Abstract
The origin and the evolution of toxin–antitoxin (TA) systems remain to be uncovered. TA systems are abundant in bacterial chromosomes and are thought to be part of the flexible genome that originates from horizontal gene transfer. To gain insight into TA system evolution, we analyzed the distribution of the chromosomally encoded ccdO157 system in 395 natural isolates of Escherichia coli. It was discovered in the E. coli O157:H7 strain in which it constitutes a genomic islet between two core genes (folA and apaH). Our study revealed that the folA–apaH intergenic region is plastic and subject to insertion of foreign DNA. It could be composed (i) of a repetitive extragenic palindromic (REP) sequence, (ii) of the ccdO157 system or subtle variants of it, (iii) of a large DNA piece that contained a ccdAO157 antitoxin remnant in association with ORFs of unknown function, or (iv) of a variant of it containing an insertion sequence in the ccdAO157 remnant. Sequence analysis and functional tests of the ccdO157 variants revealed that 69% of the variants were composed of an active toxin and antitoxin, 29% were composed of an active antitoxin and an inactive toxin, and in 2% of the cases both ORFs were inactive. Molecular evolution analysis showed that ccdBO157 is under neutral evolution, suggesting that this system is devoid of any biological role in the E. coli species.
BACTERIAL chromosomes and plasmids harbor, often in multiple copies, addiction modules also known as toxin–antitoxin (TA) systems (for a review, see Gerdes et al. 2005). They generally consist of two genes: a toxin-encoding gene whose product affects the bacterial metabolism (replication or translation) and an antitoxin-encoding gene whose product binds to the toxin and counteracts its activity. The antitoxin is constantly degraded by an ATP-dependent protease while the toxin is a stable protein. This property renders the cell addicted to antitoxin production and therefore to the TA genes. This has been well documented for plasmid-encoded TA systems. Their biological role is to help maintain plasmids in growing bacterial populations by killing plasmid-free daughter bacteria (addiction phenomenon or postsegregational killing, PSK) (Gerdes et al. 1986; Yarmolinsky 1995). The function(s) of chromosomally encoded TA systems, however, still remains unclear. While it was proposed that chromosomally encoded mazEF and relBE systems of Escherichia coli K-12 are stress response modules, it was recently shown that the five canonical TA systems of E. coli K-12 (including relBE and mazEF) do not confer any selective advantage under a variety of stress conditions (Christensen et al. 2003; Kolodkin-Gal and Engelberg-Kulka 2006; Tsilibaris et al. 2007). This raises the distinct possibility that chromosomally encoded TA systems might be devoid of function, at least in the global stress response. Nevertheless, recent studies have highlighted a variety of physiological processes involving chromosomally encoded TA systems such as developmental programming (Nariya and Inouye 2008), persistence (Keren et al. 2004), stabilization of large genomic fragments (Szekeres et al. 2007), and anti-addiction (Saavedra De Bast et al. 2008). Thus, the functions of chromosomally encoded TA systems are likely to be quite diverse, depending on the type of TA systems, their genomic location (plasmid, genomic island, chromosomal core), and their host species.
Seven families of TA systems have been defined on the basis of the homology of the toxin proteins (Pandey and Gerdes 2005). One of the striking features of these systems is their wide distribution in bacterial and archeal genomes. Most of the genomes that have been sequenced do contain TA systems (Pandey and Gerdes 2005; Sevin and Barloy-Hubler 2007) although to various extents. Some of the TA families are highly prevalent (such as relBE and vapBC) while others appear to be less represented (phd-doc and ccd) (Pandey and Gerdes 2005). The ccd family is mostly confined to γ-proteobacteria and only a small number of chromosomally encoded homologs have been identified and studied so far (Wilbaux et al. 2007; Saavedra De Bast et al. 2008).
A chromosomally encoded homolog of the ccdF system of the E. coli F plasmid was previously characterized in E. coli O157:H7 EDL933 and found to be expressed and still functional (Wilbaux et al. 2007). Indeed, the ectopic expression of this CcdBO157 toxin is lethal through gyrase poisoning, while the coexpression of the CcdAO157 antitoxin relieves its toxicity. The ccdO157 system is located between the folA and apaH metabolic genes, coding, respectively, for a dihydrofolate reductase and a diadenosine tetraphosphatase. Bioinformatics analysis on E. coli genomic sequences available at the time (17 partial or complete sequences) indicated a certain degree of plasticity in this region (Wilbaux et al. 2007). In the present work, we have analyzed the folA–apaH intergenic region of a collection of 395 E. coli isolates representing the 174 known serogroups. The aim of this study is to evaluate the prevalence of the chromosomally encoded ccdO157 system within E. coli species and to gain insight into the evolution of TA systems.
MATERIALS AND METHODS
Bacterial strains:
E. coli strains used in this work, their origin, and their pathogenic or commensal nature are listed in supplemental Table S1. Three hundred ninety-five E. coli strains were obtained from various sources. Sixty-nine strains were obtained from academic hospitals (48 from the Hôpital Universitaire des Enfants Malades Reine Fabiola, Université Libre de Bruxelles, Belgium and 21 from the Academisch Ziekenhuis, Vrije Universiteit Brussel, Belgium). Three hundred six strains were obtained from collections [2 from the Collection de l'Institut Pasteur, (Paris), 130 from the Shiga Toxin-Producing Escherichia coli Center (Michigan State University), and 174 from Laboratorio de Referencia de E. coli (Universidad de Santiago de Compostela, Spain)]. Eighteen strains were obtained from stools from healthy volunteers and 2 from urine of patients with a diagnosed cystitis. Identification of these 20 strains was confirmed using the VITEK 2 (BioMérieux, Marcy l'Etoile, France). All the strains, besides those received from LREC, were serogrouped using the complete serogrouping kit from LREC. E. coli serogrouping relies on the nature of the O antigen. One hundred seventy-four different serogroups have been described.
