Abstract
A genetic linkage map of the channel catfish genome (N = 29) was constructed using EST-based microsatellite and single nucleotide polymorphism (SNP) markers in an interspecific reference family. A total of 413 microsatellites and 125 SNP markers were polymorphic in the reference family. Linkage analysis using JoinMap 4.0 allowed mapping of 331 markers (259 microsatellites and 72 SNPs) to 29 linkage groups. Each linkage group contained 3–18 markers. The largest linkage group contained 18 markers and spanned 131.2 cM, while the smallest linkage group contained 14 markers and spanned only 7.9 cM. The linkage map covered a genetic distance of 1811 cM with an average marker interval of 6.0 cM. Sex-specific maps were also constructed; the recombination rate for females was 1.6 times higher than that for males. Putative conserved syntenies between catfish and zebrafish, medaka, and Tetraodon were established, but the overall levels of genome rearrangements were high among the teleost genomes. This study represents a first-generation linkage map constructed by using EST-derived microsatellites and SNPs, laying a framework for large-scale comparative genome analysis in catfish. The conserved syntenies identified here between the catfish and the three model fish species should facilitate structural genome analysis and evolutionary studies, but more importantly should facilitate functional inference of catfish genes. Given that determination of gene functions is difficult in nonmodel species such as catfish, functional genome analysis will have to rely heavily on the establishment of orthologies from model species.
LINKAGE maps are powerful research tools for mapping quantitative trait loci (QTL) to complement marker-assisted selection in many species, including aquaculture species (Lander and Botstein 1989; Sakamoto et al. 2000; Fishman et al. 2001; Nichols et al. 2003; Hubert and Hedgecock 2004; Moen et al. 2004, 2008; Chistiakov et al. 2005; Lee et al. 2005; Gharbi et al. 2006; Liu et al. 2006; Sekino et al. 2006; Phillips et al. 2007; Sekino and Hara 2007; for a recent review, see Danzmann and Gharbi 2007). However, marker density for all aquacultured species is still low. Aquaculture genome research can greatly benefit from genome studies of model species through comparative genome analysis, transferring genome information from fully sequenced and functionally well-characterized model species to aquacultured species (Sarropoulou et al. 2008).
Comparative genome analysis can be facilitated if a draft genome sequence is available for the species of interest. This area is rapidly expanding because whole-genome sequences are becoming available from many species, including five teleost species: zebrafish (Danio rerio), fugu (Fugu rubripes), Tetraodon (Tetraodon nigroviridis), medaka (Oryzias latipes), and three-spined stickleback (Gasterosteus aculeatus). To date, no whole-genome sequence exists for any aquaculture species. Major progress, however, has been made in the generation of other genome resources for some economically important aquaculture species such as tilapia (Katagiri et al. 2005; Lee et al. 2005; Ferreira and Martins 2008), rainbow trout (Rexroad and Palti 2003; Guyomard et al. 2006, 2007), Atlantic salmon (Moen et al. 2004, 2008), gilthead seabream (Sparus aurata) (Franch et al. 2006; Senger et al. 2006; Sarropoulou et al. 2008), and the European sea bass (Dicentrarchus labrax) (Chistiakov et al. 2005; Whitaker et al. 2006).
Channel catfish (Ictalurus punctatus) is the most economically important catfish species in the United States. It is anticipated that it will become one of the most important aquaculture fish species in Asia as well. Blue catfish (I. furcatus) is also economically important because the hybrid between channel catfish and blue catfish exhibits superior performance for several commercial traits (Dunham et al. 1987, 1990, 1993; Dunham and Argue 1998; He et al. 2003). Since these interspecific hybrids are fertile, it is possible to generate synthetic breeds using introgression strategies (Liu 2003; Liu et al. 2003).
Major progress has been made in catfish genome research, particularly in the area of genome resource development, including a large number of polymorphic markers (Serapion et al. 2004a,b; Xu et al. 2006; Somridhivej et al. 2008), construction of genetic linkage maps (Waldbieser et al. 2001; Liu et al. 2003), construction and characterization of BAC libraries (e.g., Wang et al. 2007), construction of BAC contig-based physical maps (e.g., Xu et al. 2007), generation of 63,000 BAC end sequences (Xu et al. 2006; Z. Liu, unpublished data), understanding of the genomic architecture (Liu et al. 1998; Liu 2007; Nandi et al. 2007), and a large number of ESTs (Ju et al. 2000; Cao et al. 2001; Karsi et al. 2002; Kocabas et al. 2002; Li et al. 2007). However, these genome resources are as yet nonintegrated and thus underutilized due to the lack of a platform for comparative genome analysis. A gene-based linkage map would provide a start for such a platform. In addition to these traditional genome resources, the use of single nucleotide polymorphism (SNP) markers is gaining significant momentum in aquacultured species (Hayes et al. 2007; Moen et al. 2008; Wang et al. 2008).
A major utility of gene-based linkage maps is comparative genomics, which is used to assist in the understanding of genome evolution (Meyers et al. 2005; Woods et al. 2005; Mousel et al. 2006; Sasazaki et al. 2006; Sawera et al. 2006). Because gene-associated markers are conserved through a wide evolutionary spectrum of species, they have become the most desirable type of marker for comparative mapping (Martin et al. 2005; Moretzsohn et al. 2005; Smith et al. 2005; Snelling et al. 2005; Casasoli et al. 2006; Kim et al. 2006; Sasazaki et al. 2006; Sawera et al. 2006). We have previously reported the identification of a large number of EST-associated microsatellites and SNPs (He et al. 2003; Serapion et al. 2004a; Wang et al. 2008). Here we present a gene-based linkage map of the catfish genome constructed with EST-associated microsatellites and SNPs. We report map locations for a total of 331 gene-based markers including 259 microsatellites and 72 SNPs. The linkage map is composed of 29 linkage groups (LGs). Significant differences in recombination frequencies between males and females were noted. Conserved syntenies were identified between the catfish and three model fish species: zebrafish, Tetraodon, and medaka.
MATERIALS AND METHODS
Catfish resource families:
F1 interspecific hybrid catfish were made by mating channel catfish females with blue catfish males. These F1 catfish and their parents were screened prior to the 1997 spawning season to determine which matings were most informative. Backcross families were made in the spawning season of 1997 by mating the F1 fish with channel catfish (channel catfish backcross). A specific family, F1-2 × channel catfish-6, was used for this project. The resource family was reared in 1000-liter tanks until collection of blood samples for genotyping. Individuals that were sampled for genotyping were heat-branded for future identification.
Genomic DNA:
Blood samples (0.5–1 ml) were collected in a 1-ml syringe and immediately expelled into a 50-ml tube containing 20 ml of DNA extraction buffer (100 mm NaCl, 10 mm Tris, pH 8, 25 mm EDTA, 0.5% SDS, and 0.1 mg/ml freshly made proteinase K), and DNA was isolated as previously described using standard protocols (Liu et al. 1998). Briefly, the blood samples were incubated at 55° overnight and DNA was extracted twice with phenol and once with chloroform. DNA was precipitated by adding a half volume of 7.5 m ammonium acetate and 2 vol of ethanol. DNA was collected mostly by spooling onto a micropipette tip, or in some cases by brief centrifugation, and washed twice with 70% ethanol, air-dried, resuspended in TE buffer (Tris–HCl, 10 mm, EDTA, 1 mm, pH 7.5), and quantified with a spectrophotometer.
Identification of microsatellites, primers, and PCR amplification:
EST-based microsatellites were previously published (Serapion et al. 2004b). FastPCR (Kalendar 2008) was used for the design of PCR primers. A tailed primer protocol (Oetting et al. 1995; Boutin-Ganache et al. 2001) was used to amplify microsatellite alleles. The following conditions were used to amplify the microsatellites: 1× PCR buffer, 1.5 mm MgCl2, 0.2 mm each of dNTPs, 4 ng upper PCR primer, 6 ng lower PCR primer, 20 fmol labeled primer, 0.25 units of JumpStart Taq polymerase (Sigma, St. Louis), and 20 ng genomic DNA, in a total reaction volume of 5 μl. A touchdown PCR was performed with the following thermo profile: after an initial denaturation at 94° for 3 min, PCR amplification was carried out at 94° for 30 sec, 57° for 30 sec, and 72° for 30 sec for 20 cycles as the first step and at 94° for 30 sec, 53° for 30 sec, and 72° for 30 sec for 15 cycles as the second step. A final extension at 72° for 10 min was included. The PCR products were analyzed on 7% polyacrylamide sequencing gels using LI-COR automated DNA sequencers.
