested that gp41 truncation mutations affected the establishment of a productive, spreading infection in T cell lines in a cell type-dependent manner (10).
In this report, we characterize the biological and biochemical properties of an HIV-1 mutant lacking the gp41 CT. Analysis of virus replication in a range of T cell lines and in peripheral blood mononuclear cells (PBMC) and monocyte-derived macrophages (MDM) indicates that in most T cell lines and primary cells the gp41 CT is essential for the efficient establishment of a productive, spreading infection. However, the long CT is not required for virus replication in MT-4 cells. Intriguingly, we observe that the gp41 CT is essential, in a cell type-specific manner, for efficient Env incorporation into virions; the cell type dependence of this requirement matches the biological effect of gp41 truncation.
Materials and Methods
Cells, Viruses, and Plasmids.
CEM (12D-7), Jurkat, H9, MT-4, 293T, HeLa, and MAGI cells were maintained as described (25). MT-2 and SupT1 cells were cultured in RPMI medium 1640 supplemented with 10% FBS, 2 mM Gln, and antibiotics. The isolation and culture of human PBMC and primary human MDM has been described in detail (9). For infection of T cell lines, PBMC, and MAGI cells, virus was obtained by transfecting HeLa cells with the T cell line tropic molecular clone pNL4–3 (26) and the derivative containing the CTdel-144–2 mutation. For macrophage infectivity analyses, we used the macrophage-tropic molecular clone pNL(AD8) (9) and the pNL(AD8)CTdel-144–2 derivative. Because replication of NL4–3 is restricted in macrophages, this virus was included as a negative control (9). We used the following Env expression plasmids: for HIV-1 Env, pIIINL4env and pIIINL4envCTdel-144; for vesicular stomatitis virus glycoprotein (VSV-G), pHCMV-G (16) (kindly provided by J. Burns, University of California, San Diego). The luciferase-expressing clone pNLuc was constructed as described (25).
Mutagenesis and Construction of Plasmids.
The CTdel-144–2 mutation was introduced as follows: a template for oligonucleotide-directed mutagenesis was constructed by cloning the BamHI–KpnI fragment from pNL4–3 (nucleotides 8465–9005) into M13 mp18. Two back-to-back stop codons then were introduced 143 and 144 codons from the C terminus of gp41 by using the oligonucleotide 5′-GGATATTGATGATTATCG-3′ and previously reported methods (27). The mutagenized fragment then was recloned into pNL4–3 and sequenced in its entirety. The macrophage-tropic version of CTdel-144–2 was constructed by cloning the 0.4-kbp BsmI–BamHI fragment (nucleotides 8044–8465) from pNL4–3CTdel-144–2 into the macrophage-tropic molecular clone pNL(AD8) (9). The Env expression vectors pIIINL4env and pIIINL4envCTdel-144–2 were constructed by cloning the 2.5-kbp KpnI–XhoI (pNL4–3 nucleotides 6343–8887) fragment from pNL4–3 or pNL4–3CTdel-144–2, respectively, into pIIIenv3.1 (28). The di-Lys endoplasmic reticulum retrieval signal Lys-X-Lys-X-X (29) was introduced at the gp160 C terminus by converting the wild-type (wt) sequence to Lys-Lys-Lys-Leu-Leu. This was performed by site-directed mutagenesis using the oligonucleotide 5′-GGCTTGAAAAAGAAGAAATTGCTA-3′.
Transfections and Infections.
Virus stocks of NL4–3, NL(AD8), CTdel-144–2 mutant derivatives, and VSV-G pseudotypes were obtained by transfecting HeLa cells by using the calcium phosphate precipitation method (30, 31). Pseudotyped virus stocks of pNLuc were prepared by cotransfection of 293T cells with HIV-1 Env expression vectors as described (25). Transfected cell supernatants were harvested, filtered, normalized for reverse transcriptase (RT) activity, and used in infections as indicated. Infections of T cell lines, PBMC, and MDM were performed as described (9, 25). RT assays were performed as reported (9).
Metabolic Labeling and Radioimmunoprecipitation.
