Abstract
Heterochromatin renders domains of chromosomes transcriptionally silent and, due to clonal variation in its formation, can generate heritably distinct populations of genetically identical cells. Saccharomyces cerevisiae's Sir1 functions primarily in the establishment, but not the maintenance, of heterochromatic silencing at the HMR and HML loci. In several Saccharomyces species, we discovered multiple paralogs of Sir1, called Kos1–Kos4 (Kin of Sir1). The Kos and Sir1 proteins contributed partially overlapping functions to silencing of both cryptic mating loci in S. bayanus. Mutants of these paralogs reduced silencing at HML more than at HMR. Most genes of the SIR1 family were located near telomeres, and at least one paralog was regulated by telomere position effect. In S. cerevisiae, Sir1 is recruited to the silencers at HML and HMR via its ORC interacting region (OIR), which binds the bromo adjacent homology (BAH) domain of Orc1. Zygosaccharomyces rouxii, which diverged from Saccharomyces after the appearance of the silent mating cassettes, but before the whole-genome duplication, contained an ortholog of Kos3 that was apparently the archetypal member of the family, with only one OIR. In contrast, a duplication of this domain was present in all orthologs of Sir1, Kos1, Kos2, and Kos4. We propose that the functional specialization of Sir3, itself a paralog of Orc1, as a silencing protein was facilitated by the tandem duplication of the OIR domain in the Sir1 family, allowing distinct Sir1–Sir3 and Sir1–Orc1 interactions through OIR–BAH domain interactions.
SUBSTANTIAL portions of many eukaryotic genomes are silenced, blocking transcription of genes in these regions. Proteins involved in gene silencing change the structure of chromatin, in part, by post-translational modifications of histones, leading to the recruitment of heterochromatin structural proteins that recognize these modifications. Heterochromatic regions of genomes often contain repetitive DNA such as retrotransposons, and are often regions of structural importance such as centromeres and telomeres. In yeasts, heterochromatin underlies the silencing mechanism controlling genes that determine cell type and helps to preserve the integrity of the genome. Hence, perturbation of heterochromatin can lead to drastic changes in cellular behavior that, in more complex eukaryotes, can lead to cancer and other diseases (Lafon et al. 2007; Moss and Wallrath 2007).
In Saccharomyces, the silent mating loci, HML and HMR, encode genetic regulators for both mating types, yet are constitutively silenced. Unidirectional gene conversion from HML or HMR to the MAT locus in haploid cells causes a switch in mating types (Hicks et al. 1979; Kushner et al. 1979). Silencers flanking both sides of HML and HMR prevent expression of these loci. The silencers are bound by origin recognition complex (ORC) Rap1 and Abf1, which in turn recruit the Sir proteins that result in formation of silenced chromatin (Rusche et al. 2003). Sir1 protein is required primarily for the establishment of silencing at HML and HMR but not its maintenance (Mahoney and Broach 1989; Pillus and Rine 1989). Sir1 is recruited to the silencers by interaction with Orc1 (Triolo and Sternglanz 1996; Fox et al. 1997) through the Orc1 interaction region (OIR) of Sir1, located in the C-terminal half of the protein, and the bromo adjacent homology (BAH) domain of Orc1 (Gardner et al. 1999; Zhang et al. 2002; Hou et al. 2005; Hsu et al. 2005). A duplication of the OIR of Sir1, called OIR′, has been proposed to bind to the bromo-associated homology (BAH) domain in the N-terminal region of Sir3, although no direct binding of these two domains has yet been detected (Connelly et al. 2006). Thus, these two similar domains within Sir1 may act as a scaffold to bring Orc1 and Sir3 into juxtaposition. In addition to binding Orc1, the C-terminal region of Sir1 also binds the Sir4 protein (Bose et al. 2004), which is recruited to silencers through its interaction with Rap1, and presumably Sir1 as well. Sir4 is in a complex with Sir2 and Sir3 (Moazed and Johnson 1996; Moazed et al. 1997). Recruitment of the complex to the silencers allows Sir2 to deacetylate key lysines on the tails of neighboring histone H3 and H4, creating high-affinity binding sites for the Sir2, Sir3, and Sir4 complex, promoting spreading of Sir proteins across the HML and HMR loci (Rusche et al. 2003).
Sir1 allows stable heterochromatin formation by efficiently nucleating the Sir silencing complex on the E and I silencers, yet Sir1 is not absolutely required for silencing. In the absence of Sir1, a clonal population of cells achieves an equilibrium with two heritably different subpopulations of cells, one with and one without silencing at HML and HMR (Pillus and Rine 1989; Xu et al. 2006). Telomeric silencing, which is a weaker form of silencing than that at HML and HMR, requires Sir2, Sir3, and Sir4 but not Sir1 (Gottschling et al. 1990; Aparicio et al. 1991). Nevertheless, tethering of a Sir1–Gal4 fusion protein to a telomere strengthens its silencing (Chien et al. 1993).
Silencing mechanisms in yeasts range from the Sir-based mechanism of Saccharomyces cerevisiae, to the RNAi-based mechanism of Schizosaccharomyces pombe. With the exception of Sir2/Hst4, the structural proteins of heterochromatin in Saccharomyces bear little sequence similarity to the heterochromatin proteins in S. pombe. Some progress has been made toward understanding the evolution of silencing, revealing a function of Sir2 and Sir4 proteins at least through genera as diverged from S. cerevisiae as Kluyveromyces lactis (Åström and Rine 1998). Sir1 homologs have not previously been identified in any species outside of the whole-genome-duplication clade of yeasts (Bose et al. 2004; Fabre et al. 2005). In S. cerevisiae, Sir protein-based heterochromatin can be replaced by compositionally unique but functionally equivalent heterochromatin just by changing a single amino acid in the meiotic repressor Sum1 (Rusche and Rine 2001). Given that Sir-based silencing can so easily be replaced, we undertook a study of heterochromatin and silencing in closely related species in search of insights into heterochromatin formation that have been inaccessible in comparisons among more distant species.
Our analysis of the genome sequences of yeast species that contain silent mating-type cassettes at HML and HMR (Cliften et al. 2003; Kellis et al. 2003) revealed that a family of SIR1 paralogs arose some time after the evolution of silent mating cassettes. In this study we explored the evolution of Sir1 with a focus on S. bayanus. S. cerevisiae and S. bayanus diverged after the genome duplication in hemiascomycetes (Wolfe and Shields 1997) and after the appearance of Sir1. On the basis of the extent of protein sequence divergence, S. bayanus is approximately as closely related to S. cerevisiae as mouse is to human. S. bayanus encodes single orthologs of Sir2 [plus its Homolog of Sir two (Hst) orthologs], Sir3, and Sir4. However, instead of one Sir1 protein as in S. cerevisiae, S. bayanus and other Saccharomyces species contain up to four Sir1 orthologs not previously identified (Bose et al. 2004). We tested whether the role of the single Sir1 in silencing in S. cerevisiae has been subdivided into multiple paralogs in other species, or whether these paralogs have other functions, and explored the possible implications of the positions of SIR1 orthologs in the genome on their expression.
MATERIALS AND METHODS
Yeast strains, oligonucleotides, sequences, and plasmids:
S. cerevisiae and S. bayanus strains are listed in Table 1, oligonucleotide sequences are listed in supplemental Table 1, and Sir1 orthologs and paralogs accession numbers are listed in supplemental Table 2. All S. bayanus strains were derived from CBS7001. Plasmids used in this study were based on pRS316 and are listed in supplemental Table 3. S. bayanus genes were knocked out by single-step gene replacement with HPHR, KANR, S. pombe HIS5, or Candida albicans URA3 (Goldstein and McCusker 1999; Goldstein et al. 1999), and epitope tagging with FLAG tag was carried out in a similar manner.
TABLE 1.