In addition, the MG1655 (wild-type E. coli K-12) (Gentry et al. 1991) and the SG22622 (MC4100 cpsB∷lacZ ara malP∷lacIq) (from S. Gottesman) laboratory strains were used.
Media:
Luria–Bertani medium (LB) (Miller 1972), Ceria 132 synthetic medium (CM), (Glansdorff 1965), and CM supplemented with 0.1% casamino acids (CCM) were used.
DNA manipulations:
Transformations with appropriate plasmids were performed as described in Miller (1972), and most routine DNA manipulations were performed as described in Sambrook and Russell (2001).
PCR detection of the ccdO157 system:
Presence of the ccdO157 system was examined in all our E. coli isolates by PCR, using primers flanking the folA–apaH intergenic region: primer 40-bis (5′-GCAGAACTCTCACAGCTATT-3′) complementary to the 3′-end of the folA gene and primer 93 (5′-TTGTCCAGCCGTCGAACCGGC-3′) complementary to the 3′-end of the apaH gene (Figure 1). The strains presenting a PCR amplicon of 151 bp (which corresponds to a folA–apaH intergenic region of 77 bp) were further screened by PCR with primers complementary to the ccdO157 system of EDL933 to possibly detect it at different locations. For this purpose, primer 43 (5′-TTGTTCTAGAATTGTACAGGAGCACG-3′) complementary to the 5′-end of the ccdAO157 gene and primer 47 (5′-AGTCTCTGCAGTTAAATCCCGTCGAGC-3′) complementary to the 5′-end of the ccdBO157 gene were used. As an internal control, primers 16SA (5′-CCCCCTGGACGAAGACTGAC-3′) and 16SB (5′-ACCGCTGGCAACAAAGGATA-3′) complementary to 16S rRNA were used in PCR reactions.
Figure 1.—
PCR detection of the ccdO157 system in the E. coli species. The ccdO157 system is located between the folA and the apaH gene (not on scale). Its presence was detected by PCR using the P40 and P93 primers that are complementary to the ORFs that are flanking the ccdO157 system. Another set of primers (P43 and P47) complementary to the ccdAO157 and ccdBO157 ORFs was used to detect it at other potential chromosomal locations in strains that did not carry ccdO157 between folA and apaH.
Sequencing of the folA–apaH intergenic region:
To ensure a diverse sampling of the E. coli population, the folA–apaH intergenic region of 93 isolates was sequenced using primers 40-bis and 93 (see supplemental Table S1, in boldface type). The sequence of the 712-bp fragment obtained in 47 different serogroups, as well as that of the 1499-bp fragment in 20 different serogroups, and that of the unique 2778-bp fragment was analyzed. The 151-bp fragment was analyzed for the 25 serogroups that presented a high variability in PCR size. In the case of the 712-bp fragments, the DNA sequence was translated and compared to the CcdAO157 and CcdBO157 protein sequences from the E. coli O157:H7 EDL933 reference strain using BLASTP. For the 1499- and 2778-bp fragments, the ORFs were determined using ORFinder.
Construction of the ccdA and ccdB expression plasmids:
pBAD33-derivative plasmids:
The variants of the chromosomally encoded ccdBO157 gene were amplified from a boiled colony from the various serotypes using the primers 5′-CcdBO157-XbaI (5′-GCTCTAGAAGGAGGTAGCGATGCAATTTACGG-3′) and 3′-CcdBO157-PstI (5′-AGTCGCTGCAGCTAGAAGCTCCGGTAC-3′). The variants of the F-plasmid-encoded ccdBF gene were amplified using OLI 70 (5′-TCTAGAAGGAGGGTGAAATGCAGTTTAAGG-3′) and OLI 71 (5′-AGTCGCTGCAGTTATATTCCCCAGAAC-3′). The 5′-primers (CcdBO157-XbaI and OLI 70) carried a canonical Shine–Dalgarno (SD) sequence (in boldface type). The PCR products were digested with XbaI and PstI (in italics) and ligated downstream of the PBAD promoter of the pBAD33 vector cut with the same restriction enzymes. The recombinant plasmids were sequenced. The different variants were named according to the serotype of the strain.
pKK223-3-derivative plasmids:
The variants of the chromosomally encoded ccdAO157 gene were amplified on a boiled colony from the various serotypes using the primers 5′-CcdAO157-EcoRI (5′-TTGTGAATTCTATGACTGCAAAACGTACCA-3′) and 3′-CcdAO157-PstI (5′-AGTCTCTGCAGCTAGAAGCTCCGGTACTC-3′). The variants of the plasmid-encoded ccdAF gene were amplified using P502 (5′-TTGTGAATTCTATGAAGCAGCGTATTACAGTGACAG-3′) with P516 (5′-AGTCTCTGCAGTCACCAGTC CCTGTTCTC-3′). The PCR products were cloned into the TOPO-XL vector (Invitrogen, Carlsbad, CA) and sequenced. The recombinant TOPO-XL plasmids were then digested with EcoRI and PstI (in italics) and the corresponding DNA fragments were ligated downstream of the lac/trc promoter in the pKK223-3 vector opened with the same restriction enzymes. The recombinant plasmids were sequenced. The different variants were named according to the serotype of the strain.
Toxicity and antitoxicity plate assays:
To test the toxicity of the cloned CcdBO157 and CcdBF variants, the corresponding pBAD33-ccdB constructs were transformed in MG1655. The resulting transformants were plated on LB plates containing chloramphenicol with or without arabinose (1%). The CcdB variants were considered to be functional (toxic) when transformants were able to grow only in the absence of arabinose.
To test the ability of the cloned CcdAO157 and CcdAF variants to counteract the toxicity of CcdBO157 and CcdBF, respectively, the corresponding pKK-ccdA constructs were transformed in MG1655 expressing the reference ccdBO157 or ccdBF genes from the pBAD33 vector. The resulting transformants were plated on LB plates containing chloramphenicol and ampicillin with arabinose (1%). Basal expression of ccdA from the pTac promoter of pKK223-3 in MG1655 is sufficient to test the antitoxicity phenotype. The CcdA variants were considered to be functional when the toxicity of CcdBO157 or CcdBF protein was counteracted, i.e., when strains coexpressing a ccdA variant with the ccdBO157 or ccdBF reference genes were able to grow in the presence of arabinose while strains expressing only the ccdBO157 or ccdBF reference gene were not.
