Abstract
Methylation of histone H3 lysine 9 (H3K9) is a key feature of silent chromatin and plays an important role in stabilizing the interaction of heterochromatin protein 1 (HP1) with chromatin. Genomes of metazoans such as the fruit fly Drosophila melanogaster generally encode three types of H3K9-specific SET domain methyltransferases that contribute to chromatin homeostasis during the life cycle of the organism. SU(VAR)3-9, dG9a, and dSETDB1 all function in the generation of wild-type H3K9 methylation levels in the Drosophila genome. Two of these enzymes, dSETDB1 and SU(VAR)3-9, govern heterochromatin formation in distinct but overlapping patterns across the genome. H3K9 methylation in the small, heterochromatic fourth chromosome of D. melanogaster is governed mainly by dSETDB1, whereas dSETDB1 and SU(VAR)3-9 function in concert to methylate H3K9 in the pericentric heterochromatin of all chromosomes, with dG9a having little impact in these domains, as shown by monitoring position effect variegation. To understand how these distinct heterochromatin compartments may be differentiated, we examined the developmental timing of dSETDB1 function using a knockdown strategy. dSETDB1 acts to maintain heterochromatin during metamorphosis, at a later stage in development than the reported action of SU(VAR)3-9. Surprisingly, depletion of both of these enzymes has less deleterious effect than depletion of one. These results imply that dSETDB1 acts as a heterochromatin maintenance factor that may be required for the persistence of earlier developmental events normally governed by SU(VAR)3-9. In addition, the genetic interactions between dSETDB1 and Su(var)3-9 mutations emphasize the importance of maintaining the activities of these histone methyltransferases in balance for normal genome function.
KEY constituents of heterochromatin include the structural protein HP1 and histone H3 methylated on lysine 9 (H3K9me) (Grewal and Elgin 2002). The formation of heterochromatin in most eukaryotes, from the unicellular fission yeast to humans, involves an ortholog of the Drosophila SU(VAR)3-9 protein, which is an H3K9-specific methyltransferase (Schotta et al. 2003b). However, most metazoan genomes encode members of at least three H3K9-specific SET domain methyltransferase families, including SU(VAR)3-9, G9a, and SETDB1 (Huisinga et al. 2006). Mammalian G9a and SETDB1 methyltransferases have important roles in euchromatic gene regulation (Schultz et al. 2002; Tachibana et al. 2002). The importance of these methyltransferases for heterochromatin formation is less well understood. Likewise, the contribution of each of these enzymes to genomewide H3K9me homeostasis is not well documented. The Drosophila melanogaster genome encodes one representative of each H3K9-specific SET methyltransferase family, namely SU(VAR)3-9, dG9a, and dSETDB1 (Stabell et al. 2006a,b; Seum et al. 2007a). We have utilized this system to study the combined function and interaction of the three enzymes and their contribution to chromatin homeostasis in a metazoan.
In most species, heterochromatin is present in the regions around centromeres and at telomeres (Dillon 2004). This is also true in Drosophila, with the additional presence of heterochromatin throughout the length of the small fourth chromosome [also referred to as the “dot” chromosome because of its appearance in mitotic nuclei (Locke and McDermid 1993)]. The heterochromatic character of the fourth chromosome, as well as pericentric regions, is apparent from cytological preparations showing enrichment for HP1 at those locations (James and Elgin 1986). H3K9me2-3 (di- and trimethylation) shares the same distribution as HP1 in Drosophila heterochromatin, and SU(VAR)3-9 is codistributed with HP1 and H3K9me2-3 at pericentromeric and fourth chromosome locations (Schotta et al. 2002). Despite this similarity in distribution, SU(VAR)3-9 is not solely responsible for heterochromatic H3K9 methylation in Drosophila; in Su(var)3-9 mutants, pericentric H3K9me is diminished, but in the fourth chromosome H3K9me appears unchanged (Schotta et al. 2002).
It has recently been noted that the dSETDB1 gene controls H3K9 methylation critical for heterochromatin formation in the fourth chromosome (Seum et al. 2007b; Tzeng et al. 2007). However, the precise developmental requirement for dSETDB1, its interaction with dG9a and SU(VAR)3-9 in the formation of heterochromatin, and the potential impact of its function for fourth chromosome gene expression have yet to be explored. To study the individual and combinatorial contributions of the H3K9 methyltransferases to heterochromatin stability and normal genome function in D. melanogaster, we have utilized a set of RNAi knockdown lines for dG9a and dSETDB1 along with available classical mutations. We find that dSETDB1 activity is critical during development, and that loss of this activity alone is more deleterious than loss of both SU(VAR)3-9 and dSETDB1 activities together.
MATERIALS AND METHODS
Fly culture and stocks:
Fly lines produced for this study include: yw;+/+;P{dSETDB1hp2101B=my+}, yw;+/+;P{dG9ahp1002=my+}, yw,P{dSETDB1hp0408=my+};+/+;Su(var)3-906, yw,P{dG9ahp2201=my+};+/+;Su(var)3-906, yw;P{Act5CGAL4=mw+}/CyO;Su(var)3-906. (For a description of the hairpin constructs P{dG9ahp1002=my+}, P{dG9ahp2201=my+}, P{dSETDB1hp2101B=my+}, and P{dSETDB1hp0408=my+} see the next section and supplemental Figure S1F.) Various combinations of these lines and the parental w;P{Act5CGAL4=mw+}/CyO were crossed to produce single- and double-mutant offspring for Western blot analysis and results displayed in Table 1. Variegating lines carrying daGAL4 were configured: yw;+/+;P{daGAL4}/TM3;P{hsp26-pt, hsp70-w} or yw,P{hsp26-pt, hsp70-w};+/+;P{daGAL4}/TM3. Variegating lines carrying dSETDB1hp were configured: yw;+/+;P{dSETDB1hp2101B=my+}/TM3;P{hsp26-pt, hsp70-w} or yw,P{hsp26-pt, hsp70-w};+/+;P{dSETDB1hp2101B=my+}/TM3. For double-mutant analysis: yw;egg235/CyO;Su(var)3-906 and for analysis of polytene chromosomes of double mutants: yw;egg235/CyOGFP;Su(var)3-906 and yw;egg1473/CyOGFP;Su(var)3-906. In all cases the genetic background yw is the same inbred line bearing the yw67c23 chromosome.
TABLE 1.
Viability of H3K9 methyltransferase knockdown mutants and combinations
| % of expected female progeny with this genotype | % of expected male progeny with this genotype | |
|---|---|---|
| dSETDB1KD | 65.2 (62/190)* | 32.4 (35/216)* |
| dG9aKD | 100 (53/96) | 100 (46/86) |
| dG9aKD;dSETDB1KD | 32.7 (46/562)* | 0 (0/369)* |
| dSETDB1KD;Su(var)3-906 | 89.3 (123/551) | 30.4 (28/368)* |
| dG9aKD;Su(var)3-906 | 100 (74/247) | 100 (47/188) |
Viability of HMT double mutants (KD) compared to single-mutant siblings. Survival of double mutants is expressed as percentage of expected survival, with respect to single-mutant siblings carrying the CyO balancer chromosome rather than the Act5CGAL4 transgene hairpin driver, assuming normal Mendelian ratios. Control crosses were performed to normalize for any lethality effects associated with the balancer chromosome itself. Hairpin lines carrying constructs dSETDB1hp2101B and dG9ahp1002 were used for the single gene knockdown and dG9ahp2201 and dSETDB1hp2101B for the dG9a/dSETDB1 double knockdown. For combination with the Su(var)3-906 mutation, dG9ahp2201 and dSETDB1hp0408 were used. *Significant deviation from expected ratio (P < 0.001).
Fly lines obtained for this study include: egg1473/CyO and egg235/CyO (provided by T. Hazelrigg), Df(2R)Dll-MP/SM6a (deleted region: 60E1–60E6; Bloomington no. 1465, supplied by T. Hazelrigg), w;P{Act5CGAL4}/CyO, w;P{daGAL4.w− }3 (Bloomington no. 8641), yw;P{ey1xGAL4=w+mC}3 (Bloomington no. 8227), P{nosGAL4= w+mC }2/CyO (Bloomington no. 4442), Su(var)3-901 (Bloomington no. 6209), Su(var)3-902 (Bloomington no. 6210), and w;Su(var)3-906. For larval selection, egg1473, egg235, and Df(2R)Dll-MP) were maintained over either CyO or CyoGFP. See supplemental Methods for detailed information on each cross.
Hairpin constructs:
To generate conditional knockdown lines for dG9a and dSETDB1, hairpin constructs were introduced into the yw67c23 strain of D. melanogaster by standard P-element transformation. Transformations were performed by Genetic Services. The construct employed is a modification of the pWIZ vector described in Lee and Carthew (2003); see supplemental Figure S1F. The white transformation marker of pWIZ was replaced by a copy of the yellow gene lacking introns under the control of its own promoter including the wing and body enhancer elements. The fragment of dG9a introduced into the pWIZ vector was amplified from genomic DNA using the following primers: forward 5′-GCT CTA GAT AAA CTC GCT GCT GAC GCA ACC AA-3′ and reverse 5′-GTC CTA GAT CAC CCA ATT CCT CCT GCT CTG TT-3′. The fragment of dSETDB1 introduced into the pWIZ vector was amplified from genomic DNA using the following primers: forward 5′-GCT CTA GAT GGC GAT TCC GCT GGT AAA CTA CT-3′ and reverse 5′-GCT CTA GAA CCG TCT CGT CCA GAT CAG CAT TT-3′. The PCR fragments were digested with XbaI and cloned sequentially into the AvrII and NheI cloning sites of pWIZ to complete the hairpin constructs.
Potential off-target effects of the two hairpin constructs were investigated computationally using custom Perl scripts. A library of all possible 19mers derived from the forward and reverse strand of the hairpin sequence was compared to all transcripts annotated in the genome (FlyBase release 5.12). Exact matches of the 19mers to the transcriptome were considered to determine off-target effects, with any gene other than the intended receiving one or more hits counting as a possible off-target. For the egg hairpin construct, no off-target matches were detected. For the dG9a hairpin construct, a total of 10 potential off-targets were detected due to the presence of several short regions of low sequence complexity in the hairpin. These off-targets each match a single 19mer (Cf2, shn, ctp, mthl1, mgl, CG1673, CG9027, and CG12347) with the exception of Hr46 and scyl, which each contain sequence complementary to three different 19mers. Hr46 defects induce distinct visual phenotypes, which were not observed here. Neither Hr46 nor scyl is thought to impact histone methylation or chromatin structure. It remains a formal possibility that changes in the expression of these genes impacts the measures of lethality shown in Table 1.