For T cell lines and HeLa cells, 5 × 106 or 2 × 106 cells, respectively, were infected with 1 ml of VSV-G-pseudotyped HIV-1 in the presence of 1 μM of the CXCR4 inhibitor T22 (32). The next day, infected cells were plated in 5 ml (2 ml for HeLa cells) of Cys-free RPMI medium supplemented with 10% FBS and 1 μM T22 in 25-cm2 flasks. After addition of [35S]Cys (500 μci), the cultures were incubated for 16 h at 37°C. Preparation of cell lysates, pelleting of labeled virions in an ultracentrifuge, and immunoprecipitation of cell- and virion-associated proteins with AIDS patient sera (HIV-1 neutralizing sera; obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, catalogue nos. 1983 and 1984) have been described (33). Quantitative analysis of bands visualized by radioimmunoprecipitation was performed with a FujiX BAS2000 Bio-image analyzer. Metabolic labeling and radioimmunoprecipitation of MDM was performed as described (34).
Western Blot Analysis.
Proteins were separated by 10% SDS/PAGE and transferred to poly(vinylidine difluoride) membranes (Millipore) by using a semidry blotter. For biotinylation experiments, a passive diffusion method (35) was used. Membranes were incubated with one of the following primary antibodies: a rabbit anti-gp120 antibody (a kind gift of K. Strebel, National Institutes of Health), the anti-gp41 mAb T32 (36) (a kind gift of P. Earl, National Institutes of Health), or AIDS patient sera. Subsequently, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia). Antigens recognized by antibodies were detected by enhanced chemiluminescence (Amersham Pharmacia).
Cell Surface Biotinylation.
Cell-surface proteins were biotinylated essentially as described (37) except that 0.25 mg/ml of sulfo-N-hydroxysuccinimide-biotin was used. After biotinylation, lysates immunoprecipitated with AIDS patient sera or precipitated with neutravidin-agarose beads (Pierce) were subjected to Western blotting with rabbit anti-gp120 polyclonal antibody as described above.
Flow Cytometric Analysis of HIV-1 Env Surface Expression.
CEM (12D-7) cells (106) that were uninfected or infected with VSV-G-pseudotyped HIV-1 were incubated for 1 h at 4°C with 12.5 μg/ml of the T32 anti-gp41 mAb. After incubation with this primary antibody, cells were washed twice with PBS supplemented with 1% FBS and incubated with FITC-conjugated goat anti-mouse IgG for 30 min at 4°C. After incubation with secondary antibody, the cells were washed once with Dulbecco's PBS supplemented with 1% FBS and once with PBS and resuspended in 1% formaldehyde in PBS. Cell-surface expression of gp41 was analyzed with a FACScan flow cytometer (Becton Dickinson).
Single-Cycle, Luciferase-Based Infectivity Assay.
The pNLuc molecular clone (25) was cotransfected into 293T cells with HIV-1 Env expression vectors (pIIINL4env or pIIINL4envCTdel-144–2). Stocks were normalized for RT activity and used to infect MT-4 or CEM (12D-7). Luciferase activity was measured 2 days postinfection as described (25).
Results
Replication of a gp41 CT Truncation Mutant Is Cell-Type Dependent.
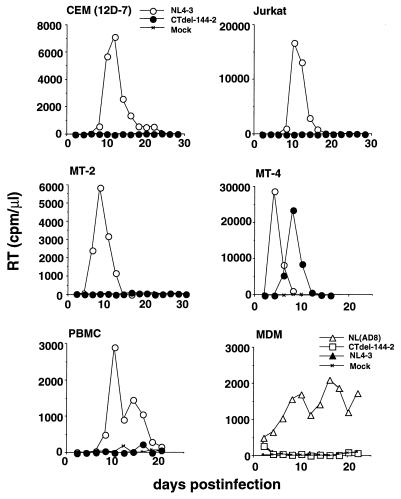
To characterize the role of the gp41 CT in HIV-1 replication, we constructed a version of the infectious pNL4–3 HIV-1 molecular clone in which two stop codons were introduced into the sequence encoding the amino acids 144 and 143 residues from the C terminus of gp41. This mutant molecular clone, pNL4–3(CTdel-144–2), thus expresses a truncated Env glycoprotein lacking all but 6 aa of the gp41 CT. We observed that the CTdel-144–2 mutant readily established a productive, spreading infection in MT-4 cells, consistent with the results of Wilk et al. (6) using a similar mutant. In marked contrast, the gp41 CT deletion mutation blocked detectable virus replication in the other T cell lines tested: CEM (12D-7), Jurkat, and MT-2 (Fig. 1), H9, and SupT1 (data not shown).
