Abstract
Detecting and localizing selective sweeps on the basis of SNP data has recently received considerable attention. Here we introduce the use of hidden Markov models (HMMs) for the detection of selective sweeps in DNA sequences. Like previously published methods, our HMMs use the site frequency spectrum, and the spatial pattern of diversity along the sequence, to identify selection. In contrast to earlier approaches, our HMMs explicitly model the correlation structure between linked sites. The detection power of our methods, and their accuracy for estimating the selected site location, is similar to that of competing methods for constant size populations. In the case of population bottlenecks, however, our methods frequently showed fewer false positives.
ADVANCES in genotyping and sequencing technology have made it feasible to scan a large amount of DNA sequence data for genes that underwent a recent event of positive selection. Since selective sweeps affect the patterns of genetic variation also in some neighborhood of the selected locus, such scans are also known as “hitchhiking mapping” (Maynard Smith and Haigh 1974). The patterns that are usually searched for in a population genetic sample are a lower number of segregating sites (Kaplan et al. 1989), a skewed site frequency spectrum with many low-frequency and high-frequency derived alleles (Braverman et al. 1995; Fay and Wu 2000), and a modified linkage disequilibrium structure (Kim and Nielsen 2004; McVean 2006; Stephan et al. 2006; Pfaffelhuber et al. 2008).
Several statistical methods have been developed to detect signatures of selective sweeps. Most of them rely on the computation of some summary statistics of the data at each locus; see, for instance, Teshima et al. (2006) for a review on this strategy. This approach, however, implies a loss of information and brings the difficult question of the best summary statistics to use. An important improvement has been achieved by Kim and Stephan (2002), who proposed to compute the maximum composite likelihood of the observed allele frequencies for a family of models involving a selective sweep and to compare it with the composite likelihood of the same data under a model without selective sweep. With this approach, the full site frequency spectrum information is used, while also taking into account the spatial pattern of genetic diversity that is shaped by recombination. However, this method is sensitive to demographic effects, because the likelihoods are derived under a model that assumes a constant population size. To overcome this problem, Nielsen et al. (2005) proposed to estimate a background frequency spectrum from the data, rather than computing it under the hypothesis of a constant size population. Jensen et al. (2005) developed a goodness-of-fit test in the same spirit. Both methods provide an efficient control of the rate of false positives in the case of an expanding population, but still fail to do so in a wide range of bottleneck scenarios (Jensen et al. 2005; Williamson et al. 2007). As an alternative, Li and Stephan (2005, 2006) proposed to estimate the demographic history of the population under study and to compute the likelihoods using simulations that took into account this demographic information. To also use information concerning the linkage disequilibrium pattern, Kim and Nielsen (2004) proposed to compute the composite likelihood using two-locus haplotype data instead of single-locus allele data. They observed, however (see also Jensen et al. 2007), that the improvement compared to the original method of Kim and Stephan (2002) was not significant. To date, no method achieves both the high power of the original method of Kim and Stephan (2002) and a reasonable robustness against past bottleneck events.
The above-mentioned composite-likelihood methods make the simplifying assumption that the allele frequencies observed at two close sites are independent random variables. These random variables are indirectly correlated, because the distance from the selected site is taken into account when computing their distributions. However, by assuming that the probability distribution of allele frequencies is a deterministic function of the distance to the sweep, composite-likelihood methods do not capture the stochasticity of the genealogies along the sequence and the correlation pattern resulting from this stochasticity. This issue is addressed by our alternative approach based on hidden Markov models (HMMs). Hidden Markov models allow the hidden state, which should be thought of as a representation of the genealogy, to evolve stochastically along the sequence. Although the genealogy of a sample is not strictly Markovian along the sequence (Wiuf and Hein 1999), the approximation seems reasonable, given that most of the information about the genealogy at one site is provided by the genealogy at the previous site (assuming an arbitrary direction on the sequence). This assumption was successfully used, for instance, by Husmeier and Wright (2001) for detecting recombination events and by Hobolth et al. (2006) for inferring speciation times and ancestral population sizes between human, chimp, and gorilla. In addition, HMMs in general benefit from very efficient estimation and prediction algorithms and are already widely used in bioinformatics for sequence analysis (Durbin et al. 1998; Siepel and Haussler 2004).
Here, we introduce HMM methods for the detection of selective sweeps. Using computer simulations, we show that these methods have a high power for detecting selective sweep events in a population of constant size. Compared to methods that have a similar power in the case of a constant size population, we also show that they often have similar or lower rates of false positives due to demographic events such as population expansions or bottlenecks.
METHODS
Suppose we have a sample consisting of n aligned DNA sequences of length L taken from the same population. Using these data, we wish to determine whether a selective sweep has occurred in the corresponding chromosomal region. For i = 1, … , L, let Yi = 1, … , n − 1 be the number of derived alleles at site i, assuming an infinite site model. Whether an allele is ancestral or derived can be determined by looking at an outgroup. To take benefit from the lower polymorphism level expected in the vicinity of the selected locus, we also include the nonsegregating sites in the model. In our simulations, we set Yi to 0 for those sites, corresponding to the output produced by the software we used to simulate sequences [SelSim (Spencer and Coop 2004)]. In practice, distinguishing between the states Yi = 0 and Yi = n would be an option that might lead to a slight increase in performance and could easily be implemented.
Furthermore, if no reliable outgroup data are available, Yi could easily be defined as the smaller of the counts of the two alleles, thus using the folded frequency spectrum. We do not, however, consider this case in the present study.
In our model, Yi = 0,…, n − 1 is the observed state at site i. The hidden state Xi indicates whether site i has been affected by selection. While Xi should ideally represent the genealogy of the sample at site i, and thus have an infinite number of possible values, we chose to focus on a simplified model with only three hidden states: “neutral,” “intermediate,” and “sweep.” A site is in a sweep state when it is very close to the selected locus. Its site frequency spectrum is strongly influenced by the sweep. The intermediate state applies to those loci that are only slightly influenced by the sweep because of their larger distance to the selected locus. One could obviously consider models with more hidden states, but such models did not lead to a noticeable improvement of our results.
The assumption that X = X1,…, XL is a first-order Markov chain and that the Yi's are independent conditionally on X (the distribution of Yi thus depends only on Xi) leads to a HMM. Its full description requires us to specify the following components: (i) the transition matrix between hidden states, (ii) the probability distribution of the initial hidden state X1, and (iii) the emission probabilities, i.e., the probability distribution of Yi given a hidden state Xi.
We put the following constraints on the transition matrix:
The probability that the hidden chain moves directly from the neutral state to the sweep state or the reverse is set to zero. This is because we expect a gradual shift in the site frequency spectrum along the sequence and do not want to detect a random and localized loss of diversity.
From the intermediate state, the transition probabilities to the neutral state and to the sweep state are the same.
The probability of staying in the same state must be nearly equal among states.