List of strains
| Name | Species (alias) | Genotype |
|---|---|---|
| JRY4621 | S. cerevisiae | MATα sir1∷LEU2 can1-100, his3-111 leu2-3,112, lys2Δ, trp1-1, ura3-1 |
| CBS7001 | S. bayanus | MATa/MATα prototroph |
| JRY8145 | S. bayanus | MATaho∷NAT, leu1-1 |
| JRY8146 | S. bayanus | MATα ho∷NAT, leu1-1 |
| JRY8147 | S. bayanus | MATaho∷NAT, ade2-2, his3-1, lys2-5, ura3-1 |
| JRY8148 | S. bayanus | MATα ho∷NAT, ade2-2, his3-1, lys2-5, ura3-1 |
| JRY8149 | S. bayanus | MATaho∷NAT, his3-1, lys2-5, ura3-1 |
| JRY8150 | S. bayanus | MATα ho∷NAT his3-1, lys2-5, ura3-1 |
| JRY8151 | S. bayanus | MATaho∷NAT, ade2-2, his3-1, lys2-5, trp-1, ura3-1 |
| JRY8152 | S. bayanus | MATα ho∷NAT, ade2-2, his3-1, lys2-5, trp-1, ura3-1 |
| JRY8153 | S. bayanus | MATaho∷NAT, his3-1, lys2-5, trp-1, ura3-1 |
| JRY8154 | S. bayanus | MATα ho∷NAT, his3-1, lys2-,5 trp-1, ura3-1 |
| JRY8155 | S. bayanus | JRY8153 SIR1-FLAG∷KAN |
| JRY8157 | S. bayanus | JRY8153 KOS1-FLAG∷KAN |
| JRY8159 | S. bayanus | JRY8153 KOS2-FLAG∷KAN |
| JRY8161 | S. bayanus | JRY8153 KOS3-FLAG∷KAN |
| JRY8165 | S. bayanus | JRY8149 sir1∷HPH |
| JRY8166 | S. bayanus | JRY8150 sir1∷HPH |
| JRY8169 | S. bayanus | JRY8149 kos1∷HPH |
| JRY8170 | S. bayanus | JRY8149 kos2∷HPH |
| JRY8173 | S. bayanus | JRY8149 kos2∷HPH |
| JRY8174 | S. bayanus | JRY8150 kos2∷HPH |
| JRY8177 | S. bayanus | JRY8149 kos3∷HPH |
| JRY8178 | S. bayanus | JRY8150 kos3∷HPH |
| JRY8181 | S. bayanus | JRY8153 sir1∷HIS5, kos1∷KAN, kos2∷HPH. kos3∷URA3 |
| JRY8182 | S. bayanus | JRY8154 sir1∷HIS5, kos1∷KAN, kos2∷HPH. kos3∷URA3 |
| JRY8185 | S. bayanus | JRY8153 sir1∷HIS5, kos1∷KAN, kos2∷HPH |
| JRY8186 | S. bayanus | JRY8154 sir1∷HIS5, kos1∷KAN, kos2∷HPH |
| JRY8189 | S. bayanus | JRY8153 sir1∷HIS5, kos1∷KAN, kos3∷URA3 |
| JRY8190 | S. bayanus | JRY8154 sir1∷HIS5, kos1∷KAN, kos3∷URA3 |
| JRY8193 | S. bayanus | JRY8153 sir1∷HIS5, kos2∷HPH, kos3∷URA3 |
| JRY8194 | S. bayanus | JRY8154 sir1∷HIS5, kos2∷HPH, kos3∷URA3 |
| JRY8197 | S. bayanus | JRY8153 kos1∷KAN, kos2∷HPH. kos3∷URA3 |
| JRY8198 | S. bayanus | JRY8154 kos1∷KAN, kos2∷HPH. kos3∷URA3 |
| JRY8201 | S. bayanus | JRY8153 sir1∷HIS5, kos1∷KAN |
| JRY8202 | S. bayanus | JRY8154 sir1∷HIS5, kos1∷KAN |
| JRY8205 | S. bayanus | JRY8153 sir1∷HIS5, kos2∷HPH |
| JRY8206 | S. bayanus | JRY8154 sir1∷HIS5, kos2∷HPH |
| JRY8209 | S. bayanus | JRY8153 kos1∷KAN, kos3∷URA3 |
| JRY8210 | S. bayanus | JRY8154 kos1∷KAN, kos3∷URA3 |
| JRY8213 | S. bayanus | JRY8153 sir1∷HIS3, kos3∷URA3 |
| JRY8214 | S. bayanus | JRY8154 sir1∷HIS3, kos3∷URA3 |
| JRY8217 | S. bayanus | JRY8153 kos1∷KAN, kos3∷URA3 |
| JRY8218 | S. bayanus | JRY8154 kos1∷KAN, kos3∷URA3 |
| JRY8221 | S. bayanus | JRY8153 kos2∷HPH, kos3∷URA3 |
| JRY8222 | S. bayanus | JRY8154 kos2∷HPH, kos3∷URA3 |
| JRY8237 | S. bayanus | JRY8150 matα∷URA3, hml:HIS5 |
| JRY8238 | S. bayanus | JRY8150 matα∷URA3, hml:HIS5, sir4∷KAN |
| JRY8239 | S. bayanus | JRY8150 matα∷URA3, hml:HIS5, sir1∷HPH |
| JRY8240 | S. bayanus | JRY8150 matα∷URA3,hml∷HIS5, kos1∷HPH |
| JRY8241 | S. bayanus | JRY8150 matα∷URA3, hml∷HIS5, kos2∷HPH |
| JRY8242 | S. bayanus | JRY8150 matα∷URA3, hml∷HI5S, kos3∷HPH |
| JRY8243 | S. bayanus | JRY8149 hmra1:URA |
| JRY8244 | S. bayanus | JRY8149 hmra1:URA3, sir4∷KAN |
| JRY8245 | S. bayanus | JRY8149 hmra1:URA3, sir1∷HPH |
| JRY8246 | S. bayanus | JRY8149 hmra1:URA3, kos1∷HPH |
| JRY8247 | S. bayanus | JRY8149 hmra1:URA3, kos2∷HPH |
| JRY8248 | S. bayanus | JRY8149 hmra1:URA3, kos3∷HPH |
| JRY8249 | S. bayanus | JRY8149 mata∷HIS5, hmra1:URA |
| JRY8250 | S. bayanus | JRY8149 mata∷HIS5, hmra1:URA3, sir4∷KAN |
| JRY8251 | S. bayanus | JRY8149 mata∷HIS5, hmra1:URA3, sir1∷HPH |
| JRY8252 | S. bayanus | JRY8149 mata∷HIS5, hmra1:URA3, kos1∷HPH |
| JRY8253 | S. bayanus | JRY8149 mata∷HIS5, hmra1:URA3, kos2∷HPH |
| JRY8254 | S. bayanus | JRY8149 mata∷HIS5, hmra1:URA3, kos3∷HPH |
| JRY8255 | S. bayanus | JRY8149 hmra1:URA3, sir1∷KAN, kos1∷HPH |
| JRY8257 | S. bayanus | JRY8149 hmra1:URA3, sir1∷HIS, kos2∷HPH |
| JRY8259 | S. bayanus | JRY8149 hmra1:URA3, sir1∷HYG,kos3∷HIS |
| JRY8261 | S. bayanus | JRY8149 hmra1:URA3, kos1∷KAN, kos2∷HPH |
| JRY8263 | S. bayanus | JRY8149 hmra1:URA3, kos1∷KAN, kos3∷HPH |
| JRY8265 | S. bayanus | JRY8149 hmra1:URA3, kos2∷HPH, kos3∷HIS |
| JRY8269 | S. bayanus | JRY8153 sir1∷HYG, kos3∷KAN |
| JRY8273 | S. bayanus | JRY8153 kos1∷HYG, kos3∷KAN |
| JRY8277 | S. bayanus | JRY8153 kos2∷HYG, kos3∷KAN |
| JRY8279 | S. bayanus | JRY8153 SIR3-FLAG |
| JRY3352 | S. cerevisiae | MATa, hmr∷TRP1 sir1∷LEU2, leu2-3,112, his3-11, lys2Δ, trp1-1, ura3-1, can1-100 |
| JRY8951 | S. cerevisiae | MATa, hmlα2∷ADE2 sir1∷LEU2, leu2-3,112, his3-11, lys2Δ, trp1-1, ura3-1, can1-100 |
Zygosaccharomyces rouxii KOS3 was sequenced on a plasmid clone isolated from a genomic DNA library of strain CBS 732. The S. bayanus KOS3 sequence in GenBank (accession no. AACG02000101) has an error that results in a frameshift in the ORF. We sequenced the gene, identified the error, and deposited the corrected sequence (accession no. EU880229). The S. bayanus SIR4 sequence in GenBank (accession nos. AACA01000411 and AACA01000334) was on two contigs. Therefore we cloned and sequenced the complete gene (accession no. FJ472632).
Sequence analysis:
Sir1 family protein sequences were aligned by Clustal W. All species contained one gene with substantially greater similarity to S. cerevisiae Sir1 than the paralogs, with E-values between 9.5e-272 and 2.6e-206 via BLASTp (Figure 1); the E-values from comparison of Sir1 to Kos proteins were between 3.7e-25 and 2e-9.