Figure 1.
Replication kinetics of viruses containing the CTdel-144–2 mutation. Virus stocks, obtained by transfection of HeLa cells with the indicated molecular clones, were normalized for RT activity and used to infect T cell lines [CEM (12D-7), Jurkat, MT-2, and MT-4], primary PBMC, or MDM. RT activity was monitored in the culture supernatant over time. T cell line and PBMC infections were performed with the T cell line tropic molecular clone NL4–3 expressing wt or CTdel-144–2-mutant Env; MDM infections were performed with the macrophage-tropic molecular clone NL(AD8) expressing wt or CTdel-144–2-mutant Env. NL4–3, which fails to productively infect MDM, was included in the macrophage infections as a negative control. The results are representative of at least two independent experiments.
To extend these results to primary cell types that constitute natural targets for HIV-1 infection in vivo, we tested the ability of CTdel-144–2 to replicate in phytohemagglutinin-stimulated human PBMC and MDM (Fig. 1). To perform the macrophage experiments, we introduced the CTdel-144–2 mutation into the macrophage-tropic pNL4–3 derivative pNL(AD8) (9, 33). The CTdel-144–2 mutation blocked the establishment of a spreading infection in both of these primary cell types. Thus, truncation of the gp41 CT abolishes virus replication in the majority of T cell lines examined and in primary PBMC and MDM.
The CTdel-144–2 Mutation Does Not Block an Early Stage of the Virus Replication Cycle.
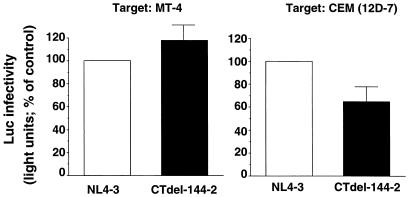
To determine the efficiency of the early events in virus replication in cells that were “nonpermissive” for gp41 truncation, we used an env-minus pNL4–3 derivative, pNLuc, which encodes the luciferase gene in the place of nef (25). 293T cells, which were permissive for the gp41 truncation (data not shown), were cotransfected with pNLuc and vectors expressing either the wt NL4–3 Env glycoprotein or the CTdel-144–2 mutant derivative. The pseudotyped virions released from the transfected cells were harvested, normalized for RT activity, and used to infect T cell lines. Two days postinfection, cells were lysed and luciferase activity was determined. This assay provides a quantitative measure of virus infectivity in a single round of infection (i.e., in the absence of virus spread). The CTdel-144–2 mutant showed more than 60% of wt infectivity both in MT-4 and CEM (12D-7) cells (Fig. 2). Similar results were observed in Jurkat cells (data not shown). These results indicate that the block imposed by truncation of the gp41 CT is not at an early stage of the virus replication cycle.
Figure 2.
Effect of the CTdel-144–2 mutation on infectivity in single-round assays. Relative infectivies of wt (NL4–3) and CTdel-144–2 in MT-4 and CEM (12D-7) cells. Virus preparations were obtained by cotransfecting 293T cells with a luciferase-expressing molecular clone (pNLuc) and HIV-1 Env expression vectors (pIIINL4env or pIIINL4envCTdel-144–2). Stocks were normalized for RT activity and used to infect MT-4 or CEM(12D-7). Luciferase activity was measured 2 days postinfection (Materials and Methods). Data presented are averages of at least two assays, ±SD.
The CTdel-144–2 Mutation Markedly Reduces Levels of gp120 in Virions Produced by Nonpermissive T Cell Lines and MDM.