If one state has a higher probability, we indeed observed that the most likely hidden chain was generally staying always in this state. According to these constraints, we choose a transition matrix of the form
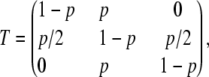 |
where Tj,k denotes the transition probability from state j to state k. The index j = 1 refers to the neutral state, j = 2 refers to the intermediate state, and j = 3 refers to the sweep state. The larger p is chosen, the more sweeps tend to be detected. We therefore use the parameter p to calibrate the false positive rate of the HMM under the null model of no sweep (see the Detecting selection from a sample section). For any value of p, the Markov chain X has the stationary distribution (π1, π2, π3) = (0.25, 0.5, 0.25), where πj = P(Xi = j). We assume that X1 is drawn from this stationary distribution, which ensures that the probability of detecting a sweep will not depend on the position of this sweep within the sequence.
If one has relevant information about the proportion of the analyzed sequence influenced by selection, one might want to modify the transition matrix to obtain a stationary distribution reflecting this information.
We considered different strategies for computing the emission matrix, leading to the following HMMs:
HMMA: The emission probabilities in all hidden states are computed according to the formulas by Kim and Stephan (2002).
HMMB: The emission probabilities are determined using the approach described by Nielsen et al. (2005).
HMMB-SEG: The same model as HMMB, except that only segregating sites are considered.
More details about these models can be found below. Two further models are also discussed in the supplemental material.
Kim and Stephan's (2002) approach (HMMA):
The emission probabilities in this model are taken from Equations 3–5 derived by Kim and Stephan (2002). These formulas approximate the probability of observing k derived alleles (k = 1, …, n − 1) in a sample of size n, both for neutral sites and for sites linked to a selected site. They assume that the sweep has just occurred and that the population size has been constant over time. The probabilities depend on the scaled mutation rate θ = 4Nμ, where N is the effective population size and μ the mutation rate per site and generation. In the case of a site linked to a selected site, the expression also depends on the quantity C = 1 − (1/α)R/s, where s is the selection intensity at the selected site, α = 2Ns is the scaled selection intensity, and R is the recombination rate between the selected site and the neutral site.
To determine the emission probabilities in our HMM, we therefore have to choose the value of θ and, in the sweep and intermediate states, the value of C. Consistent with previous results (Watterson 1975), we used Watterson's estimator of θ. Since the selection intensity and the per site recombination rate are rarely known in practice, the choice of C is more difficult. Furthermore the value of R appearing in the formula for C depends on the distance from the selected site that is modeled by our three hidden states. This leads to the rough guideline that the parameter C should be chosen smaller in the sweep state than in the intermediate state, because R is smaller in the sweep state. Without additional information, we propose to choose the parameter C to optimize the prediction ability of our HMM. According to our simulations, choosing C according to a rather weak selection pattern (i.e., a rather large value of C) under the sweep state tends to improve the sensitivity of the HMM. If the emission patterns in the three hidden states are too similar, on the other hand, the ability of the method to discriminate between neutral and sweep samples turns out to be low. We therefore chose C by optimizing the detection power in the case of a selective sweep with weak selection intensity (α = 100); for more details see the Power analysis section. We obtained C = 0.101 for the sweep state and C = 0.411 for the intermediate state. Given that α = 100, these values correspond to R/s = 0.02 (sweep) and R/s = 0.1 (intermediate). According to our results, this choice leads to a good overall detection power (see results).
Nielsen et al.'s (2005) approach (HMMB):
As in Kim and Stephan (2002), Nielsen et al. (2005) obtain an approximate formula for the probability qk* of observing k derived alleles in a sample of size n (k = 1, …, n − 1) after a selective sweep. However, in contrast to Kim and Stephan (2002), each probability qk* is expressed as a function of q = (q1, …, qn−1), where qk is the probability of observing k derived alleles in a sample of size n just before the sweep. In practice, the background probability distribution q can be estimated by the frequency spectrum in the observed sample. Since we do not know which sites are close to a selected site, the frequency spectrum is estimated from the whole sequence. Therefore the approach will be conservative, if a substantial part of the sequences is affected by selection. The probability distribution q* after the sweep also depends on the additional parameter a that quantifies the relative influence of both the recombination with the selected locus and the selection intensity. Low values of a correspond to severe hitchhiking effects.
Adapting this approach to our setting, the emission probabilities for the neutral state of our HMM can be taken from the frequency spectrum of the observed sample and the emission probabilities in the intermediate and sweep states can be computed from Equation 6 in Nielsen et al. (2005). As with the parameter C discussed above, the values of a are chosen to optimize the predictive ability of the HMM. This led to a = 0.7 for the intermediate state and a = 0.2 for the sweep state.
Segregating sites (HMMB-SEG):
SweepFinder (Nielsen et al. 2005) was developed for analyzing SNP data and does not include information about nonsegregating sites. To better compare the composite-likelihood approach and the HMM approach, we studied a variant of HMMB where only the segregating sites are included in the observed sequence. In this SNP model, Yi cannot be 0, and the emission probabilities for Yi = 1, …, n − 1 are calculated conditional on 0 < Yi < n, to sum to 1. The distance between two consecutive segregating sites is still taken into account by taking Td as the transition matrix between two sites that are d bases away from each other. The per-site transition matrix T is chosen as before. What is not accounted for by this model is that a segregating site is less likely to be found in an intermediate state than in a neutral state, and even more so in a sweep state.
Detecting selection from a sample:
To detect selection from a sample using one of our HMM methods, we first compute a sequence of frequencies of the derived allele. We then run the Viterbi algorithm (implemented in the statistical toolbox of MATLAB), to predict the sequence of hidden states X = X1, …, XL from the sequence of observed states Y = Y1, …, YL. If at least one site in the sequence has the predicted sweep state, we conclude that a sweep has occurred in the history of the sample. The presence of sites with the predicted intermediate state does not provide sufficient evidence for a sweep.
The type I error of our method can be defined as the probability of detecting a sweep when analyzing a neutral sample. We control this type I error at level 5% by selecting an appropriate coefficient p in the transition matrix. For a given test sample, p is chosen as follows: (i) Assuming a neutral evolution scenario and a population of constant size, we use ms (Hudson 2002) to simulate a set of samples of the same size n and the same length L as the test sample; (ii) for an initial value p0, we analyze these neutral samples as described above and estimate the type I error of the method by the percentage of the samples for which a sweep was detected; and (iii) if this initial error rate is greater (resp. lower) than 5%, we iteratively decrease (resp. increase) p until the error rate becomes lower (resp. greater) than 5%. We also stop the algorithm if p becomes >0.1 (even if the error is still <5%), because we do not want the Markov chain to move too fast from one state to another, which would not be consistent with our idea of modeling the correlation between close sites. For p = 0.1, the expected number of sites the chain stays in the same hidden state is equal to 10. The quantity dp that is added or subtracted to p at each iteration is small compared to p0. The value of p returned by this procedure is then used for the analysis of the test sample.
