Abstract
The rDNA arrays in Drosophila contain the cis-acting nucleolus organizer regions responsible for forming the nucleolus and the genes for the 28S, 18S, and 5.8S/2S RNA components of the ribosomes and so serve a central role in protein synthesis. Mutations or alterations that affect the nucleolus organizer region have pleiotropic effects on genome regulation and development and may play a role in genomewide phenomena such as aging and cancer. We demonstrate a method to create an allelic series of graded deletions in the Drosophila Y-linked rDNA of otherwise isogenic chromosomes, quantify the size of the deletions using real-time PCR, and monitor magnification of the rDNA arrays as their functions are restored. We use this series to define the thresholds of Y-linked rDNA required for sufficient protein translation, as well as establish the rate of Y-linked rDNA magnification in Drosophila. Finally, we show that I-CreI expression can revert rDNA deletion phenotypes, suggesting that double-strand breaks are sufficient to induce rDNA magnification.
THE genes that encode three of the four RNA components of ribosomes, the 28S (sometimes called 26S), 18S, and 5.8/2S rRNAs, are found in repeated arrays of cistrons on the X and Y chromosomes of Drosophila melanogaster (Wellauer and Dawid 1977; Tautz et al. 1988). Each ribosomal RNA gene array contains ∼100–300 copies of the multigene cistron (Tartof 1973; Long and Dawid 1980), although the number may vary within laboratory or wild strains (Lyckegaard and Clark 1989; Clark et al. 1991; Averbeck and Eickbush 2005). The individual cistrons within an array, as well as the arrays on the two sex chromosomes, are redundant, since only a subset are required to supply the demands of normal protein synthesis (Ritossa and Atwood 1966; Gersh 1968). Deletions within the arrays are without phenotype unless extreme enough to limit the total number of copies of cistron in the cell to < ∼200 (Ritossa 1968; Tartof 1973).
The rDNA arrays are volatile, as the number of cistrons per array varies from generation to generation (Ritossa 1968; Tartof 1974) or within the cells of an individual (Cohen et al. 2003, 2005). Wild-type arrays are seen to reduce through somatic development and deleted X-linked arrays to magnify during mitosis and meiosis (de Cicco and Glover 1983; Hawley and Tartof 1985), retaining an average of ∼200 copies in each array. Measurements of array sizes have been accomplished genetically (e.g., Marcus et al. 1986), which has the benefit of revealing individual cell information, but at low resolution and only in specific tissues, or molecularly by using membrane hybridization analyses, which necessitate the averaging of array size over many individuals or many tissue types (Tartof 1973; Lyckegaard and Clark 1989).
The rDNA array is arguably the best-understood locus controlled by epigenetic regulation (McStay and Grummt 2008) and the best-characterized repeated gene locus, yet many aspects of its size, structure, and regulation have been beyond experimental manipulation. Many studies have sought to investigate the biology of the rDNA loci but, with few exceptions (Robbins 1981, 1996), most have suffered from an inability to cause specific, graded, and easily induced damage to the locus. To probe rDNA biology, we developed a facile and reproducible system for rDNA cistron deletion. The I-CreI homing endonuclease cleaves a degenerate consensus, which appears in both Chlamydomonas and Drosophila rDNA (Seligman et al. 1997; Maggert and Golic 2005).
In this report, we demonstrate that deletions within the rDNA arrays can be induced by exposure to I-CreI. We used genetic tests to initially identify deletions to a length less than that necessary to serve as the sole source of rRNA in the cell. We developed a reliable real-time polymerase chain reaction assay to quantify the amount of rDNA on these chromosomes, establishing an allelic series of otherwise isogenic Y chromosomes. Using this series, we defined thresholds of Y-linked rDNA array size required for protein synthetic demands. Despite being kept as stocks with wild-type X-linked rDNA arrays, these Y-linked arrays magnified in size. A second exposure to I-CreI induced large magnifications that rapidly restored deleted rDNA arrays. Our work establishes methods for generating and characterizing mutations of the rDNA and expands our understanding of rDNA magnification of Y-linked rDNA arrays.
MATERIALS AND METHODS
Fly stocks and husbandry:
The Y10A chromosome is y+Yw+, Dp(1;Y) y+, P{w+=RSw}10A (Maggert and Golic 2005). The first exon of the white+ gene in RSw is flanked by FRT sequences (Golic and Golic 1996). A chromosome with FLP-induced loss of white+ is referred to as Y10B. Prior to using either Y10A or Y10B for these experiments, we crossed single males to females for three generations prior to our experiments. The X chromosome is y1 w67c23. The I-CreI-expressing line is P{v+t1.8=hs-I-CreI.R}2A, v1/Y; Sb/TM6b, Ubx (Maggert et al. 2008), obtained from the Bloomington Drosophila Stock Center. The attached-X chromosome is C(1)DX, y1 f1 bb0 (Lindsley and Zimm 1992). Flies were raised on cornmeal molasses agar at 25° and 80% humidity.