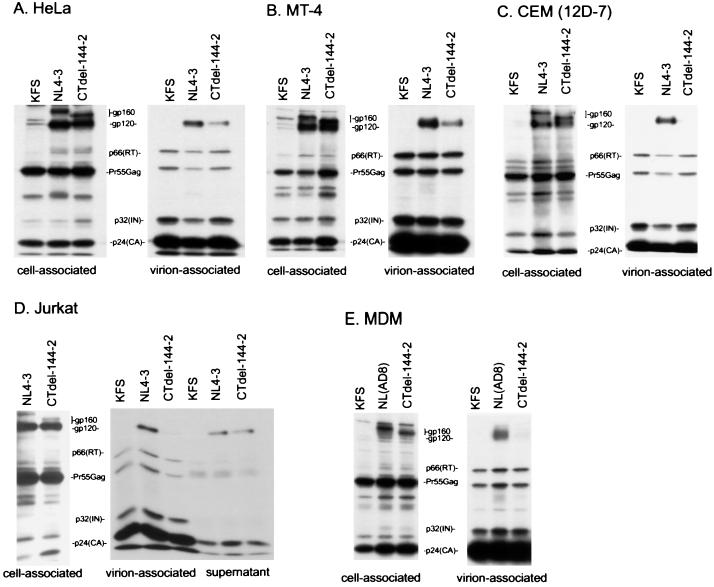
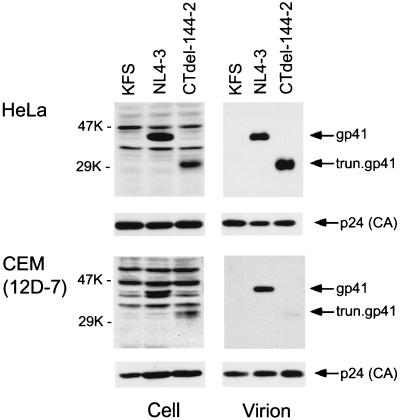
To identify the defect caused by gp41 truncation in nonpermissive T cell lines, we examined the effect of the CTdel-144–2 mutation on Env incorporation. Because T cell lines generally cannot be transfected with efficiencies required for biochemical analysis of Env incorporation, we used a high-level, transient HIV-1 expression system based on pseudotyping with VSV-G (38, 39). HeLa cells were cotransfected with a VSV-G expression vector (16) and either wt pNL4–3 or the CTdel-144–2 mutant derivative. The env-minus molecular clone pNL4–3KFS (9) was used as a negative control. VSV-G-pseudotyped virus stocks were harvested and used at high multiplicity of infection to infect a variety of target cells, including HeLa, MT-4, CEM (12D-7), and Jurkat. One day postinfection, cells were metabolically labeled overnight with [35S]Cys, virions were pelleted in an ultracentrifuge, and cell and virion lysates were prepared and immunoprecipitated with AIDS patient sera. In some assays, virion-free gp120 released into the medium was recovered from the supernatant of the ultracentrifuge spin by immunoprecipitation. In HeLa and MT-4 cells, the CTdel-144–2 mutation had a relatively minor impact on Env incorporation; gp120 was detected at a level that was reduced only 3-fold relative to wt (Fig. 3 A and B). In contrast, gp120 levels in virions produced from nonpermissive cell lines [e.g., CEM (12D-7) and Jurkat] and primary MDM were reduced more than 10-fold by the gp41 truncation (Fig. 3 C–E). As observed previously, the gp120 derived from MDM appeared as a broad band because of modification with lactosaminoglycans (34).
Figure 3.
Radioimmunoprecipitation analysis of the wt (NL4–3) and the CTdel-144–2 mutant. HeLa (A), MT-4 (B), CEM (12D-7) (C), or Jurkat (D) cells were infected with VSV-G-pseudotyped KFS (Env-minus), NL4–3, or CTdel-144–2 and metabolically labeled overnight with [35S]Cys. Virion-associated material was obtained by pelleting the infected cell supernatant in an ultracentrifuge; lysates derived from cell- and virion-associated material were immunoprecipitated with AIDS patient sera (see Materials and Methods). In Jurkat cells (D), virus-free supernatant obtained from the ultracentrifuge spin also was immunoprecipitated. (E) Radioimmunoprecipitation analysis of MDM infected with the VSV-G-pseudotyped, macrophage-tropic molecular clone NL(AD8) expressing wt or CTdel-144–2-mutant Env. Radioimmunoprecipitation was performed as described above except metabolic labeling was performed with [35S]Met. The positions of the Env precursor gp160, the mature surface glycoprotein gp120, p66(RT), the Gag precursor Pr55Gag, p32(IN), and p24(CA) are indicated. The results are representative of at least two independent experiments.