Note that the simulation of samples under neutrality described at step i requires the specification of θ and ρ. In practice, some estimates of these parameters may be found in the literature for the population where the test sample is from. If this is not the case, we suggest estimating θ and ρ directly from the test sample.
One alternative prediction approach, based on the estimation of the posterior probabilities of the sweep state at each sequence position, is briefly covered in the supplemental material.
Power analysis:
We performed computer simulations to assess the ability of the various HMM methods to detect a selective sweep (Tables 1–3). We considered different values of the scaled selection intensity α and the time τ since the fixation of the positively selected allele. The per site scaled mutation rate θ and the per site scaled recombination rate ρ = 4Nr were chosen as 0.005 and 0.02, respectively. While most of the classical tests for detecting natural selection require data from independent loci, the proposed HMM methods can be used for analyzing DNA sequences of any length. It seems thus natural to take advantage of this feature to analyze rather long sequences. Here we considered sequences of length 100 kb and a constant sample size n = 30. The influence of L and n on the power of the HMMs is covered in supplemental Table S3.
TABLE 1.
Prediction ability of the HMM methods and of composite-likelihood methods
| Method
|
|||||
|---|---|---|---|---|---|
| Selection strength | HMMA | HMMB | HMMB-SEG | CLsw | SF |
| Detection power | |||||
| α = 300 | 0.98 | 0.98 | 0.84 | 1.00 | 0.94 |
| α = 500 | 1.00 | 0.99 | 0.94 | 1.00 | 0.98 |
| Average no. of sweep windowsa | |||||
| α = 300 | 1.23 | 1.13 | 1.13 | — | — |
| α = 500 | 1.44 | 1.16 | 1.18 | — | — |
| Average length of the largest sweep window (kb)a | |||||
| α = 300 | 4.52 | 6.19 | 5.97 | — | — |
| α = 500 | 6.20 | 8.64 | 8.32 | — | — |
| Proportion of the largest sweep windows including the selected sitea | |||||
| α = 300 | 0.96 | 0.98 | 0.81 | — | — |
| α = 500 | 0.97 | 0.99 | 0.85 | — | — |
| Average distance from the largest sweep window to the selected site (kb)ab | |||||
| α = 300 | 0.97 | 1.10 | 1.68 | 0.69 | 3.20 |
| α = 500 | 1.15 | 1.45 | 1.92 | 0.90 | 2.40 |
Power to detect a simulated recent selective sweep event (τ = 0.001) is shown. HMMA, three-state model; HMMB, three-state model with estimated background emission probabilities; HMMB-SEG, the same as HMMB but with segregating sites only; CLsw, Kim and Stephan's (2002) method; SF, SweepFinder (Nielsen et al. 2005). n = 30, L = 100 kb. Type I error (percentage of falsely detected sweeps using neutral samples) is 5%. —, irrelevant.
Among those replicates where at least one sweep is detected.
Computed from the center of the sweep window.
TABLE 2.
Prediction patterns with multiple sweep windows
| No. of sweep windows
|
|||
|---|---|---|---|
| 1 | 2 | ≥3 | |
| Proportion | 0.79 | 0.19 | 0.02 |
| Average length of the largest sweep window (kb) | 4.60 | 4.16 | 4.53 |
| Average length of the second largest sweep window (kb) | — | 1.54 | 2.35 |
| Average length of the third largest sweep window (kb) | — | — | 1.31 |
| Largest distance from a sweep window to the selected site (kb)a | 0.92 | 6.78 | 16.47 |
| Distance from the largest sweep window to the selected site (kb)a | 0.92 | 1.11 | 1.62 |
| Proportion of the largest sweep windows that include the selected site | 0.97 | 0.92 | 0.89 |
Detailed results obtained with HMMA (three-state model) for the sweep scenario of Table 1 with α = 300 are shown. Averages over the replicates where at least one sweep window was detected are displayed.
Computed from the center of the sweep window.
TABLE 3.
Influence of the age of the sweep
| Age of the sweep
|
|||
|---|---|---|---|
| Selection strength | τ = 0.001 | τ = 0.01 | τ = 0.1 |
| Detection power | |||
| α = 300 | 0.98 | 0.99 | 0.89 |
| α = 500 | 0.99 | 1.00 | 0.97 |
| Average no. of sweep windowsa | |||
| α = 300 | 1.13 | 1.12 | 1.16 |
| α = 500 | 1.16 | 1.20 | 1.28 |
| Average length of the largest sweep window (kb)a | |||
| α = 300 | 6.19 | 6.21 | 5.03 |
| α = 500 | 8.64 | 8.49 | 7.03 |
| Proportion of the largest sweep windows including the selected sitea | |||
| α = 300 | 0.98 | 0.98 | 0.87 |
| α = 500 | 0.99 | 0.97 | 0.93 |
| Average distance from the largest sweep window to the selected site (kb)ab | |||
| α = 300 | 1.10 | 1.08 | 1.31 |
| α = 500 | 1.45 | 1.50 | 1.60 |
Performances of HMMB (three-state model with estimated background emission probabilities) for several values of τ and α are shown. n = 30, L = 100 kb. Type I error is 5%.
Among those replicates where at least one sweep is detected.
Computed from the center of the sweep window.
For a given set of parameter values, we simulated 400 samples under selection using SelSim (Spencer and Coop 2004).
To calibrate the transition matrix, neutral samples were simulated with the true θ and ρ. Since these true parameters will be usually unknown in practice, we also tried to estimate them independently for each test sample. We tried this approach for one set of parameter values and estimated ρ using PHASE (version 2.1) (Li and Stephens 2003; Crawford et al. 2004) and θ, using Watterson's estimator. This led to very similar results as in the case of known parameter values (not shown). This might have been expected, because these estimators are generally accurate when applied to populations with constant size. Assuming known values for θ and ρ considerably reduces the computation time, because the same 200 neutral samples (and thus the same p) can then be used for the 400 test samples.
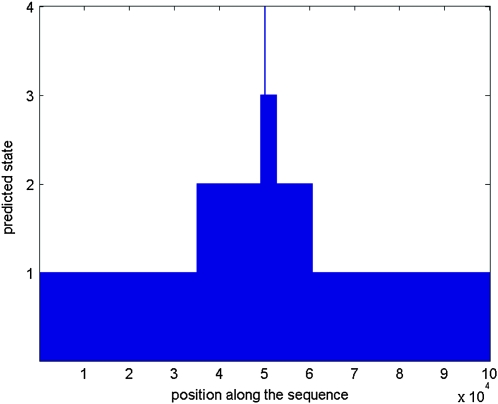
We estimated the power of a method by the percentage of test samples for which a sweep was detected. To also investigate the accuracy of the estimated sweep position, we introduce the notion of a “sweep window,” by which we mean a set of consecutive sites with the predicted hidden sweep state. When analyzing a sample, we recorded the number of these windows as well as their length and position. Because of the Markov structure of the hidden state sequence and the fact that the probability of moving from one hidden state to another is very low (otherwise the type I error of the methods would always be very high), we generally observe only few sweep windows in one sequence. For a sample that is simulated under selection, we ideally want to get only one sweep window of relatively small length and including the selected site (see Figure 1). Our simulation results (see the next section) actually show that this was in most cases the pattern that we observed.