Toxicity and antitoxicity liquid assays:
Strains carrying the toxin-expressing plasmids and/or the antitoxin-expressing plasmids were grown overnight (ON) at 37° in CCM supplemented with glucose (0.4%) and the appropriate antibiotics. ON cultures were diluted in the same medium to an optical density (OD) at 600 nm of 0.01 and grown at 37° to an OD600 of 0.1–0.2. The cultures were centrifuged at 4000 rpm for 10 min at room temperature. The bacterial pellets were resuspended in CCM, prewarmed at 37°, and supplemented with glycerol (0.4%) and the appropriate antibiotics. Arabinose was then added (0.25 or 1%), and the cultures were grown at 37°. Samples were removed at the indicated time, diluted in MgSO4 (10 mm), and plated on CCM plates supplemented with glucose (0.4%) and the appropriate antibiotics.
Construction of the O51 ΔccdO51 strain:
The pKOBEG plasmid (Chaveroche et al. 2000) was transformed to E. coli O51 and used as described at the website http://www.pasteur.fr/recherche/unites/Ggb/3SPCRprotocol.html. The kanamycin resistance cassette of pKD4 was amplified by PCR with the following primers: P1 (5′-ATACTAGACGTATAAATTGTACAGGAGCACGATATCGTGTAGGCTGGAGCTGCTTC-3′) and P6 (5′-AAAGATATGGGTGAGGGAGAGGCGGCCGCGTCTTAACATATGAATATCCTCCTTAG-3′). Deletion of the ccdO51 system was constructed by following the method described in reference Datsenko and Wanner (2000). The deletion and the flanking regions were checked afterward by DNA sequencing.
Simpson index:
To measure the diversity of the CcdBO157 and CcdAO157 sequences, the Simpson index was calculated as
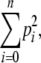 |
with n being the total number of different variants of each gene and p their frequencies. The value of the Simpson index is between 0 (maximum diversity) and 1 (no diversity, one variant) (Simpson 1949).
Natural selection measurements:
To evaluate the nature and magnitude of natural selection acting on the ccdBO157, folA, and apaH genes, the dN/dS ratio (ω) was calculated. We were unable to carry such an analysis for ccdAO157 due to the small number of variants. For the 14 variants of the chromosomally encoded CcdBO157 found in this study, an amino acid-based nucleotide alignment of the variants was built, along with 13 CcdBF plasmid-encoded homologs, using the online version of MAFFT v6 (Katoh et al. 2002; Katoh and Toh 2008). The following plasmids were considered: the pIP1206, pSE11, pETEC-74, pETEC-80, and pO113 E. coli plasmids; the pBS512-211 and pCP301 Shigella plasmids; and the pCVM29188, pCVM291882, pOU1115, pOU7519, and pOU1113 from various Salmonella isolates. The folA and apaH genes from the 47 serogroups (isolates from which the ccdO157 system was sequenced) were sequenced. The FolAFOR (5′-GCACCAGTCGACGACGGTTTAC-3′) and P47 primers and ApaREV (5′-CTTTCAGCATCGACATTCCCG-3′) and P43 primers were used for folA and apaH, respectively. The apaH sequence was complete (846 bp) while we analyzed ∼86% of the sequence of floA [from base pair 58 to the end (420 bp)].
The folA and apaH alignments were made manually. Twenty-four and 38 variants were identified for folA and apaH, respectively.
Phylogenetic analyses were carried out with the maximum-likelihood method as implemented in the online version of Bootstrap RAxML (Stamatakis et al. 2008) available on the CIPRES portal (http://www.phylo.org/sub_sections/portal/). Since we wanted to compare ω for plasmid and chromosomally encoded ccdB sequences, we used PAML v4.1 (Yang 1997, 2007) with the branch model. This model assumes nonindependent ratios for each branch, the user being allowed to specify which branches should have which rates. We fixed one rate (ω1) for the plasmid sequences, another (ω2) for the chromosome ones, and a last (ω0) for all the internal branches. The folA and apaH ratios were calculated using the same model, with for each analysis one rate (ω1) for the sequences and one rate (ω0) for all the internal branches. To obtain the variance on the dN/dS values, we computed the pairwise ω-ratios for each couple of sequences using DNA Master (http://cobamide2.bio.pitt.edu/computer.htm). These ratios were weighted according to the inverse of the distance between the two sequences. The weighted pairwise ω allowed us to calculate the variance on the dN/dS ratios.
RESULTS
Size variability of the folA–apaH intergenic region within the E. coli species:
In E. coli O157:H7 EDL933, the ccdO157 system is located between the folA and the apaH genes (Wilbaux et al. 2007). To evaluate the general prevalence of this system in E. coli species, PCR was used to probe the folA and apaH intergenic region (IR) of 395 E. coli isolates. Interestingly, four different lengths of PCR amplicons were obtained: (i) a fragment of 151 bp, which corresponds to the folA–apaH intergenic region in MG1655 K-12; (ii) a fragment of 712 bp, which corresponds to the ccdO157 system in the O157:H7 EDL933 strain; (iii) a fragment of 1499 bp; and (iv) a fragment of 2778 bp (Figure 2). The actual sizes of the IR are shown in Table 1. The isolates presenting the 151-bp fragment were further screened by PCR with primers specific to the ccdO157 system and not the ccdF system (primers 43 and 47, see materials and methods). No amplification was obtained, suggesting that these isolates did not carry the ccdO157 system at another chromosomal location and/or on a plasmid.