SNP markers:
A total of 384 EST-derived SNP markers were genotyped as described in Wang et al. (2008) using the F1-2 × channel catfish-6 hybrid catfish.
Genotyping:
For microsatellites, after running through the LI-COR automated sequencers, genotypes were called by recording the amplified fragment sizes (in base pairs) in a Microsoft Excel spreadsheet. The fragment sizes were determined by using labeled size markers (LI-COR). Loci that did not show any polymorphism were recorded as nonpolymorphic. The complex loci and parental type microsatellites were also recorded. A chi-square goodness-of-fit test was used to assess the departures from the expected Mendelian segregation patterns. Genotype configurations of markers were categorized into three expected segregation types when null-allele segregation was allowed: 1:1:1:1-ratio type (♀ × ♂: AB × CD or AB × AC), 1:1 ♀ type (AB × AA or CC), and 1:1 ♂ type (AA or CC × AB). All statistical analyses described below were completed using JoinMap 4.0 (Kyazma B.V., Wageningen, The Netherlands) with the cross-pollinating coding scheme, which handles the data containing various genotype configurations with unknown linkage phases (Sekino et al. 2006). Segregation data from expected 1:1:1:1-type markers into 1:1 ♀- and 1:1 ♂-type markers were partitioned by creating maternal and paternal data sets using JoinMap 4.0 to perform linkage analysis for each sex (Jacobs et al. 1995; Viruel et al. 1995). This option in JoinMap 4.0 creates maternal and paternal genotypes by converting genotypes from 1:1:1:1-ratio type (♀ × ♂: AB × CD or AB × AC) to 1:1 ♀ type (AB × AA or CC) and to 1:1 ♂ type (AA or CC × AB).
Linkage analysis:
Linkage between markers, recombination rate (Θ), and map distances was calculated using the Kosambi mapping function. Significance was tested by JoinMap, which tests for independence of segregation using LOD scores. LOD scores were generated using the G2 statistic, which was multiplied by 0.5 × log10e to convert into a normalized LOD scale. Significance was determined at a LOD threshold of 3.0, and a threshold Θ of 0.6 was set to detect suspect linkage possibly resulting from allele-coding errors. Six individuals were omitted from the analyses because they were missing too many genotypes.
Markers were linearly aligned in each linkage group, converting the recombination rates into the Kosambi's map distance (in centimorgans). The position of markers was developed using a sequential map buildup (Stam 1993). With this method, the most informative pair of markers was selected, followed by sequential addition of other markers. The “ripple” was performed after adding each marker. The best fitting position of an added marker was examined on the basis of the goodness-of-fit test (chi-square) for the resulting map, which is the normalized difference in chi-square value before and after adding the marker. Mean chi-square contribution values were used to determine if genotyping errors were suspected. Suspect loci were manually regenotyped. When a marker generated a negative map distance, or a large jump in goodness-of-fit, the marker was removed, and map construction was continued as a first-round map. After the first-round marker ordering, the previously removed markers were added back and again subjected to the goodness-of-fit testing. In this manner, the marker ordering was continued to a third round until an optimum order of markers was found.
Genome size and coverage:
Genome length from the linkage map was calculated according to Hubert and Hedgecock (2004). Telomeric regions were added to the map distance by adding 2x to the length of each linkage group (Fishman et al. 2001), where x is the average spacing between markers, which was calculated by dividing the total length of all linkage groups by the number of markers minus the number of linkage groups (29).
Comparative genome analysis:
The ESTs containing the microsatellites or SNPs used for linkage mapping were used as queries for BLAST searches to locate their genomic location in zebrafish, Tetraodon, and medaka genome sequences (E-value < e−10). The chromosomal locations and linkage groups of the microsatellites and SNPs were recorded. Putative conserved syntenies were identified when the genes were located in the same chromosome and the same linkage group. The distances among genes on the same chromosome of zebrafish, Tetraodon, and medaka are given in base pairs, whereas the distances among markers on the linkage groups of catfish are given in centimorgans.
RESULTS AND DISCUSSION
EST-derived microsatellite markers:
As summarized in Table 1, a total of 992 EST-derived microsatellites were used for PCR analysis. Nine of these loci were later found to represent duplicate ESTs of the same genes and therefore were removed; of the 983 remaining microsatellite loci, 450 were unsuccessful in PCR. A total of 533 EST-derived microsatellites were successful in PCR amplifications. Of these 533 successful amplifications, 120 were not polymorphic. One hundred three of the remaining 413 microsatellites could not be scored, mostly due to duplicated gene loci and non-Mendelian segregation patterns, leaving a total of 310 microsatellite markers for linkage mapping analysis. The overall success rate from the identification of the EST-derived microsatellites to successful genotyping was 31.3%. This low success rate was attributed to several major factors related to the nature of the microsatellites. A large fraction (45.4%) of EST-derived microsatellites failed in PCR amplification. The major reason for failures at this step was most likely caused by the involvement of introns. In spite of the efforts to limit the PCR product size to <300 bp for genotyping using automated sequencers, the unknown involvement of introns could have made the PCR amplification impossible or the size of amplicons was too large to be analyzed on the automated sequencers. Approximately 12% of the microsatellites were nonpolymorphic in the resource family. In addition, amplification of duplicated gene loci made it impossible to call the genotypes in almost 10% of the EST-derived microsatellites. Overall, EST-derived microsatellites had a much lower success rate as compared to microsatellites identified from genomic survey sequences, e.g., BAC end sequences (Xu et al. 2006; Somridhivej et al. 2008). The advantage of representing genes by microsatellites is severely limited by this low success rate. However, such problems can be alleviated using full-length cDNAs or draft genome sequences for accurate predictions of intron locations. As discussed by Massault et al. (2008), such gene-based maps should not only facilitate QTL analysis in aquaculture, but also set the foundation for orthology establishment, thus enabling functional inference of genes in aquaculture species where direct functional genomics work is difficult.
TABLE 1.
EST-derived microsatellites and SNPs and their performance in genotyping analysis
| Category | Microsatellites | SNPs |
|---|---|---|
| Total no. of markers | 992 | 384 |
| Duplicated EST contigs | 9 | 0 |
| No. of failed markers | 450 | 118 |
| Nonpolymorphic markers | 120 | 143 |
| Markers not scored due to gene duplication | 103 | 0 |
| Validated in mapping family | 310 | 125 |
| Over five parent–parent–children genotyping errors | NA | 27 |
| Markers used for linkage mapping analysis | 310 | 98 |
NA, not applicable.
EST-derived SNP markers:
Wang et al. (2008) described the factors that are significant for validation of EST-derived SNPs. One hundred eighteen of 384 SNP markers failed, and 125 were polymorphic in the mapping family. Twenty-seven of these polymorphic SNP markers were not included in the mapping analysis because they contained a P-P-C error value (parent–parent–children genotyping error) >5, resulting in a total of 98 SNPs for the linkage mapping. These markers were subjected to BLAST searches against the zebrafish, medaka, and Tetraodon nigrovirides cDNA databases in ENSEMBL with genome location information. A total of 72 SNPs of 98 SNP markers with significant hits were associated with linkage groups at a minimum LOD score of 3.0 while 26 of them remained unassigned.
Segregation of markers and linkage analysis:
One hundred sixteen (28%) of the 408 markers exhibited a segregation ratio of 1:1:1:1, serving as the most useful markers segregating in codominant fashion. Two hundred ninety-two (72%) of the 408 markers were segregating with a ratio of 1:1 ♀ type (AB × AA or CC) and 1:1 ♂ type (AA or CC × AB). Eighteen markers showing a significant level of distorted segregation were excluded from initial map construction, but 9 of them were later added back manually to the linkage groups using the “strongest cross-link” feature of the software. The remaining 9 markers could not be assigned to any linkage groups at the threshold LOD score. Two hundred seventy-six markers were organized in linkage groups at an initial LOD score of 15.0. Using the “strongest cross-link” feature in JoinMap 4.0, initially ungrouped and excluded markers were assigned to groups to which they have linkage with a minimum LOD score of 3.0. Similarly, markers in smaller groups were also assigned to the groups that have the strongest linkage with a minimum LOD score of 3.0. No suspect linkages were detected. As the DNA for grandparents was not available, the linkage phase of the mapping family was unknown. Therefore, we first obtained the genotypes of the polymorphic microsatellite markers of the female and the male separately to construct sex-specific linkage maps. The software made the best estimate of the linkage phases. A total of 331 EST-derived microsatellites and SNPs were mapped on the combined map.