Induction and screen for deletions:
Flies were allowed to lay eggs for 2–3 days, and larvae to develop for 1 more day. Second and third instar larvae were heat-shocked in circulating water baths at 36°. In experiments involving Y10A, larvae were heat-shocked on 2 successive days, each treatment lasting 45 min. In experiments involving Y10B, larvae were heat-shocked on 1 day for 45 min.
Heat-shock-induced expression was monitored by underrepresentation of I-CreI bearing male progeny in relation to P{v+t1.8=hs-I-CreI.R}2A, v1/y1 w67c23 siblings and by cuticle or eye defects indicating expression-induced cell lethality (Maggert and Golic 2005). X–Y translocation chromosomes were identified as sterile yellow males and yellow+ females and were excluded from analysis.
Real-time polymerase chain reaction:
Primers AGCCTGAGAAACGGCTACCA and AGCTGGGAGTGGGTAATTTACG amplify 63 nucleotides of the 18S gene in the 35S rDNA. After confirming single melting curve kinetics using an ABI Step-One real-time polymerase chain reaction machine (Applied Biosystems) running Step-One v1.0 software, we used the Power SYBR Green master mix (Applied Biosystems) reagent, 500 nm primers, and 10 ng nucleic acid with 40 cycles alternating between 95° for 3 sec and 60° for 30 sec. DNA samples were prepared using a modified procedure from K. Dobie (Gloor et al. 1993; Dobie et al. 2001). The organic extractions were followed with ether extraction, rather than ethanol precipitation, which produced 1–2 μg total nucleic acid/fly. Amplification data were processed by determining the point at which fluorescence first crossed a threshold of 10 standard deviations above the average of all previous cycles (“no amplification”) of fluorescence from each extract, as determined by the Step-One software. Extracts were run in triplicate (occasionally quadruplicate) identical samples. Samples in discordance with the other samples (a threshold cycle with a difference of >2 standard errors of the mean) were interpreted as errors in reaction or reaction preparation and were excluded. Fewer than fifty of ∼5000 total samples were discarded using this criterion. tRNAK-CTT genes were amplified using primers CTAGCTCAGTCGGTAGAGCATGA and CCAACGTGGGGCTCGAAC to generate a 63-nucleotide product. Cycle differences between rDNA and tRNA genes (“ΔCT”) were compared to the same measurement from DNA pooled from a large population (∼200) of adult flies or larvae bearing chromosome Y10B (“ΔΔCT”), generating the percentage of wild-type rDNA quantity. Adult DNA was used for rDNAbb lines, and larval DNA was used for rDNAbb-l lines. The same pooled Y10B preparations of DNA were used for all experiments.
We present either standard deviation (with pooled root-sum errors) if individuals are compared to other individuals or standard errors of the mean (with pooled root-squared-sum errors) if array size from individuals is shown.
Cytology and photography:
Photographs of adult flies were taken using a Nikon D2H camera attached to a Nikon SMZ-1500 microscope. Neuroblast spreads were prepared following the protocol of S. Pimpinelli, S. Bonaccorsi, L. Fanti, and M. Gatti (Sullivan et al. 2000).
RESULTS
Creation of rDNA deletions using I-CreI expression:
To create an allelic series of deletions within the rDNA, we devised an easily employed system to induce varying degrees of damage to isogenic target chromosomes. We chose to make a deletion series of the Y chromosome because of the ease with which this chromosome is manipulated in Drosophila (Bridges 1916). Since the X-linked rDNA array has been the primary target for previous analysis of rDNA mutation (summarized in Lindsley and Zimm 1992; Ashburner et al. 2005), information on the Y-linked rDNA would produce the additional benefit of revealing novel information or supporting the generality of the work on the X-linked array.
Variability in rDNA array size likely exists even within populations derived from a common ancestor (Averbeck and Eickbush 2005). We suspected that small deletions would reveal little useful information; however, larger deletions had the potential to reveal some features of rDNA biology, including transcriptional regulation and magnification. Hence, we sought to generate deletions that removed enough of the rDNA cistrons to manifest a phenotype.
In the first experiment, we used Y chromosome Y10A, which contains an active white+ transgene near the telomere of the short arm (Golic et al. 1998), and a translocation between the X and the tip of the long arm to introduce a yellow+ marker. For the second experiment, we used a related Y chromosome, Y10B, which differs only in the absence of the promoter and first exon of the white+ gene, rendering it white−. We generically refer to either chromosome as Y10, meaning either Y10A or Y10B.