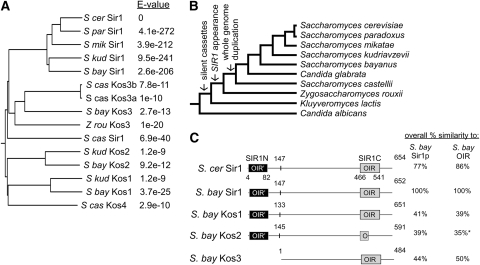
Figure 1.—
SIR1 family represented a rapidly evolving family of paralogs that diverged after the whole-genomewide duplication. (A) N-J bootstrap tree of SIR1 paralogs with E-value of alignment with S. cerevisiae Sir1. (B) Evolutionary tree of several Saccharomyces species and other yeast (Wolfe 2006). (C) Representation of S. cerevisiae Sir1 protein and paralogs from S. bayanus. Sir1 protein from S. bayanus was aligned with Sir1 from S. cerevisiae and with paralogous sequences from S. bayanus, called Kos1–3 (Kin of Sir1). The OIR is boxed. The amino-terminal duplication of the OIR is represented by a solid box labeled OIR′. Amino acid similarity to the full-length S. bayanus Sir1 protein and to its OIR was determined by BLAST. *Kos2 contained significant gaps in the OIR alignment compared to S. cerevisiae OIR. The similarity to S. bayanus OIR was based only on the partial alignment.
Growth and transformation of S. bayanus:
S. bayanus strain CBS7001 was used in this study. Standard S. cerevisiae yeast media and lithium acetate transformation conditions were used, except cells were heat-shocked for 5 min during transformations and subsequently grown at 25°. The coding region for the FLAG epitope was fused at the 3′ end of each Sir1 orthologous ORF in S. bayanus.
Site-directed mutagenesis:
M1R, M25R, and M25A mutations were made in S. cerevisiae SIR1 in pRS316 (Gardner and Fox 2001). Independently isolated mutant plasmids from Quick-change PCR (Stratagene) were sequenced to confirm the point mutation and transformed into the sir1∷LEU2 strain (JRY4621). S. cerevisiae SIR1 was mutagenized to create M1R (pJR2793), M25R (pJR2794-5), and M25A (pJR2796-7).
Immunoblots:
A S. cerevisiae sir1 strain (JRY4621) was transformed with pRS316 with S. cerevisiae SIR1-HA (pJR2793-7) and grown in supplemented minimal medium lacking uracil (CSM −Ura selective medium). Sir1-3xHA was immunoprecipitated from whole-cell extracts with anti-HA resin and detected on an immunoblot with anti-HA antibody (Sharp et al. 2002, 2003). S. bayanus Sir1 paralogs tagged with the FLAG epitope were immunoprecipitated from whole-cell extracts with anti-FLAG M2 resin and detected by immunoblotting with rabbit anti-FLAG antibody (Sigma).
Chromatin immunoprecipitation:
Cultures were grown in rich medium (YPD) to mid-log phase, and 50 OD600 units of cells with FLAG-tagged Sir1, Kos1, Kos2, Kos3, or Sir3, and were treated with 1% formaldehyde for 2 hr. Chromatin immunoprecipitation was then carried out as in Kuras and Struhl (1999). The co-immunoprecipitated DNA was amplified with primers specific to the predicted E and I elements of HML and HMR from S. bayanus. Values shown are fold enrichment from biological triplicates relative to the actin gene, ACT1.
Quantitative-reverse transcription PCR:
Total RNA was isolated by hot phenol extraction (Ausubel et al. 1995) from cultures at mid-log phase. Once precipitated, 10 μg of RNA were treated with 4 μl of Invitrogen RNAse-free DNAse for 20 min at room temperature and then precipitated in ethanol. The resulting 2 μg of RNA was then converted to cDNA using Invitrogen SuperScript III First-Strand kit. The 10 ng of cDNA was amplified in triplicate using specific primers in Finnzyme SYBR Green on a Stratagene MX3000 real-time PCR system. ACT1 mRNA was used for normalization.
Mating assay:
Cells were grown overnight in YPD and then diluted to 1 × 107 cells/ml and then serially diluted threefold onto minimal medium either along with 1 × 107 cells/ml of a mating-type-tester strain or spotted onto a lawn of tester strain. This method allowed semiquantitative measurement of relative mating strength of both mating types. Plates were photographed after 3 days at 25°. To test for cross-species complementation by S. cerevisiae SIR1 in MATa S. bayanus, mutants were transformed with empty plasmid or plasmid encoding S. cerevisiae SIR1. Cells were grown on selective medium overnight and then replica plated onto YM and the MATα mating-type tester.
Shmoo assay:
A line of MATα cells were streaked onto solid YPD and incubated for 2 hr. Then 50–100 small unbudded freshly grown MATa cells were micromanipulated next to the MATα cells. The majority of MATa cells either arrested division and formed a shmoo or continued division, forming a bud, which upon the completion of cell division resulted in a pair of shmoon (plural of shmoo). A small fraction of cells arrested division without forming a shmoo and hence were ambiguous with respect to HML silencing.
RESULTS
The availability of sequenced genomes from closely related yeast species allowed us to examine the evolution of proteins with roles in heterochromatic silencing in the Saccharomyces sensu stricto species. On the basis of the best matches from reciprocal BLAST analysis, the Sir2, Sir3, and Sir4 genes of S. cerevisiae each had one obvious ortholog in each of the sensu stricto species. For Sir2 the extent of amino acid sequence divergence was characteristic of the genome as a whole (82% identical between S. bayanus and S. cerevisiae), whereas Sir4 and Sir3 were more diverged than the other silencing proteins (43 and 59% identical, respectively; O. Zill and J. Rine, unpublished results). In contrast to these three silencing proteins, the Sir1 family has undergone dramatic expansions and contractions in the different species.
Organization of SIR1 homologs in Saccharomyces species:
The gene in each species with the highest degree of similarity to S. cerevisiae's SIR1 was designated SIR1, with its paralogs referred to as KOS for kin of Sir1. We found SIR1 orthologs in S. paradoxus, S. mikatae, S. kudriavzevii, S. bayanus, and S. castellii (Figure 1A). The SIR1 genes from these yeast species all contained the C-terminal OIR (Gardner et al. 1999) which binds the BAH domain of Orc1 (Bose et al. 2004) and the N-terminal duplication of the OIR, called OIR′ (or SIR1N) (Connelly et al. 2006). The Sir1's from the different species were between 77 and 58% identical to S. cerevisiae Sir1 across the entire protein sequence (Table 2). SIR1 was not found in C. glabrata (Figure 1B), which contains silenced mating-type cassettes and shared a common ancestor with S. cerevisiae after the whole-genome duplication (Butler et al. 2004; Conant and Wolfe 2006). The Sir1 found in S. castellii was the most divergent ortholog (Bose et al. 2004).
TABLE 2.
Pairwise alignment of Sir1 orthologs and paralogs
| S cer Sir1 | S kud Sir1 | S bay Sir1 | S cas Sir1 | S kud Kos1 | S bay Kos1 | S kud Kos2 | S bay Kos2 | S bay Kos3 | S cas Kos3a | S cas Kos3b | Z rou Kos3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S par Sir1 | 77 | 67 | 61 | 23 | 22 | 22 | 21 | 22 | 23 | 21 | 22 | 23 |
| S mik Sir1 | 67 | 63 | 61 | 21 | 23 | 22 | 21 | 22 | 21 | 22 | 22 | 23 |
| S kud Sir1 | 63 | |||||||||||
| S bay Sir1 | 58 | 66 | ||||||||||
| S cas Sir1 | 22 | 24 | 24 | |||||||||
| S kud Kos1 | 22 | 21 | 22 | 23 | ||||||||
| S bay Kos1 | 23 | 21 | 22 | 24 | 65 | |||||||
| S kud Kos2 | 18 | 20 | 21 | 22 | 21 | 21 | ||||||
| S bay Kos2 | 22 | 20 | 21 | 21 | 26 | 25 | 62 | |||||
| S bay Kos3 | 25 | 19 | 21 | 21 | 22 | 22 | 19 | 20 | ||||
| S bay Kos3a | 25 | 21 | 23 | 21 | 24 | 20 | 19 | 21 | 37 | |||
| S cas Kos3b | 25 | 20 | 23 | 21 | 24 | 21 | 19 | 21 | 37 | 98 | ||
| Z rou Kos3 | 22 | 21 | 26 | 22 | 21 | 22 | 20 | 21 | 29 | 29 | 26 | |
| S cas Kos4 | 24 | 23 | 25 | 24 | 26 | 26 | 24 | 23 | 22 | 23 | 23 | 23 |
Sequence identity and similarity of Sir1 orthologs and Kos paralogs. All Sir1 family members from S. cerevisiae, S. kudriavzevii, S. bayanus, S. castellii, and Z. rouxii were aligned by BLASTp. Numbers shown represent percentage of identity. Sequences with identity >50% are in boldface type and between 26% and 29% are in italics.