The genetic linkage map:
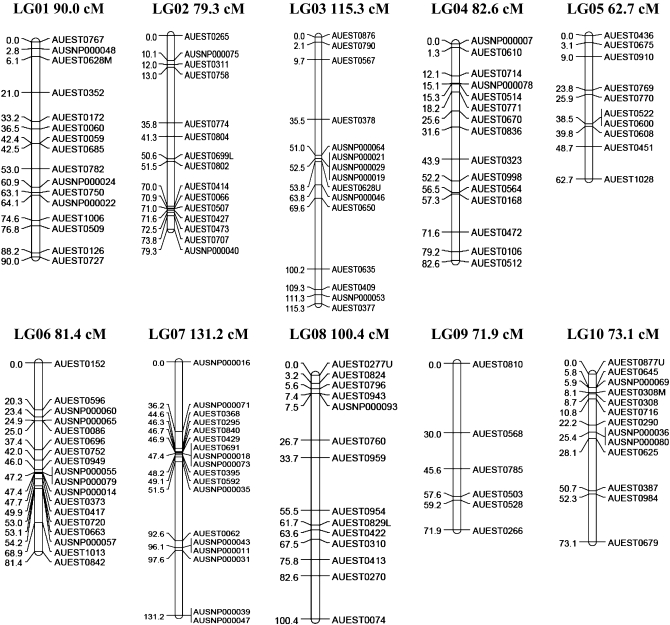
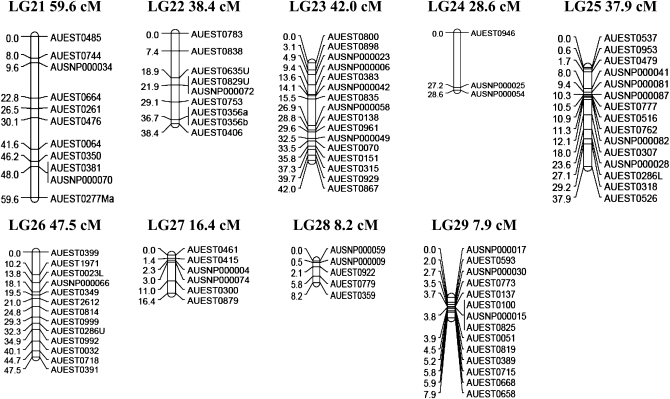
A linkage map for channel catfish was constructed with 331 markers composed of 259 type I microsatellites and 72 type I SNP markers. The linkage map contains 29 linkage groups (Figure 1) with 3–18 markers/linkage group; the number of linkage groups is consistent with expectations from the 29 haploid chromosomes of catfish. The largest linkage group contained 15 loci and spanned almost 131.2 cM, while the smallest linkage group contained 14 loci and spanned 7.9 cM. The linkage map covers a genetic distance of 1811 cM with an average of one marker every 6.0 cM. An additional 348.0 cM for the telomeric regions increased the estimated genome size to 2159.0 cM.
Figure 1.—
Linkage map of catfish constructed from EST-derived microsatellites and SNPs. Location of a locus relative to the neighboring locus (LOD = 3) is indicated on the left side of each linkage group in centimorgans, and names of the loci are indicated on the right side of each linkage group.
Two linkage maps were previously published:
One linkage map was constructed using AFLP markers (Liu et al. 2003), and the other was constructed using microsatellite markers (Waldbieser et al. 2001). While the current EST-based map is not comparable with an AFLP-based map because of the dominant nature of AFLPs, the current map had similar genome coverage to the previously published microsatellite map. The number of mapped markers was similar and the mapped genome size was similar as well, suggesting similar recombination frequencies within the intraspecific and interspecific mapping populations. However, a direct comparison of the two mapping populations should be conducted using a common set of microsatellite markers.
Of the 408 polymorphic markers, 358 were segregating female markers and 150 were segregating male markers. Linkage mapping analysis using JoinMap allowed the mapping of 315 female segregating markers into 29 linkage groups, while 43 markers were ungrouped. Similarly, of the 150 segregating male markers, 123 were mapped into 27 groups (5 of them were 2-point linkage groups), while 27 were ungrouped. Clearly, in the case of the male map, the markers were not sufficient to cover all the chromosomes (N = 29). The male and female linkage groups with shared markers are shown in supplemental Figure 1. The female map spanned 2009.8 cM with an average marker spacing of 6.15 cM. The male map spanned 761.0 cM with an average marker spacing of 4.95 cM. Clearly, the number of polymorphic markers was much larger in the female than in the male parent; this was at least in part due to the greater levels of polymorphism between channel catfish and blue catfish than within channel catfish. The female parent used for the resource family production was an F1 hybrid catfish made from mating a channel catfish female with a blue catfish male, whereas the male parent was a channel catfish.
Differences in recombination between sexes:
The sexes show substantial differences in recombination rate, both in general and for specific pairs of linked markers. In general, there is less recombination and genetic distance in the male linkage map (supplemental Figure 1). When common informative markers were selected (90 loci; 18 linkage groups), a higher recombination rate was observed in the female map (supplemental Figure 1, Table 3). Summing up the map distances for common markers for each LG resulted in a total length of 1891.2 and 3403.2 cM in the male and female maps, respectively. Thus, the ratio of female:male recombination rates for shared markers was 1.6:1. In 5 of the 22 linkage groups, recombination frequency was larger in the male than in the female, and the ratio of male-to-female recombination frequency in these 22 linkage groups varied greatly from 0.3 to 4.7. In contrast, in 17 of the 22 linkage groups, the recombination frequency was greater in the female than in the male with a female-to-male recombination frequency ratio of 0.2 to 3.4 (Table 2).
TABLE 2.
Comparison of male and female recombination rates in linkage groups with two or more shared markers
| Sex-specific linkage groups | Total shared markers | Cumulative distance in female | Cumulative distance in male | Female:male ratio |
|---|---|---|---|---|
| F1_M38 | 2 | 0.00 | 1.93 | ND |
| F2_M25 | 2 | 9.62 | 28.03 | 0.3 |
| F4_M7 | 5 | 84.12 | 84.02 | 1.0 |
| F6_M6 | 5 | 83.10 | 42.90 | 1.9 |
| F7_M1 | 2 | 7.59 | 2.23 | 3.4 |
| F7_M3 | 6 | 229.20 | 131.70 | 1.7 |
| F8_M1 | 4 | 85.50 | 66.70 | 1.3 |
| F9_M11 | 3 | 43.50 | 29.10 | 1.5 |
| F10_M21 | 2 | 53.40 | 19.20 | 2.8 |
| F11_M5 | 3 | 7.20 | 29.10 | 0.2 |
| F16_M11 | 2 | 28.80 | 12.70 | 2.3 |
| F17_M15 | 4 | 115.30 | 53.70 | 2.1 |
| F17_M30 | 2 | 3.80 | 9.20 | 0.4 |
| F18_M13 | 4 | 58.30 | 49.50 | 1.2 |
| F19_M16 | 3 | 30.10 | 27.40 | 1.1 |
| F20_M12 | 5 | 7.20 | 33.60 | 0.2 |
| F21_M8 | 6 | 505.40 | 308.00 | 1.6 |
| F23_M4 | 6 | 440.00 | 221.90 | 2.0 |
| F30_M17 | 4 | 143.00 | 47.50 | 3.0 |
| F31_M2 | 9 | 1465.30 | 683.90 | 2.1 |
| F33_M17 | 2 | 0.00 | 7.10 | ND |
| F33_M18 | 2 | 2.80 | 1.80 | 1.6 |
| Total/average | 83 | 3403.23 | 1891.21 | 1.6 |
A differential recombination rate was reported for a number of species. In several teleost species, recombination appears to be reduced in males. For example, the female:male recombination ratios are 8.26:1 in the Atlantic salmon (Johnson et al. 1987; Moen et al. 2004, 2008), 3.25:1 in rainbow trout (Sakamoto et al. 2000), and 1.48:1 in the European sea bass (Chistiakov et al. 2005). Our finding here of a recombination rate of 1.6 (female) to 1 (male) is in line with the general pattern found in other teleost fish. However, because the recombination rates are related to specific markers corresponding to specific chromosome regions, species, and specific matings, they are expected to be variable when additional markers are analyzed in catfish.