Viability analysis:
Deviation from Mendelian ratios and sex differences in survival were evaluated with the chi-square test. Significance levels for differences in lethality between single and double mutants were determined by comparing the proportions of survivors between the two populations using a Z-statistic.
Monitoring position effect variegation:
To be able to study white variegation, we employed a yellow reporter instead of white to mark our hairpin expressing transgenic constructs (see above) and used these in conjunction with non-white-marked GAL4 drivers. All assays of position effect variegation (PEV) phenotypes were carried out using flies raised in vials in a humidified 25° incubator. For pigment assays, three samples of five flies (three females and two males) were selected at random from aged populations (3–4 days) of each genotype tested. The samples were prepared and analyzed by spectrophotometry using previously described methods (Brower-Toland et al. 2007). Images of fly eyes were captured from the same aged population. All test crosses were performed in both directions to exclude maternal and paternal effects. For single-mutant effects, comparison was with the progeny of wild-type (yw67c23) flies mated with flies carrying the variegating insertion of interest. For double-mutant effects, comparison of progeny was with parental lines (flies collected from the same culture and at the same time parents were selected).
Polytene chromosome staining:
Polytene chromosomes were prepared and stained using standard methods (Stephens et al. 2004). Antibodies were used at the following dilutions: anti-HP1 (C1A9, 1:25), anti-H3K9me1 (Upstate, 1:500), anti-H3K9me2 (Upstate, 1:25), anti-H3K9me3 (Upstate, 1:50), anti-SXL (University of Iowa Hybridoma Bank, affinity purified, 1:20), and anti-SU(VAR)3-9 (G. Reuter, 1:10).
Western blotting:
Where not otherwise specified, samples were isolated from the indicated tissues by hypotonic extraction of homogenized tissues in TE (10 mm Tris-Cl, 1 mm EDTA, pH 8.0) with 1.0 mm DTT and protease inhibitor cocktail for 30 min on ice, followed by addition of SDS loading buffer (final concentration 50 mm Tris-Cl, 2% SDS, 0.1% bromophenol blue, 10% glycerol, 100 mm β-mercaptoethanol), centrifugation to remove insolubles, and incubation at 100° for 15 min. Samples were electrophoresed on 18% SDS/PAGE gels following the method of Laemmli (1970), transferred to nitrocellulose, and probed with antibodies in TBS + 0.05% Tween with 2.5% nonfat dried milk using standard methods (Sambrook and Russell 2001). Antibodies were used at the following dilutions: anti-H2B (Upstate, 1:5000), anti-H3K9me1 (Upstate, 1:2000), anti-H3K9me2 (Upstate, 1:1000), and anti-H3K9me3 (Upstate, 1:500). Western analysis was repeated at least three times, and representative images are shown.
Reverse transcriptase PCR:
Oligo(dT) primed synthesis of cDNAs was carried out using total RNA from the indicated genotypes and developmental stages. Amplification of target cDNAs was accomplished by PCR with the following primer sets: RpL32Fwd 5′-CGAGCTCGCCGCAGTAAAC-3′, RpL32Rev 5′-CTTCATCCGCCACCAGTCG-3′; dG9aFwd 5′-AACAGAGCAG GAGGAATTGGGTGA-3′, dG9aRev 5′-TGAAGTTCGCATCCACCAGAGGAA-3′; dSETDB1Fwd 5′-CCAGTGACTATGTGCAT GAAGTTCC-3′, dSETDB1Rev 5′-AATCACGCGGGTACCAAATGG-3′; and SU(VAR)3-9Fwd 5′-AATCAAGCGGGCCCAATTTG TACG-3′, SU(VAR)3-9Rev 5′-CGCACACAAATTCACCCTTAC GCA-3′.
Microarray analysis:
Total RNA samples were prepared from fly heads of the following genotypes: yw;+/CyO, yw;egg235/CyO, yw;+/CyO;Su(var)3-906, yw;egg235/CyO;Su(var)3-906. Each sample was isolated in triplicate (resulting in three biological replicates) and hybridized independently along with an additional replicate of the wild type (yw; +/CyO) controlling for technical variability in the hybridization process. cDNA preparation and hybridization to Affymetrix Drosophila 2.0 expression microarrays was carried out by the Siteman Cancer Center Multiplexed Gene Analysis Core Facility at Washington University according to the manufacturer's recommended protocols. Resulting data were analyzed using the Partek Genomic Suite. Normalized mean probe signals called present on the arrays were compared to obtain fold differences between genotypes. Gene lists were compiled for fold differences with a significance of P ≤ 5 × 10−6.
A second analysis of the microarray data was carried out using the Bioconductor packages affy and limma in R on the log2-transformed signal intensities (Gentleman et al. 2004). The density estimates for the two technical replicates agree very well (data not shown), therefore only data from one of the two technical replicates was used in the final data analysis. The RMA method was employed to carry out background correction, quantile normalization, and summarization of the gene expression levels. The empirical Bayes method was used to identify statistically differentially expressed genes for the following six comparisons: comparisons between each of the three mutants and the control, comparisons between the double mutant and each of the two single mutants, and the comparison between the double mutant and the average of the two single mutants. To correct for multiple comparisons (total number of tests was 18,952 × 6), Benjamini and Hochberg's false discovery rate (FDR) controlling procedure was applied to the P-values at significance level α = 0.05 (Benjamini and Hochberg 1995).
The microarray data can be found in the GEO (Gene Expression Omnibus) database, accession number no. GSE14756.
For gene ontology (GO) analyses, GOToolBox (http://burgundy.cmmt.ubc.ca/GOToolBox/) software was used.
RESULTS
dSETDB1 mutants exhibit multiple phenotypes arising after the larval stage:
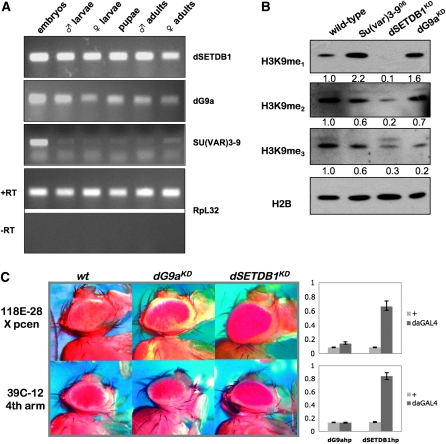
Previous studies have shown that the SU(VAR)3-9 enzyme is essential for the production of wild-type levels of methylated H3K9 in Drosophila embryos (Kuhfittig et al. 2001; Schotta et al. 2002, 2003b; Rudolph et al. 2007). We observed that SU(VAR)3-9 mRNA was present throughout the life cycle of Drosophila, but is most abundant in embryos (Figure 1A). The expression levels of dG9a and dSETDB1, the other two H3K9-specific methyltransferases, were more constant throughout development than those of SU(VAR)3-9 (Figure 1A). To study the contributions of these two enzymes to methyl-H3K9 metabolism and heterochromatin formation and maintenance we produced transgenic flies expressing double-stranded RNA identical to portions of the dSETDB1 and dG9a transcripts under control of the GAL4 UAS system (Lee and Carthew 2003). The resulting knockdown alleles are referred to as dSETDB1KD and dG9aKD throughout this article, to distinguish them from classical alleles. These hairpin transgenes effectively reduced target gene expression as shown in supplemental Figure S1. For the alleles most commonly used in our work the transcript reduction is >80%; due to the remaining activity, the knockdown alleles should be considered hypomorphs.
Figure 1.—
In contrast with dG9a, dSETDB1 is a major H3K9 methyltransferase that is required for heterochromatin formation in Drosophila. (A) H3K9 methyltransferases differ in their expression patterns during Drosophila development. RT–PCR assays with primer sets specific for each methyltransferase sequence are shown in the top three panels; RpL32 amplification, both with and without reverse transcriptase, is provided at the bottom as an input control. All samples are from wild-type OR flies. Embryos, 6- to 18-hr mixed embryos; larvae, third instar. (B) dSETDB1, dG9a, and SU(VAR)3-9 all participate in methyl-H3K9 metabolism in vivo. Duplicate Western blots of wild-type vs. HMT mutant lysates from adult flies were probed with antibodies specific for mono-, di- and trimethyl H3K9; anti-H2B was used as loading control. Knockdown was achieved by using the Act5CGAL4 driver with dSETDB1hp2101B or dG9ahp1002. Numbers at the bottom of each blot represent the normalized band intensity expressed as fold difference from wild type. (C) dSETDB1KD is a potent suppressor of variegation, while dG9aKD is not. PEV assays shown compare Mod(var) effects of dG9aKD vs. dSETDB1KD using representative fourth chromosome (39C-12) and pericentric (118E-28) reporters. Knockdown was achieved by using the daGAL4 driver with dSETDB1hp2101B or dG9ahp1002. To the right of the eye images, data from quantitative eye pigment assays are shown, confirming the Su(var) effect. Shown in light gray are values from flies carrying the indicated hairpin construct and the reporter in the absence of the driver, while the dark gray bars illustrate the pigment level in flies that carry the reporter, the hairpin construct, as well as the driver. Y-axis, fraction of wild-type pigment. Error bars represent standard deviations.
Knockdown of dG9a or dSETDB1 throughout development with an ACT5C-GAL4 driver produced organism-level defects only in the case of dSETDB1 (Table 1 and data not shown). No defects were observed with dG9a knockdown, consistent with recently published observations derived from targeted knockout of dG9a (Seum et al. 2007a). dSETDB1KD animals exhibited several visible phenotypes in larval, pupal, and adult stages. Mutant larvae were more sluggish in their feeding and wandering behavior than wild-type counterparts (data not shown). At the end of pupal development, when the adult flies eclose from the pupal case, dSETDB1KD animals often failed to complete this process, and newly eclosed mutants often failed to complete wing expansion. Adult females and males with dSETDB1 defects were underrepresented in crosses resulting in dSETDB1KD (Table 1), and both dSETDB1KD males and females had shorter life spans, often dying within 2 days of eclosion. Adult dSETDB1KD females were less fecund, laying greatly reduced numbers of eggs, resulting in fewer progeny per parent than wild-type females. Adult males were more severely impacted by dSETDB1KD, demonstrated by their significantly lower survival (P < 0.001; Table 1). These results demonstrate that dSETDB1 function is critically important for normal development.