We crossed heat-shocked P{v+t1.8=hs-I-CreI.R}2A, v1/Y10 males en masse to virgin y1 w67c23 females (Figure 1, generation 1) and collected male offspring. Individual male progeny were each crossed separately. The number of translocation chromosomes (Table 1) confirmed that I-CreI was expressed and that damage occurred to the X- and Y-linked rDNA arrays and was subsequently repaired. We reasoned that, within the chromosomes that we collected by isolating individual male progeny, we would find an allelic series of rDNA deletions.
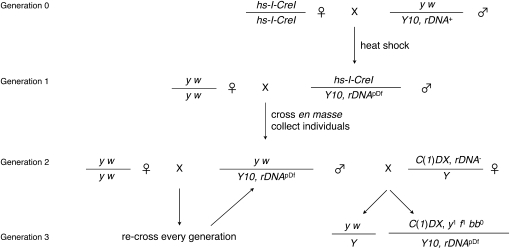
Figure 1.—
Genetic cross and screen for Y, rDNA deletions. In generation 0, females harboring a heat-shock-inducible I-CreI nuclease are mated to males with a recently isogenized yellow+-marked Y chromosome. Males were heat-shocked as larvae and crossed to a common yellow white stock en masse in generation 1. Collecting individual males in generation 2 allowed us to sample independent I-CreI-induced rDNA events of the Y chromosomes. Males were crossed to both fresh yellow white females to establish a stock and to C(1)DX females, whose compound-X chromosome lacks rDNA, to determine if damage to the rDNA had occurred. Damage could be assessed as an altered female-to-male ratio (at an extreme, 0:1) or as bobbed female phenotypes (in generation 3). In every subsequent generation, males were backcrossed to the maternal genotype (yellow white) to maintain the stock and C(1)DX females to retest the rDNA array. Genetic nomenclature: hs-I-CreI is P{v+t1.8=hs-I-CreI.R}2A, v1; y w is y1 w67c23; Y10 is y+Yw+, Dp(1;Y) y+, P{w+=RSw}10A, or y+Yw−, Dp(1;Y) y+, P{w−=RSw−}10B; and C(1)DX is C(1)DX, y1 f1 bb0.
TABLE 1.
Derived Y,rDNADf chromosomes
| Screen | Parental Y chromosome | Chromosomes screened | Altered sex ratio | T(X;Y) | Y,rDNAbb | Y,rDNAl |
|---|---|---|---|---|---|---|
| 1 | Y10A | 560 | 32 | 12 | 0 | 7 |
| 2 | Y10B | 600 | 60 | 6 | 9 | 16 |
| Total | 1160 | 92 | 18 | 9 | 23 |
I-CreI-induced deletions within the rDNA of two related Y chromosomes. Y10A and Y10B differ only by a white+ transgene. Progeny that displayed altered sex ratios and translocation chromosomes [T(X;Y)] were not pursued. Deficiencies of the rDNA (Y,rDNAbb and Y,rDNAl) are described in the text.
A genetic test for rDNA deletion:
Since the I-CreI-mediated damage to the Y chromosomes is specific to the rDNA array, we could easily monitor the extent of damage genetically by making the potentially damaged Y chromosome the sole source of rDNA to the organism. Compound chromosome C(1)DX, y1 f1 bb0 (C(1)DX) contains no rDNA genes (Lindsley and Zimm 1992), and so we replaced the normal Y chromosome in a C(1)DX/Y stock with the rDNA potential Deficiency (Y10, rDNApDf) chromosomes from our study (Figure 1). We expected large deletions to be inviable and moderate deletions to be subviable or express a bobbed phenotype.
We tested 1160 individual Y10, rDNApDf chromosomes using this assay and identified 23 Y chromosomes incapable of supplying sufficient rRNA for survival (1.9%), 9 Y chromosomes that expressed a majority penetrant (>50% of flies showed the phenotype) bobbed cuticular phenotype (0.7%), and 92 more that exhibited a sex ratio significantly different from unity (7.9%) (see Table 1 for summary). Each potential reduction chromosome was retested; those with no or bobbed female progeny again produced similar female progeny upon retest, but those that showed altered sex ratios showed normal ratios upon retest. Lethality and the bobbed phenotypes were thus reliable indicators of rDNA deletion; however, the use of subviability (and consequent sex-ratio distortion) was not.