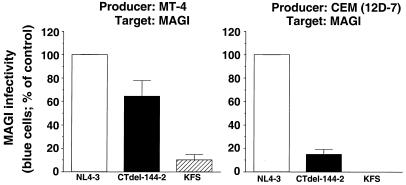
To test the impact of the Env incorporation phenotypes described above on virus infectivity, permissive (MT-4) or nonpermissive [CEM (12D-7)] cells were infected with VSV-G-pseudotyped NL4–3 or NL4–3CTdel-144–2. The infectivity of virus produced in these T cell lines then was measured in the MAGI assay. The results demonstrated that CTdel-144–2 virus produced from MT-4 cells displayed an infectivity that was approximately 60% that of wt. In contrast, the CTdel-144–2 mutation reduced infectivity approximately 10-fold when virus was produced in CEM (12D-7) cells (Fig. 4). These results suggest that the replication defect imposed by the CTdel-144–2 mutation in nonpermissive cells can be attributed to impaired Env incorporation.
Figure 4.
Relative MAGI infectivity of virions derived from MT-4 (Left) or CEM (12D-7) (Right) cells. Virus preparations were obtained by cotransfecting HeLa cells with HIV-1 molecular clones (pNL4–3, pNL4–3CTdel-144–2, or pNL4–3KFS) and a VSV-G expression vector. Stocks were normalized for RT activity and used to infect MT-4 and CEM (12D-7) cells. Two days postinfection, the infectivity of NL4–3, NL4–3CTdel-144–2, or NL4–3KFS virus recovered from the T cell line supernatant was determined by MAGI assay (Materials and Methods). Data presented are averages of at least two assays, ±SD.
The Low Levels of gp120 in CTdel-144–2 Mutant Virions Produced by Nonpermissive Cells Are Not Caused by Increased gp120 Shedding.
The data presented in Fig. 3 do not exclude the possibility that the apparent Env incorporation defect imposed by the CTdel-144–2 mutation is caused by increased shedding of gp120 from virions after incorporation. To address this possibility, we infected cells with VSV-G-pseudotyped virions as described above and examined the levels of cell- and virion-associated gp41 by Western blot analysis using an anti-gp41 mAb. In parallel, the amount of viral Gag proteins present in cell and virion lysates was determined by blotting with AIDS patient sera. In virions derived from HeLa cells, comparable levels of full-length and truncated gp41 were observed, confirming the lack of a major Env incorporation defect in HeLa cells (Fig. 5 Upper). In striking contrast, CTdel-144–2 virions produced in CEM (12D-7) cells contained a barely detectable amount of gp41 despite the fact that cell-associated levels of wt and mutant gp41 were comparable (Fig. 5 Lower). We also immunoprecipitated the soluble (virion-free) gp120 released from the surface of Env-expressing, nonpermissive cells; levels of gp120 released from wt- and mutant-Env expressing cells were comparable (Fig. 3D, supernatant). These results demonstrate that truncation of the gp41 CT severely impairs Env incorporation in nonpermissive cells.
Figure 5.
Analysis of gp41 incorporation by Western blotting. Cell and virion lysates were prepared from HeLa or CEM (12D-7) cells infected with VSV-G-pseudotyped KFS, NL4–3, or CTdel-144–2. Samples were transferred to poly(vinylidine difluoride) membranes and blotted with an anti-gp41 mAb (T32) and reprobed with AIDS patient sera to detect p24 (CA) (Materials and Methods). The positions of full-length and truncated (trun.) gp41 and p24 (CA) are shown. The results are representative of duplicate experiments.
The CTdel-144–2 Mutation Does Not Reduce Cell Surface Env Expression in Nonpermissive Cells.