Clough et al. (2007) recently isolated a number of recessive alleles of the gene encoding dSETDB1 referred to as “eggless” (egg) because of the female oogenesis phenotype. Egg homozygous individuals or those with mutations over a small deficiency [Df(2R)Dll-MP] recapitulated the traits of dSETDB1KD individuals (described above), including motility defects, eclosion failure, wing expansion defects, and female infertility. All combinations of dSETDB1 mutant alleles resulted in some lethality in females, and, to a much greater extent, adult males (Table 2). This difference in lethality between males and females is statistically significant for the three allelic combinations tested (P < 0.01). Similar effects of egg mutations on adult longevity and male viability have been observed by E. Clough and T. Hazelrigg (personal communication). The lethality observed in individuals deficient for dSETDB1 occurred primarily after the L2/L3 stage, since mutant larvae (−/−) from crosses of heterozygous parents (balanced over a GFP-marked chromosome) were observed at the expected frequency (data not shown). Significantly decreased viability of adult dSETDB1 mutants demonstrated that dSETDB1 encodes a product essential during metamorphosis (Table 2; P < 0.001 for all mutant combinations with the exception of egg235/Df(2R)Dll-MP females). These observations are very much in contrast with those of the Su(var)3-906 allele which, like all mutant Su(var)3-9 alleles isolated, is not homozygous lethal (Tschiersch et al. 1994). In fact, Su(var)3-906 can be maintained as a homozygous stock indefinitely, although homozygotes are less healthy and fecund (data not shown).
TABLE 2.
Viability of homozygous progeny from crosses between heterozygous egg parents in wild-type and Su(var)3-906 backgrounds
| % of expected female progeny with this genotype | % of expected male progeny with this genotype | |
|---|---|---|
| egg1473 | 57.6 (27/142)** | 2.1 (1/141)** |
| egg1473/Df(2R)Dll-MP | 68.6 (43/190)* | 13.3 (7/159)** |
| egg235/Df(2R)Dll-MP | 89.2 (43/146) | 27.8 (9/98)** |
| egg1473;Su(var)3-906 | 97.1 (33/103) | 11.4 (4/106)** |
dSETDB1(egg) mutant viability expressed as percentage of expected progeny homozygous for the indicated dSETDB1 allele, or carrying that allele over a deficiency, by comparison with heterozygous siblings. *Significant deviation from expected ratio (P < 0.01); **significant deviation from expected ratio (P < 0.001).
Consistent with their function as H3K9 methyltransferases, ubiquitous knockdown of dG9a and dSETDB1 using the Act5CGAL4 driver and the respective hairpin construct decreased total H3K9 methylation levels in adult flies. Compared to bulk H3K9 methylation in wild-type and Su(var)3-906 homozygous mutants, we found that knockdown of dSETDB1 (dSETDB1KD) dramatically reduced total mono-, di- and trimethyl forms (10–30% of adult wild-type levels), while dG9aKD had an impact primarily on trimethylation (Figure 1B). Similar results were obtained using the daGAL4 driver (data not shown). Su(var)3-9 mutants showed modest reductions in di- and trimethylation (∼60% of adult wild-type levels), but in these flies H3K9me1 accumulated to levels approximately twofold higher than seen in the wild-type adults (Figure 1B). Thus, the more severe phenotype was associated with lower overall levels of H3K9 methylation in dSETDB1 mutant flies.
Knockdown mutation confirms that dSETDB1, in addition to SU(VAR)3-9, is necessary for normal heterochromatin formation in Drosophila:
The deficiencies observed in H3K9 methylation upon knockdown of dG9a or dSETDB1 function suggest a possible role in heterochromatin formation. To test this hypothesis for our knockdown mutations, we used a PEV assay with a white eye color reporter, where increased variegation indicates increased heterochromatin formation, while a decrease of variegation (and increase of red eye color) indicates decreased heterochromatin formation. Thus, we combined the H3K9 methyltransferase RNAi knockdown lines with a set of variegating reporters, including three pericentric reporters (118E-28 [X], 39C-2 [2L] and 118E-10 [4]) and two reporters in heterochromatin on the banded arm of the fourth chromosome (39C-12 and 118E-25-5). With one exception (118E-28, which has two copies), all reporter lines bear only one copy of the hsp26-pt, hsp70-w transgene, inserted by P transgenesis, which has been mapped by in situ hybridization and/or inverse PCR (Wallrath and Elgin 1995; Sun et al. 2000).
Driving knockdown of dSETDB1 in the presence of representative variegating insertions on the fourth chromosome and in pericentric heterochromatin with the ubiquitous daughterless driver (daGAL4) resulted in strong suppression of the variegating phenotype at both locations (Figure 1C). The UAS-dSETDB1 hairpin construct functioned to knock down expression of dSETDB1 regardless of insertion site, and knockdown potency was generally correlated with its activity as a suppressor of variegation on the fourth chromosome (supplemental Figure S1, A–C). In contrast with dSETDB1KD, knockdown of dG9a produced no change in silencing of a fourth chromosome reporter (39C-12; Figure 1C) and acted only as a very weak suppressor of pericentric silencing (118E-28; Figure 1C), regardless of hairpin insertion site (supplemental Figure S1, D and E), consistent with the lack of a visible phenotype noted in recent reports (Seum et al. 2007a; Kato et al. 2008). These findings demonstrate that the knockdown mutations generated here give phenotypes in each instance consistent with earlier reports using classical alleles.
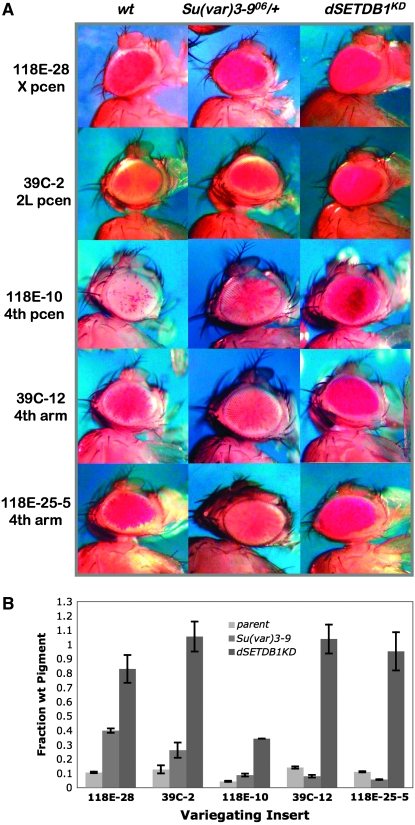
Mutant alleles of the Su(var)3-9 gene, which produces the third H3K9-specific methyltransferase, are potent suppressors of PEV when present with a variegating rearrangement of the white gene or with hsp70-w reporters in pericentric heterochromatin (Tschiersch et al. 1994; Haynes et al. 2007). SU(VAR)3-9 has been described as the main chromocenter-specific histone methyltransferase (HMT) in Drosophila (Schotta et al. 2003b). When we compared the dominant suppression effects of Su(var)3-906 with the suppression produced by dSETDB1KD, we found that whereas dSETDB1KD resulted in dramatic derepression (approximately six- to eightfold) of reporters in the fourth chromosome arm, Su(var)3-906 had no such effect in this domain and appears to have increased silencing of such fourth chromosome reporters (Figure 2 and Table 3). This finding is consistent with reports that Su(var)3-906 acts not as a suppressor but as a weak dominant enhancer of PEV with reporters located in the fourth chromosome arm (Haynes et al. 2007). This enhancement of PEV has been suggested to be the consequence of redistribution of the heterochromatin components made available upon loss of pericentric targets (Haynes et al. 2007). Since the putative enhancement effect seen in this case was subtle, we repeated the assay at a greater level of sensitivity, using three different mutant alleles of Su(var)3-9. Each of the mutant alleles tested displayed a statistically significant decrease in eye pigmentation of a variegating fourth chromosome arm reporter in contrast with the increase seen for a pericentric reporter (supplemental Figure S2). However, when tested with pericentric X and second chromosome reporters, both dSETDB1KD and Su(var)3-906 consistently produced robust suppression effects (Figure 2 and Table 3).
Figure 2.—
The knockdown system verifies that dSETDB1 and SU(VAR)3-9 have opposing roles in fourth chromosome heterochromatin formation, but act similarly in pericentric heterochromatin. (A) dSETDB1KD (5–25% of wild-type expression, see supplemental Figure S1) is a suppressor of variegation for insertions throughout the Drosophila genome. Su(var)3-906 is a dominant suppressor of pericentric variegation on all chromosomes including the fourth but acts as a dominant enhancer of variegation on reporters in the banded region of the fourth chromosome. For the dSETDB1 knockdown the daGAL4 driver was combined with dSETDB1hp2101B. (B) Quantification of pigment content of all populations represented in A. Values for the “parent” control are shown in lightest shade of gray, for Su(var)3-906 lines in the medium shade of gray, and for dSETDB1KD lines in dark gray. Note the distinct E(var) effect of Su(var)3-906 at variegating reporters in the arm of the fourth chromosome, and the large Su(var) effect of dSETDB1KD at all sites (refer to Table 3 for numerical values). Error bars are standard deviation (n = 3). Parent refers to the reporter in a yw background.
TABLE 3.
Fold change in pigment values resulting from suppression of variegation by HMT mutations
| Chromosomal location | Fold change (± SD)
|
||
|---|---|---|---|
| Reporter | dSETDB1KD | Su(var)3-906/+ | |
| 118E-28 | X, pericentric | 7.8 ± 0.9 | 3.7 ± 0.1 |
| 39C-2 | 2L, pericentric | 6.6 ± 0.7 | 2.3 ± 0.5 |
| 118E-10 | 4, pericentric | 7.5 ± 1.0 | 2.0 ± 0.2 |
| 39C-12 | 4, arm | 8.6 ± 0.8 | −2.6 ± 0.1 |
| 118E-25-5 | 4, arm | 9.7 ± 0.2 | −2.3 ± 0.1 |
PEV modification by HMT mutations. Fold change in pigmentation in dominant modification assays upon knockdown of dSETDB1 expression (5–25% transcript quantities relative to wild type; dSETDB1hp2101B driven by daGAL4) or with the Su(var)3-906 allele (dominant effect). Mean fold change from the parental pigmentation is indicated with standard deviation from the mean; negative values indicate fold enhancement of PEV.