Surprisingly, most species contained additional paralogs of Sir1. The paralogs of SIR1 within each species were designated KOS for Kin of Sir1, followed by a number. The Kos proteins encoded by three genes shared obvious similarity to Sir1 in the C-terminal OIR domain (Figure 1C). The Kos proteins were distinguished from S. cerevisiae Sir1 protein either by their weaker similarity to the Sir1-defining N-terminal OIR′ of S. cerevisiae or in the case of Kos3, by the absence of an OIR′. Nevertheless the N termini of Kos1, Kos2, and Kos4 all had similar lengths to Sir1 and aligned to the OIR′ region of Sir1 (supplemental Figure 1). The S. bayanus genome encoded the most divergent set of Sir1-related proteins, one Sir1 and three Kos proteins.
Phylogenetic analysis (Figure 1A) revealed that the Sir1, Kos1, Kos2, Kos3, and Kos4 proteins define five clades, all approximately equidistant from each other (Table 2). The distribution of genes among species was variable: among the sensu stricto species, Sir1 is present in all five species but Kos1 is present in only two species. This pattern implies multiple gene losses during evolution. S. castellii lacked KOS1 and KOS2 but had another gene, KOS4, a KOS3 paralog discussed later.
Origin and evolution of the Sir1 family from the Kos3 founder:
S. bayanus, S. castellii, and Z. rouxii contained a further SIR1-related gene, designated KOS3, which was much shorter than the other SIR1 and KOS genes. The predicted Kos3 protein lacked the first 145 amino acids corresponding to the OIR′ domain of Sir1 (Figure 1C and supplemental Figure 1) (Connelly et al. 2006). However, the N termini of the Kos3 proteins were strongly conserved relative to each other (supplemental Figure 1). The remainder of the Kos3 proteins were 22–25% identical to Sir1 proteins and were slightly more similar than the other Kos1 and Kos2 proteins to S. cerevisiae Sir1 (Table 2). S. castellii contained two nearly identical KOS3 genes (KOS3a and KOS3b), encoding proteins differing in only five amino acids. Among the Sir1 and Kos paralogs, the Kos3 proteins were the most diverged between species.
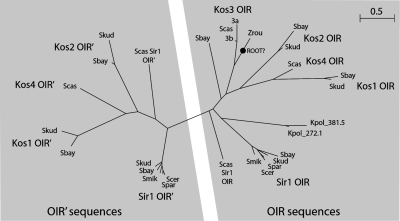
Z. rouxii KOS3 was apparently the only member of the Sir1/Kos family in a species that did not undergo the whole genome duplication. The Z. rouxii genome has not yet been completely sequenced, but shotgun coverage to approximately 1× depth on two different strains (Butler et al. 2004; Gordon and Wolfe 2008) did not reveal any additional members of the family in this species. This observation raised the possibility that Z. rouxii Kos3 was an outgroup to all the other sequences in our analysis and therefore that Kos3 represented the ancestral gene. By this view, other family members originated from it by duplication and diversification. It was interesting that Kos3 contains only a single OIR domain. To investigate the timing of the OIR tandem domain duplication relative to the gene duplication and speciation events, we constructed a phylogenetic tree of the OIR and OIR′ domains themselves (Figure 2). In this analysis we included OIR-like domains from two proteins of K. polysporus, a postwhole-genome-duplication species that lacked full-length Sir1/Kos homologs but which contained two proteins in which an OIR domain was fused to a putative helicase similar to S. cerevisiae Yrf1 helicase. The OIR and OIR′ domains are short, making reliable phylogenetic analysis difficult, but the unrooted tree (Figure 2) showed a clear separation of OIR domains from OIR′ domains, which indicated that the two-domain structure originated only once during evolution. This topology confirmed that the divergent N termini of Kos1, Kos2, and Kos4 indeed contained OIR′ sequences. The single OIR of Kos3 clustered with the clade of C-terminal OIRs. When we rooted the tree on the Z. rouxii branch as suggested above, the tree's branch lengths implied massive acceleration of the rate of sequence evolution of both the OIR and the OIR′ domains after the genome and domain duplications.
Figure 2.—
Phylogenetic tree of OIR and OIR′ domains. A possible position for the root, based on the species phylogeny, is marked. The domains (94- to 123-residues long) were aligned using MUSCLE (Edgar 2004). The tree was constructed by maximum likelihood using PHYML (Guindon and Gascuel 2003) using the JTT substitution model and four rate classes. Branch lengths are drawn to scale, indicating the number of amino acid substitutions per site. Bootstrap support for the branch separating OIR and OIR′ domains was 94% (100 replicates).
The variation in the number of Sir1 paralogs in the different species was striking. The most parsimonious interpretation, given the species tree, was that the expansion of the Sir1 family occurred prior to the ancestor of the sensu stricto species, but that many gene copies were later eliminated, including loss of the entire set of KOS genes on the branch leading to S. cerevisiae, S. paradoxus, and S. mikatae. Furthermore, considering the existence of KOS3 in Z. rouxii and of paralogs in S. castellii, Sir1 and its paralogs were all presumably lost in C. glabrata.
A revision of the Sir1 protein primary structure:
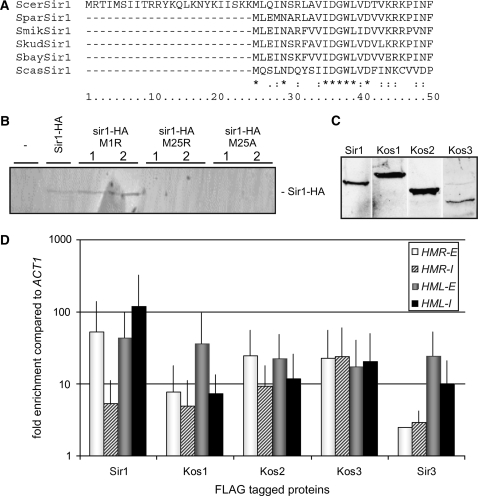
Comparison of all the Sir1 and Kos proteins across the Saccharomyces species suggested that the sequence of Sir1 of S. cerevisiae as represented in the Saccharomyces Genome Database (SGD) and earlier publications (Stone et al. 1991; Gardner et al. 1999; Gardner and Fox 2001; Hou et al. 2005; Hsu et al. 2005; Connelly et al. 2006) contains a 24 amino acid N-terminal extension compared to most other species. The S. cerevisiae SIR1 ORF contains three in-frame methionine codons, 1 (Met1), 5 (Met5), and 25 (Met25) residues into the ORF, the latter of which corresponded to the first methionine codon in the Sir1 ORFs of other species (Figure 3A). Given that some Saccharomyces species had multiple Sir1 paralogs, the three in-frame methionine codons in the putative N-terminal extension of S. cerevisiae suggested that it might produce two Sir1 proteins of differing length from the same gene by translating from different start codons. We changed two potential start codons, Met1 and Met25, to arginine or serine codons by site-directed mutagenesis of a C-terminally epitope-tagged version of S. cerevisiae SIR1 (SIR1-3xHA) (Gardner and Fox 2001) and found that HA-tagged Sir1 of identical mobility was detected from cells expressing the wild-type ORF and the Met1-Arg mutant ORF. No Sir1-3xHA protein was detected either in cells carrying a vector control or in cells with a SIR1 gene in which the Met25 codon was mutated to arginine or serine codon (Figure 3B). Therefore, amino acids 1–24 of S. cerevisiae's Sir1 ORF were not translated into protein from S. cerevisiae SIR1 mRNA. S. paradoxus Sir1 ORF, which is slightly longer at the N terminus than the revised Sir1 ORF of S. cerevisiae, also contains a methionine codon at the position corresponding to the start codon of Sir1 from all other species. We inferred that the N-terminal extension in this species was an annotation artifact, and a corrected sequence was included in supplemental Figure 1.
Figure 3.—
Expression, translation, and silencer localization of S. cerevisiae SIR1 and the four paralogs of SIR1 in S. bayanus. (A) Clustal W alignment of N terminus of Sir1 orthologs as provided from the Saccharomyces Genome Database. (B) Immunoblot of S. cerevisiae Sir1 immunoprecipitated from yeast expressing C-terminally HA-tagged SIR1 or mutated sir1 from a plasmid. (C) Immunoblot of FLAG-tagged Sir1 and Kos proteins immunoprecipitated. Kos1 migrated anomalously slower than Sir1. Kos1 had a calculated pI of 7.25 compared to 5.55 of Sir1, 5.2 of Kos2, and 7.9 of Kos3. (D) Chromatin immunoprecipitations of FLAG-tagged S. bayanus Sir1 paralogs at HML and HMR silencer.