Figure 1.—
Predicted hidden states along the sequence obtained from a typical HMM run for a sample of length 100 kb. “Neutral” states have value 1, “intermediate” states have value 2, and “sweep” states have value 3. The true selected site is indicated by the vertical line.
Scenarios with variable population size:
The expansion and bottleneck scenarios studied in Tables 4 and 5 were simulated using ms, with the same values of n, L, θ, and ρ as in the power analysis. We simulated 100 test samples for each scenario and analyzed them with HMMA and HMMB. To account for the fact that the estimators of ρ and θ are usually biased for populations with varying size, we did not use the true values of these parameters for adjusting the transition matrix of the HMMs. For each test sample, we instead estimated ρ and θ from the data and simulated 100 independent neutral samples on the basis of these values.
TABLE 4.
False positives in the case of a population expansion
| HMMA | HMMB | CLsw | SF | |
|---|---|---|---|---|
| Error rate | 0.00 | 0.01 | 0.02 | 0.00 |
False positive rate due to population growth (population size increased by a factor 10, N/2 generations ago) is shown. HMMA, three-state model; HMMB, three-state model with estimated background emission probabilities; CLsw, Kim and Stephan's (2002) method; SF, SweepFinder (Nielsen et al. 2005). n = 30, L = 100 kb. Type I error is 5% for simulations with constant population size.
TABLE 5.
False positive rates under bottlenecks
| Onset time
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
t = 0.002
|
t = 0.02
|
t = 0.2
|
||||||||||
| Depth (severity) | HMMA | HMMB | CLsw | SF | HMMA | HMMB | CLsw | SF | HMMA | HMMB | CLsw | SF |
| Duration δ = 0.04 | ||||||||||||
| 0.5 (0.08) | 0.02 | 0.02 | 0.01 | 0.02 | 0.07 | 0.06 | 0.04 | 0.08 | 0.05 | 0.02 | 0.08 | 0.05 |
| 0.1 (0.4) | 0.22 | 0.38 | 0.33 | 0.26 | 0.40 | 0.60 | 0.57 | 0.48 | 0.29 | 0.32 | 0.53 | 0.31 |
| 0.02 (2) | 0.33 | 0.25 | 0.93 | 0.73 | 0.28 | 0.13 | 1.00 | 0.60 | 0.14 | 0.04 | 0.43 | 0.29 |
| Duration δ = 0.2 | ||||||||||||
| 0.5 (0.4) | 0.10 | 0.13 | 0.12 | 0.10 | 0.12 | 0.15 | 0.18 | 0.14 | 0.11 | 0.08 | 0.16 | 0.10 |
| 0.1 (2) | 0.17 | 0.07 | 0.48 | 0.46 | 0.21 | 0.04 | 0.80 | 0.33 | 0.04 | 0.01 | 0.19 | 0.12 |
| 0.02 (10) | 0.00 | 0.00 | 0.14 | 0.02 | 0.00 | 0.00 | 0.63 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 |
False positive rates obtained with HMMA, HMMB, CLsw, and SweepFinder for various population bottleneck scenarios are shown. Parameters are n = 30, L = 100 kb. Type I error is 5% for simulations with constant population size. Time is in units of 2N generations. The depth of a bottleneck is the factor by which the population size is reduced.
HMMs and composite-likelihood methods:
As a comparison to our HMMs, the simulated samples were also analyzed with the composite-likelihood methods of Kim and Stephan (2002) (implemented in CLsw) and of Nielsen et al. (2005) (implemented in SweepFinder). HMMA is related to CLsw, because its emission matrix is computed from Kim and Stephan's (2002) formulas. Similarly, the emission matrices in HMMB and HMMB-SEG, and the composite likelihood in SweepFinder, are computed from the empirical frequency spectrum combined with Nielsen et al.'s (2005) formulas.
The statistical model behind SweepFinder and CLsw is, however, quite different from the one used for our HMMs. These methods scan a large number of positions in the sequence and look for the one that is the most likely to be a selected site. (For the analysis with SweepFinder, we took a step of 40 bases between two positions. For CLsw, the moves between positions are stochastic and handled by an optimization algorithm, starting from a set of 16 locations equally spaced along the sequence.) At each position, a composite-likelihood value, measuring the evidence for a sweep, is computed. Unlike our proposed hidden Markov models, the method takes into account the exact distance from the hypothetical selected site when computing the contribution of a site to the likelihood. On the other hand, the correlation between close sites is taken into account more realistically in a hidden Markov context. One other important point is that HMMA, HMMB, and CLsw use the information from all the sites, while HMMB-SEG and SweepFinder consider only the segregating sites.
With Clsw and SweepFinder, a selective sweep is detected when the maximum composite-likelihood value exceeds a given threshold. As for the parameter p in our HMMs, this threshold is adjusted to control the type I error of the method. In our simulation study, the same neutral samples were used to adjust the likelihood thresholds of Clsw and SweepFinder, and the parameters p of the HMMs.
RESULTS
We present the most significant results of our simulation study. We first compare the efficiency of the different HMMs for detecting the presence of a selective sweep and estimating the position of the selected site. We also investigate the influence of the strength of selection and of the age of the sweep on the results. The last section deals with the problem of robustness against demographic events such as population growth or bottlenecks.
Comparison of the HMM methods and influence of the sweep intensity:
The results obtained with our proposed HMMs for recent selective sweeps (τ = 0.001) of intensity α = 300 or 500 are provided in Table 1.
For these parameters, all our proposed methods were able to detect the sweep event with high power. Among the samples showing evidence for selection, the average number of sweep windows was close to one, indicating that in most of the cases there was exactly one sweep window along the sequence. In the rare case when multiple sweep windows were detected, we mostly observed one “large” sweep window and one or more “small” windows (an example is provided in Table 2). In most cases, the large window was located very close to the sweep and included the selected site. Consequently, we considered only the largest sweep window for the results shown in Tables 1 and 3.
For the scenarios of Table 1, all methods were able to estimate the location of the selected site accurately. The average size of the sweep window was relatively small (between 4.52 and 8.64 kb depending on the scenario and the method). For HMMA and HMMB, the sweep window included the selected site in at least 96% of the replicates. Any sweep window returned by HMMA or HMMB can consequently be viewed as a confidence interval of level almost 1 for the position of the selected site. Interestingly, the position of the selected site could be well estimated by the center of the sweep window.