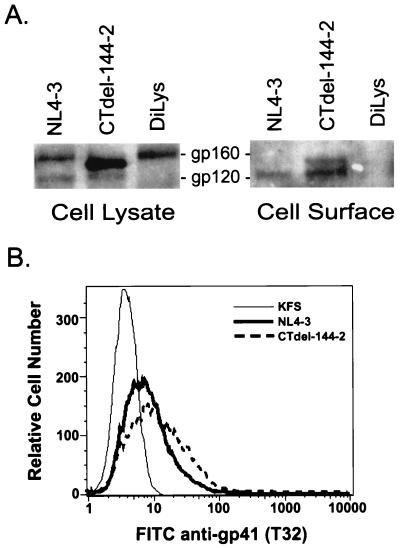
Because reduced expression of the truncated Env on the surface of nonpermissive cells could explain the very low levels of gp120 and gp41 on virions produced from these cells, it was necessary to directly measure cell-surface Env expression. Levels of wt and CTdel-144–2 Env on the cell surface were determined by using CEM (12D-7) cells infected with VSV-G-pseudotyped virions. Cells were biotinylated with sulfo-N-hydroxysuccinimide-biotin; lysates were immunoprecipitated with AIDS patient sera (Fig. 6A Left) or incubated with neutravidin-agarose beads (Fig. 6A Right). Immunoprecipitated and neutravidin-precipitated proteins were subjected to Western blotting with an anti-gp120 polyclonal antibody. Levels of gp120 at the surface of wt- and CTdel-144–2-Env expressing cells were comparable. The Western blotting procedure used in this experiment detects gp160 more efficiently than the immunoprecipitation analysis used in Fig. 3 (compare Fig. 3C with Fig. 6A). As a result, and in agreement with previous studies using similar biotinylation methods (37), some gp160 is detected at the cell surface. The levels of wt and mutant gp160 (as well as gp120) at the cell surface were comparable in a repeat experiment (data not shown). Thus, we conclude that the CTdel-144–2 mutation affected the cell-surface expression of neither gp120 nor gp160. To verify that our biotinylation assays were specific for cell-surface proteins we performed three controls: (i) cells expressing a mutant form of HIV-1 Env containing a di-Lys endoplasmic reticulum retrieval signal (29) at the C terminus were biotinylated. The di-Lys signal previously has been shown to prevent cell surface expression of HIV-1 Env (40, 41), (ii) before biotinylation, Env-expressing cells were treated with brefeldin A, which traps glycoproteins in the endoplasmic reticulum (42), and (iii) immunoprecipitated and neutravidin-agarose-precipitated proteins were subjected to Western blotting with AIDS patient sera. The viral Gag proteins (e.g., Pr55Gag and p24), which are not exposed at the cell surface, should not be detected in the neutravidin-agarose-precipitated (cell-surface) samples. All three controls confirmed that proteins that were expressed abundantly intracellularly but not at the cell surface were not biotinylated (Fig. 6A, diLys lane; and data not shown), thus indicating that the biotinylation conditions used were specific for proteins exposed at the cell surface.
Figure 6.
Analysis of cell-surface Env expression. (A) CEM (12D-7) cells were infected with VSV-G-pseudotyped NL4–3, CTdel-144–2, or DiLys. Cells were biotinylated with sulfo-N-hydroxysuccinimide-biotin. Cell lysates were immunoprecipitated with AIDS patient sera (Left) or incubated with neutravidin-agarose beads (Right). Proteins immunoprecipitated with AIDS patient sera or precipitated with neutravidin-agarose beads were subjected to Western blotting with an anti-gp120 polyclonal antiserum. The results are representative of at least two experiments. (B) Cell-surface immunostaining with an anti-gp41 mAb (T32) was performed by using CEM (12D-7) cells infected with VSV-G-pseudotyped KFS, NL4–3 or CTdel-144–2. Fluorescence intensity was determined by FACScan. Percentage of antibody-positive cells for NL4–3- and CTdel-144–2-infected cells was 29% and 45%, respectively. Data presented are representative of triplicate experiments.