Figure 2.—
Diversity of the folA–apaH region in the E. coli species. PCR reactions were carried out on E. coli isolates using P40 and P93. Lane 1, molecular weight marker (SMART ladder, Eurogentec), size is indicated in base pairs; lane 2, MG1655; lane 3, O157 (2886-75, STEC Center); lane 4, O153 (LREC); lane 5, O163 (LREC); lane 6, O86 (LREC); lane 7, negative control. The size (base pairs) of the different amplicons is indicated.
TABLE 1.
Diversity of the folA–apaH intergenic region among 395 E. coli isolates belonging to 174 serogroups
| Size (bp)
|
No. of serogroups (n = 174) | No. of isolates (n = 395) | No. of fragments sequenced (n = 93) | |
|---|---|---|---|---|
| PCR | IR | |||
| 151 | 77 | 135 | 196 | 25 |
| 712 | 638 | 47 | 137 | 47 |
| 1499 | 1425 | 20 | 61 | 20 |
| 2778 | 2704 | 1 | 1 | 1 |
The PCR amplicon (PCR) and actual intergenic region (IR) sizes are indicated.
Our collection is composed of the 174 different E. coli serogroups that have been described so far. Although most of them are represented by only one isolate, 54 different serogroups contained at least 2 independent isolates. Table 1 shows the variability of the folA–apaH IR among the 174 serogroups. The 151-bp fragment was detected in isolates belonging to 135 different serogroups, while the 712-bp fragment was present in 47 different serogroups. The 1499-bp fragment was less prevalent and was detected in 20 different serogroups, while the 2778-bp fragment was detected in only one serogroup (O163). This shows that independent isolates from the same serogroup may display a different IR (29 over 174 serogroups). For instance, 14 serogroups contained isolates presenting a fragment of 151 or 712 bp, while 19 contained only isolates presenting the 151-bp fragment and 9 only those at 712 bp (data not shown). Moreover, the variability of the folA–apaH IR region among one serogroup was very high for some serogroups and low for others, and this variability was not correlated to the number of isolates per serogroup. Serogroups such as O153 and O23 contained isolates presenting the 151-, 712-, and 1499-bp PCR fragments (see Figure 3 for O153) and the O111 serogroup contained 18 isolates presenting the 1499-bp fragment and 14 presenting the 151-bp fragment (Table 2). Other serogroups display no variability among the different isolates (Table 2). For instance, the 19 isolates of serogroup O55 all harbored the 712-bp fragment.
Figure 3.—
Diversity of the folA–apaH region within the O153 serogroup. PCR reactions were carried out on three O153 isolates using P40 and P93. Lane 1, molecular weight marker (SMART ladder, Eurogentec), size is indicated in base pairs; lane 2, negative control; lane 3, HC45; lane 4, MG1655; lane 5, HC71; lane 6, O153 (LREC). The size (base pairs) of the different amplicons is indicated.
TABLE 2.
Diversity of the folA–apaH intergenic region among isolates belonging to the same serogroups
| Serogroup | Total no. of isolates | No. of isolates with an IR of 151 bp | No. of isolates with an IR of 712 bp | No. of isolates with an IR of 1499 bp |
|---|---|---|---|---|
| O2 | 10 | — | 10 | — |
| O26 | 19 | — | — | 19 |
| O55 | 19 | — | 19 | — |
| O111 | 32 | 14 | — | 18 |
| O128 | 15 | 10 | — | 5 |
| O157 | 24 | 1 | 23 | — |
Serogroups represented by at least 10 isolates are indicated.
All together, these results show that the folA–apaH IR is highly diversified and is most likely a region that allows integration of foreign DNA. No correlation between the presence of a ccdO157 system and the strain serogroup could be highlighted.
The plasticity of the folA–apaH intergenic region:
As shown in Table 1, 135 serogroups contained isolates showing a folA–apaH IR of 77 bp, 47 serogroups contained the 638-bp ccdO157 system (see below), 20 serogroups contained isolates displaying an IR of 1425 bp, and one isolate had an IR of 2704 bp. Figure 4A shows the IRs of various sizes and their content. The 77-bp IR was sequenced for 25 serogroups and was found to be identical to that of the MG1655 strain. This IR is composed of a repetitive extragenic palindromic (REP) sequence (Wilbaux et al. 2007). The 1425-bp fragments were sequenced for 20 isolates and were highly similar. We identified a small sequence that presents identity with the 3′ region of ccdAO157 and two putative ORFs (Figure 4, A and B). The sequenced E. coli B7A, E. coli E110019, and E. coli SMS-3-5 strains also present the 1425-bp IR. ORF2 presents identity with hypothetical proteins from other bacterial species, i.e., E. albertii TW07627 and Bacteroides uniformis ATCC 8492. ORF3 shows identities with a large number of hypothetical proteins found in bacteria, fungi, and amoebas and one in a sea urchin. The 2704-bp IR corresponds to that of 1425 bp with the insertion of an IS621 in the sequence presenting identity with ccdAO157 (nt 83).
Figure 4.—
Plasticity of the folA–apaH IR within the E. coli species. (A) Representation of the diversity of the folA–apaH IR. The strains and the size (base pairs) of the different folA–apaH intergenic regions (IR) are indicated. The folA and apaH genes are represented in black. The ccdAO157 and ccdBO157 genes are represented in white and dark gray, respectively (indicated as A and B). A′ represents the 3′-terminal part of the ccdAO157 gene. ORF2 (129 amino acids) is represented in light gray, ORF3 (199 amino acids) in white, and IS621 in stippled white. (B) Sequence alignment of the seq1 sequence and CcdAO157. The amino acid sequences were aligned with CLUSTALW. Symbols: asterisk, identical amino acids; colon, strongly similar amino acids; period, weakly similar amino acids. The total number of amino acids for each protein is in parentheses.