On the basis of the PEV data, it appears that dG9a has little or no impact on constitutive heterochromatic domains. However, the knockdown allele is a hypomorph (supplemental Figure S1, D and E). If only very little dG9a is required for function, a dG9a role in heterochromatin formation might not be detected. The absence of such a role for dG9a suggested by the PEV data is supported by the fact that dG9a is not associated with heterochromatic regions of the genome on polytene chromosome preparations (Stabell et al. 2006b; Seum et al. 2007a).
These results suggest that dSETDB1 functions both in pericentric and in fourth chromosome heterochromatin formation and/or maintenance, while SU(VAR)3-9 function is restricted to pericentric heterochromatin. Consistent with this broad role in heterochromatin formation, loss of dSETDB1 function was deleterious, resulting in high lethality rates, unlike loss of dG9a function, which did not act as a strong Su(var) and was not by itself deleterious (Table 1). The dramatic suppression of variegation of fourth chromosome reporters upon dSETDB1 knockdown confirmed reports that the enzyme has a unique function in this chromosome (Seum et al. 2007b; Tzeng et al. 2007). In contrast, the antithetical effect of several Su(var)3-9 mutant alleles on two of these reporters confirmed reports that Su(var)3-9 does not participate in heterochromatin formation in the distal arm of the fourth chromosome (Schotta et al. 2002).
Loss of dSETDB1 and SU(VAR)3-9 together is less deleterious than loss of dSETDB1 alone:
Suppression of PEV at pericentric insertions by either Su(var)3-906 or dSETDB1KD was sometimes less dramatic than suppression of reporters in the fourth chromosome arm where dSETDB1KD acts alone (see Figure 2). This result implies cooperation: since pericentric heterochromatin utilizes the function of both dSETDB1 and SU(VAR)3-9, diminishing the function of both HMTs should result in even greater suppression of pericentric heterochromatin formation. Loss of dG9a appears to have little impact on normal heterochromatin formation (Figure 1C), but it may function redundantly to compensate for defects in Su(var)3-9 or dSETDB1 mutants.
To test this prediction, we generated fly lines that when crossed produced nearly complete loss of function of any two of these three H3K9 HMTs. Double-mutant offspring displayed defects in viability that differed from single-mutant offspring. Knocking down dSETDB1 and dG9a function together was more deleterious than either alone and resulted in male and female sterility, suggesting that some compensation occurs in the single mutants (P < 0.001; Table 1 and data not shown). However, combining either of these two knockdown mutations with the Su(var)3-906 mutation had no such effect. dG9aKD;Su(var)3-906 mutant offspring displayed no significant change in viability when compared with dG9a mutants alone, although females were sterile (Table 1 and data not shown). Strikingly, concomitant loss of dSETDB1 and SU(VAR)3-9 function resulted in greater viability than dSETDB1 loss of function alone (P < 0.001; Table 1).
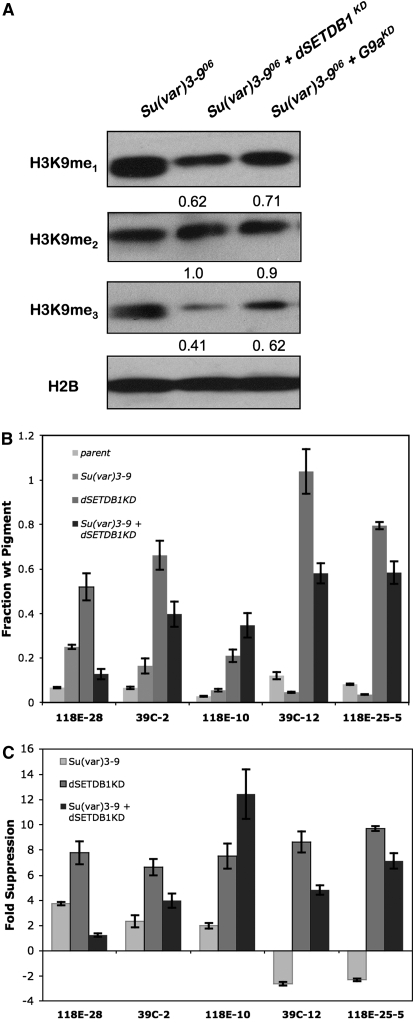
Evaluation of bulk H3K9 methylation in these double mutants by comparison with Su(var)3-906 homozygous mutants showed that the buildup of the H3K9me1 substrate in Su(var)3-9 mutants was relieved by 30–40% when the function of either dSETDB1 or dG9a was also diminished (Figure 3A, samples from adults). Su(var)3-906 homozygous mutants with dSETDB1KD showed less H3K9me1 than dG9aKD;Su(var)3-906 double mutants, consistent with the almost complete loss of H3K9me1 observed in dSETDB1KD flies (see Figure 1B). Virtually no difference in H3K9me2 was observed in these double mutants, but combinations of the Su(var)3-906 homozygous mutation with knockdown of either of the other two methyltransferases had the greatest impact (reductions of 40–60%) on H3K9me3 (Figure 3A). These results indicate that all three H3K9 methyltransferases contribute in significant ways to the bulk metabolism of H3K9 methylation. Furthermore, both dSETDB1 and dG9a activities may provide monomethyl substrate to SU(VAR)3-9, raising questions about the way their activities are coordinated.
Figure 3.—
A complex interaction between loss of dSETDB1 and of SU(VAR)3-9 impacts heterochromatin formation, with the double mutant showing a less severe phenotype. (A) dSETDB1 and dG9a act with SU(VAR)3-9 to produce methyl-H3K9 in vivo. Western blot showing quantities of H3K9 methylation in double mutants (adults). Values at the bottom of each blot express the fraction of mono-, di-, or trimethylation in double mutants by comparison with homozygous Su(var)3-906. Knockdown of dG9a and dSETDB1 was achieved by combining the Act5CGAL4 driver with hairpins dG9ahp2201 or dSETDB1hp0408. (B) Loss of silencing in dSETDB1KD lines is partially restored by depletion of SU(VAR)3-9, except at pericentric fourth chromosome heterochromatin. Pigment values for dominant suppression by Su(var)3-906/+, dSETDB1KD and dSETDB1KD;Su(var)3-906/+ are compared with parental (no HMT mutant) values. Knockdown of dSETDB1 was achieved by combining the daGAL4 driver with hairpin dSETDB1hp2101B. Error bars represent standard deviations (n = 3). (C) Comparison of fold suppression changes from the parental pigment values are expressed for single and double mutants on the basis of the data from B. Enhancement of PEV (reduced pigmentation) is expressed as a negative fold change.
Analysis of variegating eye phenotypes with these mutant combinations provides additional insight into the interactive functions of the three Drosophila histone methyltransferases. Loss of both dG9a and SU(VAR)3-9 function modestly increased suppression of variegation at second and fourth chromosome pericentric insertions over loss of SU(VAR)3-9 activity alone (data not shown). The simultaneous loss of dG9a and dSETDB1 function was not significantly different than loss of dSETDB1 alone by this assay (data not shown), although there was a significant increase in lethality (P < 0.001; Table 1). The ability of dSETDB1KD to reduce silencing at reporters was significantly modified in combination with a Su(var)3-9 loss-of-function mutation. In contrast with the predicted cooperation between dSETDB1 and SU(VAR)3-9 and the predicted increase in suppression of variegation in pericentric regions, suppression of variegation by dSETDB1KD in pericentric X and 2L and in the arm of chromosome four was actually decreased when flies also lacked SU(VAR)3-9 function (Figure 3, B and C).
Cooperation between these two Su(var)'s occurred only at the pericentric fourth chromosome reporter 118E-10, which exhibits a greater loss of silencing in a double-mutant dSETDBKD; Su(var)3-906 than as a consequence of either mutation independently (Figure 3, B and C). These results indicate a bivalent nature for fourth chromosome pericentric chromatin by contrast with chromatin that is distinctly controlled either by pericentric or fourth chromosome heterochromatin-forming mechanisms. The reason for this is not clear, as this domain otherwise generally behaves as pericentric heterochromatin.
Region-specific changes in heterochromatin structure resulting from HMT mutations:
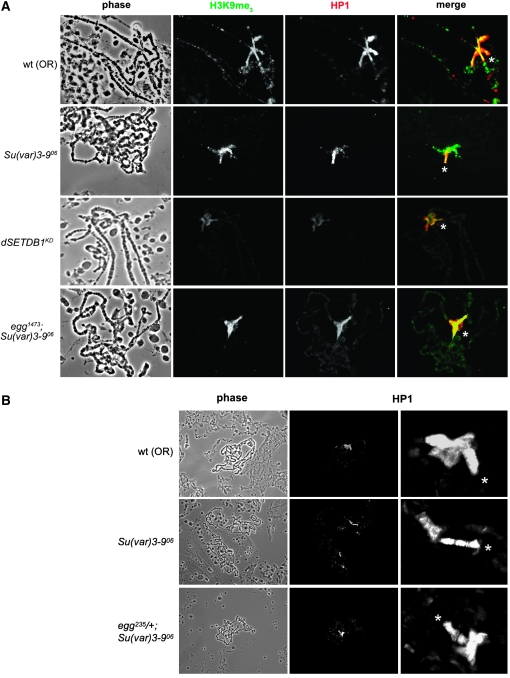
To visualize H3K9 methylation defects in Su(var)3-9 and dSETDB1 mutant flies, we examined polytene chromosomes of larvae bearing mutations in the dSETDB1 and Su(var)3-9 genes. For this purpose we utilized the aforementioned classical alleles of the dSETDB1 gene (egg1473 and egg235) as well as a deficiency [Df(2R)Dll-MP] that covers the dSETDB1 locus in addition to the knockdown alleles (Clough et al. 2007). dSETDB1KD mutant chromosomes exhibited dramatic reduction in H3K9 trimethylation in the fourth chromosome in contrast with the pericentric chromatin, and absence of methylation and HP1 in the prominent pattern seen at cytological region 31 in the wild-type chromosomes (Figure 4A) as previously reported for other alleles (Seum et al. 2007b). [Note while using a shorter exposure time very little staining of H3K9me3 and HP1 is evident on the fourth chromosome, longer exposures indicate that residual amounts of HP1 are still present on chromosome four (data not shown).] Using the same experimental conditions, we observed the opposite pattern for H3K9me3 with Su(var)3-9 homozygous mutant chromosomes: partial loss of pericentric methylation but retention of H3K9me3 on the fourth chromosome (Figure 4A). We also noted robust fourth chromosome HP1 staining in Su(var)3-9 homozygous mutant chromosomes, which appears to be in excess of what is seen in the wild type. In the wild type, HP1 levels on chromosome four and the chromocenter appear very similar, while the Su(var)3-906 chromosomes show a much stronger HP1 staining on chromosome four compared to the chromocenter. In addition, the fourth chromosome morphology was altered in these mutants, most often appearing elongated and constricted, with HP1 bands more well defined in the polytene chromosomes (Figure 4, A and B). As previously suggested, this configuration may arise as an effect of surplus heterochromatin components liberated from pericentric heterochromatin by loss of SU(VAR)3-9 function (Haynes et al. 2007). Liberated heterochromatin components may thereby become available to the fourth chromosome heterochromatin forming mechanism, increasing the heterochromatic character of chromosome four. Consistent with this hypothesis, we observed dominant enhancement of silencing of variegating fourth chromosome reporters by several Su(var)3-9 mutant alleles (supplemental Figure S2).