With α = 300, the sweep signal is weaker than with α = 500. The detection power was consequently slightly lower for all methods. On the other hand, the location of the sweep was more accurately estimated for smaller α. In particular, (i) the average number of sweep windows was closer to one; (ii) the largest sweep window was smaller, while it still included the selected site with a very high probability; and (iii) the distance between the center of the largest sweep window and the true position of the selected site was reduced. In our simulation study, we always used the same value of C with HMMA and the same value of a with HMMB, whatever the value of α in the test samples (which seems reasonable since the true value of α is generally unknown in practice). These choices imply that in all our HMMs the emission pattern in the sweep state is expected in the vicinity of the selected site for a sweep with weak selection intensity. This emission pattern is even more likely to be found in samples generated under a sweep with strong selection intensity, but it is generally found farther from the selected site. This explains the higher power and the lower accuracy in locating the selected site observed for larger α. Taking smaller values of C with HMMA and a with HMMB would provide a higher accuracy in estimating the position of the selected site for test samples generated with α = 500. But it would also lead to lower power, especially for the simulations with α = 300 (data not shown).
The performance of the different HMMs is shown in Table 1. First, it appears that HMMB-SEG is not as efficient as the other methods. This is consistent with previous results by Kim and Stephan (2002) and Kim and Nielsen (2004), who reported that the density of segregating sites along the sequence provided useful information. Second, HMMB is slightly more reliable than HMMA. It returned on average fewer sweep windows, which more often included the selected site. On the other hand, these windows were a bit larger than with HMMA, and their center was also farther from the sweep.
Comparison to other methods:
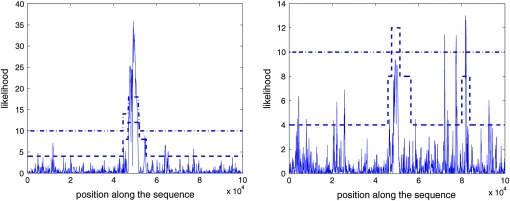
The results obtained with the composite-likelihood methods of Kim and Stephan (2002), implemented in the software CLsw, and of Nielsen et al. (2005), implemented in the software SweepFinder, are also presented in Table 1. Among all methods, CLsw was the most powerful and the one providing the most accurate estimations of the selected site location. It performed slightly better than HMMA. Among the methods that use an estimated background frequency spectrum, HMMB had a higher power than SweepFinder and the position of the selected site was more accurately estimated by HMMB. The estimate of the selected site position provided by SweepFinder was >20 kb away from the true position in 3.95% of the samples for which a sweep was detected, while this never happened with HMMB. The likelihood curve obtained for one such sample is provided in Figure 2. While still estimating the sweep position more accurately, HMMB-SEG was a bit less powerful than SweepFinder. This suggests that the better power obtained with HMMB compared to SweepFinder comes from the additional information used by HMMB.
Figure 2.—
Likelihood curves obtained by SweepFinder (solid lines) and predicted hidden sequences obtained by HMMB (dashed lines) and HMMB-SEG (dashed-dotted lines) for two samples of size n = 30 and length L = 100 kb generated with α = 300 and τ = 0.001. For SweepFinder, the 95% threshold was 10.00 in this case. The states “neutral,” “intermediate,” and “sweep” are represented, respectively, by the values 4, 8, and 12 for HMMB and by the values 10, 14, and 18 for HMMB-SEG. The true selected site is in the middle of the segment, at position 50 kb. Large differences between SweepFinder and HMMB, as for sample 2, were observed in ∼4% of the simulated samples.
Influence of the age of the sweep:
In all our HMMs, the computation of the emission probabilities in the sweep state is based on the assumption of a very recent sweep. The ability to detect older sweeps was tested using simulation scenarios with τ = 0.01 and τ = 0.1. We report the results of these simulations in Table 3, focusing on HMMB. With τ = 0.01, the quality of the predictions was almost the same as with τ = 0.001. With τ = 0.1, the results obtained with α = 300 were affected. Both the detection power and the accuracy of localization decreased. Interestingly, the results obtained with α = 500 remained very good even with τ = 0.1. Furthermore, the difference between HMMB and HMMB-SEG was larger with τ = 0.1 than with τ = 0.001 (not shown). Using the density of segregating sites seems important to detect older sweeps, for which there is only a small excess of high-frequency derived alleles.
Impact of population demography:
As outlined in several studies (e.g., Jensen et al. 2005; Teshima et al. 2006; Thornton and Jensen 2007; Wiehe et al. 2007), demographic events such as population expansions or bottlenecks are often falsely detected as selective sweep events, because their effects on the frequency spectrum can be very similar. To investigate the influence of such demographic events, we first studied a neutral scenario where the population size was increased by a factor 10 at time t = 0.25. For each test sample, we estimated ρ and θ from the data and simulated independent neutral samples on the basis of these values to adjust the type I errors of the different methods. As expected, ρ and θ were underestimated. The average 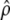 was 0.015 (ρ = 0.02) and the average
was 0.015 (ρ = 0.02) and the average 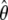 was 0.002 (θ = 0.005).
was 0.002 (θ = 0.005).
The rates of falsely detected sweeps obtained with the different methods for this expansion scenario are given in Table 4. The good performance of HMMA and CLsw may seem surprising, given that the frequency spectrum of an expanding population shares two common properties with the one of a selection event: the low number of segregating sites and the high number of low-frequency derived alleles. Two reasons actually explain that no sweep was detected by HMMA and CLsw for the samples of Table 4. First, the emission probabilities in HMMA, and the likelihoods in CLsw, are computed using the estimated θ, which is based on the observed number of segregating sites, and therefore the low level of polymorphism in the data is not detected as an evidence for selection. Second, a relatively high proportion of high-frequency derived alleles is expected close to a selected site, but is not found for an expanding population. Confirming the results of Nielsen et al. (2005), SweepFinder did not detect selection in the samples of Table 4. This is due to the homogeneity of the frequency spectrum of an expanding population along the sequence. For the same reason, HMMB also performed very well.
In the samples simulated under population expansion, θ was generally more strongly underestimated than ρ. Consequently, in the neutral samples used for adjusting the transition matrix of the HMMs, the numbers of derived alleles at two consecutive segregating sites were weakly correlated, which reduces the probability of observing several consecutive segregating sites that all exhibit a sweep pattern. A false detection rate of 5% among these samples would require us to set a rather high transition probability p between hidden states. But using high transition probabilities between hidden states could lead us to detect very short sweep windows. To make the method more robust against random fluctuations of the frequency spectrum along the sequence, an upper bound on p turned out to be helpful. In our simulations, the bound p ≤ 0.1 provided satisfactory results.