Variants of the ccdO157 system:
Table 1 shows that 137 isolates, spread over 47 serogroups, display a folA–apaH IR of 638 bp, indicating the presence of the ccdO157 system within the IR. Of each serogroup, the IR of one such isolate was sequenced (in boldface type in supplemental Table S1) and found indeed to correspond to the ccdO157 system or subtle variants of it. Table 3 shows the amino acid sequences of the corresponding CcdA and CcdB proteins. The 47 antitoxin proteins presented very few variations, except in the case of CcdAO138. Overall, seven classes of alleles could be identified. The most prevalent one was identical to the ccdAO157 gene of the O157:H7 EDL933 reference strain (37/47 isolates). Five classes, representing 9/47 isolates, presented single point variations (S76R, A54T, D72E, T47I, or T34M). The capacity of 1 representative protein of each class to antagonize the toxic activity of the reference CcdBO157 protein was tested, using the antitoxicity plate assay (see materials and methods). The single point variations did not affect the capacity of the CcdA variants to antagonize CcdBO157 activity (data not shown, Table 3). The last class (1 isolate) presented a frameshift mutation caused by a 1-nt deletion. This led to a major modification of the carboxy terminus of CcdAO138 that affects the antitoxic activity of this variant. Indeed, when coexpressed with CcdBO157, this protein was unable to restore viability, showing that CcdAO138 is inactive (Figure 5A). This result was expected since it has been shown that the carboxy-terminal domain of CcdAF is responsible for the antitoxin activity (Bernard and Couturier 1991).
TABLE 3.
The variants of CcdAO157 and CcdBO157 proteins
| Strain serogroup | No. of isolates | Antitoxin activity | Amino acid sequence |
|---|---|---|---|
| CcdA variants | |||
| 157 (EDL933 reference strain) | — | + | MTAKRTTQSVTVTVDRELVNRARDAGLNMSATLTVALNAELKKHAATRWREENAEAIAALNQLADETGCFSDEYRSF------------------------------------------------------------------------------------------------------ 77 |
| 2/4/5/6/11/15/16/17/22/23/24/25/33/45/51/52/55/58/81/83/85/86/89/112/114/115/120/134/139/142/145/150/153/157/161/166/176 | 37 | + | MTAKRTTQSVTVTVDRELVNRARDAGLNMSATLTVALNAELKKHAATRWREENAEAIAALNQLADETGCFSDEYRSF------------------------------------------------------------------------------------------------------- 77 |
| 44/133 | 2 | + | MTAKRTTQSVTVTVDRELVNRARDAGLNMSATLTVALNAELKKHAATRWREENAEAIAALNQLADETGCFSDEYRRF------------------------------------------------------------------------------------------------------- 77 |
| 1/7/75/79 | 4 | + | MTAKRTTQSVTVTVDRELVNRARDAGLNMSATLTVALNAELKKHAATRWREENTEAIAALNQLADETGCFSDEYRSF------------------------------------------------------------------------------------------------------- 77 |
| 171 | 1 | + | MTAKRTTQSVTVTVDRELVNRARDAGLNMSATLTVALNAELKKHAATRWREENAEAIAALNQLADETGCFSEEYRSF-------------------------------------------------------------------------------------------------------77 |
| 119 | 1 | + | MTAKRTTQSVTVTVDRELVNRARDAGLNMSATLTVALNAELKKHAAIRWREENAEAIAALNQLADETGCFSDEYRSF------------------------------------------------------------------------------------------------------- 77 |
| 102 | 1 | + | MTAKRTTQSVTVTVDRELVNRARDAGLNMSATLMVALNAELKKHAATRWREENAEAIAALNQLADETGCFSDEYRSF------------------------------------------------------------------------------------------------------- 77 |
| 138 | 1 | − | MTAKRTTQSVTVTVDRELVNRARDAGLNMSAPLRLRSMLNLKNMQQHVGVKRTQKLSLR-------------------59 |
| Strain serogroup | No. of isolates | Toxic activity | Amino acid sequence |
| CcdB variants | |||
| 157 (EDL933 reference strain) | — | + | MQFTVYRSRSRNAAFPFVIDVTSDIIGVINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 44/55/157/171 | 4 | + | MQFTVYRSRSRNAAFPFVIDVTSDIIGVINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 24/33/52/112/114/145/176/115 | 8 | + | MQFTVYRSRSRNAAFPFVIDVTSDIIGVINRRIVIPLTPIERFIRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 11/58/161 | 3 | + | MQFTVYRSRSRNAAFPFVIDVTSDIIGEINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 133 | 1 | + | MQFTVYHSRSRNAAFPFVIDVTSDIIGEINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 85/89 | 2 | + | MQFTVYRSRGRNAAFPFVIDVTSDIVGEINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 2/45/83 | 3 | − | MQFTVYRSRGRNAAFPFVIDVTSDIVGEINRRIVIPLTPI------------------------------------------------------- 40 |
| 81 | 1 | + | MQFTVYHSRGRNAAFPFVIDVTSDIVGEINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLLKGALDFMLDGI----------------------------------------------------------------- 104 |
| 1/4/5/6/16/22/25/75/79/120/134/142/150 | 13 | + | MQFTVYRSRGRNAAFPFVIDVTSDIIGEINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 102 | 1 | NT | MQFTVYRSRGRNAAFPFVIDVTSDIIGEINRRIVIPLTPIERFSRIRPPERLNPILLLVDGKEYVLMTHETATVPVNALGTKF----------------------------------------------------------------------------------------------------- 83 |
| 138/153 | 2 | − | MQFTVYRSRGRNAAFPFVIDVTSDIIGEINRRIVIPLTPIERFIRIRPPERLNTILLLVDGKEYVLMTHETATVPVNALGTKFCDASAHRTLIKGALDFMLDGI------------------------------------------------------------------ 104 |
| 166 | 1 | − | MQFTVYRSRGRNAAFPFVIDVTSDIIGEINRRIVIPLTPIERFIRIRPPERLNTILLLVDGKEYVLMTHETATVPVNALGTKF----------------------------------------------------------------------------------------------------- 83 |
| 17/15 | 2 | − | MQFTVYRSRGSNAAFPFVIDVTSDIIGEINRSIVIPLTPIERFIRIRPPERLNTILLLVDGS------------------------- 62 |
| 23/51/86/119/139 | 5 | − | MQFTVYRSRGRNAAFPFVIDVTSDIIGEINRRIVIPLTPIERFIRIRPPERLNTILLLVDGS------------------------ 62 |
| 7 | 1 | NT | MQFTVYRSRGRNAAFPFVIDVTSDIIGEINRRIVIPLTPIE------------------------------------------------------ 41 |
NT, not tested; +, active for either toxicity or antitoxicity; −, not active for either toxicity or antitoxicity. Amino acid variation as compared to amino acid sequence of the reference CcdAO157 and CcdBO157 proteins is indicated in italics. The CcdA and CcdB variants that have been tested experimentally are represented in boldface type.