We wanted to confirm that the lethality phenotypes that we observed were due to reduction of the rDNA, so we crossed males from eight of our identified Y10, rDNApDf chromosomes (three from Y10A and five from Y10B) to females of the genotype In(1)sc4sc8/FM7a, BS. In(1)sc4sc8, like C(1)DX, lacks rDNA. Half of the male progeny of the cross were expected to express a Bar phenotype (FM7a/Y) and half to be non-Bar [In(1)sc4sc8/Y10B, rDNApDf], unless the rDNA was removed, in which case the non-Bar class of males would be absent or express a bobbed phenotype. We found strict concordance between the C(1)DX/Y10, rDNApDf lethality or bobbed phenotypes and the In(1)sc4sc8/Y10B, rDNApDf lethality or bobbed phenotypes (data not shown), indicating that the lethality is linked to the Y chromosome and most likely due to rDNA deletion, and not to the induction of other genomic alterations that interacted with the C(1)DX background to produce lethality. We confirmed that the only cytologically visible alteration to the chromosome structures was in Y-linked band h20, the location of the rDNA locus (data not shown).
Since we interpreted our results to mean that all of the identified lines possessed significant reduction of the rDNA, these chromosomes from hereon will be referred to as Y10B, rDNAl or Y10B, rDNAbb (or, generically, Y10B, rDNADf), consistent with established nomenclature for bobbed-lethal or bobbed alleles with reduced rDNA copy number.
Molecular test and quantification of rDNA:
The genetic test for rDNA array size relies on active rDNA cistrons. We sought a method to quantify the size of the deletions irrespective of genetic activity and so developed a real-time (quantitative) polymerase chain reaction (qPCR) to measure the copy number of rDNA template.
To normalize the rDNA qPCR amplification rate, we chose a denominator that fulfilled several criteria. First, we needed a normalizing DNA sequence with a high copy number, which would make the quantification of rDNA robust despite fluctuations in DNA yield from individual flies. Second, we needed a sequence that did not vary between individuals within a population or between strains so that our results would be easily comparable without performing cumbersome crosses to establish isogeny. Third, since many tandem-repeat (arrayed) DNAs are eliminated during development (Cohen et al. 2003, 2005), we wanted to find a dispersed repeat. For these reasons, we chose the high-copy-number tRNAK-CTT. Although lysine is encoded by three anticodons, the CTT isotype is most common, and all 15 tRNA genes of this isotype (in a haploid genome) have identical sequence (Schattner et al. 2005). Amplification of 30 tRNAK-CTT genes (in a diploid genome) then was used as a denominator in our calculations. We are aware that not all tRNA genes may be equally amplified in our reaction, so absolute values of rDNA copy number may be inaccurate; however, the copy number of rDNA (on Y10B or an unrelated unmarked Y), by these calculations, is ∼290 (Figure 2, 95% confidence interval of 270–315 copies), in agreement with population studies performed by many labs using other techniques (Tartof 1973; Shermoen and Kiefer 1975; Long and Dawid 1980; Lyckegaard and Clark 1989). We are cautious about comparing absolute copy numbers of rDNA derived from different techniques and so report quantification relative to our Y10 chromosomes.
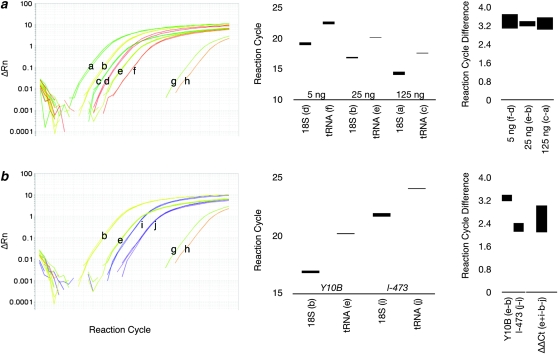
Figure 2.—
Real-time qPCR to measure rDNA content. (a) Traces of qPCR reactions to amplify the 18S rRNA and tRNAK genes at three different concentrations of DNA extracted from C(1)DX/Y10B females. Traces show triplicate reactions set in parallel; each triplet is labeled with a letter (a–f), which corresponds to the data in the accompanying graphs. g and h are no-template controls. Bar graphs show the average ± standard error of the mean ranges for amplification cycles (Ct). The rightmost graph shows the difference between rDNA and tDNA threshold cycles (ΔCt) with ranges equal to root pooled squares of standard errors. (b) Traces of qPCR reactions to amplify 18S and tRNAK genes from wild type (Y10B, traces b and e) and a rDNA deficiency chromosome (l-473, traces i and j). b, e, g, and h are the same traces as in a. Accompanying graphs show Ct for these reactions: the rightmost graph shows ΔCt as a measure of the rDNA/tDNA copy number, and ΔΔCt shows the difference between l-473 and Y10B in Ct, corrected (by the tDNA measurement) for DNA concentration. The difference in rDNA copy number is 2ΔΔCt. The y-axes are either cycles of qPCR or differences in cycles between different samples.