Localization of Sir1 and Kos proteins at the silencers:
Because S. bayanus contained the most diverse set of SIR1 and KOS genes (Figure 1A), we investigated their roles in formation of silenced chromatin. As a first step, we created a series of C-terminal FLAG-tagged versions of S. bayanus Sir1, Kos1, Kos2, and Kos3, all of which were functional in complementing the phenotype of null alleles of the corresponding genes (see below), and evaluated their expression by immunoblotting with anti-FLAG antibody (Figure 3C). As for S. cerevisiae Sir1 protein (Gardner and Fox 2001), the S. bayanus Sir1 and Kos proteins were expressed at low levels and could be visualized only by immunoblotting of immunoprecipitated samples. There was a surprising difference in gel migration of Sir1, Kos1, and Kos2, all of which had similar calculated molecular weights.
Having established that all four paralogs were translated, we assayed their localization at the S. bayanus silencers. In S. cerevisiae, Sir1 binds to the E and I silencers of HML and HMR. The potential occupancy of the Sir1/Kos proteins of S. bayanus at the E and I silencers of HML and HMR was determined by chromatin immunoprecipitation (ChIP). As in S. cerevisiae, E and I silencers of S. bayanus each contain a match to the S. cerevisiae ARS consensus sequence, which serves as binding site for the ORC complex, and binding sites for Abf1 and Rap1 (Teytelman et al. 2008). All four paralogs were found at all silencers, with rather high levels of enrichment relative to the ACT1 control locus. Sir1, Kos1, and Kos2 showed preference for E elements, whereas Kos3 was found at similar levels at all four silencers (Figure 3D).
Role of Sir1 and Kos proteins in silencing HMR and HML:
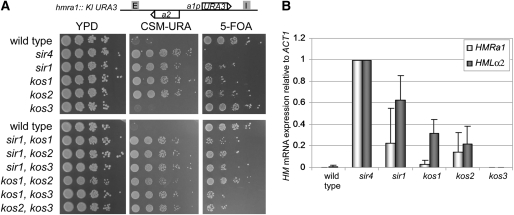
Given the presence of all Sir1 paralogs at HML and HMR, we evaluated whether they shared the silencing function found in the single Sir1 protein of S. cerevisiae by several assays. In the first assay we used a strain of S. bayanus with the K. lactis URA3 gene integrated in place of the HMRa1 ORF, placing URA3 under the control of the a1 promoter. At this position, URA3 is fully silenced in otherwise wild-type S. bayanus cells (O. Zill, personal communication). The hmra1∷Kl URA3 reporter strain was crossed with strains containing a deletion of SIR1, KOS1, KOS2, or KOS3 genes. sir1, kos1, and kos2 single mutants grew on minimal medium lacking uracil and grew poorly on medium containing 5-FOA (Figure 4A). kos3 mutants were indistinguishable from wild type. Therefore SIR1, KOS1, and KOS2 each contributed to silencing of HMR, but in no case did loss of one of these genes lead to full derepression.
Figure 4.—
Derepression of HML and HMR loci in S. bayanus mutants of SIR1 paralogs. (A) Derepression of hmra1∷Kl URA3 in S. bayanus allowed growth on CSM −Ura, and repression allowed growth on 5-FOA media. (B) Quantitative reverse transcription (QRT)–PCR of HMLα2 mRNAs from S. bayanus lacking the HMR and MAT loci, and of HMRa1 from cells lacking HML and MAT loci, normalized to levels in sir4 cells.
Given that at least three of these genes contributed to silencing HMR, we tested whether their contributions were additive or otherwise by measuring the effect of all possible sir1/kos double mutants on hmra1∷Kl URA3 expression. The most significant insight from the double-mutant analysis was that kos2, kos3 double mutant had a greater silencing defect at HMR as it grew less on 5-FOA medium than either single mutant, revealing partially overlapping roles of Kos2 and Kos3 in silencing HMR (Figure 4A). In summary Sir1 paralogs were at the HMR silencer, and contributed to silencing of HMRa1∷Kl URA3 reporter, although the contribution of Kos3 was masked by the contribution of Kos2.
To provide an independent assessment of the role of S. bayanus SIR1 and KOS genes in silencing, we used quantitative reverse transcription (QRT)–PCR to measure HML and HMR silencing. This analysis required care to avoid complications from the autoregulation of mating-type genes by mating type itself. Specifically, compared to haploid cells, MATa/α diploids downregulate a1 mRNA 50% and α2 mRNA 15% in S. bayanus and in S. cerevisiae (J. E. G. Gallagher and O. Zill, unpublished data). To circumvent the complications from autoregulation, S. bayanus sir1/kos mutant strains were made that contained no MAT locus and retained either HMR or HML, but not both. Expression from sir4 mutants containing no MAT locus and only HML or HMR as a source of mating-type genes provided the benchmark for complete derepression.
HMLα2 was derepressed in sir1 and kos mutants relative to the parental control, but less than in the sir4 mutant (Figure 4B). Of the sir1/kos mutants, the sir1 mutant exhibited the most derepression, with kos1 and kos2 mutants expressing similar levels of HMLα2. In sir1, kos1, and kos2 mutants, HMRa1 was derepressed to a lesser extent than in a sir4 mutant, whereas there was no expression of HMRa1 or HMLα2 in kos3 mutants (Figure 4B), as seen above for the hmral∷Kl URA3 reporter.
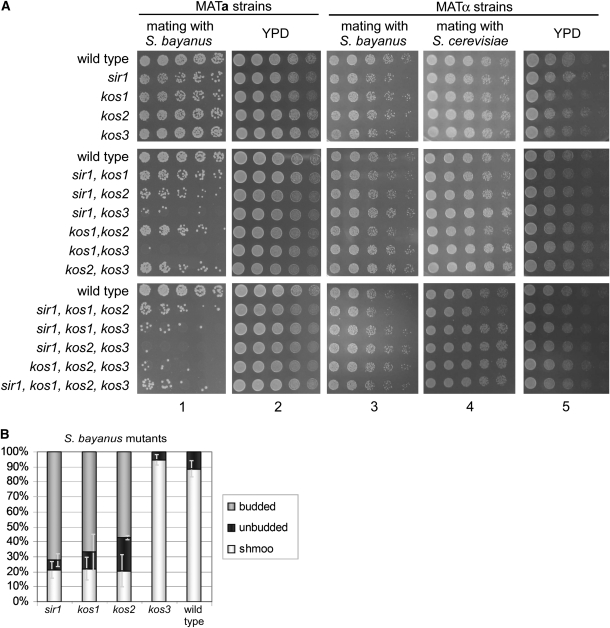
A comprehensive survey of mating defects in single, double, triple, and quadruple mutants of sir1 and kos genes was determined by mating tests to S. bayanus MATa and MATα tester strains. With respect to HML, single sir1 and kos1 MATa mutants displayed a slightly reduced level of mating compared to wild type, kos2, or kos3 mutants (Figure 5A, column 1), indicative of derepression of HMLα. Stronger mating defects were obvious in all double mutants that included the kos3 null mutation. The sir1 kos1 double mutant was more similar to the corresponding single mutants than any other double mutants were to their corresponding single mutants. Formally, it would appear that Sir1 and Kos1 were jointly required to perform a common function in silencing HML. In contrast, these analyses indicated that the contribution of Kos3 to HML silencing was the most dissimilar to the contributions of the other paralogs, since cells lacking both Kos3 and one other paralog were the most different from the corresponding single mutants. All single and multiple mutants had approximately the same plating efficiency as wild type. Thus, no combination of SIR1 and KOS gene mutations had a measurable effect on viability under these conditions.
Figure 5.—
Mating efficiencies of S. bayanus single, double, triple, and quadruple mutants of SIR1 paralogs. (A) S. bayanus MATa strains mated to a lawn of S. bayanus MATα mating-type tester cells (column 1). S. bayanus MATα strains were mated to a lawn of S. bayanus MATa mating-type tester strain (column 3) and to a S. cerevisiae mating-type tester (column 4). Growth of mutants on YPD is shown in columns 2 and 5, at the same dilutions as used on the mating tester plates. (B) Efficiency of shmoo formation was measured for MATa strains with single mutants of SIR1 paralogs in the presence of α-factor. The percentage of cells that formed a shmoo is open and those that budded are shaded. Cells that remained small and failed to bud are solid. The standard deviation of cells that formed shmoon is shown to the left within each bar and the standard deviation of cells that budded is shown to the right.
With respect to HMR, only slight mating defects were evident in mutant MATα cells with combinations of mutations in the paralogs (Figure 5A, column 3). The modest mating defects in these MATα cells indicated derepression of HMR. This result qualitatively mirrored the QRT–PCR and hmral∷Kl URA3 reporter strain. The parental S. bayanus strain contained a nonsense mutation in the bar1 gene, which leads to hypersensitivity of MATa stains to α-factor (Zill and Rine 2008). To rule out any possible unknown influence of exaggerated cell-cycle arrest of MATa tester cells, we repeated the mating assay with a S. cerevisiae MATa mating-type tester lawn and found the same lack of strong derepression of HMR (Figure 5A, column 4). Hence all four genes from the SIR1 family contributed to HML and HMR silencing, although no single assay was sufficient to reveal all the subtleties in the phenotype.