Figure 5.—
Properties of the ccdO51 and ccdO138 variants. (A) The CcdAO138 protein is not able to counteract the toxicity of CcdBO157. SG22622/pKK223-3/pBAD33-ccdBO157 (triangle), SG22622/pKK-ccdAO138/pBAD33-ccdBO157 (square), and SG22622/pKK-ccdAO157/pBAD33-ccdBO157 (diamond) were grown in the presence of 1% arabinose. (B) The CcdBO51 and CcdBO138 variants are not toxic. SG22622/pBAD33 (diamond), SG22622/pBAD33-ccdBO51 (square), SG22622/pBAD33-ccdBO138 (circle) and SG22622/pBAD33-ccdBO157 (triangle) were grown in the presence of 1% arabinose. In A and B serial dilutions of the cultures were plated at regular time intervals on CCM plates without arabinose and incubated overnight at 37°. Values correspond to the mean of results of three independent experiments and SDs are indicated. (C) The CcdAO51 protein is expressed in E. coli O51 and counteracts the toxicity of CcdBO157 expressed in trans. O51 (triangle) and O51ΔccdO51 (square) containing the pBAD33 vector (solid symbols) or the pBAD33-ccdBO157 plasmid (open symbols) were grown in the presence of 0.25% arabinose. At regular time intervals, serial dilutions of the cultures were plated on CCM plates without arabinose and incubated overnight at 37°. Values correspond to the mean of results of three independent experiments and SDs are indicated.
Interestingly, the CcdB toxin proteins were much more diversified than the antitoxins. Among the 47 serogroups, 14 classes of alleles could be identified. Note that, as mentioned earlier, only one isolate for each serogroup was sequenced and tested (47 isolates). One class was composed of 4 isolates presenting sequence identical to the ccdBO157 gene of the O157:H7 EDL933 reference strain. The 2 most prevalent classes represented 13/47 and 8/47 isolates, and the corresponding alleles harbored either two variations (S10G and V28E) or one variation (S44I), respectively. The toxic activity of at least one representative CcdB protein of each class was tested using the toxicity plate assay (see materials and methods) and was comparable to that of the CcdBO157 protein. Four classes, together representing 7 isolates (7/47), contained from one to five variations (V28E, RH7-V28E, S10G-I26V-V28E, and RH7-S10G-I26V-V28E-I93L). At least one representative of each class was assayed for toxicity using the toxicity plate assay. Interestingly, expression of these variants led to cell killing, showing that the variations did not affect the toxic activity of the CcdB proteins. Of the 7 classes remaining, 1 is composed of 2 isolates (representing serogroups O138 and O153) containing four variations in their ccdB gene (S10G-V28E-S44I-P54T). The CcdBO138 protein (from serogroup O138) was assayed for toxicity and surprisingly shown not to affect viability upon ectopic expression (Figure 5B). The comparison of the variations among the different classes suggests that the P54T mutation is responsible for abolishing the toxic activity of the CcdBO138 variant. The 6 last classes, representing 13/47 isolates, were composed of truncated proteins. Deletions of the carboxy-terminal region were caused by amber mutations at various locations (E41, R42, E63, and C84). Interestingly, all these truncated proteins contained several point variations that were also found in the full-length variants that were still toxic (S10G, I26V, V28E, and S44I) or not (R7H and P54T). Two of these classes (7/47 isolates) contained one extra variation (K62S), while another class (1/47 isolates) contained two more variations (R11S and R32S). One representative of 4 of these 6 classes was tested and was shown to be nontoxic, using the plate toxicity assay (data not shown). Figure 5B shows that ectopic overexpression of CcdBO51 in the liquid toxicity assay did not lead to the loss of viability. These truncated variants of CcdBO157 are thus inactive and it is likely to be the case for CcdBO7 and CcdBO102 since the active site of the CcdBO157 is located at the caboxy terminus of the protein (Bahassi et al. 1995; Wilbaux et al. 2007). The corresponding antitoxins were functional (Table 3).
These results showed that while the vast majority of the ccdO157 system or variants of it were composed of an active antitoxin (46/47), the proportion of active toxins was much smaller (31/45 tested). Thus, 69% of the variants (31/45) were composed of an active toxin and antitoxin, 29% (13/45) were composed of an active antitoxin and an inactive toxin, and in 2% (1/45) of the cases both ORFs are inactive. The Simpson index measurements showed that the antitoxin sequences are homogeneous (0.63) while the toxin sequences show a high level of diversity (0.14).
Evolution of the ccdO157 system and its flanking regions:
To evaluate the nature of the selective pressures acting on the ccdO157 system, the dN/dS ratio for the CcdBO157 proteins was measured and compared to that of 13 CcdBF homologs found on plasmids (see materials and methods). The dN/dS for the plasmid-encoded toxin proteins reflected a negative selection (0.11 ± 0.0035), showing that these sequences are very constrained. On the contrary, the dN/dS value for the chromosomally encoded toxins was close to 1 (0.9 ± 0.1665), indicating a neutral selection. This analysis could not be performed for ccdAO157 due to the small number of variants, indicating that the antitoxin sequences are highly constrained. The evolution of the folA and apaH genes flanking the ccdO157 system appeared to be also constrained (0.03 ± 0.0004 and 0.32 ± 0.0166, respectively).