Identification of potentially conserved syntenies:
The use of EST-derived microsatellites and gene-derived SNP markers in this work provided opportunities to compare the similarities of the genome organization in catfish with those of its closely related species such as zebrafish where whole-genome sequences are available. To identify potential conserved syntenies between the catfish and the model fish genomes, EST sequences containing the mapped microsatellites and SNPs were used as queries for BLAST analysis against the zebrafish, medaka, and Tetraodon cDNA databases in ENSEMBL with genome location information. Of the 331 loci mapped, 131 had significant hits when searched against the zebrafish cDNA database, 139 had significant hits against the medaka cDNA database, and 130 had significant hits against the Tetraodon cDNA database. As summarized in Table 3, a total of 29 conserved syntenic blocks were identified between the linked catfish EST-derived microsatellites and SNPs and the physically linked zebrafish genes. Eight of these syntenic blocks contained four or more markers mapped to linkage groups 8, 9, 10, 11, 25, 27, and 29. The largest syntenic block contained eight markers mapped to LG10. In three syntenic blocks, a linear syntenic relation was evident with distances between the mapped markers being proportional to the distances of the genes on the zebrafish chromosomes. For example, the three loci AUEST0074, AUSNP000270, and AUSNP000093 were mapped to LG12 spanning ∼100 cM, while the genes homologous to these ESTs span almost 50 million base pairs in the zebrafish genome in a linear fashion. Similarly, the three loci AUSNP000042, AUEST0070, and AUSNP000151 were mapped to LG25 spanning ∼20 cM, while the genes homologous to these ESTs span almost 22 million base pairs in the zebrafish genome in a linear fashion. However, for most of the identified syntenic blocks, the gene/marker order and orientation may not be the same (Table 3). The conservation of marker/gene positions was the highest between catfish and zebrafish, consistent with their phylogenetic relationships (Xu et al. 2006; Steinke et al. 2006). The overall annotation rate of the 331 mapped ESTs was lower than that of the average ESTs (>50%; Li et al. 2007), largely because of the location of microsatellites being associated mostly with 5′- or 3′-UTRs.
TABLE 3.
Identification of putative conserved syntenies between the catfish and zebrafish genomes
| Locus | GID/contig | ENSEMBL ID | Chromosome | Chromosome location (bp) | E-value | Linkage group | Map location (cM) |
|---|---|---|---|---|---|---|---|
| AUEST0368 | CF262908 | ENSDART00000024945 | 1 | 37,727,657 | 0.74 | 29 | 44.6 |
| AUSNP000047 | Ctg_3078 | ENSDART00000054230 | 1 | 22,493,244 | 1.00 | 29 | 131.2 |
| AUEST0074 | BM028055 | ENSDART00000081134 | 1 | 4,593,682 | 0.84 | 12 | 100.4 |
| AUEST0270 | CF262276 | ENSDART00000103588 | 1 | 5,640,711 | 0.27 | 12 | 82.6 |
| AUSNP000093 | Ctg_3139 | ENSDART00000100195 | 1 | 55,725,417 | 1.00 | 12 | 7.5 |
| AUEST0137 | BM495657 | ENSDART00000101881 | 1 | 31,938,503 | 0.23 | 9 | 3.7 |
| AUEST0106 | BM495226 | ENSDART00000021158 | 2 | 12,139,259 | 0.29 | 6 | 79.2 |
| AUEST0168 | BM438559 | ENSDART00000055792 | 2 | 2,187,626 | 0.15 | 6 | 57.3 |
| AUEST0290 | CB937768 | ENSDART00000048277 | 2 | 35,593,551 | 0.16 | 16 | 22.2 |
| AUEST0154 | BE469707 | ENSDART00000012487 | 2 | 17,181,829 | 0.25 | 13 | 11.3 |
| AUEST0023L | BM496054 | ENSDART00000036997 | 2 | 40,658,639 | 0.42 | 19 | 13.8 |
| AUSNP000009 | Ctg_0027 | ENSDART00000087086 | 2 | 25,356,938 | 0.18 | 5 | 0.5 |
| AUEST0002 | BM438455 | ENSDART00000016407 | 3 | 35,368,255 | 0.25 | 11 | 27.9 |
| AUEST0671 | CK413397 | ENSDART00000055360 | 3 | 28,793,198 | 0.68 | 23 | 37.7 |
| AUSNP000026 | Ctg_1273 | ENSDART00000074561 | 3 | 41,753,186 | 0.44 | 13 | 54.3 |
| AUEST0838 | BE469511 | ENSDART00000080075 | 3 | 13,504,775 | 1.00 | 26 | 7.4 |
| AUSNP000072 | Ctg_4221 | ENSDART00000004305 | 3 | 13,516,452 | 1.00 | 26 | 21.9 |
| AUSNP000042 | Ctg_2754 | ENSDART00000030890 | 3 | 23,436,798 | 1.00 | 25 | 14.1 |
| AUEST0070 | AF267989 | ENSDART00000055675 | 3 | 19,486,003 | 0.71 | 25 | 33.5 |
| AUEST0151 | BM027834 | ENSDART00000046995 | 3 | 1,374,688 | 0.77 | 25 | 35.8 |
| AUEST0824 | BE468808 | ENSDART00000039572 | 4 | 16,948,230 | 0.20 | 12 | 3.2 |
| AUEST0149 | BM497130 | ENSDART00000066929 | 4 | 21,411,838 | 0.28 | 22 | 31.6 |
| AUEST0704 | CK413527 | ENSDART00000000020 | 4 | 15,491,685 | 0.32 | 22 | 0.0 |
| AUEST0679 | CK413500 | ENSDART00000051554 | 5 | 14,064,654 | 0.48 | 16 | 73.1 |
| AUSNP000036 | Ctg_2405 | ENSDART00000024676 | 5 | 62,066,731 | 1.00 | 16 | 25.4 |
| AUEST0286L | CF262064 | ENSDART00000074117 | 5 | 70,268,448 | 0.26 | 10 | 27.1 |
| AUEST0516 | CK412782 | ENSDART00000051236 | 5 | 30,026,995 | 0.30 | 10 | 10.9 |
| AUEST0526 | CK412855 | ENSDART00000038929 | 5 | 68,419,340 | 0.31 | 10 | 38.0 |
| AUEST0537 | CK412946 | ENSDART00000051135 | 5 | 38,197,794 | 0.44 | 10 | 0.0 |
| AUSNP000041 | Ctg_2749 | ENSDART00000089992 | 5 | 12,496,309 | 0.19 | 10 | 8.0 |
| AUSNP000081 | Ctg_5550 | ENSDART00000080919 | 5 | 12,171,859 | 0.61 | 10 | 9.4 |
| AUSNP000082 | Ctg_0743 | ENSDART00000023554 | 5 | 51,276,820 | 1.00 | 10 | 12.2 |
| AUSNP000087 | Ctg_1741 | ENSDART00000050949 | 5 | 51,428,185 | 1.00 | 10 | 10.3 |
| AUEST0767 | BM497034 | ENSDART00000041882 | 6 | 26,178,677 | 0.56 | 28 | 0.0 |
| AUSNP000048 | Ctg_3079 | ENSDART00000064904 | 6 | 26,868,281 | 1.00 | 28 | 2.8 |
| AUEST0010 | BE469169 | ENSDART00000019845 | 6 | 27,616,006 | 0.67 | 15 | 32.4 |
| AUEST0078 | BM495047 | ENSDART00000083670 | 6 | 37,763,322 | 0.11 | 14 | 70.6 |
| AUSNP000023 | Ctg_1244 | ENSDART00000073780 | 6 | 3,537,041 | 0.36 | 25 | 4.9 |
| AUEST0835 | BE469419 | ENSDART00000018503 | 6 | 9,055,562 | 1.00 | 25 | 15.5 |
| AUSNP000058 | Ctg_3435 | ENSDART00000004656 | 6 | 13,058,847 | 1.00 | 25 | 26.9 |
| AUEST0929 | BM494953 | ENSDART00000065502 | 6 | 3,510,301 | 0.84 | 25 | 39.7 |