Analysis of silencing at the single-cell level:
Conventional patch mating tests, most reporter assays, and QRT–PCR assays of gene expression evaluate the average phenotype of millions of cells, which can mask interesting variation at the single-cell level. Because of the mild mating defect in single mutants in S. bayanus, we evaluated potential defects in silencing at the single-cell level, by testing the ability of MATa S. bayanus mutants to respond to α-factor. MATa yeast in which HMLα is silenced form shmoon in the presence of α-factor. In contrast, loss of HMLα silencing confers α-factor resistance. Shmoo formation was decreased to <20% for sir1, kos1, and kos2 single mutants, whereas wild-type and kos3 mutants were indistinguishable (Figure 5B).
Cross-species complementation of SIR1 paralogs:
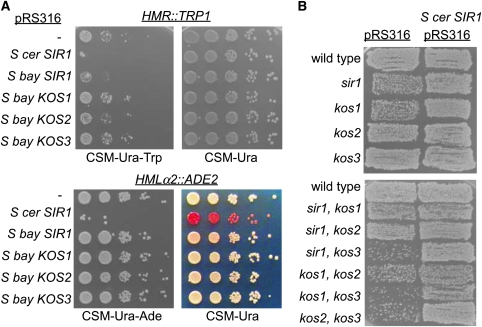
We tested the ability of the SIR1 orthologs of S. bayanus to complement the silencing defect of a S. cerevisiae sir1 mutant. Each SIR1/KOS gene with its promoter and terminator was amplified from genomic DNA and cloned into a CEN/ARS vector (pRS316 with the URA3 marker). The plasmids were transformed into a S. cerevisiae MATa sir1 mutant containing the hmr∷TRP1 reporter (Sussel and Shore 1991) and 10-fold serial dilutions on CSM −Ura −Trp were used to measure silencing of hmr∷TRP1. By this assay, S. bayanus SIR1 increased silencing of the hmr∷TRP1 in S. cerevisiae sir1 cells ∼10-fold and expression of KOS2 increased silencing ∼5-fold, whereas expression of KOS1 or KOS3 had no effect (Figure 6A). We independently assayed complementation by S. bayanus SIR1 and KOS genes with an hmlα2∷ADE2 reporter. While S. cerevisiae colonies expressing SIR1 were red and did not grow on medium lacking adenine, cells with S. bayanus SIR1 were pink on YPD (an intermediate silencing phenotype) and grew on CSM −Ura −Ade media. In contrast, S. bayanus KOS1, KOS2, or KOS3 had no ability to support silencing of HML in S. cerevisiae.
Figure 6.—
Cross-species complementation of SIR1 paralogs in S. bayanus and S. cerevisiae. (A) S. cerevisiae sir1 yeast contain the reporter hmr∷TRP1 or hmlα2∷ADE2, complemented by S. bayanus SIR1 paralogs expressed from pRS316 plasmids. (B) Patch mating tests of MATa S. bayanus sir1 or kos mutants carrying a plasmid with S. cerevisiae SIR1.
Patch mating assays were used to determine whether S. cerevisiae SIR1 could complement the silencing defect of sir1/kos mutants of S. bayanus (Figure 1C). Rescue by S. cerevisiae SIR1 of the mating defects in the S. bayanus double mutants revealed that S. cerevisiae Sir1 retained most of the Sir1/Kos functions in silencing. Thus, a function of SIR1/KOS genes was conserved between S. cerevisiae and S. bayanus, with the extent of complementation not directly proportional to the extent of sequence similarity.
Telomere position effect on the regulation of KOS gene expression:
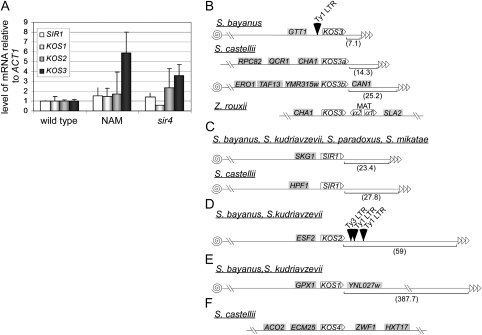
In general, the map position of genes in the Saccharomyces genome has given few clues as to their function or regulation. However, the map position of some SIR1 family members in S. bayanus and other species suggested the potential for a previously unrecognized form of heterochromatin regulation. S. cerevisiae and S. bayanus each contain 16 chromosomes, with only five translocations and three inversions distinguishing the species (Fischer et al. 2000). Thus, there is sufficient conservation in gene order that we could infer the chromosomal location of the SIR1 and KOS genes of S. bayanus and other species on the basis of the identities of flanking genes even though the genome of three other species is not yet fully assembled.
On the basis of chromosome coordinates from S. cerevisiae, we estimated that SIR1, KOS1, KOS2, and KOS3 genes of S. bayanus were 23, 389, 61, and 7 kb pairs, respectively, from their nearest telomeres. The telomere proximity of the KOS3 gene suggested it may be subject to telomeric position effect (TPE). To explore this possibility, the expression of the SIR1/KOS genes was measured in cells treated with nicotinamide (NAM), a competitive inhibitor of Sir2-dependent deacetylases, and in cells carrying a mutation in SIR4 (Figure 7A). Expression of SIR1, KOS1, and KOS2 was not significantly altered in nicotinamide-treated cells. In contrast, KOS3 was derepressed almost sixfold in cells treated with nicotinamide, and 3.5-fold in sir4 cells. KOS1 and KOS2 expression were only slightly affected in sir4 mutants. Thus KOS3 was one member of the SIR1 family that was itself, notably regulated by heterochromatin, presumably in the form of a telomere position effect.
Figure 7.—
Telomere-position-effect regulation of, and genomic organization of, SIR1 and KOS paralogs in Saccharomyces and Zygosaccharomyces. (A) Transcriptional regulation of SIR1 and KOS genes in S. bayanus. Levels of mRNA of SIR1 paralogs were measured by QRT–PCR from cells treated with nicotinamide (NAM) or from cells containing a sir4 mutation relative to ACT1 mRNA and then normalized to wild-type cells. (B–F) Chromosomes are depicted as a line, with telomeres as triangles and centromeres as spirals. On the basis of genes encoded on the same contig and using sequence from S. cerevisiae genome, each paralog's distance in kilobases from the telomeres was predicted as indicated in parentheses below the chromosome. The positions of S. cerevisiae Ty elements are labeled above the chromosome as triangles. The names of each flanking gene is in a shaded box. (B) Predicted locations of KOS3 paralogs in S. bayanus, S. castellii, and Z. rouxii. (C) Predicted locations of SIR1 in S. bayanus, S. kudriavzevii, S. paradoxus, S. mikatae, and S. castellii. (D) Predicted locations of KOS2 paralogs in S. bayanus and S. kudriavzevii. (E) Predicted locations of KOS1 paralogs in S. bayanus and S. kudriavzevii. (F) Predicted locations of KOS4 in S. castellii.
The genome organization of the SIR1 family:
To determine whether the telomere position of KOS3 in S. bayanus was evolutionarily conserved and to gain insights into how SIR1 family members were gained or lost, we extrapolated the position and genome organization of the SIR1 gene family in the Saccharomyces genera (Figures 7, B–F).
The genomic location of KOS3 differed in S. bayanus, S. castellii, and Z. rouxii. The telomere proximity of KOS3 in S. bayanus was recapitulated in S. castellii, in which KOS3 was duplicated, forming KOS3a and KOS3b (Figure 7B). Many S. castellii chromosomes were subject to rearrangements and gene loss (Cliften et al. 2006) reducing the number of chromosomes from 16 to 9 (Vaughan-Martini et al. 1993). Therefore, positioning in this species by synteny to S. cerevisiae was less reliable. With this caveat, KOS3a was on the same contig as CHA1 and QCR2, which in S. cerevisiae are near telomeric sequences of chromosome III (CHA1 is 15 kb from telomeric sequences) and XVI (QCR2 is 22 kb from telomeric sequences). Genes on the contig with S. castellii's KOS3b were between 20 kb (YMR135w) and 5 kb (ERO1) from telomeric sequences from both arms of chromosome XIII of S. cerevisiae. In Z. rouxii, KOS3, the presumptive founder of the SIR1 family, was not telomeric but was found beside the mating-type locus (J. L. Gordon and K. H. Wolfe, unpublished results; GenBank accession no. AM989983), apparently resulting from a chromosomal rearrangement that involved a recombination between the MAT locus and a telomeric silent cassette.