Expression of the ccdO51 system in E. coli O51:
A quite significant proportion of systems (29%) are constituted of an inactive toxin and an active antitoxin. To better characterize this type of variant, the expression of such a system was analyzed. For that purpose, we deleted the ccdO51 system in E. coli O51. Strains O51 and O51ΔccdO51 were transformed with the pBAD-ccdBO157 plasmid. The viability of both strains was measured upon expression of the CcdBO157 toxin. Figure 5C shows that the viability of the O51ΔccdO51 strain was more strongly affected than that of the O51 strain by ectopic overexpression of CcdBO157. This shows that CcdAO51 is still induced from its natural location at a basal level that is sufficient to at least partially counteract the toxic activity of CcdBO157 produced in trans.
DISCUSSION
Our work revealed that the intergenic region between the folA and apaH genes is a hot spot for integration of exogenous mobile genetic elements. In several E. coli isolates that have been fully sequenced, this region is composed of a REP sequence (Wilbaux et al. 2007). REP sequences are noncoding, short (21–65 nt) palindromic sequences detected in bacterial chromosomes in multiple copies in intergenic regions and between genes from the same operon (Gilson et al. 1984; Stern et al. 1984). REPs have been shown to be the integration site for various insertion sequences (Clement et al. 1999; Choi et al. 2003; Wilde et al. 2003; Tobes and Pareja 2006). Among the 395 E. coli isolates that were tested in this work, (i) 49.6% (196/395) had the REP sequence in the folA–apaH IR; (ii) 34.7% (137/395) had the ccdO157 system or variants of it; (iii) 15.4% (61/395) contained two hypothetical ORFs, one of them being largely represented even in eukaryotic cells, and a 3′ remnant of the ccdAO157 gene; and (iv) 0.3% (1/395) contained an IS621 inserted in this remnant sequence. No sign of REP-like sequence or inverted repeat was detected in the ccdAO157 remnant. How the ccdO157 system was inserted at that location remains also unclear since no transposase gene or repeats are detectable (Wilbaux et al. 2007). We can hypothesize that it originated from a larger composite transposon that was trimmed with evolution. Unfortunately, we were unable to retrace the evolutionary history of that region and to determine whether one or several insertion/deletion events have occurred. However, comparing the sequences of the ccdO157 system and its variants did provide insights about its evolution. Thirty percent of the variants of the chromosomally encoded ccdO157 system are composed of an inactive toxin. A similar survey, although less extensive, was carried out for the ccdF plasmid-encoded system within the same isolate collection (data not shown). The variants of the CcdBF toxin that were sequenced and tested were toxic, indicating that the plasmid-encoded systems are under a stronger selective pressure to retain their addiction function. This was confirmed by molecular evolutionary analysis of the CcdBF-like toxins encoded by various plasmids.
It is likely not to be the case for the chromosomal ones since a significant proportion of the toxin variants were inactive. The dN/dS ratio showed that the CcdBO157-like toxins were under neutral selection, suggesting that this system might be devoid of any biological role. Interestingly, the evolution of ccdAO157-like antitoxins and of the flanking apaH and folA genes appears to be much more constrained. Inactivation of the toxin gene prior to the antitoxin gene presumably constitutes the first and safer step of TA systems degradation. Moreover, at least in the case of E. coli O51, the regulatory regions remain unchanged since this strain was resistant to a moderate expression of the CcdBO157 toxin in trans. The case of ccdO138 reinforces the hypothesis that toxin inactivation arises first. Indeed, the inactive CcdBO138 variant is coupled either to the inactive CcdAO138 as in the case of the ccdO138 variant or to an active antitoxin as in the case of the ccdO153 variant. Thus, the situation at present strongly indicates a decay of the ccdO157 system. An alternative hypothesis is that the antitoxin might play an anti-addictive role as shown for the ccdEch system (Saavedra De Bast et al. 2008) although not against CcdBF-like toxin since CcdAO157 antitoxin does not protect against ccdF addiction (Wilbaux et al. 2007). Therefore, we are in favor of a degeneration of the chromosomally encoded ccdO157 system.
The role of chromosomally encoded TA systems remains controversial. The well-studied mazEF and relBE systems of E. coli K-12 were reported to be induced under stress conditions (e.g., amino acid starvation, antibiotic treatments, and heat shock) although the outcome of their induction is quite different and controversial: induction of mazEF was shown to lead to programmed cell death, while that of relBE induces a bacteriostatic state (Gerdes et al. 2005; Engelberg-Kulka et al. 2006). Moreover, in competition experiments between the wild-type strain and a strain devoid of 5 TA systems (among them mazEF and relBE) under stress conditions, no selective advantage of the wild-type strain was evident (Tsilibaris et al. 2007), further underscoring the discrepancy with the stress response model. Our present observations add to this and indicate that the chromosomally encoded ccdO157 system might actually be devoid of any biological role in the E. coli species although the ccdO157 variants are composed in 69% of the cases of active ORFs (a toxin that when ectopically overexpressed inhibits colony formation and an antitoxin that relieves the toxin toxicity).
A still striking and not yet understood observation is the abundance of TA systems in bacterial chromosomes (Pandey and Gerdes 2005; Sevin and Barloy-Hubler 2007; Guglielmini et al. 2008). Although no extensive comparative bioinformatics analysis has been carried out, there is indication that TA systems are part of genomic islands (Pandey and Gerdes 2005; Magnuson 2007). As for the ccdO157 system, they might constitute an “islet” by themselves. This indicates that they disseminate and invade chromosomes through horizontal gene transfer. We propose that some TA systems might be maintained in bacterial chromosomes without conferring any selective advantage to the host, but only due to the interdependence of the toxin and antitoxin ORFs (selfish view). In time, genetic drift might lead to the appearance of inactive toxin mutants, which could then be selected for if sporadic toxin induction proves disadvantageous for the cell. Unless chromosomally encoded TA systems become accommodated in regulatory networks, such as developmental programming (Nariya and Inouye 2008) and persistence (Keren et al. 2004), TA systems would gradually degenerate and eventually disappear.