By titrating the template, we determined that the amount of template is free to vary over a concentration range of at least 25-fold and that the denominator tRNAK-CTT is able to normalize the signal to a relative copy number (Figure 2). We have been able to detect template rDNA from as few as five genome equivalents (data not shown), a total template rDNA copy number of <2000, and a tRNAK-CTT copy number of ∼150; however, our analyses of Y10B, rDNADf presented here were done with 10–20 ng of total nucleic acid to ensure that we were well within the range of linear sensitivity.
To make the reduced rDNA array unique within the genome, we crossed y1 w67c23/Y10B, rDNADf single males to C(1)DX/Y females. Female progeny are of two types: C(1)DX/y1 w67c23 (triplo-X) metafemales that die late in development and are identifiable by their yellow phenotype (Lindsley and Zimm 1992) and C(1)DX/Y10B, rDNADf females that have only Y-linked rDNA and are identifiable by their yellow+ phenotype. Flies devoid of rDNA still possess rRNA by virtue of maternally loaded RNAs and ribosomes and, in our hands, survived to late larvae or early pupae stages. Hence, we were able to purify DNA from C(1)DX/Y10B, rDNADf larval, pupal, or adult females whose only rDNA was the Y-linked array.
The results of our analyses of all Y10B, rDNADf are presented in Figure 3a, which shows quantification from multiple (three to seven) C(1)DX/Y10B, rDNADf female siblings from single fathers in the second generation after being isolated as independent stocks (Figure 1, generation 3). The ranges shown are pooled standard errors from replicate reactions using DNA from three to eight individuals of generation 2. These ranges include experimental error and standard deviation of the population analyzed, sorted by mean after the reference pool of Y10B. We used wild-type (Y10B) reference DNA preparations separately for adults and larvae.
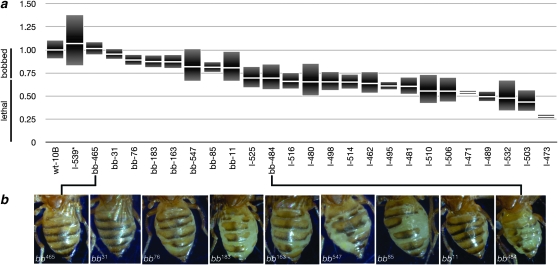
Figure 3.—
Y, rDNADf allelic series. qPCR was used to measure rDNA array size in the alleles generated in this work. (a) 10B is the progenitor chromosome. The remainder are the recovered alleles, sorted by average size. Data are presented as the average ± standard error of the mean. Chromosome names indicate their phenotype (wt, wild-type; bb, bobbed; l, lethal) as the sole source of rDNA in the organism, as well as their allele number. The approximate range that corresponds to those phenotypes is indicated at the left of the graph. The y-axis is ratio of wild-type Y10B chromosome rDNA content. (b) Bobbed flies were photographed and are presented in order of decreasing rDNA array size (taken from a), which correlates with an increasing severity of the bobbed phenotype.
As expected, the rDNA arrays that are largest among our allelic series express a bobbed phenotype. Figure 3b shows the abdomens of surviving C(1)DX/Y10B, rDNADf females carrying bobbed alleles of the rDNA. Moreover, we saw a correlation between the deletion size and the expressivity of the bobbed phenotype. More extensive deletion of the rDNA caused bobbed-lethal phenotypes. Hence, two transitions are defined by this graph: the wild-type to bobbed transition, and the bobbed to lethal transition.
The wild-type to bobbed transition occurred at ∼90% of the hemizygous (Y-linked) rDNA cistrons, or ∼260 copies. Although we expected to define such a transitional rDNA size, we were surprised that, in our studies of the Y chromosome, this transition was higher than in other studies that investigated the X-linked rDNA bobbed threshold. The X rDNA locus required a deletion to ∼50–80% of the wild-type size, or to ∼150–200 cistrons, to produce a bobbed phenotype (Tartof 1973; Terracol and Prud'Homme 1986). This difference may be due to the disparate chromosomes used in these studies, to differences in the proportion of intact and R1- and R2-interrupted cistrons (Lyckegaard and Clark 1989; Averbeck and Eickbush 2005), or to differences in the quantification techniques used in each study.