SIR1 was at the same chromosomal location in S. cerevisiae, S. bayanus, S. paradoxus, S. mikatae, and S. kudriavzevii, ∼23 kb pairs from the telomere (Figure 7C). The SIR1 of S. castellii was flanked by genes whose orthologs in S. cerevisiae are from telomere-proximal regions of chromosomes X and XV. Therefore, SIR1 was likely to be near a telomere in S. castellii, similarly to other species.
KOS2 from S. bayanus and S. kudriavzevii mapped to the telomere-proximal region of chromosome XVI, to the right of ESF2, ∼59 kb pairs from the telomere (Figure 7D). In the genomes of S. cerevisiae, S. paradoxus, and S. mikatae, which did not have KOS2 genes, there was a transposon at the corresponding region to the right of ESF2, suggesting a mechanism by which KOS2 may have been lost from these closely related species.
KOS1 from S. bayanus and S. kudriavzevii, the only two species to share this family member, was between GPX1 and YNL027w of S. cerevisiae, on the left arm of chromosome XI, 389 kb pairs from the closest telomere (Figure 7E). There were no obvious genomic clues to the origin or loss of KOS1 from Saccharomyces genomes.
S. castellii contained a highly diverged Kos protein, designated Kos4p, which was 47% similar to Kos1 and Kos2 proteins from S. kudriavzevii and S. bayanus. The contig containing S. castellii KOS4 also had genes whose orthologs in S. cerevisiae were from subtelomeric regions common to numerous chromosomes (Figure 7F). From the analysis of the chromosomal locations of KOS genes, we found the highest diversity in genes and gene order in the paralogs near the telomeres in all species.
DISCUSSION
Of the genes involved in heterochromatin formation in Saccharomyces species, the SIR1 gene family stands out due to its late appearance relative to mating-type cassettes, its divergence in gene number, the locations of these genes in genomes, and in at least one case, its mode of regulation. This study of the four S. bayanus Sir1 paralogs revealed roles for all of the paralogs in silencing, provided evidence for the divergence and subspecialization of their roles, and inspired a model for the evolution of this protein family.
The entire SIR1 family contributed to silencing in S. bayanus:
Using strains with null alleles of SIR1, KOS1, KOS2, or KOS3, and strains with combinations of these null alleles, we established that all four genes contribute to silencing HML and HMR by several independent assays. Thus, S. bayanus uses a family of Sir1 paralogs to accomplish what S. cerevisiae does with its single Sir1. Interestingly, the various assays were necessary in combination to reveal the subtleties of how each paralog contributed to silencing at each locus. HML and HMR were at least partially derepressed by null alleles of the SIR1, KOS1, or KOS2 paralogs. A null allele in KOS3 had no effect on its own on the silencing of HML or HMR.
The analysis of double mutants by the mating-based silencing assay was instrumental in revealing a relationship among the contributions of the Sir1 paralogs to silencing. In particular, double-mutant combinations of sir1, kos1, and kos2 were, to a first approximation, about as defective in silencing HML as was each single mutant on its own. Hence, it would appear that these three genes were jointly required to provide a common contribution to silencing. However, when any of these three single mutations was tested in combination with kos3, the silencing defect was much more pronounced than in any single mutant alone. Thus Kos3 contributed a different and complementary function to silencing HML.
Site of action of the Sir1 family members:
The effect of all the Sir1 paralogs on HML and HMR expression was likely to be a direct effect of those proteins acting at those loci. By ChIP analysis, all the Sir1 paralogs localized to both the E and I silencing elements of both HML and HMR. In S. cerevisiae, the recruitment of the single Sir1 species to silencers occurs through direct interaction between Sir1 and Orc1. It is presumed that there is a single Orc1 in the ORC complex and a single ORC complex bound to the ARS consensus sequence common to all silencers. Thus one would expect a single Sir1 ortholog molecule recruited to a silencer by direct binding of its OIR to the BAH domain of the single Orc1. However, because our data revealed that all four Sir1 orthologs were at each silencer, there would likely be other OIR (and OIR′) domains at silencers with the potential to interact with other proteins with BAH domains. Sir3 is an obvious candidate, and the potential for multivalent interactions among Sir3 with multiple silencer-binding Sir1 orthologs could serve to enhance the efficiency of establishing silencing, explaining in part the very weak silencing defects of mutations in individual SIR1 orthologs in S. bayanus. Of course, the multiple OIR and OIR′ domains would have the potential to recruit other proteins with BAH domains, such as Rsc1 and Rsc2. Clearly the nature of the superstructure assembled at S. bayanus silencers is worthy of deeper exploration.
In addition to silencers, S. cerevisiae's Sir1 protein is found at centromeres where it interacts with chromatin assembly factor I to promote centromere function (Sharp et al. 2003). It was possible that the dual functions of S. cerevisiae Sir1 were segregated into different paralogs in S. bayanus. However, our on-going work has shown that all four Sir1 paralogs in S. bayanus are resident at centromeres (J. E. G. Gallagher, unpublished data), suggesting that this subspecialization was not the reason for retaining multiple paralogs in S. bayanus.
Transcriptional regulation of KOS3 by its genomic location:
Three of the Sir1 paralogs from S. bayanus, S. kudriavzevii, and S. castellii are encoded by genes inferred to be within a modest distance of telomeres, with KOS3 being the closest, and potentially vulnerable to telomere position effects. Indeed, treatment of wild-type S. bayanus cells with nicotinamide, a competitive inhibitor of the Sir2 class of histone deacetylases, resulted in a sixfold increase of KOS3 expression. As expected, KOS3 was also derepressed in sir4 cells. Thus a gene encoding a protein that assists in the assembly of heterochromatin was itself regulated by heterochromatin, presumably through a telomere position effect. At this point it has not been possible to unambiguously determine whether the SIR1 paralogs contribute to telomere position effect because of the poor assembly of subtelomeric sequences in S. bayanus.
Evolution of the SIR1 family:
Prior to the detection of the KOS3 ortholog in Z. rouxii, it appeared as if the Sir1 family arose after the whole-genome duplication (Butler et al. 2004). However, we are now able to position the appearance of Sir1 some time after the evolution of the mating cassettes, but before the genome duplication. By this model, KOS3 would be the founding member of the family, with its loss in the lineage leading to C. glabrata. The internal duplication of the OIR in all SIR1 orthologs, but missing from KOS3 orthologs, implied an early duplication of KOS3, followed by a partial intragenic duplication of the OIR in one of the resulting genes. In this model, this gene with the duplicated OIR would have led to the other SIR1 family members. The evolution of the other remaining family members could have been facilitated by their telomere proximity and unequal crossing over in these regions.
The genome duplication in the Saccharomyces lineage occurred once, and duplicated gene blocks were lost both before and after speciation events (Langkjaer et al. 2003). If the last common ancestor of the Saccharomyces clade (Figure 1A) had four Sir1 paralogs, then there must have been multiple loss events leading to S. cerevisiae and its closest neighbors. Recombination among the σ- and two δ-transposons that occupy the position of KOS2 in species lacking it offered one mechanism for its loss. Because KOS1 was the only SIR1 paralog that was neither subtelomeric nor near a transposable element, some event must have moved this paralog from the subtelomeric birthplace of its orthologs. The proximity of some paralogs to telomeres suggested that the expansion and contraction of the family may be facilitated by the genomic churning in these neighborhoods, as suggested by the recent elaboration of KOS3a and KOS3b in S. castellii. One of the driving influences of gene duplication is the opportunity for neofunctionalization and diversification. However, at least in the case of the Sir1/Kos family of S. bayanus, all the members retained at least the function of silencing. Whether they have gained new functions remains to be determined.
Implications for the evolution of silencing:
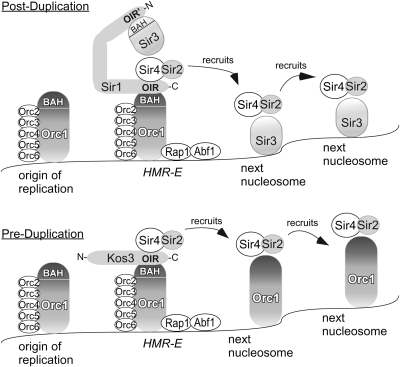
ORC1 and SIR3 are paralogs created by the whole-genome duplication (Scannell et al. 2007). Hence silencing of HML and HMR in preduplication species had only one of these two proteins to work with. Since ORC1 is essential and SIR3 is not, we designated ORC1 as the ancestor. There is substantial experimental support indicating that the OIR region of the Sir1 protein family interacts with the BAH domain of Orc1 (Bell et al. 1995; Triolo and Sternglanz 1996; Gardner et al. 1999; Zhang et al. 2002; Hou et al. 2005; Hsu et al. 2005; Connelly et al. 2006). Hence in the ancestral species, Kos3 was likely responsible for the recruitment of Sir2/Sir4 to the silencer. In these species it would seem possible that Orc1 may have served the structural role of Sir3 in heterochromatin in addition to its role in replication (Figure 8). There is indirect evidence that in S. cerevisiae the N-terminal OIR′ domain in Sir1 interacts with the BAH domain of Sir3, allowing Sir1 to act simultaneously as a recruitment agent for bringing Sir proteins to the silencer, and potentially as a scaffold to help assemble a Sir protein complex (Connelly et al. 2006). The affinity of Sir3 for deacetylated nucleosome tails would then provide a mechanism of spreading additional Sir complexes. This model provides an explanation for how the ancestral Kos3, with only a single OIR domain, could still support silencing, and suggests that Orc1 in preduplication species may have some undiscovered link to histone H3 and H4 tails. A limitation of this model is that it offers little insight into why S. bayanus would need four paralogs to accomplish what S. cerevisiae, and presumably Z. rouxii, accomplish with one.