Figure 4.—
The fourth chromosome appears hypercondensed in Su(var)3-9 mutant salivary glands; this is reversed in Su(var)3-9; dSETDB1 double mutants. (A) Localization of heterochromatin components on polytene chromosomes from wild-type, Su(var)3-906 homozygous mutant, dSETDB1 mutant (dSETDB1hp2101B driven by daGAL4), and egg1473;Su(var)3-906 double homozygous mutant chromosomes. Retention of H3K9me3 (green in merged image) and superabundance of HP1 (red) on the fourth chromosome (asterisk) is observed in Su(var)3-906 mutants. A broader distribution of HP1 is observed in egg1473;Su(var)3-906 double-mutant salivary glands. (B) Heterozygosity for egg235 reduces condensation of chromosome four in Su(var)3-906 mutants. Polytene chromosomes comparing HP1 localization in wild-type, Su(var)3-906, and egg235/+;Su(var)3-906 lines. Close-up views of the chromocenter are provided at the right of each image showing each set of chromosomes with the fourth chromosome indicated by an asterisk. Side-by-side comparisons (generated by squashing the different salivary glands together on the same slide) are provided in supplemental Figure S3.
To further investigate the interaction of dSETDB1 and SU(VAR)3-9, and the effect of their mutations on chromosome four chromatin structure, we created Drosophila lines containing balanced hypomorphic alleles of egg in a Su(var)3-9 mutant background. These crosses produced offspring that were homozygous for the Su(var)3-906 mutant chromosome and heterozygous for the egg mutation. Su(var)3-9 null, egg heterozygous flies could be maintained indefinitely with fecundity and viability only slightly less than the wild type. The Su(var)3-906 homozygous mutant background also increased survival rates for egg homozygous mutants in the case of the dSETDB1 allele egg1473 (P < 0.05; Table 2) and improved survival of the dSETDB1KD lines (P < 0.001; Table 1).
Chromosomes from salivary glands of larvae homozygous for both HMT mutations were distinctly mutant in appearance: they were less condensed, the chromocenter was ill defined, and the HP1 signal across the euchromatic arms appeared more general (Figure 4A). The HP1 and H3K9me3 localization patterns and chromosome morphology observed on egg/+;Su(var)3-906 double-mutant chromosomes provided some insight into the improved viability of Su(var)3-906 mutants when they had only one wild-type copy of dSETDB1. These chromosomes appear to have less overall H3K9 methylation than Su(var)3-906 or egg mutant chromosomes, but immunofluorescence associated with HP1 was distributed more evenly across the chromocenter, being present both in pericentric chromatin and on the fourth chromosome at similar intensities (Figure 4B and supplemental Figure S3). In the egg235/+;Su(var)3-906 larvae, the fourth chromosome no longer appeared elongated and instead of the pronounced HP1 banding present in Su(var)3-906 homozygous mutant chromosomes, it was difficult to discriminate distinct bands; the chromosome often appeared broader and shorter than in the wild type (Figure 4B and supplemental Figure S4). Overall, mutant chromosome phenotypes illustrated the spatially distinct but overlapping functions of dSETDB1 and SU(VAR)3-9 in genome maintenance and suggest that chromosome four morphology more closely resembles the wild-type in the double mutant as contrasted with the Su(var)3-906 homozygous lines.
Developmental timing of dSETDB1 function in heterochromatin formation:
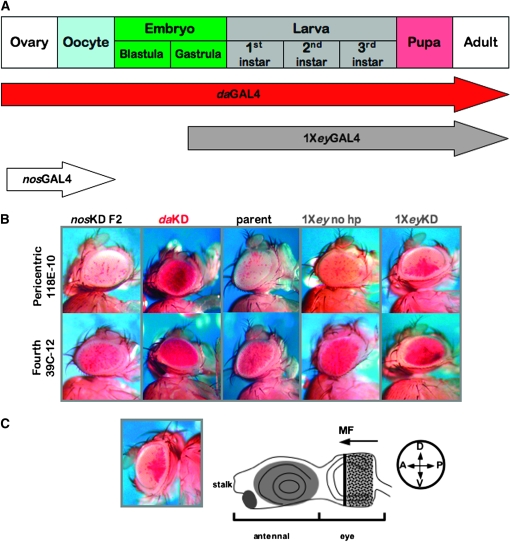
Several observations hint at a complex temporal relationship between the two methyltransferase activities SU(VAR)3-9 and dSETDB1. SU(VAR)3-9 activity is utilized for formation of heterochromatin in the earliest stages of development (Kuhfittig et al. 2001; Rudolph et al. 2007). Although dSETDB1 functions in the germarium of the female ovary (Clough et al. 2007; Yoon et al. 2008), SU(VAR)3-9 is the primary H3K9 methyltransferase functioning in later oogenesis and immediately following fertilization (Yoon et al. 2008). In particular, the occurrence of many dSETDB1KD phenotypes in later development led us to consider the developmental timing of dSETDB1 function. To narrow the developmental window for the functional requirement of dSETDB1 in heterochromatin formation, we utilized GAL4 drivers, one expressed during egg production and in early embryogenesis (nosGAL4) and one expressed from late embryogenesis in the eye anlagen and developing eye tissue (eyGAL4) (Figure 5A). First, we tested the effect of dSETDB knockdown in the maternal germline and early embryo using the nosGAL4 driver. Although it decreased egg production (data not shown), nosGAL4 driven knockdown of dSETDB1 in the maternal germline had no effect on the variegating status of fourth chromosome or pericentric reporters in the offspring, arguing against an irreversible requirement for a very early function of dSETDB1 contributed through oocyte chromatin or by maternally loaded embryonic dSETDB1 in the initial stages of heterochromatin formation (Figure 5B). (The efficacy of nosGAL4 in driving knockdown is shown in supplemental Figure S4.) We note that a similar experiment, in which female dSETDB1daKD virgins bearing strongly suppressed variegating reporters at each location tested were backcrossed to males bearing the same reporters in the wild-type background, produced offspring exhibiting no suppression of PEV (data not shown). The ubiquitous expression pattern of daughterless is known to include strong expression in all cells of the ovary (Cummings and Cronmiller 1994).
Figure 5.—
dSETDB1 contributes to fourth chromosome heterochromatin formation during metamorphosis. (A) Schematic illustration of drivers spanning the development of Drosophila used to assess impact of dSETDB1 knockdown on PEV at different developmental times. (B) dSETDB1 is required for heterochromatin formation and/or maintenance after oogenesis but prior to late pupal development. Representative eye phenotypes resulting from knockdown of dSETDB1 in the ovary and early embryo (nosGAL4) are contrasted with parental phenotypes and ubiquitous knockdown of this methyltransferase enzyme (daGAL4). Knockdown of dSETDB1 in eye tissue lineages (eyGAL4) results in an unusual pattern of suppression. [Note that unlike the other drivers, eyGAL4 is marked with a copy of white; this results in the low level background pigmentation shown (1Xey, no hp) but does not interfere with the assay.] dSETDB1hp2101B was used for these experiments. The reporter was line 39C-12 (insertion in the banded region of chromosome 4) or 118E-10 (insertion in the pericentric heterochromatin, chromosome 4) as indicated. (C) The wedge-shaped pattern of hsp70-w expression (depicted at left; 118E-10) displayed when knockdown is driven by 1XeyGAL4 indicates that dSETDB1 is specifically required for the stability of heterochromatin during morphogenetic furrow (MF) progression in the eye imaginal disc (illustrated at right), which occurs during late larval development. D, dorsal; V, ventral; A, anterior; and P, posterior.
We were able to pinpoint a developmental window during which dSETDB1 function is required to produce/maintain fourth chromosome heterochromatic silencing using a GAL4 driver under the control of the 200-bp eye-specific enhancer of eyeless (1XeyGAL4). In contrast with siblings bearing only the driver, dSETDB1eyKD individuals showed dramatic suppression of variegation at both fourth chromosome reporters tested (Figure 5B). Interestingly, the pigmentation in suppressed variegating eyes displayed a distinctive pattern reflecting late third instar development of the eye: loss of silencing exhibited a gradient from posterior to anterior ommatidia. This posterior-anterior pattern mirrors morphogenetic furrow progression in the eye imaginal disc, which also occurs from posterior to anterior (Figure 5C). Additional variegating pericentric reporters exhibited the same type of eye phenotypes, raising the possibility that dSETDB1 functions during the time of morphogenetic furrow progression. If that is the case, dSETDB1 functions during the same, late, developmental window in pericentric heterochromatin as in fourth chromosome heterochromatin on the basis of posterior-anterior patterns observed in PEV studies (data not shown). The eye pigmentation patterns observed with each reporter tested bore a striking resemblance to patterns displayed in the eye imaginal disc by a variegating lacZ reporter (Lu et al. 1996, 1998); however those patterns are observed before the furrow has completely crossed the imaginal disc. In summary, the results of the knockdown PEV studies argue that dSETDB1 plays a critical role in maintaining heterochromatin during development.