We also studied the impact of a bottleneck on the predictions and considered several bottleneck scenarios. The scenarios were chosen by varying (i) the depth d of the bottleneck, i.e., the factor by which the population size is reduced (we assume the population size to be the same before and after the bottleneck); (ii) the duration δ of the bottleneck, i.e., the time during which the population size is reduced; and (iii) the onset time t, i.e., the time since the bottleneck ended. The range of values we chose for these parameters is consistent with those used in recent studies on Drosophila melanogaster (Haddrill et al. 2005; Thornton and Andolfatto 2006; Wiehe et al. 2007) or on non-sub-Saharan human populations (Teshima et al. 2006). As for the samples simulated under population expansion, ρ and θ were estimated from the data for each bottleneck sample.
The percentage of replicates where a sweep was (wrongly) detected is presented in Table 5. In addition to the bottleneck parameters d, δ, and t, we indicate for each scenario the severity γ = δ/d of the bottleneck as defined in Wiehe et al. (2007). The results obtained with the HMMs and the composite-likelihood methods are qualitatively similar to the ones in (Wiehe et al. 2007): The error rates were in general low both for very small (γ = 0.08) and very large (γ = 10) severities, but they were higher for intermediate severities (0.4 or 2). Among the bottlenecks with intermediate severity, the ones with t = 0.2 caused fewer errors than the ones with t = 0.02 and in most cases fewer errors than the ones with t = 0.002 (but see line 2). It is natural to observe lower detection rates in the case of old and less severe bottlenecks, because they have only a small effect on the frequency spectrum. On the other hand, severe bottlenecks do produce a very strong effect on the frequency spectrum, but their effect is quite similar along the sequence, as in a population expansion scenario. The results obtained in such cases can thus be linked to the ones presented in Table 4. Recent bottlenecks with intermediate severity are the most difficult to distinguish from sweeps because they cause a very large variance in the frequency spectrum along the sequence (Jensen et al. 2005), with some segments where this spectrum may appear as that of a selective sweep when compared to the global spectrum. Consistent with our results, Williamson et al. (2007) also found that SweepFinder was conservative for bottlenecks with depth 0.01, anti-conservative for bottlenecks with depth 0.1, and almost unaffected for bottlenecks with depth 0.5.
Besides this overall pattern, large differences were often observed between the different methods. In all the bottleneck scenarios considered here, HMMA had similar or lower false positive rates than CLsw. The difference between the two methods was particularly large for bottleneck scenarios with depth <0.5 (rows 2, 3, 5, and 6 in Table 5). Note that CLsw could be made more robust by using the goodness-of-fit test proposed by Jensen et al. (2005). As shown by the authors, the false positive rates obtained with severe bottlenecks could be substantially reduced using this method. But the use of this correction would also result in a lower detection power. In the scenario with α = 300 studied in Table 1, we indeed found that the detection power, equal to 1.00 without the correction, would fall to 0.73 with the correction. HMMB performed equally well or better than SweepFinder, except for two scenarios. The difference between the two methods was sometimes large, for instance, for scenarios with severity 2 (Table 5, rows 3 and 5).
For the scenarios with severity 0.4 shown in row 2 of Table 5, 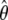 was large compared to
was large compared to 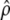 , which implied that the observations between consecutive segregating sites were highly correlated. The value of p used with the HMMs was quite small in this case. The lower false positive rates obtained by HMMA compared to CLsw may thus be related to the fact that the correlation between sites is taken into account more directly in the HMM framework. For the scenarios with severity >2 (lines 3, 5, and 6 in Table 5), the restriction p ≤ 0.1 turned out to be helpful for avoiding very short artificial sweep windows, as in the case of the expanding population scenario.
, which implied that the observations between consecutive segregating sites were highly correlated. The value of p used with the HMMs was quite small in this case. The lower false positive rates obtained by HMMA compared to CLsw may thus be related to the fact that the correlation between sites is taken into account more directly in the HMM framework. For the scenarios with severity >2 (lines 3, 5, and 6 in Table 5), the restriction p ≤ 0.1 turned out to be helpful for avoiding very short artificial sweep windows, as in the case of the expanding population scenario.
We also looked at sweeps within a bottleneck background and used the software developed by Jensen et al. (2007) to simulate scenarios where a sweep of intensity α = 300 occurred during a bottleneck. The power obtained by the different methods is shown in Table 6. We considered two of the bottleneck scenarios in Table 5, both with t = 0.002 and d = 0.1. The first scenario was the one with severity 0.4 and depth 0.1, where both the HMMs and the composite-likelihood methods had high rates of false positives, and where p was small for the HMMs. We can see in Table 6 that all the methods had reasonably high power in this case. The highest ratio between true positives and false positives was obtained with HMMA (∼4). For the second scenario, the HMMs had almost no power. One way to increase the power in this case would be to relax the constraint p ≤ 0.1. However, we do not recommend this, since the false positive rate when no sweep occurred would then also increase. We note that also CLsw had almost as many false positives as true positives in this case.
TABLE 6.
Detection power vs. false positives for sweeps occurring within a bottleneck
| Method
|
||||
|---|---|---|---|---|
| Bottleneck scenario | HMMA | HMMB | CLsw | SF |
| δ = 0.04 | 0.84/0.22 (= 3.82) | 0.75/0.38 (= 1.97) | 0.92/0.33 (= 2.79) | 0.66/0.26 (= 2.54) |
| δ = 0.2 | 0.04/0.17 (= 0.23) | 0.02/0.07 (= 0.29) | 0.65/0.48 (= 1.35) | 0.23/0.46 (= 0.50) |
Power of HMMA, HMMB, CLsw, and SweepFinder for detecting a sweep of intensity α = 300 that occurred during a bottleneck and false positive rate obtained for the same bottleneck scenario without sweep are shown. The bottleneck scenarios considered here were chosen from among the ones presented in Table 5, with t = 0.002 and d = 0.1. To distinguish true sweep signals from false ones, we count as true signals only those sweep windows whose distance from the actual selected site is <10 kb. The false positive rates are taken from Table 5. The results are displayed in the form “power”/“false positive rate” (= ratio).
In all the scenarios of Table 5 with severity ≤0.4, HMMA produced fewer false positives than HMMB. This may seem surprising, given that with HMMB the background frequency spectrum is estimated from the data, which is supposed to make the method conservative. We note, however, that this is true only if the effects of demography on the frequency spectrum are similar to the ones of positive selection. This is typically the case for a population expansion, but not for a bottleneck, for which the overall frequency spectrum is actually modified in the “opposite” direction (toward an excess of intermediate-frequency derived alleles) to that for a sweep within a neutral background.
Consequently, the emission probabilities in the sweep state of HMMB correspond to a weaker sweep signal than the ones in the sweep state of HMMA, which explains the higher rate of false positives obtained with HMMB. However, this general idea does not explain the results for the scenarios with severity >2, where HMMB often had fewer false positives than HMMA. Finally, the higher power of HMMA compared to HMMB in the bottleneck scenario with severity 0.4 (Table 6) was not expected, given that for this scenario the rate of false positives was lower with HMMA. While little is known on the combined effects of bottlenecks and sweeps, this result shows that the frequency spectrum observed around a positively selected site can be similar for a population with constant size and for a population that went through a bottleneck of intermediate severity.