| AUSNP000074 | Ctg_4582 | ENSDART00000056319 | 6 | 53,324,672 | 0.25 | 17 | 3.0 |
| AUEST0051 | BM425105 | ENSDART00000003898 | 6 | 14,085,842 | 0.20 | 9 | 3.9 |
| AUEST0608 | BM496609 | ENSDART00000052318 | 7 | 24,276,447 | 0.54 | 20 | 39.8 |
| AUEST0769 | BM496763 | ENSDART00000100149 | 7 | 29,092,776 | 0.17 | 20 | 23.8 |
| AUEST0152 | BE212675 | ENSDART00000075757 | 7 | 34,233,565 | 0.72 | 27 | 0.0 |
| AUEST0918 | BM439064 | ENSDART00000074463 | 7 | 24,815,798 | 1.00 | 21 | 13.6 |
| AUEST0805 | CB936968 | ENSDART00000052539 | 7 | 32,892,667 | 1.00 | 8 | 36.0 |
| AUSNP000083 | Ctg_0030 | ENSDART00000027000 | 7 | 2,534,611 | 1.00 | 8 | 21.3 |
| AUEST0027 | BM438274 | ENSDART00000062702 | 8 | 26,857,785 | 0.45 | 8 | 24.7 |
| AUEST0376 | CF262727 | ENSDART00000099025 | 8 | 28,749,949 | 0.38 | 8 | 17.3 |
| AUEST0945 | BM495553 | ENSDART00000083790 | 8 | 33,771,883 | 0.23 | 8 | 28.8 |
| AUSNP000032 | Ctg_1974 | ENSDART00000099708 | 8 | 22,251,295 | 0.41 | 8 | 24.4 |
| AUSNP000060 | Ctg_3638 | ENSDART00000022074 | 9 | 41,386,855 | 1.00 | 27 | 23.4 |
| AUEST0100 | AF396747 | ENSDART00000100386 | 9 | 34,893,437 | 1.00 | 9 | 3.8 |
| AUSNP000015 | Ctg_0867 | ENSDART00000101338 | 9 | 21,792,056 | 1.00 | 9 | 3.8 |
| AUSNP000017 | Ctg_0881 | ENSDART00000006948 | 9 | 27,291,927 | 1.00 | 9 | 0.0 |
| AUSNP000030 | Ctg_1591 | ENSDART00000101985 | 9 | 15,176,111 | 1.00 | 9 | 2.7 |
| AUSNP000061 | Ctg_3648 | ENSDART00000100022 | 10 | 25,016,652 | 0.58 | 1 | 60.3 |
| AUEST0086 | BM028141 | ENSDART00000104260 | 11 | 9,753,944 | 0.64 | 27 | 25.0 |
| AUEST0373 | CF262754 | ENSDART00000035560 | 11 | 26,287,029 | 0.12 | 27 | 47.7 |
| AUEST0696 | CK413701 | ENSDART00000087597 | 11 | 31,179,554 | 0.47 | 27 | 37.4 |
| AUSNP000014 | Ctg_0865 | ENSDART00000104360 | 11 | 5,177,192 | 1.00 | 27 | 47.5 |
| AUSNP000065 | Ctg_3707 | ENSDART00000030103 | 11 | 9,704,698 | 1.00 | 27 | 25.0 |
| AUEST0017 | BE468998 | ENSDART00000026017 | 11 | 33,770,768 | 0.26 | 15 | 21.0 |
| AUSNP000085 | Ctg_1136 | ENSDART00000103368 | 11 | 27,142,612 | 0.48 | 15 | 33.4 |
| AUSNP000033 | Ctg_2272 | ENSDART00000054788 | 12 | 35,429,324 | 0.71 | 23 | 11.6 |
| AUEST0265 | CF262438 | ENSDART00000022684 | 13 | 38,174,389 | 0.74 | 3 | 0.0 |
| AUEST0758 | CB938230 | ENSDART00000043312 | 13 | 38,246,633 | 0.10 | 3 | 13.0 |
| AUSNP000040 | Ctg_2695 | ENSDART00000057774 | 13 | 4,030,776 | 0.53 | 3 | 79.3 |
| AUEST0678 | CK413486 | ENSDART00000101853 | 13 | 14,053,231 | 1.00 | 21 | 62.8 |
| AUSNP000054 | Ctg_3193 | ENSDART00000102941 | 13 | 407,677 | 0.24 | 24 | 28.6 |
| AUSNP000019 | Ctg_1050 | ENSDART00000061001 | 14 | 55,809,901 | 1.00 | 2 | 52.5 |
| AUSNP000016 | Ctg_0878 | ENSDART00000061001 | 14 | 55,809,901 | 1.00 | 29 | 0.0 |
| AUSNP000035 | Ctg_2393 | ENSDART00000039660 | 14 | 22,980,719 | 1.00 | 29 | 51.5 |
| AUSNP000071 | Ctg_4192 | ENSDART00000079608 | 14 | 18,466,098 | 1.00 | 29 | 36.2 |
| AUSNP000073 | Ctg_4466 | ENSDART00000023540 | 14 | 38,067,562 | 0.42 | 29 | 47.4 |
| AUEST0302 | CF261566 | ENSDART00000020961 | 14 | 53,554,821 | 0.11 | 4 | 70.1 |
| AUSNP000068 | Ctg_3792 | ENSDART00000105389 | 14 | 53,613,423 | 0.91 | 4 | 70.4 |
| AUEST0472 | BM496853 | ENSDART00000063783 | 15 | 35,442,248 | 0.55 | 6 | 71.6 |
| AUSNP000078 | Ctg_5097 | ENSDART00000019330 | 15 | 9,862,693 | 1.00 | 6 | 15.1 |
| AUEST0377 | CF262687 | ENSDART00000020363 | 16 | 20,391,653 | 0.40 | 2 | 115.3 |
| AUSNP000007 | Ctg_4213 | ENSDART00000081477 | 16 | 3,876,888 | 1.00 | 6 | 0.0 |
| AUEST0436 | CK414043 | ENSDART00000081259 | 16 | 5,590,055 | 0.33 | 20 | 0.0 |
| AUEST0220U | CB937920 | ENSDART00000081649 | 16 | 2,253,445 | 0.52 | 11 | 47.9 |
| AUEST0936 | BM495325 | ENSDART00000049323 | 16 | 7,467,887 | 0.50 | 11 | 3.7 |
| AUSNP000038 | Ctg_2583 | ENSDART00000058965 | 16 | 24,388,825 | 1.00 | 11 | 18.9 |
| AUSNP000052 | Ctg_3158 | ENSDART00000058945 | 16 | 25,554,914 | 1.00 | 11 | 19.9 |
| AUEST0032 | BM424544 | ENSDART00000058385 | 16 | 43,054,577 | 0.40 | 19 | 40.1 |
| AUSNP000066 | Ctg_3768 | ENSDART00000078310 | 16 | 22,069,295 | 0.88 | 19 | 18.1 |
| AUSNP000034 | Ctg_2379 | ENSDART00000064739 | 17 | 2,086,675 | 1.00 | 7 | 9.6 |
| AUSNP000070 | Ctg_4140 | ENSDART00000064633 | 17 | 6,482,607 | 0.28 | 7 | 48.0 |
| AUEST0132 | BM424646 | ENSDART00000053440 | 18 | 45,420,729 | 0.23 | 11 | 40.6 |
| AUEST0288 | CB937073 | ENSDART00000032151 | 18 | 21,353,197 | 0.62 | 21 | 61.5 |
| AUEST0771 | BM496501 | ENSDART00000052556 | 19 | 12,650,517 | 0.11 | 6 | 18.2 |
| AUEST0067 | BM497044 | ENSDART00000062518 | 19 | 267,404 | 0.35 | 4 | 94.1 |
| AUSNP000010 | Ctg_0286 | ENSDART00000023156 | 19 | 40,605,399 | 1.00 | 13 | 54.0 |
| AUEST0138 | BM028228 | ENSDART00000104083 | 19 | 14,992,257 | 0.44 | 25 | 28.8 |
| AUSNP000049 | Ctg_3094 | ENSDART00000052421 | 19 | 15,217,159 | 1.00 | 25 | 32.5 |
| AUEST0266 | CF262406 | ENSDART00000058527 | 20 | 20,006,535 | 0.32 | 18 | 71.9 |
| AUEST0406 | BM439180 | ENSDART00000032393 | 20 | 24,132,370 | 0.92 | 26 | 38.4 |
| AUSNP000021 | Ctg_1207 | ENSDART00000053208 | 21 | 36,366,077 | 0.92 | 2 | 52.5 |
| AUSNP000046 | Ctg_3032 | ENSDART00000040598 | 21 | 4,298,522 | 1.00 | 2 | 63.8 |
| AUSNP000053 | Ctg_3179 | ENSDART00000055325 | 21 | 39,207,385 | 1.00 | 2 | 111.3 |
| AUEST0052 | BM495288 | ENSDART00000020174 | 21 | 31,406,378 | 0.60 | 21 | 47.2 |
| AUEST0635U | BM496516 | ENSDART00000015576 | 21 | 44,368,280 | 0.34 | 26 | 18.9 |
| AUEST0451 | CK424102 | ENSDART00000063133 | 22 | 9,212,561 | 0.37 | 20 | 48.7 |
| AUEST0417 | BM494174 | ENSDART00000048775 | 22 | 20,284,919 | 1.00 | 27 | 49.9 |
| AUSNP000055 | Ctg_3226 | ENSDART00000062618 | 22 | 13,542,614 | 1.00 | 27 | 47.2 |
| AUSNP000079 | Ctg_5104 | ENSDART00000076082 | 22 | 35,077,720 | 1.00 | 27 | 47.2 |
| AUEST0210 | CB939628 | ENSDART00000059140 | 22 | 7,599,621 | 0.59 | 15 | 0.0 |
| AUSNP000088 | Ctg_1769 | ENSDART00000092082 | 22 | 33,774,628 | 0.78 | 15 | 28.8 |
| AUEST0509 | CK412708 | ENSDART00000085054 | 23 | 30,669,614 | 0.68 | 28 | 76.8 |