Gene misregulation resulting from H3K9 methyltransferase defects:
Although it represents only a small fraction of the Drosophila genome, a number of fourth chromosome genes encode components essential for proper development. Among its >80 genes, the fourth chromosome encodes several essential genes including pan (dTCF) and lgs (dBCL9), two components of the wg/WNT signaling pathway, zfh2, a zinc finger protein required for normal wing development, ribosomal protein S3a (RpS3a), mutations which produce female infertility, and a host of other nonessential genes. The enhanced PEV and unusual fourth chromosome morphology evident in Su(var)3-9 null chromosomes (Figures 2–4) may be indicative of misregulation of such essential genes, leading to compromised health and reproductive vigor. An additional reduction in the amount of dSETDB1 activity relieves this hyperheterochromatinization induced by the homozygous Su(var)3-9 mutation as assayed cytologically with polytene chromosomes (Figure 4 and supplemental Figure S3). Conversely, dSETDB1 knockdown is less deleterious in a Su(var)3-9 mutant background (Figure 3 and Table 1). We hypothesized that this relief occurs because of a more balanced distribution of heterochromatin components, which results in restoration of more balanced fourth chromosome gene expression, leading to the improved viability observed in dSETDB1/+;Su(var)3-9 double mutants.
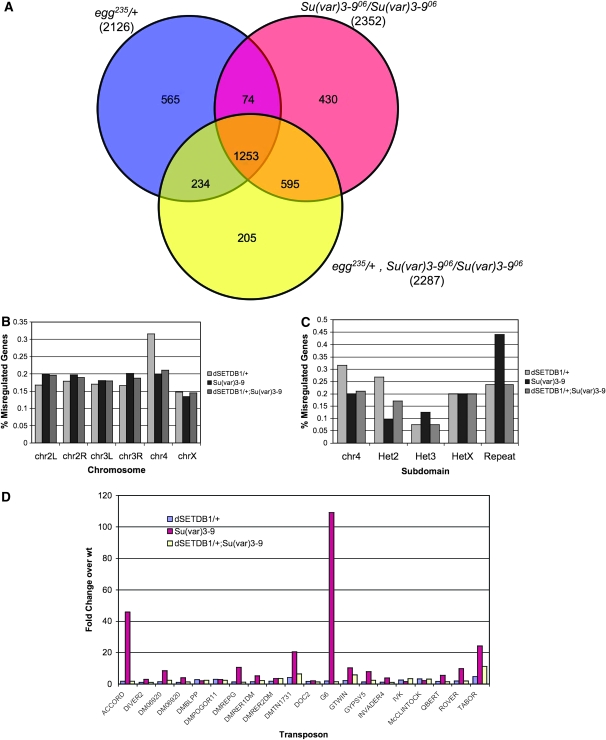
To test this hypothesis we compared expression levels of fourth chromosome genes in four genetic backgrounds: wild-type, egg235/+, Su(var)3-906 homozygous mutant, and the double-mutant egg235/+;Su(var)3-906 by microarray analysis. Results from this analysis showed a complex pattern of misregulation. We carried out two independent analyses of the microarray data (see materials and methods), the results of which were very similar. The results presented here use those transcripts that were significantly misregulated (up or down) in both analyses and met a significance cutoff of P < 5 × 10−6 in the Partek analysis. Generally, we found (not unexpectedly) that a large number of genes were affected by mutations in the HMTs dSETDB1 and SU(VAR)3-9. On the basis of the above criteria, 2126 transcripts were misregulated in the egg235/+ genotype compared to the wild type. A slightly larger number of genes, 2352, were significantly different from wild type in the Su(var)3-906 homozygous mutant, and in the double mutant, 2287 transcripts were misregulated in relation to wild type (see Figure 6A). While the three lines examined all affected approximately the same number of genes, they did not affect the same transcripts. Approximately half of the transcripts affected in each line were shared between all lines (1253). In addition to the shared misregulated genes, each strain examined uniquely affected some genes that were not misregulated in the other two strains. The double mutant exhibited the smallest number of uniquely misregulated genes (205), while the egg heterozygote had the most (565; Figure 6A).
Figure 6.—
Global changes in gene expression occur in histone methyltransferase mutant Drosophila. (A) Venn diagram illustrating overlap of misregulated genes in Su(var)3-906 (red) and egg235/+ (blue) single mutants as well as the egg235/+; Su(var)3-906 (yellow) double mutant. At the bottom of each genotype, the total number of misregulated genes detected is given. RNA prepared from fly heads. (B) Bar chart depicting the percentage of misregulated genes in each mutant background compared to wild type by chromosome. A high proportion of fourth chromosome genes are misregulated as a consequence of the dSETDB1 mutation. (C) Bar chart depicting the percentage of misregulated genes in each heterochromatin domain. (D) Bar chart illustrating transposon reactivation in each mutant background based on increased expression levels compared to wild type. Loss of SU(VAR)3-9 activity has the more pronounced effect on transposon expression; this is frequently reversed in the double-mutant line.
In all three HMT mutant genotypes, the average level of misregulation was greater for downregulated than for upregulated genes. This discrepancy was most extreme in the case of the Su(var)3-906 homozygous line, where the average level of downregulation was 11.9-fold, while the average level of upregulation was only 2.4-fold (data not shown). In addition to the bias in the level of misregulation between up- and downregulated genes, there was also a bias in the prevalence of up- and downregulation among the three genotypes examined. For the Su(var)3-906 mutant, there were 884 significantly upregulated genes, but 1468 significantly downregulated genes. The same 1:2 ratio was observed in the egg/+; Su(var)3-9 double mutant, with 885 genes significantly upregulated and 1402 genes significantly downregulated. Interestingly, in the egg235/+ single-mutant line, an ∼1:1 ratio was observed between significantly up- and downregulated genes (1035:1091).
We found that misregulated fourth chromosome genes were disproportionately represented in dSETDB1 heterozygotes (Figure 6B). Comparing gene expression in all heterochromatin domains, we found that the percentage of fourth chromosome genes misregulated in dSETDB1 mutants was still disproportionately larger, with 31.6% of fourth chromosome genes showing significant changes in expression compared with an average of 16.6 ± 1.1% in the other chromosomes (Figure 6C). While overall, there were similar numbers of genes significantly up- and downregulated in egg235/+ individuals, the distribution for the fourth chromosome did not follow this trend. Of the 30 misregulated genes on chromosome 4, 27 or 90% were upregulated, while only three were downregulated. This bias for upregulation is also seen in other regions of the genome generally considered heterochromatic, namely chromosome U (the nonmapped regions of the Drosophila genome), repeats, and the pericentric regions of chromosomes 2, 3, and X. The X chromosome also showed a slight bias toward upregulation (183 vs. 111 genes), while on the other chromosome arms downregulation was most frequently observed in response to the egg235 mutation (supplemental Figure S5). These observations are consistent with the importance of dSETDB1 in modification of the fourth chromosome and argue that the resulting H3K9 methylation downregulates fourth chromosome genes.
In the Su(var)3-906 homozygotes, the fourth chromosome and the other heterochromatic regions of the genome also exhibited a misregulation profile different from the euchromatic chromosome arms. While on the euchromatic arms of chromosomes 2 and 3 the majority of misregulated genes were downregulated, in heterochromatic portions of the genome upregulation was seen in 63 of 73 cases. The X chromosome was intermediate between these two extremes, containing 121 upregulated and 148 downregulated genes. Two genes downregulated on chromosome four are noteworthy, JYalpha and ATPsyn-beta. These genes defy the commonly seen upregulation pattern on chromosome four in both HMT single-mutant strains examined.
The trends in misregulation patterns described above for Su(var)3-906 and egg235 mutants also held true for the egg235/+; Su(var)3-906 double mutant. In heterochromatic regions of the genome upregulation was predominantly observed compared to wild type, while downregulation was prevailing in all other regions with the exception of the X chromosome where both up- and downregulation were equally likely to occur. The disproportionate impact on fourth chromosome genes seen in the egg235/+ samples is reduced in the double mutant, which has 19 misregulated fourth chromosome genes compared to the 30 seen in the double mutant, and 19 seen in the Su(var)3-9 mutant.
Gene ontology (GO) annotation analysis of the misregulated genes revealed that in all three HMT mutant lines examined a wide variety of genes were affected. The affected gene products were associated with various cellular components such as membranes, intracellular organelles, and protein complexes, to name a few. Compared to the genome as a whole, the annotated fraction of the significantly misregulated genes represented on the microarray were enriched and depleted for several classes of cellular components. Specifically, membrane-associated proteins and proteins associated with intracellular organelles were overrepresented in the set of misregulated genes in all three HMT mutant lines, while proteins localized to extracellular regions were underrepresented. Similarly, a wide variety of molecular functions were represented among the genes found to be misregulated in this study. Most noteworthy were the overrepresentation of genes associated with the GO term “oxidoreductase activity” and the underrepresentation of genes characterized as possessing “nucleic acid binding,” “helicase activity,” and, in two lines [Su(var)3-906 single and egg235/+; Su(var)3-906 double mutant], the class with “trancription regulator activity.”
Another class of genes noticeably affected by the HMT mutations was composed of repeats and transposable elements (Figure 6C). Several transposable elements were derepressed in each genotype, although to a much greater extent in Su(var)3-9 homozygotes. Of the transposable elements represented on the microarray, 44% exhibited marked upregulation in Su(var)3-906 homozygotes compared to 24% in egg235 heterozygotes (Figure 6D). Notably, in the egg235/+; Su(var)3-9 double-mutant background, many transposable elements exhibited significant reduction in expression when compared to Su(var)3-906 (Figure 6, C and D). Reduction in transposon activation induced by the Su(var)3-9 defect in the dSETDB1/+;Su(var)3-906 mutant line may be linked to the redistribution of heterochromatin components to pericentric heterochromatin observed in polytene chromosomes of larvae having the same genotype (Figure 4B and supplemental Figure S3). Similarly, the number of misregulated fourth chromosome genes was reduced in the double-mutant compared to the egg235/+ line. These results do not, however, immediately suggest an explanation of the rescue of dSETDB1 mutant effects in the Su(var)3-906 mutant background in any more than general terms. Consideration of the small set of genes whose expression was restored to wild type in the double-mutant background when compared to expression in the egg235/+ mutant was not informative, as these were not clearly biased to any functional class (data not shown). These results emphasize the complexity of genomic regulation governed by dSETDB1 and SU(VAR)3-9, where altering the balance of these two activities has effects on gene expression that are difficult to predict.