The cell-surface expression data presented in Fig. 6A were corroborated by immunostaining Env-expressing cells with an anti-gp41 mAb and subjecting the cells to flow cytometric analysis (Fig. 6B). Consistent with the biotinylation data, cell-surface expression of wt and mutant Env glycoproteins was comparable. Taken together, these results demonstrate that the Env incorporation defect imposed by the CTdel-144–2 gp41 truncation in nonpermissive cells does not result from reduced cell-surface Env expression.
Discussion
No clear consensus has emerged from previous studies regarding whether the long gp41 CT tail is required for infectivity, Env incorporation, both, or neither. A limitation of the previous studies, including our own, was that biochemical analyses were performed in cell lines that can be readily transfected (e.g., COS, CV-1, or HeLa), rather than in cells that are targets for productive HIV-1 infection. To address this issue, and to establish a link between biochemical and biological properties of gp41 CT truncation mutants, we used a high-level, transient HIV-1 expression system based on pseudotyping with VSV-G. This system provided high-level expression of wt and gp41 truncation mutants in a range of cell types including T cell lines and primary MDM. Using this system, we were able to demonstrate that in permissive HeLa and MT-4 cells, CTdel-144–2 mutant Env incorporation was reduced only 3-fold relative to wt (Fig. 3 A and B), whereas gp120 levels in virions produced from nonpermissive T cell lines [e.g., CEM (12D-7) and Jurkat] and primary MDM were reduced more than 10-fold (Fig. 3 C–E).
What mechanism governs Env incorporation into virions in permissive and nonpermissive cell types? Several models could account for the cell-type differences observed in this study. (i) The gp41 CT may promote Env recruitment to the site of virus assembly via interactions with host factors. Differences in the expression or localization of such putative host factors could explain cell type-specific effects of gp41 CT truncation. (ii) The Gag/Env interaction, and the virion incorporation process itself, may be facilitated by host factor(s) whose expression differs between permissive and nonpermissive cells. (iii) Env glycoproteins lacking the CT may be specifically excluded from incorporation in cells that are nonpermissive for gp41 truncation, and (iv) permissive cells may express higher levels of Env at the cell surface than nonpermissive cells, making Env incorporation in nonpermissive cells more dependent on active recruitment into virions via gp41 CT interactions with MA and/or host factors.
There are numerous precedents for the suggestion that the gp41 CT engages in interactions with host factors. For example, the gp41 CT is apparently responsible for basolateral targeting of virus release in polarized epithelial cells (21, 38, 43); a membrane-proximal, Tyr-based signal mediates rapid Env internalization via interactions with the AP-2 clathrin complex (44); and membrane-proximal residues in the gp41 CT prevent Env localization at sites of VSV budding (45). Finally, simian immunodeficiency virus passaged in human cells spontaneously acquires mutations that prematurely truncate the gp41 CT; passaging of virus expressing the truncated gp41 in cells of monkey origin reselects for a full-length CT (46, 47). The relationship between host factors responsible for these effects, and those involved in Env incorporation, remains to be determined.
This study demonstrates that the long CT of gp41 is required, in a cell type-dependent manner, for HIV-1 Env incorporation into virions. These results, in part, provide an explanation for the evolution of the unusually long CT of lentiviral Env glycoproteins.
Acknowledgments
We thank A. Ono, D. Demirov, and R. Willey for helpful suggestions and critical review of the manuscript. We acknowledge J. Burns for pHCMV-G; P. Earl for the anti-gp41 antibody, T32; and K. Strebel for rabbit anti-gp120 antiserum. The following reagents were obtained through the National Institutes of Health AIDS Research Reference and Reagent Program: MAGI cells (from M. Emerman) and HIV-1 neutralizing sera (from L. Vujcic).