DISCUSSION
We introduced a new approach, based on HMMs, for detecting selective sweeps using DNA sequence data. This approach makes use of the full frequency spectrum information and of the spatial structure of the diversity along the sequence. In contrast to composite-likelihood methods, it also models the stochasticity of the correlations between sites along the sequence. We considered two different ways of computing the emission probabilities of the HMM, using either Kim and Stephan's (2002) or Nielsen et al.'s (2005) approach.
We evaluated the statistical properties of our methods using computer simulations. As illustrated in Tables 1 and 3, both HMMA and HMMB have a very high power for detecting sweeps occurring in a population with constant size. They are powerful even for rather weak selection intensities (α = 300) and quite old sweeps (τ = 0.1). In most of the replicates, the estimated sweep location is very accurate. We also found that both HMMA and HMMB are robust against population growth and against bottleneck scenarios with both weak (0.08) and strong (10) severity (Table 5). For bottleneck scenarios with intermediate severity (0.4 and 2), HMMA and HMMB often produced a considerable proportion of false positives, but generally fewer than the composite-likelihood methods of Kim and Stephan (2002) and Nielsen et al. (2005). HMMA performed better than HMMB for bottleneck scenarios with severity ≤0.4, with both fewer false positives and more true positives (Table 6). Given that both methods performed equally well in Tables 1–4, we would rather recommend using HMMA if we had to choose between both. However, for some bottleneck scenarios with severity 2, HMMB had fewer false positives.
The performance of the HMM approach in comparison to composite-likelihood methods depends on the parameters used. In our simulations, SweepFinder was a bit more powerful than HMMB-SEG (Table 1), but with α = 300 the estimation of the selected site position was less accurate. In some samples where HMMB-SEG could not detect a sweep, SweepFinder did detect it but at a quite wrong position (Figure 2, sample 2). Clsw also performed slightly better than HMMA for detecting selection in a population of constant size. In contrast to HMMA, however, Clsw had a very high rate of false positives under several bottleneck scenarios, up to 1.00 in some cases. For the most difficult bottleneck scenarios, HMMA led to both much fewer false positives and much fewer true positives than CLsw (cf. Table 6, row 2). In other cases, HMMA had a better ratio between true and false positives than Clsw (cf. Table 6, row 1). Overall, it seems that the significant increase of robustness offered by HMMA is more important than the loss of power, which is small for constant size populations. For completeness, we note that Jensen et al.'s (2005) correction also provided a great gain in robustness in the case of bottleneck scenarios, but led also to a considerable loss of power for constant size populations (only 73% power in the scenario of Table 1 with α = 300).
In addition, the results of Tables 5 and 6 suggest the HMMs perform better in the scenarios where the correlation between sites is strong, which is probably due to the fact that the correlation between sites is directly modeled via the Markov structure of the hidden states in a HMM framework. This may thus provide a criterion for deciding when these methods should be preferred over other existing methods. In general, it also seems that the HMM approach is more flexible, because it does not assume that the frequency spectrum is a deterministic function of the distance to the sweep. Hence, it should be less affected by heterogeneities in the recombination rates, for instance, the hot-and-cold spots of recombination, which have been described for humans (Jeffreys et al. 2001; Gabriel et al. 2002).
The good results obtained by HMMA and HMMB are partly due to the fact that they use all the sites, not only the segregating ones. In our simulations, HMMB performed clearly better than HMMB-SEG. Given that we estimate θ using Watterson's estimator, the additional information provided by the nonsegregating sites is not related to the average diversity level, but to its spatial variation along the sequence (Kim and Stephan 2002). In the composite-likelihood framework, Kim and Stephan (2002) and Kim and Nielsen (2004) had already observed that using all the sites was a clear advantage. However, Nielsen et al. (2005) pointed out that the usual SNP data sets did not correctly represent the genomic diversity and focused on segregating sites while taking the ascertainment process into account. With the recent advent of high-throughput sequencing, such ascertainment problems will disappear in many cases so we think that methods that combine information from the frequency spectrum and from the diversity level provide promising tools for future analysis.
Even if the HMMs proposed here led to a decrease in the rate of false positives in some bottleneck scenarios, they do not solve the problem completely. Indeed, several bottleneck scenarios with intermediate severities (0.4 and 2) resulted in a high rate of false positives both with the HMM and with the composite-likelihood approach. Such bottleneck scenarios are very difficult to keep apart from selective sweeps, because they induce a high spatial variation in both the diversity level and the site frequency spectrum along the sequence. An alternative approach for such scenarios would be to use the linkage disequilibrium information. Jensen et al. (2007) studied a bottleneck scenario that was very similar to one where our HMMs had a lot of false positives, i.e., with depth d = 0.1 and duration δ = 0.05 (Jensen et al.'s Figure 2C), and showed that it could be well distinguished from a sweep scenario with α = 500 (Jensen et al.'s Figure 2A). They used a statistic introduced by Kim and Nielsen (2004) and aimed at identifying a linkage disequilibrium structure that is typical for a sweep site.
One alternative way of controlling the errors resulting from demography would be of course to use samples simulated under the “true” demographic scenario for choosing the appropriate value of p in the transition matrix. This would be possible, if the true demographic scenario were known from previous studies, or if it can be inferred by other methods. However, we note that it will be difficult to predict, whether the errors made when estimating the true demographic scenario result in a too conservative or a too liberal test. Indeed, the results of Table 5 suggest that different bottleneck parameters can lead to very different detection patterns.
In our models, the sweep state is characterized by both a low diversity level and an excess of high-frequency derived alleles. This pattern is actually expected in two regions flanking the selected site, but not very close to the selected site (Fay and Wu 2000). One may thus wonder if, in our HMMs, an alternative sweep signal would be a pair of two close sweep windows, rather than one single sweep window. However, the results of Tables 1 and 3 do not support this idea, since in most of the samples only one sweep window was detected.
This could in principle be changed by increasing p, but we observed that the average number of sweep windows increases only slowly with p (data not shown). Consistent with these results, we also observed that a three-state HMM, where the sweep state had essentially no diversity (as in the central region of a sweep), and where the intermediate state had an excess of high-frequency derived alleles [as in the flanking regions (Fay and Wu 2000)], did not perform as well as our current models. One reason might be that, for weak selection, the central region of the sweep is too small. Combining the information from the frequency spectrum and the diversity level seems to be a strength of our HMMs, which makes them powerful even for noisy signals, such as those produced by older sweeps (Table 3).