While the bioinformatics approach constitutes a powerful tool for detecting TA systems, we should consider the possibility that some of them might be pseudogenes. An integrated approach of bioinformatics and experimental characterization will certainly provide valuable information about the evolution of TA systems.
Acknowledgments
We thank Régis Hallez, Abram Aertsen, and Manuel Saavedra De Bast for their comments on the manuscript. We are grateful to Régis Hallez and Damien Geeraerts for their assistance in cloning toxin and antitoxin genes and Michel Milinkovitch for providing advice for molecular evolution analysis. We thank Denis Piérard (Academisch Ziekenhuis Vrije Universiteit Brussel, Brussels), Anne Vergison (Hôpital Universitaire des Enfants Malades Reine Fabiola, Université Libre de Bruxelles, Belgium), and Beth Whittam (STEC Center, Michigan State University) for providing us with the E. coli strains and Jean-Marc Ghigo and Christophe Beloin (Institut Pasteur, Paris) for the pKOBEG plasmid. This work was supported by the European Union (Combating Resistance to Antibiotics; Specific Targeted Research Project, LSH-2004-2.1.2-2), by the Fonds de la Recherche Scientifique Médicale (3.4510.02), and by the Fonds Brachet. At the time of this work, M.W. was supported by the Fonds pour la formation à la Recherche dans l'Industrie et dans l'Agriculture and the European Union (CRAB STREP LSH-2004-2.1.2-2).
References
- Bahassi, E. M., M. A. Salmon, L. Van Melderen, P. Bernard and M. Couturier, 1995. F plasmid CcdB killer protein: ccdB gene mutants coding for non-cytotoxic proteins which retain their regulatory functions. Mol. Microbiol. 15 1031–1037. [DOI] [PubMed] [Google Scholar]
- Bernard, P., and M. Couturier, 1991. The 41 carboxy-terminal residues of the miniF plasmid CcdA protein are sufficient to antagonize the killer activity of the CcdB protein. Mol. Gen. Genet. 226 297–304. [DOI] [PubMed] [Google Scholar]
- Chaveroche, M. K., J. M. Ghigo and C. d'Enfert, 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28 E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S., S. Ohta and E. Ohtsubo, 2003. A novel IS element, IS621, of the IS110/IS492 family transposes to a specific site in repetitive extragenic palindromic sequences in Escherichia coli. J. Bacteriol. 185 4891–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S. K., K. Pedersen, F. G. Hansen and K. Gerdes, 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332 809–819. [DOI] [PubMed] [Google Scholar]
- Clement, J. M., C. Wilde, S. Bachellier, P. Lambert and M. Hofnung, 1999. IS1397 is active for transposition into the chromosome of Escherichia coli K-12 and inserts specifically into palindromic units of bacterial interspersed mosaic elements. J. Bacteriol. 181 6929–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko, K. A., and B. L. Wanner, 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka, H., S. Amitai, I. Kolodkin-Gal and R. Hazan, 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2 e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry, D., H. Xiao, R. Burgess and M. Cashel, 1991. The omega subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J. Bacteriol. 173 3901–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, K., P. B. Rasmussen and S. Molin, 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83 3116–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, K., S. K. Christensen and A. Lobner-Olesen, 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3 371–382. [DOI] [PubMed] [Google Scholar]
- Gilson, E., J. M. Clement, D. Brutlag and M. Hofnung, 1984. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 3 1417–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glansdorff, N., 1965. Topography of cotransducible arginine mutations in Escherichia coli K-12. Genetics 51 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini, J., C. Szpirer and M. C. Milinkovitch, 2008. Automated discovery and phylogenetic analysis of new toxin-antitoxin systems. BMC Microbiol. 8 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., and H. Toh, 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9 286–298. [DOI] [PubMed] [Google Scholar]
- Katoh, K., K. Misawa, K. Kuma and T. Miyata, 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren, I., D. Shah, A. Spoering, N. Kaldalu and K. Lewis, 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186 8172–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal, I., and H. Engelberg-Kulka, 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188 3420–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson, R. D., 2007. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189 6089–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J., 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nariya, H., and M. Inouye, 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132 55–66. [DOI] [PubMed] [Google Scholar]
- Pandey, D. P., and K. Gerdes, 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra De Bast, M., N. Mine and L. Van Melderen, 2008. Chromosomal toxin-antitoxin systems may act as antiaddiction modules. J. Bacteriol. 190 4603–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. Russell, 2001. Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sevin, E. W., and F. Barloy-Hubler, 2007. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 8 R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, E. H., 1949. Measurement of diversity. Nature 163 688. [Google Scholar]
- Stamatakis, A., P. Hoover and J. Rougemont, 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57 758–771. [DOI] [PubMed] [Google Scholar]
- Stern, M. J., G. F. Ames, N. H. Smith, E. C. Robinson and C. F. Higgins, 1984. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37 1015–1026. [DOI] [PubMed] [Google Scholar]
- Szekeres, S., M. Dauti, C. Wilde, D. Mazel and D. A. Rowe-Magnus, 2007. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 63 1588–1605. [DOI] [PubMed] [Google Scholar]
- Tobes, R., and E. Pareja, 2006. Bacterial repetitive extragenic palindromic sequences are DNA targets for insertion sequence elements. BMC Genomics 7 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilibaris, V., G. Maenhaut-Michel, N. Mine and L. Van Melderen, 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189 6101–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbaux, M., N. Mine, A. M. Guerout, D. Mazel and L. Van Melderen, 2007. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. J. Bacteriol. 189 2712–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde, C., F. Escartin, S. Kokeguchi, P. Latour-Lambert, A. Lectard et al., 2003. Transposases are responsible for the target specificity of IS1397 and ISKpn1 for two different types of palindromic units (PUs). Nucleic Acids Res. 31 4345–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky, M. B., 1995. Programmed cell death in bacterial populations. Science 267 836–837. [DOI] [PubMed] [Google Scholar]