| AUSNP000022 | Ctg_1221 | ENSDART00000104618 | 23 | 15,796,134 | 1.00 | 28 | 64.1 |
| AUSNP000024 | Ctg_1260 | ENSDART00000081215 | 23 | 7,812,918 | 1.00 | 28 | 60.9 |
| AUEST0066 | BM027884 | ENSDART00000025414 | 23 | 20,600,486 | 0.94 | 3 | 70.9 |
| AUSNP000003 | Ctg_0621 | ENSDART00000077539 | 23 | 33,339,203 | 1.00 | 15 | 17.4 |
| AUSNP000004 | Ctg_1691 | ENSDART00000009337 | 23 | 21,056,564 | 1.00 | 17 | 2.3 |
| AUEST0814 | CB937452 | ENSDART00000066630 | 24 | 23,314,688 | 0.14 | 19 | 24.8 |
| AUEST0062 | BE469322 | ENSDART00000039485 | 25 | 28,735,776 | 0.65 | 29 | 92.6 |
| AUSNP000011 | Ctg_0381 | ENSDART00000021006 | 25 | 6,532,907 | 1.00 | 29 | 96.1 |
| AUSNP000018 | Ctg_1045 | ENSDART00000064204 | 25 | 13,669,875 | 1.00 | 29 | 47.4 |
| AUSNP000043 | Ctg_2875 | ENSDART00000005627 | 25 | 32,398,863 | 0.22 | 29 | 96.1 |
| AUEST0043 | AF063836 | ENSDART00000073566 | 25 | 18,547,487 | 1.00 | 4 | 36.6 |
| AUEST0126 | BM028849 | ENSDART00000018751 | Unassigned | 32,254 | 0.12 | 28 | 88.2 |
| AUEST0685 | CK413603 | ENSDART00000092525 | Unassigned | 919 | 1.00 | 28 | 42.5 |
| AUEST0716 | CK413890 | ENSDART00000082614 | Unassigned | 123,432 | 0.21 | 16 | 10.8 |
| AUSNP000069 | Ctg_4051 | ENSDART00000013310 | Unassigned | 171,790 | 1.00 | 16 | 5.9 |
| AUEST0242 | CF261514 | ENSDART00000097310 | Unassigned | 29,480 | 0.49 | 13 | 0.0 |
| AUEST0064 | AF410785 | ENSDART00000098979 | Unassigned | 34,198 | 0.23 | 7 | 41.7 |
| AUEST0476 | BM496810 | ENSDART00000098973 | Unassigned | 169,711 | 1.00 | 7 | 30.1 |
| AUEST0383 | CF262296 | ENSDART00000053700 | Unassigned | 2,733 | 0.28 | 25 | 13.6 |
Marker loci were named with the prefix AUEST for gene-associated microsatellites and AUSNP for gene-associated SNPs. GID is the GenBank identifier for the gene (accession numbers for ESTs); for SNPs, the contig number from which the SNP was identified is given. The ENSEMBL ID column is the sequence ID of the zebrafish genome sequence homologous to the specific locus. The E-value column describes the similarity between the catfish gene and the zebrafish gene, but is encoded: e.g., 0.74 = e−74; 1 ≤ e−100; 0.27 = e−27, etc.
A total of 21 conserved syntenic blocks was identified between the linked catfish EST-derived microsatellites and SNPs and the physically linked Tetraodon genes (supplemental Table 1). The largest syntenic block contained five markers mapped to linkage group 9. Only one syntenic block with three markers (AUEST0767 AUSNP000048, AUSNP000024) located on LG28 had a linear syntenic relationship with chromosome 11 of Tetraodon. In the case of medaka, 29 conserved syntenic blocks were identified (supplemental Table 2). Five of these syntenic blocks contained four or more markers mapped to linkage groups 9, 10, 11, 27, and 29.
The evolutionary syntenic conservation appeared to be relatively low between the catfish genome and the genomes of the three model fish species. In spite of the identified conserved syntenic blocks, the extent to which the syntenies were conserved was limited in most cases. For example, the five markers on chromosome 1 of the zebrafish mapped to two linkage groups in catfish; the 6 genes on chromosome 2 of the zebrafish mapped to five different linkage groups with only a couple of markers linked together in catfish; and the 8 genes on chromosome 3 of the zebrafish mapped to five linkage groups (Table 3). This indicates that, among the fish genomes, much chromosome breakage and many rearrangements occurred during evolution. However, in a few instances, the syntenic conservation was extensive. For example, of the 10 genes on zebrafish chromosome 5, 8 were mapped to linkage group 10, and the other 2 were mapped to linkage group 16; of the 7 genes on zebrafish chromosome 11, 5 were mapped to linkage group 27, and the other 2 were mapped to linkage group 15 (Table 3). These findings are consistent with our previous findings that high levels of conservation were found within small genomic regions, whereas high levels of large-scale genome reshuffling were evident when comparing the genomes of catfish and zebrafish (Wang et al. 2007). These results indicate that comparative genome analysis is highly efficient when dealing with small genome segments for which conserved syntenies have been identified. Therefore, many smaller conserved syntenies in catfish may need to be used when comparing zebrafish or other model fish species for which whole-genome sequences are available. Such findings also strongly support the need to produce the whole-genome sequence of catfish for the purpose of genome evolution studies. Catfish is an economically important member of a large order of Siluriformes from which no whole-genome sequence is available.
This study represents a first-generation linkage map constructed by using EST-derived microsatellites and SNPs, laying the ground for large-scale comparative genome analysis in catfish. We previously reported a large number of BAC end sequences (XU et al., 2006) and their associated microsatellites (Somridhivej et al. 2008). Further expansion of this linkage map using physical-map-anchored polymorphic markers should enhance comparative mapping, thereby transferring genome information from model species to catfish. In spite of the apparent high levels of chromosome rearrangements between the catfish and zebrafish genomes, comparative mapping is still of great value, not only for the understanding of genome organization and genome evolution, but also for the understanding of genome functions. Given that determination of gene functions is very difficult in nonmodel species such as catfish, functional genome analysis will have to rely heavily on the establishment of orthologies from model species, such as zebrafish, to infer functions. Such comparative genomics information will be valuable in narrowing down suggestive candidate genes around significant QTL, which are expected to be easily found by use of such a dense linkage map.