The transition from bobbed to lethal occurred in ∼65% of the hemizygous (Y-linked) level of rDNA. This is ∼190 copies according to our calculations and is again higher than previous studies that indicated as few as 114 copies of X-linked rDNA are sufficient for viability (Terracol and Prud'Homme 1986).
One chromosome, Y10B, rDNAl-539, carried a lethal allele of the rDNA despite showing an array size larger than Y10B, rDNA+. We do not know why ample rDNA would not supply sufficient rRNA, but consider that the cistrons may have been damaged during I-CreI-induced damage, magnification may have occurred using inactive R1- or R2-interrupted cistrons as template, the copies on the chromosome may be epigenetically inactive, or some other explanation may account for this (Terracol and Prud'Homme 1981; Terracol 1987).
Evidence for rDNA magnification:
We initially established stocks of three of the seven rDNAl deletions derived from Y10A, assuming that they would be stable as stocks containing an X chromosome with a normal rDNA array, since it is generally accepted that magnification of the Y requires special circumstances. Instead, we found that, upon retest after seven generations as a stock, two of the stocks had reverted and produced bobbed-viable and wild-type individuals despite the presence of a fully functional X-linked array. X-linked rDNA magnification is a well-characterized phenomenon (Marcus et al. 1986), yet we were surprised to see that the new Y10A, rDNAl chromosomes exhibited this phenotype after so few generations without obvious selection. Leonard Robbins (1981) showed that many deletion alleles of the rDNA are stable once generated, while ours are not. Both those alleles and ours rely on creating damage specifically to the rDNA arrays, however by different means. Komma and colleagues showed that certain Y chromosomes are capable of magnification even when the cell possesses sufficient rDNA (Komma and Endow 1986; Komma et al. 1993). This feature is not understood but has been shown to reside on some Y chromosomes; it is possible that our chromosome possessed this ability prior to being reduced.
We confirmed that the suppression of the lethal phenotypes in Y10A,rDNAl-3-revertant and Y10A,rDNAl-39-revertant did not map to the X chromosome or large autosomes (data not shown). We interpreted these results to indicate that the Y-linked rDNA array had increased in size, rather than the stock accumulating modifiers of rDNA expression, which has been shown as an alternate means of rDNA magnification (Marcus et al. 1986).
To observe magnification as it occurred on our chromosomes, we outcrossed males from each Y chromosome stock to virgins of a common y1 w67c23 stock to prevent the accumulation of modifiers that could affect rDNA expression. At every generation, we also crossed sibling males to C(1)DX, y1 f bb0/Y or C(1)DX, y1 f bb0/Y, BS females to genetically assess the status of the rDNA array size in female offspring. This is represented by the recursive recross in generation 3 of Figure 1.
We chose to monitor six chromosomes over four generations. For each generation, we extracted DNA and measured rDNA content from 4 to 10 pupae or adults, which allowed us to investigate bobbed and bobbed-lethal lines. The results are presented in Figure 4.
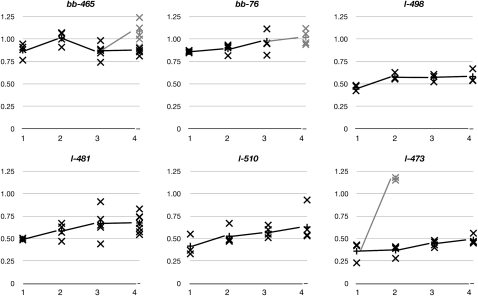
Figure 4.—
rDNA arrays undergo slow gradual magnification as well as sporadic fast magnification. Six chromosomes were monitored every generation by selecting four to eight individuals for rDNA array size measurement. The data for each individual are shown (x's), as well as the average of the population (connected by lines). Solid data points indicate individuals with the same phenotype (lethal or bobbed) as previous generations, and shaded data points are from individuals whose phenotype changed (to wild type for bb-465 or to bobbed for l-473). The x-axes are the successive generations after establishment as stock. The y-axes are the ratios to wild-type Y10B chromosome rDNA content. For clarity, standard errors are not depicted.
We observed progressive rDNA magnification by monitoring individuals every generation. All lines showed an average array size increase every generation. This gradual increase was varied, but was ∼5% (5.6% ± 1.4%) of the wild-type rDNA array size when averaged for all individuals between the first and fourth generations, or 7.2% ± 5.6% for each generation, excluding those that underwent large increases (e.g., Y10B, rDNAbb−473) or decreases (e.g., generation 2 to generation 3 for Y10B, rDNAbb−465). These results underscore the variation in the rDNA magnification amount, but indicate a trend toward progressively larger arrays. Consistent with this, five of the six chromosomes (all but Y10B, rDNAbb−473) showed an increasing coefficient of variance (ratio of standard deviation to average) in each generation. Although some individuals showed arrays that dropped in size compared to the previous generation's average, these were less frequent than were instances where the array magnified. This value, the addition of ∼15 copies of rDNA per generation, is essentially identical to that observed for the X chromosome (Tartof 1973).