As for most sweep detection methods (but see Hudson et al. 1987; Schlötterer 2002), we have made the implicit assumption of a homogeneous mutation rate along the entire genomic region surveyed. Hence, a sequence block with lower mutation rate, or a block subject to strong purifying selection, could eventually lead to a signature of a selective sweep. Future studies will be required to determine if the observed heterogeneity in mutation rates poses a significant problem for our HMM methods.
The HMMs that we presented here have only three hidden states (neutral, intermediate, and sweep) and thus do not fully capture the continuity of the frequency spectrum and of the diversity level along the sequence. We point out that we do not consider these models as a good representation of the reality, but only as good prediction tools. It is well known in statistics that the most sophisticated models are usually not the most efficient for prediction purposes. For sweep detection, we actually observed that a HMM with only two states, neutral and sweep, performed almost as well as a HMM with three states (supplemental material). We consequently do not think that increasing the number of hidden states would significantly improve our results.
Many of the sweeps that have already been detected in natural populations have stronger selection intensities than those we used in the simulations we presented. Intensities up to a few thousand have been reported, for instance, in D. melanogaster (Li and Stephan 2006). Since all methods are equally able to detect very strong sweeps, our focus has been on weak sweeps, which are more challenging. In addition, the results in Table 1 indicate that our HMM methods can also easily identify stronger sweeps, because the power increases with α. Due to the larger genomic region affected by a strong selective sweep, however, the precise identification of the target of selection is more difficult for stronger than for weaker selection. This could actually be solved by a second run of the HMM method, using a more skewed emission pattern in the sweep and intermediate states. Alternatively, prior information concerning the population or organism of interest could also be used when selecting the model parameters.
In this report, we focused on the most widely studied case of a hard sweep, in which one beneficial mutation arises and subsequently sweeps through the population until it reaches fixation. Recently, it has been suggested that soft sweeps, which involve either recurrent mutations or selection from standing variation, are another scenario that should be considered (Przeworski et al. 2005; Pennings and Hermisson 2006). We anticipate that the HMM methods introduced here could be modified to account for different selection regimes.
Overall, the HMM approach provides high power for detecting selective sweeps and is often more robust than other existing methods with comparable power.
Acknowledgments
We thank C. Vogl for general discussions and helpful comments on the manuscript. M. H. Quang provided some data-processing routines. K. Thornton kindly helped us with the simulations combining selection and bottleneck. Finally, we are grateful to two anonymous reviewers for useful and insightful comments on the manuscript. This work was financially supported by grants from the Wiener Wissenschafts-, Forschungs- und Technologiefonds and the Fonds zur Förderung der Wissenschaftlichen Forschung (P19467-B11).
References
- Braverman, J., R. Hudson, N. Kaplan, C. Langley and W. Stephan, 1995. The hitchhiking effect on the site frequency spectrum on DNA polymorphisms. Genetics 140 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, D., T. Bhangale, N. Li, G. Hellenthal, M. Rieder et al., 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36 700–706. [DOI] [PubMed] [Google Scholar]
- Durbin, R., S. Eddy, A. Krogh and G. Mitchison, 1998. Biological Sequence Analysis. Cambridge University Press, Cambridge, UK.
- Fay, J., and C.-I. Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, S., S. Schaffner, H. Nguyen, J. Moore, J. Roy et al., 2002. The structure of haplotype blocks in the human genome. Science 296 2225–2229. [DOI] [PubMed] [Google Scholar]
- Haddrill, P., K. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophilia melanogaster populations. Genome Res. 15 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth, A., O. Christensen, T. Mailund and M. Schierup, 2006. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3(2): e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18 337–338. [DOI] [PubMed] [Google Scholar]
- Hudson, R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husmeier, D., and F. Wright, 2001. Detection of recombination in DNA multiple alignments with hidden Markov models. J. Comp. Biol. 8 401–427. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A., L. Kauppi and R. Neumann, 2001. Intensely punctate meiotic recombination in the class ii region of the major histocompatibility complex. Nat. Genet. 29 217–222. [DOI] [PubMed] [Google Scholar]
- Jensen, J., Y. Kim, V. Bauer DuMont, C. Aquadro and C. Bustamante, 2005. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J., K. Thornton, C. Bustamante and C. Aquadro, 2007. On the utility of linkage disequilibrium as a statistic for identifying targets of positive selection in nonequilibrium populations. Genetics 176 2371–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, N., R. Hudson and C. Langley, 1989. The hitchhiking effect revisited. Genetics 123 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and R. Nielsen, 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and W. Stephan, 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and W. Stephan, 2005. Maximum-likelihood methods for detecting recent positive selection and localizing the selected site in the genome. Genetics 171 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and W. Stephan, 2006. Inferring the demographic history and rate of adaptative substitution in Drosophila. PLoS Genet. 2(10): e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N., and M. Stephens, 2003. Modeling linkage disequilibrium, and identifying recombination hotspots using SNP data. Genetics 165 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23 23–35. [PubMed] [Google Scholar]
- McVean, G., 2006. The structure of linkage disequilibrium around a selective sweep. Genetics 175 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R., L. Williamson, Y. Kim, M. Hubisz, A. Clark et al., 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings, P., and J. Hermisson, 2006. Soft sweeps iii: the signature of positive selection from recurrent mutation. PLoS Genet. 2(12): e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffelhuber, P., A. Lehnert and W. Stephan, 2008. Linkage disequilibrium under genetic hitchhiking in finite populations. Genetics 179 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski, M., G. Coop and J. Wall, 2005. The signature of positive selection on standing genetic variation. Evolution 59(11): 2321–2323. [PubMed] [Google Scholar]
- Schlötterer, C., 2002. A microsatellite-based multilocus screen for the identification of local selective sweeps. Genetics 160 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A., and D. Haussler, 2004. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comp. Biol. 11 413–428. [DOI] [PubMed] [Google Scholar]
- Spencer, C., and G. Coop, 2004. Selsim: a program to simulate population genetic data with natural selection and recombination. Bioinformatics 20(18): 3673–3675. [DOI] [PubMed] [Google Scholar]
- Stephan, W., Y. Song and C. Langley, 2006. Hitchhiking effect on linkage disequilibrium between linked neutral loci. Genetics 172 2647–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima, K., G. Coop and M. Przeworski, 2006. How reliable are empirical genomic scans for selective sweeps. Genome Res. 16 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K., and P. Andolfatto, 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K., and J. Jensen, 2007. Controlling the false-positive rate in multilocus genome scans for selection. Genetics 175 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Wiehe, T., V. Nolte, D. Zivkovic and C. Schlötterer, 2007. Identification of selective sweeps using a dynamically adjusted number of linked microsatellites. Genetics 175 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, S., M. Hubisz, A. Clark, B. Payseur, C. Bustamante et al., 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3(6): e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf, C., and J. Hein, 1999. Recombination as a point process along sequences. Theor. Popul. Biol. 55 248–259. [DOI] [PubMed] [Google Scholar]