DISCUSSION
Results presented here demonstrate the importance of the H3K9 methyltransferases dSETDB1, dG9a, and SU(VAR)3-9 for wild-type function of the Drosophila genome. Our expression analysis (Figure 1A), as well as the work by others, shows that the three Drosophila HMTs have different developmental profiles and distinct roles (Stabell et al. 2006a,b; Seum et al. 2007b; Yoon et al. 2008). While some transcript was detected in all tissues assayed for the three HMTs, their transcript levels differ. dSETDB1 plays an important part in oogenesis (Clough et al. 2007; Yoon et al. 2008), SU(VAR)3-9 functions prominently during early embryogenesis (Kuhfittig et al. 2001; Schotta et al. 2002, 2003b; Rudolph et al. 2007), while dG9a's expression pattern is more uniform, possibly being increased in ovaries (Stabell et al. 2006b). Interestingly, given dSETDB1/egg's role in the female germline, impaired dSETDB1 function causes greater lethality in males than in females, indicating either the existence of a second yet unknown function for dSETDB1 in males or that males inherently are more sensitive to disturbance of dSETDB1-mediated H3K9 methylation. The presence of the heterochromatic Y chromosome may increase the sensitivity of males to the perturbations in heterochromatic silencing; males frequently show a greater loss of silencing in response to Su(var) mutations.
Western analysis of histone H3 methylation levels in adult flies from Drosophila lines impaired in dG9a, dSETDB1, or SU(VAR)3-9 function corroborates the interpretation of different roles for these HMTs. Mutations in the three HMTs have different effects on the methylation states of H3K9 (Figure 1B), with SU(VAR)3-9 apparently responsible for significant amounts of dimethylation, dG9a affecting di-and especially trimethylation, and dSETDB1 having the greatest impact on all three methylation states, indicating that it might be responsible for the bulk of H3K9me1 in adult flies. Evidence from the literature supports the idea of varying specificities of the three Drosophila HMTs and also suggests that the effect on global H3K9 methylation states observed depends greatly on the developmental state and/or tissue assayed (Schotta et al. 2002, 2003a; Mis et al. 2006; Clough et al. 2007; Seum et al. 2007b; Tzeng et al. 2007; Yoon et al. 2008). For example, on the basis of our findings, dSETDB1 mutations show the greatest disturbance of the various methylation patterns in adult samples, while in early embryos SU(VAR)3-9 has the greatest effect (Mis et al. 2006).
PEV analysis as well as the study of polytene chromosomes sheds further light on the functional differences between dG9a, dSETDB1, and SU(VAR)3-9. In the PEV assay, loss of function of any of these HMTs has an effect on pericentric insertions, with SU(VAR)3-9 and dSETDB1 showing a much stronger effect than dG9a (Figures 1C and 2). For fourth chromosome reporters, dSETDB1 loss of function has the strongest impact, consistent with the observation on polytene chromosomes that it, not SU(VAR)3-9, is responsible for most H3K9me in this genomic region (Figure 4 and Seum et al. 2007b; Tzeng et al. 2007). In contrast, loss of SU(VAR)3-9 results in enhancement of variegation for reporters in the fourth chromosome arm (Figure 2 and supplemental Figure S2, and Haynes et al. 2007), possibly due to increased accumulation of heterochromatin proteins leading to an altered chromosome structure (Figure 4). The variegating fourth chromosome reporter exhibiting a twofold suppression of variegation by Su(var)3-906 (118E-10; Table 3) presents an interesting case, since it is located within pericentric chromatin rather than residing in the banded portion of the chromosome (Wallrath and Elgin 1995). Su(var)3-906 had a two- to fourfold suppression effect on pericentric insertions located in the X, second, and fourth chromosomes (Table 3), further substantiating the idea that SU(VAR)3-9 functions primarily at pericentric sites. This finding is supported by the observation that loss of SU(VAR)3-9 has a major impact on expression of repetitious elements, most often resulting in upregulation, and many of these elements are located in the pericentric regions of the genome (Figure 6). In contrast, dSETDB1KD resulted in uniformly drastic losses of silencing of pericentric reporters and multiple chromosome four reporters. We conclude that dSETDB1 functions generally in pericentric chromatin and uniquely on the chromosome four arm. Interestingly, in this case as well as in others previously published, the extent to which a given mutation impacts PEV is highly dependent on the exact reporter/mutation combination used. For example, Mis et al. (2006) report a much stronger Su(var) effect for mutation in the dG9a gene with a Sb reporter, while Seum et al. (2007b) fail to detect an effect of dSETDB1 mutations on pericentric reporters with their system.
Taken together, the expression profile of the three D. melanogaster HMTs, the H3K9 histone methylation defects in mutants, and the PEV data, illustrate the subfunctionalization of these enzymes. In all assays, the three enzymes exhibit clear differences, and mutant phenotypes are very distinct. However, especially given the stage dependency of some assay results, no simple model for the interaction between the enzymes emerges from the accumulated data. Rather, at this point, all data hint at the complexity of a system that is yet to be fully understood.
Analyses using dSETDB1 knockdown lines with distinct developmental profiles allowed for the definition of a developmental window during which depletion of dSETDB1 results in an impact on heterochromatin formation/maintenance as assayed by white PEV (Figure 5). dSETDB1 knockdown during the very early stages (nosGAL4 driver) of fly development did not impact PEV. The heterochromatin-specific function of dSETDB1 is critical during the time when developmental changes involved in metamorphosis are initiated in the late third larval instar (and perhaps earlier in larval development), indicating that this enzyme plays an important role as a heterochromatin maintenance factor (Figure 5). This result is complimentary to recent work which shows that SU(VAR)3-9 is the key H3K9 methyltransferase acting in heterochromatin formation in late oogenesis and early zygotic development (Yoon et al. 2008). Our results suggest that dSETDB1 function is required subsequent to these early events to maintain heterochromatin. dSETDB1 function also continues to be required in oogenesis for the development of germ cells from their progenitors (Clough et al. 2007), again indicating the complex temporal patterns of HMT activity. Additional studies will be required to elucidate the mechanism by which SU(VAR)3-9 and dSETDB1 negotiate the handoff of their functions in heterochromatin initiation and maintenance at different stages in the life cycle of the fly.
Our results also highlight the importance of dSETDB1 for maintenance of the peculiar chromatin structure found on the small fourth chromosome (Haynes et al. 2004). On the basis of various lines of evidence, the fourth chromosome can be considered a distinct chromatin domain (for a recent review see Riddle and Elgin 2006; Riddle et al. 2009). We have confirmed that dSETDB1, unlike SU(VAR)3-9, is uniquely required for H3K9 methylation and normal variegating silencing in the banded portion of the fourth chromosome. Given the dependence of the fourth chromosome on dSETDB1 for H3K9 methylation, the developmental timing of dSETDB1 function observed suggests that fourth chromosome heterochromatin may be considered a unique form of HP1-dependent facultative heterochromatin rather than constitutive heterochromatin. Recently, Tzeng et al. (2007) have shown that dSETDB1 interacts with POF, which binds the polytene fourth chromosome in a banded pattern similar to HP1 deposition (Johansson et al. 2007a). POF and HP1 functions are required for normal fourth chromosome gene expression, and at a higher level of resolution their pattern of binding reflects the distribution of expressed exons on the fourth chromosome, on top of a high level overall in comparison to the other chromosome arms (Johansson et al. 2007b). POF and HP1 are thought to act in an opposing manner on gene expression, constituting a fine-tuning system (Johansson et al. 2007b). Given the interaction of POF and dSETDB1, it is likely that dSETDB1 shares the same distribution and is in this way responsible for the H3K9 methylation pattern observed on the fourth chromosome.
The complexity of the system responsible in Drosophila for maintaining H3K9me homeostasis is further illustrated by the results from the study of mutants lacking more than one HMT activity. Curiously, the combination of mutations in dSETDB1 and Su(var)3-9 leads to less severe phenotypes than what is observed for dSETDB1 mutations alone. This “compensation” is seen for viability (Table1), H3K9me levels (Figure 3), as well as chromosome four morphology (Figure 4B). dSETDB1 functions to maintain constitutive pericentric heterochromatin on all chromosomes tested (Figures 2 and 3), where it cooperates with SU(VAR)3-9. Given the developmental timing of its function, dSETDB1 is likely to act at sites initially established by SU(VAR)3-9, and may function as the primary maintenance methyltransferase specific for H3K9 in heterochromatin in later stages of development (Figure 1). Our results demonstrating reduced Su(var) impact of dSETDB1 mutations in a Su(var)3-9 mutant background (Figure 3) may indicate that the developmentally prior deposition of SU(VAR)3-9 or associated marks limits the activity of dSETDB1. Our observation that heterochromatin components are more broadly distributed through pericentric heterochromatin in double-mutant chromosomes than in Su(var)3-9 mutant chromosomes is also consistent with a role for SU(VAR)3-9 in limiting the sites of activity of dSETDB1 in wild-type Drosophila. Alternatively, the loss of SU(VAR)3-9 might induce some compensatory activity that then mitigates the loss of dSETDB1 as well.
Last, we have demonstrated the genomewide effects of altered H3K9me levels on gene expression by microarray analysis. On the basis of the hypothesis that along with POF and HP1 (Johansson et al. 2007a), dSETDB1 participates in regulating normal fourth chromosome gene expression, we have looked for effects of dSETDB1 depletion on gene regulation on the fourth chromosome and throughout the Drosophila genome. Indeed, dSETDB1 knockdown results in misregulation of a large number of genes, particularly impacting the fourth chromosome, while having less impact on expression from repetitious sequences than does depletion of SU(VAR)3-9 (Figure 6). Interestingly, our results indicate that on the fourth chromosome as well as in other heterochromatic regions of the genome (supplemental Figure S5), the majority of genes affected by the egg235/+ genotype are upregulated, while downregulation is more prevalent in other regions of the genome (Figure 6 and supplemental Figure S5). An interesting possibility to consider is that the downregulation we observed in most regions of the genome with dSETDB1 loss of function is connected to the H3K9 methylation seen over the bodies of some genes in genomewide ChIP studies (for examples, see Yasuhara and Wakimoto 2008). This finding of prevalent upregulation stands in contrast with data from Tzeng et al. (2007) from a similar experiment comparing dSETDB1 homozygous mutants to wild type. This set of experiments uses third instar larvae as starting material (whereas our data are produced using RNA from adult heads) and finds that most fourth chromosome genes are downregulated in response to dSETDB1 loss of function. The result is surprising, given the report from Johansson et al. (2007a) indicating that loss of HP1 (a partner of H3K9me) results in upregulation of transcription on the fourth chromosome in first instar larvae. However, this may simply reflect the complexity of the system.