Magnification is not constant, but may instead increase by small steps and spend some generations remaining steady at average size. Line Y10B, rDNAbb-498 is a lethal allele of bobbed and shows a slow increase in rDNA content. The initial increase was the largest (11.6%), and the rate slowed for the subsequent two generations (0.5% over two generations). Since we expect that magnification affects every chromosome by differing amounts, this may be an artifact of small sample size. With a small sample size, it is possible to select the less-common individuals whose rDNA arrays either have decreased in size or have not changed appreciably. Over the individuals scored in the subsequent generation, this would appear as decreasing or stable rDNA arrays, despite the majority of the population increasing in size. A trend of magnification is consistent with our results from the other lines that we analyzed, notably Y10B, rDNAl-481, Y10B, rDNAl-510, and Y10B, rDNAl-473, that rDNA size varies between generations and an increase in size is more common than a decrease, leading to a gradual and steady increase in the population.
Line Y10B, rDNAbb−465 magnified to a size that overlapped with wild type, although all flies were bobbed. In the subsequent generation, the average size decreased again, but was still within the previously defined bobbed range (Figure 3). In the fourth generation, two classes of flies were seen: those that were bobbed and those that had phenotypically reverted to wild type (shaded in Figure 4). As predicted, those that were wild type in appearance had arrays that had increased more than those that remained bobbed and were 108% ± 17% of the wild-type quantity of rDNA.
Line Y10B, rDNAbb-76 was also originally identified as a bobbed reduction. For the next two generations, bb-76 showed the same expressivity of phenotype. In the third generation, however, the C(1)DX/Y10B-rDNAbb-76 females were notably less bobbed, and by the following generation, all female progeny had normal cuticles (shaded data points in Figure 4). This corresponds to the generation in which the average array size reached 101% ± 8% of wild-type level, near the transition that we had defined by analyzing the allelic series of initial deficiencies (Figure 3).
We cannot distinguish between somatic pseudo-magnification, which our assay measures, and germline magnification, since the germline is a small fraction of the genomes measured in whole animals. However, it is likely that germline magnification contributes since the average array size grows in subsequent generations. This is particularly evident in those cases in which a chromosome (e.g., Y10B, rDNAbb-465 and Y10B, rDNAbb-76) reverts to wild type and all progeny in that stock do so.
Against a backdrop of steady increase, we also saw two large increases, similar to what is observed for the X-linked rDNA arrays (Hawley and Tartof 1985; Endow and Komma 1986). Line Y10B, rDNAl-473 produced two bobbed flies in the second generation, which corresponded to the two Y chromosomes that had very large increases in rDNA size (shaded data points in Figure 4). This increase (to 117% ± 2%) was more than two times the size of the progenitor Y chromosome (36% ± 11%), a dramatic example of a magnification event that cannot be explained simply by a single unequal sister chromatin exchange.
Induction of magnification by I-CreI expression:
Our results indicate that double-strand breaks are sufficient to induce reduction in rDNA copy number and that natural processes are then able to magnify the arrays toward their original size. It has been proposed that magnification might rely on double-strand breaks, since flies mutated for genes involved in double-strand break repair are unable to magnify their arrays (Marcus et al. 1986). We could test this assertion using a second I-CreI.
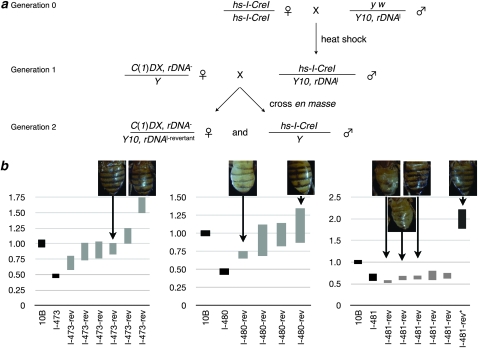
We crossed three lethal deletions from our allelic series to females carrying an I-CreI transgene and heat-shocked for 1 hr to induce I-CreI expression (Figure 5). Male progeny were crossed en masse to C(1)DX/Y females (generation 1 of Figure 5). Progeny of that cross were expected to be solely males, consistent with the phenotype of these Y10B, rDNAl chromosomes. If, however, I-CreI expression induced magnification, we expected to obtain revertant females of genotype C(1)DX/Y10B, rDNAl-revertant. The results are presented in Table 2.