Acknowledgments
We thank Rex Dunham and his students as well as Renee Beam, Karen Veverica, Esau Arana, and Randell Goodman for their excellent work in the production and maintenance of fish used in this study. We appreciate the support of the Auburn University Department of Fisheries and Allied Aquacultures, College of Agriculture, and the Vice President for Research and their matching funds to U. S. Department of Agriculture (USDA) National Research Initiative (NRI) Equipment Grants (2005-35206-15274). This project was supported by grants from the USDA NRI Animal Genome Tools and Resources Program (award no. 2006-35616-16685 and partially award no. 2003-35205-12827).
References
- Boutin-Ganache, I., M. Raposo, M. Raymond and C. F. Deschepper, 2001. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques 31 24–26. [PubMed] [Google Scholar]
- Cao, D., A. Kocabas, Z. Ju, A. Karsi, P. Li et al., 2001. Transcriptome of channel catfish (Ictalurus punctatus): initial analysis of genes and expression profiles of the head kidney. Anim. Genet. 32 169–188. [DOI] [PubMed] [Google Scholar]
- Casasoli, M., J. Derory, C. Morera-Dutrey, O. Brendel, I. Porth et al., 2006. Comparison of quantitative trait loci for adaptive traits between oak and chestnut based on an expressed sequence tag consensus map. Genetics 172 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov, D. A., B. Hellemans, C. S. Haley, A. S. Law, C. S. Tsigenopoulos et al., 2005. A microsatellite linkage map of the European sea bass Dicentrarchus labrax L. Genetics 170 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzmann, R. G., and K. Gharbi, 2007. Linkage mapping in aquaculture species, pp. 139–167 in Aquaculture Genome Technologies, edited by Z. Liu. Blackwell Publishing, Oxford.
- Dunham, R. A., and B. J. Argue, 1998. Seinability of channel catfish, blue catfish, and their F-1, F-2, F-3, and backcross hybrids in earthen ponds. Prog. Fish-Culturist 60 214–220. [Google Scholar]
- Dunham, R. A., R. O. Smitherman and R. K. Goodman, 1987. Comparison of mass selection, crossbreeding, and hybridization for improving growth of channel catfish. Progressive Fish-Culturist 49 293–296. [Google Scholar]
- Dunham, R. A., R. E. Brummett, M. O. Ella and R. O. Smitherman, 1990. Genotype environment interactions for growth of blue, channel and hybrid catfish in ponds and cages at varying densities. Aquaculture 85 143–151. [Google Scholar]
- Dunham, R. A., C. Hyde, M. Masser, J. A. Plumb, R. O. Smitherman et al., 1993. Comparison of culture traits of channel catfish, Ictalurus punctatus, and blue catfish, I. furcatus. J. Appl. Aquaculture 3 257–267. [Google Scholar]
- Ferreira, I. A., and C. Martins, 2008. Physical chromosome mapping of repetitive DNA sequences in Nile tilapia Oreochromis niloticus: evidences for a differential distribution of repetitive elements in the sex chromosomes. Micron 39 411–418. [DOI] [PubMed] [Google Scholar]
- Fishman, L., A. J. Kelly, E. Morgan and J. H. Willis, 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch, R., B. Louro, M. Tsalavouta, D. Chatziplis, C. S. Tsigenopoulos et al., 2006. A genetic linkage map of the hermaphrodite teleost fish Sparus aurata L. Genetics 174 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi, K., A. Gautier, R. G. Danzmann, S. Gharbi, T. Sakamoto et al., 2006. A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172 2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyomard, R., S. Mauger, K. Tabet-Canale, S. Martineau, C. Genet et al., 2006. A type I and type II microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) with presumptive coverage of all chromosome arms. BMC Genomics 7 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyomard, R., S. Mauger, S. Martineau, K. Tabet-Canale and E. Quillet, 2007. Construction of a female microsatellite linkage map in rainbow trout (Oncorhynchus mykiss), a tetraploid species. Aquaculture 272 S264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, B., J. K. Laerdahl, S. Lien, T. Moen, P. Berg, et al., 2007. An extensive resource of single nucleotide polymorphism markers associated with Atlantic salmon (Salmo salar) expressed sequences. Aquaculture 265 82–90. [Google Scholar]
- He, C., L. Chen, M. Simmons, P. Li, S. Kim et al., 2003. Putative SNP discovery in interspecific hybrids of catfish by comparative EST analysis. Anim. Genet. 34 445–448. [DOI] [PubMed] [Google Scholar]
- Hubert, S., and D. Hedgecock, 2004. Linkage maps of microsatellite DNA markers for the Pacific oyster Crassostrea gigas. Genetics 168 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J. M. E., H. J. Vaneck, P. Arens, B. Verkerkbakker, B. T. L. Hekkert et al., 1995. A genetic-map of potato (Solanum tuberosum) integrating molecular markers, including transposons, and classical markers. Theor. Appl. Genet. 91 289–300. [DOI] [PubMed] [Google Scholar]
- Johnson, K. R., J. E. Wright and B. May, 1987. Linkage relationships reflecting ancestral tetraploidy in salmonid fish. Genetics 116 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, Z., A. Karsi, A. Kocabas, A. Patterson, P. Li et al., 2000. Transcriptome analysis of channel catfish (Ictalurus punctatus): genes and expression profile from the brain. Gene 261 373–382. [DOI] [PubMed] [Google Scholar]
- Kalendar, R., 2008. FastPCR: a PCR primer and probe design and repeat sequence searching software with additional tools for the manipulation and analysis of DNA and protein. http://www.biocenter.helsinki.fi/bi/programs/fastpcr.htm.
- Karsi, A., D. Cao, P. Li, A. Patterson, A. Kocabas et al., 2002. Transcriptome analysis of channel catfish (Ictalurus punctatus): initial analysis of gene expression and microsatellite-containing cDNAs in the skin. Gene 285 157–168. [DOI] [PubMed] [Google Scholar]
- Katagiri, T., C. Kidd, E. Tomasino, J. T. Davis, C. Wishon et al., 2005. A BAC-based physical map of the Nile tilapia genome. BMC Genomics 6 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. S., T. Y. Chung, G. J. King, M. Jin, T. J. Yang et al., 2006. A sequence-tagged linkage map of Brassica raga. Genetics 174 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas, A. M., P. Li, D. F. Cao, A. Karsi, C. B. He et al., 2002. Expression profile of the channel catfish spleen: analysis of genes involved in immune functions. Mar. Biotechnol. 4 526–536. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. Y., W. J. Lee, J. T. Streelman, K. L. Carleton, A. E. Howe et al., 2005. A second-generation genetic linkage map of tilapia (Oreochromis spp.). Genetics 170 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., E. Peatman, S. L. Wang, J. N. Feng, C. B. He et al., 2007. Towards the Ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs. BMC Genomics 8 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. D., X. Liu, X. M. Guo, Q. K. Gao, H. G. Zhao et al., 2006. A preliminary genetic linkage map of the Pacific abalone Haliotis discus hannai Ino. Mar. Biotechnol. 8 386–397. [DOI] [PubMed] [Google Scholar]
- Liu, Z. J., 2003. A review of catfish genomics: progress and perspectives. Comp. Funct. Genomics 4 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. J., 2007. Genomescape: characterizing the repeat structure of the genome, pp. 277–290 in Aquaculture Genome Technologies, edited by Z. J. Liu. Blackwell Publishing, Oxford, UK.