Further research will be required to determine how much of the regulatory function of dSETDB1 is indirectly mediated through its general function in chromatin homeostasis and how much has to do with its role as a cofactor at specific genes. In vertebrates, SETDB1 proteins act with the transcription intermediary factor 1β (TIF1β) to specifically repress target genes (Sripathy et al. 2006; Ivanov et al. 2007). A Drosophila ortholog of TIF1β (bonus) exists and may act with dSETDB1 in the Drosophila system as well, both to regulate genes and to maintain heterochromatic regions (Beckstead et al. 2001, 2005). dSETDB1 and SU(VAR)3-9 are both necessary for normal expression of some genes, and each also regulates a nonoverlapping set of genes. We have shown that when the gene dosage of these two methyltransferases is imbalanced, heterochromatin formation is perturbed throughout the Drosophila genome and gene expression patterns are dramatically altered.
We have also observed defects in viability and fertility when dSETDB1 or SU(VAR)3-9 function is decreased together with knockdown of the third H3K9 methyltransferase in flies, dG9a (Tables 1 and 2). Like its mammalian ortholog, dG9a is involved in euchromatic gene regulation, perhaps in conjunction with the polycomb system (Kato et al. 2008). We attribute the lethality observed upon knockdown of both dG9a and dSETDB1 to catastrophic changes in gene regulation resulting from loss of the regulatory functions of both the polycomb and HP1 systems. Loss of female fertility in dG9a; Su(var)3-9 double mutants indicates a more precise set of shared functions between these two methyltransferases that may relate to transposon control. The possibility of off-target effects contributing to the increased viability and fertility defects in double-mutant combinations with dG9aKD must be considered. This scenario is unlikely given that the dG9aKD lines show no abnormal phenotypes, and mutations in the most likely off-targets are associated with morphological defects, which we do not observe.
In summary, these results illustrate the importance of properly balancing the functions of three Drosophila H3K9 methyltransferases and better define the developmental timing of H3K9 methyltransferase dSETDB1 function. All three methyltransferases described here appear to act in concert to achieve wild-type H3K9 methylation levels in Drosophila and to support the essential functions mediated by H3K9 methylation in the nucleus.
Acknowledgments
Many thanks are due to Doug Chalker (Washington University, St. Louis) for advice and conversations essential to completion of this manuscript. We are grateful for the excellent technical assistance of Gina Volpi in preparation of polytene samples used for this work and for Wilson Leung's assistance with the Perl script. We acknowledge the kind provision of classical alleles of the dSETDB1 gene by Emily Clough and Tulle Hazelrigg (Columbia University). This work was supported by National Institutes of Health grant RO1-GM68388 (to S.C.R.E.). K.H. is supported by a Kirschstein National Service fellowship (F32GM078833).
References
- Beckstead, R., J. A. Ortiz, C. Sanchez, S. N. Prokopenko, P. Chambon et al., 2001. Bonus, a Drosophila homolog of TIF1 proteins, interacts with nuclear receptors and can inhibit betaFTZ-F1-dependent transcription. Mol. Cell 7 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead, R. B., S. S. Ner, K. G. Hales, T. A. Grigliatti, B. S. Baker et al., 2005. Bonus, a Drosophila TIF1 homolog, is a chromatin-associated protein that acts as a modifier of position-effect variegation. Genetics 169 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. [Google Scholar]
- Brower-Toland, B., S. D. Findley, L. Jiang, L. Liu, H. Yin et al., 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, E., W. Moon, S. Wang, K. Smith and T. Hazelrigg, 2007. Histone methylation is required for oogenesis in Drosophila. Development 134 157–165. [DOI] [PubMed] [Google Scholar]
- Cummings, C. A., and C. Cronmiller, 1994. The daughterless gene functions together with Notch and Delta in the control of ovarian follicle development in Drosophila. Development 120 381–394. [DOI] [PubMed] [Google Scholar]
- Dillon, N., 2004. Heterochromatin structure and function. Biol. Cell 96 631–637. [DOI] [PubMed] [Google Scholar]
- Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling et al., 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, S. I., and S. C. Elgin, 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12 178–187. [DOI] [PubMed] [Google Scholar]
- Haynes, K. A., E. Gracheva and S. C. Elgin, 2007. A distinct type of heterochromatin within Drosophila melanogaster chromosome 4. Genetics 175 1539–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, K. A., B. A. Leibovitch, S. H. Rangwala, C. Craig and S. C. Elgin, 2004. Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster. Cold Spring Harbor Symp. Quant. Biol. 69 267–272. [DOI] [PubMed] [Google Scholar]
- Huisinga, K. L., B. Brower-Toland and S. C. Elgin, 2006. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma 115 110–122. [DOI] [PubMed] [Google Scholar]
- Ivanov, A. V., H. Peng, V. Yurchenko, K. L. Yap, D. G. Negorev et al., 2007. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, T. C., and S. C. Elgin, 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, A. M., P. Stenberg, C. Bernhardsson and J. Larsson, 2007. a Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 26 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, A. M., P. Stenberg, F. Pettersson and J. Larsson, 2007. b POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet. 3 e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, Y., M. Kato, M. Tachibana, Y. Shinkai and M. Yamaguchi, 2008. Characterization of Drosophila G9a in vivo and identification of genetic interactants. Genes Cells 13 703–722. [DOI] [PubMed] [Google Scholar]
- Kuhfittig, S., J. Szabad, G. Schotta, J. Hoffmann, E. Mathe et al., 2001. pitkin(D), a novel gain-of-function enhancer of position-effect variegation, affects chromatin regulation during oogenesis and early embryogenesis in Drosophila. Genetics 157 1227–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, Y. S., and R. W. Carthew, 2003. Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30 322–329. [DOI] [PubMed] [Google Scholar]
- Locke, J., and H. E. McDermid, 1993. Analysis of Drosophila chromosome 4 using pulsed field gel electrophoresis. Chromosoma 102 718–723. [DOI] [PubMed] [Google Scholar]
- Lu, B. Y., C. P. Bishop and J. C. Eissenberg, 1996. Developmental timing and tissue specificity of heterochromatin-mediated silencing. EMBO J. 15 1323–1332. [PMC free article] [PubMed] [Google Scholar]
- Lu, B. Y., J. Ma and J. C. Eissenberg, 1998. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development 125 2223–2234. [DOI] [PubMed] [Google Scholar]
- Mis, J., S. S. Ner and T. A. Grigliatti, 2006. Identification of three histone methyltransferases in Drosophila: dG9a is a suppressor of PEV and is required for gene silencing. Mol. Genet. Genomics 275 513–526. [DOI] [PubMed] [Google Scholar]
- Riddle, N. C., and S. C. Elgin, 2006. The dot chromosome of Drosophila: insights into chromatin states and their change over evolutionary time. Chromosome Res. 14 405–416. [DOI] [PubMed] [Google Scholar]
- Riddle, N. C., C. D. Shaffer and S. C. R. Elgin, 2009. A lot about a little dot: lessons learned from Drosophila melanogaster chromosome four. Biochem. Cell Biol. 87 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, T., M. Yonezawa, S. Lein, K. Heidrich, S. Kubicek et al., 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol. Cell 26 103–115. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann et al., 2002. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J. 21 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert, R. Dorn and G. Reuter, 2003. a Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14 67–75. [DOI] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert and G. Reuter, 2003. b SU(VAR)3–9 is a conserved key function in heterochromatic gene silencing. Genetica 117 149–158. [DOI] [PubMed] [Google Scholar]
- Schultz, D. C., K. Ayyanathan, D. Negorev, G. G. Maul and F. J. Rauscher, 3rd, 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum, C., S. Bontron, E. Reo, M. Delattre and P. Spierer, 2007. a Drosophila g9a is a nonessential gene. Genetics 177 1955–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum, C., E. Reo, H. Peng, F. J. Rauscher, 3rd, P. Spierer et al., 2007. b Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet. 3 e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy, S. P., J. Stevens and D. C. Schultz, 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26 8623–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell, M., M. Bjorkmo, R. B. Aalen and A. Lambertsson, 2006. a The Drosophila SET domain encoding gene dEset is essential for proper development. Hereditas 143 177–188. [DOI] [PubMed] [Google Scholar]
- Stabell, M., R. Eskeland, M. Bjorkmo, J. Larsson, R. B. Aalen et al., 2006. b The Drosophila G9a gene encodes a multi-catalytic histone methyltransferase required for normal development. Nucleic Acids Res. 34 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, G. E., C. A. Craig, Y. Li, L. L. Wallrath and S. C. Elgin, 2004. Immunofluorescent staining of polytene chromosomes: exploiting genetic tools. Methods Enzymol. 376 372–393. [DOI] [PubMed] [Google Scholar]
- Sun, F. L., M. H. Cuaycong, C. A. Craig, L. L. Wallrath, J. Locke et al., 2000. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. USA 97 5340–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana, M., K. Sugimoto, M. Nozaki, J. Ueda, T. Ohta et al., 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch, B., A. Hofmann, V. Krauss, R. Dorn, G. Korge et al., 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng, T. Y., C. H. Lee, L. W. Chan and C. K. Shen, 2007. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc. Natl. Acad. Sci. USA 104 12691–12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath, L. L., and S. C. Elgin, 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9 1263–1277. [DOI] [PubMed] [Google Scholar]
- Yasuhara, J. C., and B. T. Wakimoto, 2008. Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet. 4 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J., K. S. Lee, J. S. Park, K. Yu, S. G. Paik et al., 2008. dSETDB1 and SU(VAR)3–9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster. PLoS ONE 3 e2234. [DOI] [PMC free article] [PubMed] [Google Scholar]