Abbreviations
- Env
envelope glycoprotein
- CT
cytoplasmic tail
- PBMC
peripheral blood mononuclear cells
- MDM
monocyte-derived macrophages
- RT
reverse transcriptase
- VSV
vesicular stomatitis virus
- VSV-G
VSV glycoprotein
- MA
matrix
- MAGI
multinuclear activation of a galactosidase indicator
- wt
wild type
References
- 1.Freed E O, Martin M A. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 2.Swanstrom R, Wills J W. In: Retroviruses. Coffin M J, Hughes S H, Varmus E H, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 263–334. [Google Scholar]
- 3.Dubay J W, Roberts S J, Hahn B H, Hunter E. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu X, Yuan X, McLane M F, Lee T H, Essex M. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilk T, Pfeiffer T, Bosch V. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 7.Cosson P. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent M J, Melsen L R, Martin A S, Compans R W. J Virol. 1999;73:8138–8144. doi: 10.1128/jvi.73.10.8138-8144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed E O, Englund G, Martin M A. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed E O, Martin M A. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger H G. J Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landau N R, Page K A, Littman D R. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lusso P, di Marzo Veronese F, Ensoli B, Franchini G, Jemma C, DeRocco S E, Kalyanaraman V S, Gallo R C. Science. 1990;247:848–852. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 14.Spector D H, Wade E, Wright D A, Koval V, Clark C, Jaquish D, Spector S A. J Virol. 1990;64:2298–2308. doi: 10.1128/jvi.64.5.2298-2308.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnier L, Ravallec M, Blanchard P, Chaabihi H, Bossy J P, Devauchelle G, Jestin A, Cerutti M. J Virol. 1995;69:4060–4068. doi: 10.1128/jvi.69.7.4060-4068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yee J K, Friedmann T, Burns J C. Methods Cell Biol. 1994;43:99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 17.Ott D E. Rev Med Virol. 1997;7:167–180. doi: 10.1002/(sici)1099-1654(199709)7:3<167::aid-rmv199>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 18.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowell J F, Stanhope P E, Siliciano R F. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 20.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M, Hoxie J A. J Cell Biol. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodge R, Gottlinger H, Gabuzda D, Cohen E A, Lemay G. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller M A, Cloyd M W, Liebmann J, Rinaldo C R, Jr, Islam K R, Wang S Z, Mietzner T A, Montelaro R C. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- 23.Chernomordik L, Chanturiya A N, Suss-Toby E, Nora E, Zimmerberg J. J Virol. 1994;68:7115–7123. doi: 10.1128/jvi.68.11.7115-7123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivas S K, Srinivas R V, Anantharamaiah G M, Compans R W, Segrest J P. J Biol Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]
- 25.Kiernan R E, Ono A, Englund G, Freed E O. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 28.Sodroski J, Goh W C, Rosen C, Campbell K, Haseltine W A. Nature (London) 1986;322:470–474. doi: 10.1038/322470a0. [DOI] [PubMed] [Google Scholar]
- 29.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham F L, Eb A J v d, Eb A J v d. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 31.Hopkins N, Besmer P, DeLeo A B, Law L W. Proc Natl Acad Sci USA. 1981;78:7555–7559. doi: 10.1073/pnas.78.12.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, et al. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freed E O, Martin M A. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willey R L, Shibata R, Freed E O, Cho M W, Martin M A. J Virol. 1996;70:6431–6436. doi: 10.1128/jvi.70.9.6431-6436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y M, Tang X B, Cimakasky L M, Hildreth J E, Yu X F. J Virol. 1997;71:1443–1452. doi: 10.1128/jvi.71.2.1443-1452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Earl P L, Broder C C, Doms R W, Moss B. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzwedel K, West J T, Hunter E. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lodge R, Lalonde J P, Lemay G, Cohen E A. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartz S R, Rogel M E, Emerman M. J Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salzwedel K, West J T, Jr, Mulligan M J, Hunter E. J Virol. 1998;72:7523–7531. doi: 10.1128/jvi.72.9.7523-7531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer T, Zentgraf H, Freyaldenhoven B, Bosch V. J Gen Virol. 1997;78:1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- 42.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owens R J, Dubay J W, Hunter E, Compans R W. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boge M, Wyss S, Bonifacino J S, Thali M. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 45.Johnson J E, Rodgers W, Rose J K. Virology. 1998;251:244–252. doi: 10.1006/viro.1998.9429. [DOI] [PubMed] [Google Scholar]
- 46.Kodama T, Wooley D P, Naidu Y M, Kestler H W d, Daniel M D, Li Y, Desrosiers R C. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirsch V M, Edmondson P, Murphey-Corb M, Arbeille B, Johnson P R, Mullins J I. Nature (London) 1989;341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]