Figure 8.—
Evolution of silencing in the pre- and postduplication hemiascomycetes. The duplication of the OIR in the KOS3 ancestor corresponded with the whole-genome duplication of the ORC1/SIR3 ancestor. A simple prediction of the model was that Orc1 replaced Sir3 in heterochromatin from pregenome duplication species. Additional details are provided in the text.
One possibility for why four Sir1 paralogs were required for silencing in S. bayanus was that the intrinsic structure of the silencer was more complex than in S. cerevisiae. This model was ruled out by the ability of the S. cerevisiae Sir1 protein to replace the silencing functions of the S. bayanus orthologs, at least to a first approximation. A second possibility was that four Sir1 paralogs might offer a flexible regulatory response, with each protein optimized for responding to varying parameters in the environment. The challenge to this model is that there are no environmental conditions known to regulate silencing in S. cerevisiae, although the possibility has not been adequately explored in S. bayanus. A third possibility was that each of these paralogs has additional roles in the cell beyond silencing of HML and HMR, and presumably beyond centromere binding, that selected for their maintenance.
Acknowledgments
We thank David Wynne, Danae Schultz, Jason Zemansky, and Jessica Cande for their contributions to the construction of S. bayanus sir1 and kos mutant strains and cloning of SIR1 paralogs. We thank Oliver Zill for S. bayanus auxotrophic, sir4, and hmra1∷Kl URA3 mutant strains and Ed Louis for the original S. bayanus prototrophic strain. We thank Laura Lombardi and Debbie Thurtle for the S. cerevisiae hmlα2∷ADE2 strain and Marita Cohn for advice about S. castellii. J.E.G.G. was supported by a National Science Foundation postdoctoral fellowship (DBI-0511799) and by a National Institutes of Health (NIH) Genomics Training grant (T32 HG 000047) and L.T. by a National Science Foundation predoctoral fellowship. Additional support was provided by a grant from the NIH (GM31105 to J.R.) and by Science Foundation Ireland (to K.H.W.).
References
- Aparicio, O. M., B. L. Billington and D. E. Gottschling, 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66 1279–1287. [DOI] [PubMed] [Google Scholar]
- Åström, S. U., and J. Rine, 1998. Theme and variation among silencing proteins inSaccharomyces cerevisiae and Kluyveromyces lactis. Genetics 148 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1995. Short Protocols in Molecular Biology. John Wiley & Sons, New York.
- Bell, S. P., J. Mitchell, J. Leber, R. Kobayashi and B. Stillman, 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83 563–568. [DOI] [PubMed] [Google Scholar]
- Bose, M. E., K. H. McConnell, K. A. Gardner-Aukema, U. Muller, M. Weinreich et al., 2004. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 24 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin et al., 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 101 1632–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, C. T., S. Buck, R. Sternglanz and D. Shore, 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75 531–541. [DOI] [PubMed] [Google Scholar]
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton et al., 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301 71–76. [DOI] [PubMed] [Google Scholar]
- Cliften, P. F., R. S. Fulton, R. K. Wilson and M. Johnston, 2006. After the duplication: gene loss and adaptation in Saccharomyces genomes. Genetics 172 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant, G. C., and K. H. Wolfe, 2006. Functional partitioning of yeast co-expression networks after genome duplication. PLoS Biol. 4 e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, J. J., P. Yuan, H. C. Hsu, Z. Li, R. M. Xu et al., 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 26 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, E., H. Muller, P. Therizols, I. Lafontaine, B. Dujon et al., 2005. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 22 856–873. [DOI] [PubMed] [Google Scholar]
- Fischer, G., S. A. James, I. N. Roberts, S. G. Oliver and E. J. Louis, 2000. Chromosomal evolution in Saccharomyces. Nature 405 451–454. [DOI] [PubMed] [Google Scholar]
- Fox, C. A., A. E. Ehrenhofer-Murray, S. Loo and J. Rine, 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276 1547–1551. [DOI] [PubMed] [Google Scholar]
- Gardner, K. A., and C. A. Fox, 2001. The Sir1 protein's association with a silenced chromosome domain. Genes Dev. 15 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, K. A., J. Rine and C. A. Fox, 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., X. Pan and J. H. McCusker, 1999. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15 507–511. [DOI] [PubMed] [Google Scholar]
- Gordon, J. L., and K. H. Wolfe, 2008. Recent allopolyploid origin of Zygosaccharomyces rouxii strain ATCC 42981. Yeast 25 449–456. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63 751–762. [DOI] [PubMed] [Google Scholar]
- Guindon, S., and O. Gascuel, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Hicks, J., J. N. Strathern and A. J. S. Klar, 1979. Transposable mating type genes in Saccharomyces cerevisiae. Nature 282 478–483. [DOI] [PubMed] [Google Scholar]
- Hou, Z., D. A. Bernstein, C. A. Fox and J. L. Keck, 2005. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc. Natl. Acad. Sci. USA 102 8489–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, H. C., B. Stillman and R. M. Xu, 2005. Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc. Natl. Acad. Sci. USA 102 8519–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., N. Patterson, M. Endrizzi, B. Birren and E. S. Lander, 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423 241–254. [DOI] [PubMed] [Google Scholar]
- Kuras, L., and K. Struhl, 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399 609–613. [DOI] [PubMed] [Google Scholar]
- Kushner, P. J., L. C. Blair and I. Herskowitz, 1979. Control of yeast cell types by mobile genes: a test. Proc. Natl. Acad. Sci. USA 76 5264–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon, A., C. S. Chang, E. M. Scott, S. J. Jacobson and L. Pillus, 2007. MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene 26 5373–5384. [DOI] [PubMed] [Google Scholar]
- Langkjaer, R. B., P. F. Cliften, M. Johnston and J. Piskur, 2003. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421 848–852. [DOI] [PubMed] [Google Scholar]
- Mahoney, D. J., and J. R. Broach, 1989. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol. Cell. Biol. 9 4621–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., and D. Johnson, 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86 667–677. [DOI] [PubMed] [Google Scholar]
- Moazed, D., A. Kistler, A. Axelrod, J. Rine and A. D. Johnson, 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, T. J., and L. L. Wallrath, 2007. Connections between epigenetic gene silencing and human disease. Mutat. Res. 618 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus, L., and J. Rine, 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59 637–647. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72 481–516. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., and J. Rine, 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15 955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell, D. R., A. C. Frank, G. C. Conant, K. P. Byrne, M. Woolfit et al., 2007. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc. Natl. Acad. Sci. USA 104 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. A., A. A. Franco, M. A. Osley and P. D. Kaufman, 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. A., D. C. Krawitz, K. A. Gardner, C. A. Fox and P. D. Kaufman, 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 17 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks and R. Sternglanz, 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 11 2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel, L., and D. Shore, 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. USA 88 7749–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman, L., M. B. Eisen and J. Rine, 2008. Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 4 e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo, T., and R. Sternglanz, 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381 251–253. [DOI] [PubMed] [Google Scholar]
- Vaughan-Martini, A., A. Martini and G. Cardinali, 1993. Electrophoretic karyotyping as a taxonomic tool in the genus Saccharomyces. Antonie Van Leeuwenhoek 63 145–156. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., 2006. Comparative genomics and genome evolution in yeasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387 708–713. [DOI] [PubMed] [Google Scholar]
- Xu, E. Y., K. A. Zawadzki and J. R. Broach, 2006. Single-cell observations reveal intermediate transcriptional silencing states. Mol. Cell 23 219–229. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., M. K. Hayashi, O. Merkel, B. Stillman and R. M. Xu, 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21 4600–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill, O. A., and J. Rine, 2008. Interspecies variation reveals a conserved repressor of α-specific genes in Saccharomyces yeasts. Genes Dev. 22 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]