- Liu, Z. J., P. Li and R. A. Dunham, 1998. Characterization of an A/T-rich family of sequences from channel catfish (Ictalurus punctatus). Mol. Mar. Biol. Biotechnol. 7 232–239. [PubMed] [Google Scholar]
- Liu, Z. J., A. Karsi, P. Li, D. F. Cao and R. Dunham, 2003. An AFLP-based genetic linkage map of channel catfish (Ictalurus punctatus) constructed by using an interspecific hybrid resource family. Genetics 165 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W. J., J. McCallum, M. Shigyo, J. Jakse, J. C. Kuhl et al., 2005. Genetic mapping of expressed sequences in onion and in silico comparisons with rice show scant colinearity. Mol. Genet. Genomics 274 197–204. [DOI] [PubMed] [Google Scholar]
- Massault, C., H. Bovenhuis, C. Haley and D. J. De Koning, 2008. QTL mapping designs for species in aquaculture. Aquaculture 285 23–29. [Google Scholar]
- Meyers, S. N., M. B. Rogatcheva, D. M. Larkin, M. Yerle, D. Milan et al., 2005. Piggy-BACing the human genome. II. A high-resolution, physically anchored, comparative map of the porcine autosomes. Genomics 86 739–752. [DOI] [PubMed] [Google Scholar]
- Moen, T., B. Hoyheim, H. Munck and L. Gomez-Raya, 2004. A linkage map of Atlantic salmon (Salmo salar) reveals an uncommonly large difference in recombination rate between the sexes. Anim. Genet. 35 81–92. [DOI] [PubMed] [Google Scholar]
- Moen, T., B. Hayes, M. Baranski, P. R. Berg, S. Kjoglum et al., 2008. A linkage map of the Atlantic salmon (Salmo salar) based on EST-derived SNP markers. BMC Genomics 9 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretzsohn, M. C., L. Leoi, K. Proite, P. M. Guimaraes, S. C. M. Leal-Bertioli et al., 2005. A microsatellite-based, gene-rich linkage map for the AA genome of Arachis (Fabaceae). Theor. Appl. Genet. 111 1060–1071. [DOI] [PubMed] [Google Scholar]
- Mousel, M. R., D. J. Nonneman and G. A. Rohrer, 2006. Rearranged gene order between pig and human in a quantitative trait loci region on SSC3. Anim. Genet. 37 403–406. [DOI] [PubMed] [Google Scholar]
- Nandi, S., E. Peatman, P. Xu, S. Wang, P. Li et al., 2007. Repeat structure of the catfish genome: a genomic and transcriptomic assessment of Tc1-like transposon elements in channel catfish (Ictalurus punctatus). Genetica 131 81–90. [DOI] [PubMed] [Google Scholar]
- Nichols, K. M., W. P. Young, R. G. Danzmann, B. D. Robison, C. Rexroad et al., 2003. A consolidated linkage map for rainbow trout (Oncorhynchus mykiss). Anim. Genet. 34 102–115. [DOI] [PubMed] [Google Scholar]
- Oetting, W. S., H. K. Lee, D. J. Flanders, G. L. Wiesner, T. A. Sellers et al., 1995. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 30 450–458. [DOI] [PubMed] [Google Scholar]
- Phillips, R. B., J. J. Dekoning, K. M. Nichols, C. E. Rexroad, S. Gahr et al., 2007. Assignment of rainbow trout linkage groups to salmonid chromosomes. Aquaculture 272 S299–S300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroad, C. E., and Y. Palti, 2003. Development of ninety-seven polymorphic microsatellite markers for rainbow trout. Trans. Am. Fish. Soc. 132 1214–1221. [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Gharbi, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarropoulou, E., D. Nousdili, A. Magoulas and G. Kotoulas, 2008. Linking the genomes of non-model teleosts through comparative genomics. Mar. Biotechnol. 10 227–233. [DOI] [PubMed] [Google Scholar]
- Sasazaki, S., T. Hinenoya, B. Lin, A. Fujiwara and H. Mannen, 2006. A comparative map of macrochromosomes between chicken and Japanese quail based on orthologous genes. Anim. Genet. 37 316–320. [DOI] [PubMed] [Google Scholar]
- Sawera, M., S. Cirera, C. B. Jorgensen, J. Gorodkin and M. Fredholm, 2006. Linkage mapping of gene-associated SNPs to pig chromosome 11. Anim. Genet. 37 199–204. [DOI] [PubMed] [Google Scholar]
- Sekino, M., and M. Hara, 2007. Linkage maps for the Pacific abalone (genus Haliotis) based on microsatellite DNA markers. Genetics 175 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekino, M., T. Kobayashi and M. Hara, 2006. Segregation and linkage analysis of 75 novel microsatellite DNA markers in pair crosses of Japanese abalone (Haliotis discus hannai) using the 5′-tailed primer method. Mar. Biotechnol. 8 453–466. [DOI] [PubMed] [Google Scholar]
- Senger, F., C. Priat, C. Hitte, E. Sarropoulou, R. Franch et al., 2006. The first radiation hybrid map of a perch-like fish: the gilthead seabream (Sparus aurata L). Genomics 87 793–800. [DOI] [PubMed] [Google Scholar]
- Serapion, J., H. Kucuktas, J. A. Feng and Z. J. Liu, 2004. a Bioinformatic mining of type I microsatellites from expressed sequence tags of channel catfish (Ictalurus punctatus). Mar. Biotechnol. 6 364–377. [DOI] [PubMed] [Google Scholar]
- Serapion, J., G. C. Waldbieser, W. Wolters and Z. J. Liu, 2004. b Development of type I markers in channel catfish through intron sequencing. Anim. Genet. 35 463–466. [DOI] [PubMed] [Google Scholar]
- Smith, J. J., D. K. Kump, J. A. Walker, D. M. Parichy and S. R. Voss, 2005. A comprehensive expressed sequence tag linkage map for tiger salamander and Mexican axolotl: enabling gene mapping and comparative genomics in Ambystoma. Genetics 171 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelling, W. M., E. Casas, R. T. Stone, J. W. Keele, G. P. Harhay et al., 2005. Linkage mapping bovine EST-based SNP. BMC Genomics 6 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somridhivej, B., S. Wang, Z. Sha, H. Liu, J. Quilang et al., 2008. Characterization, polymorphism assessment, and database construction for microsatellites from BAC end sequences of catfish: a resource for integration of linkage and physical maps. Aquaculture 275 76–80. [Google Scholar]
- Stam, P., 1993. Construction of integrated genetic-linkage maps by means of a new computer package: JoinMap. Plant J. 3 739–744. [Google Scholar]
- Steinke, D., W. Salzburger and A. Meyer, 2006. Novel relationships among ten fish model species revealed based on a phylogenomic analysis using ESTs. J. Mol. Evol. 62 772–784. [DOI] [PubMed] [Google Scholar]
- Viruel, M. A., R. Messeguer, M. C. Devicente, J. Garciamas, P. Puigdomenech et al., 1995. A linkage map with RFLP and isozyme markers for almond. Theor. Appl. Genet. 91 964–971. [DOI] [PubMed] [Google Scholar]
- Waldbieser, G. C., B. G. Bosworth, D. J. Nonneman and W. R. Wolters, 2001. A microsatellite-based genetic linkage map for channel catfish, Ictalurus punctatus. Genetics 158 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., P. Xu, J. Thorsen, B. Zhu, P. de Jong et al., 2007. Characterization of a BAC library from channel catfish Ictalurus punctatus: indications of high levels of chromosomal reshuffling among teleost genomes. Mar. Biotechnol. 9 701–711. [DOI] [PubMed] [Google Scholar]
- Wang, S., Z. Sha, T. S. Sonstegard, H. Liu, P. Xu et al., 2008. Quality assessment parameters for EST-derived SNPs from catfish. BMC Genomics 9 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker, H. A., B. J. McAndrew and J. B. Taggart, 2006. Construction and characterization of a BAC library for the European sea bass Dicentrarchus labrax. Anim. Genet. 37 526. [DOI] [PubMed] [Google Scholar]
- Woods, I. G., C. Wilson, B. Friedlander, P. Chang, D. K. Reyes et al., 2005. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 15 1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., S. Wang, L. Liu, E. Peatman, B. Somridhivej et al., 2006. Channel catfish BAC-end sequences for marker development and assessment of syntenic conservation with other fish species. Anim. Genet. 37 321–326. [DOI] [PubMed] [Google Scholar]
- Xu, P., S. L. Wang, L. Liu, J. Thorsen, H. Kucuktas et al., 2007. A BAC-based physical map of the channel catfish genome. Genomics 90 380–388. [DOI] [PubMed] [Google Scholar]