Figure 5.—
rDNA arrays undergo magnification when exposed to I-CreI. (a) Y chromosomes with previously reduced rDNA arrays were exposed to I-CreI induced by heat shock. This cross is similar to the one described in Figure 1, but here we screened for reversion of the lethal-bobbed phenotype to the bobbed or wild-type phenotype. A control cross was performed in parallel with X chromosomes without the I-CreI-expressing transgene. (b) Results of rDNA quantification. Each graph contains data showing the relative average for Y10B (defined as 1.00) and the parental chromosome prior to heat-shock induction of I-CreI, both as solid data points. Shaded data points are confidence intervals for individuals (the average of replicate qPCR reactions with standard errors of the mean), and photographs of a subset of those individuals show the bobbed phenotype. The final data point, l-481-rev*, is from the control cross, which did not express I-CreI. The large amount of rDNA is most consistent with nondisjunction producing a C(1)DX/Y10B, rDNADf/Y, rDNA+ individual.
TABLE 2.
Magnified Y,rDNAl-revertant chromosomes
| X chromosome | Y10B,rDNAl chromosome | X/Y male progeny | C(1)DX/Y10B, rDNAl-revertant female progeny (and phenotypes) |
|---|---|---|---|
| I-CreI | l-473 | 149 | 3 bb, 3 bb+ |
| I-CreI | l-480 | 196 | 1 bb, 3 bb+ |
| I-CreI | l-481 | 126 | 6 (bb) |
| X | l-473 | 111 | — |
| X | l-480 | 116 | 1 (bb+) |
| X | l-481 | 62 | 1 (bb+) |
I-CreI-induced magnifications of the rDNA of three Y chromosomes previously deleted for the rDNA. I-CreI transgene-containing and wild-type (X) chromosomes were heat-shocked; only the former expresses I-CreI to create double-strand breaks in the rDNA. X/Y progeny are normal males, while C(1)DX/Y10B, rDNAl-revertant can survive only if the rDNA magnifies. rDNA phenotypes refer to the cuticular phenotype of C(1)DX/Y10B, rDNAl-revertant females.
Sires harboring each of the three tested I-CreI-exposed Y10B, rDNAl chromosomes gave female progeny when crossed to C(1)DX females. Most of these were severely bobbed, indicating that the rDNA arrays were barely sufficient for rRNA demands, although some were bobbed+. Since these Y10B, rDNAl were not able to supply sufficient rRNA prior to I-CreI expression, the rDNA array sizes must have increased on those chromosomes. To confirm an increase in rDNA, we isolated DNA for quantification (Figure 5). Each chromosome contained magnified rDNA arrays. For chromosomes Y10B, rDNAl−473 and Y10B, rDNAl−481, the amount of magnification varied between individuals and correlated well with the expressivity of the bobbed phenotype. Some revertants of chromosome Y10B, rDNAl−481 did not show a large magnification. This may be due to this chromosome being on the threshold of lethal to bobbed, so even small magnifications would be uncovered by this assay, or because the Y10B, rDNAl−481-revertant chromosomes possess a different active-to-inactive ratio of rDNA cistrons than does the original Y10B, rDNAl−481 chromosome (Terracol and Prud'Homme 1981, 1987; Terracol 1987; Ashburner et al. 2005).
Nondisjunction in the C(1)DX/Y mothers would also produce flies that appeared as revertants that had magnified to bobbed+, since progeny would be C(1)DX/Y10B, rDNAl/Y. To identify those events in our analysis, we performed the same cross and heat shock with males of genotype y w/Y10B, rDNAl. Some sires of genotype y w/Y10B, rDNAl did give female progeny, but at a much lower rate than did I-CreI-expressing sires. Surviving females were bobbed+, indicating that they were nondisjunctional progeny. Consistent with this, those surviving females had rDNA arrays that were measured to be ∼200% of the Y10B array (Figure 5b, final data point, labeled “l-481-rev*”), far above those produced by I-CreI-expressing fathers.
DISCUSSION
The rDNA is composed of the 35S cistron repeated hundreds of times on each chromosome and is responsible for nucleating the nucleolus, pairing heterogametic sex chromosomes in male meiosis, providing rRNA for ribosome biosynthesis, and modulating protein function through sequestration. Hence the rDNA array represents a central regulator in many important aspects of nuclear biology.
The rDNA arrays are regulated such that only about one-half of the cistrons are active, a form of epigenetic regulation thought to involve histone modification; ATP-dependent chromatin remodeling; and, in some organisms, DNA methylation (McStay and Grummt 2008). The proportion of active cistrons can be manipulated by altering gene dosage of important regulators (Mayer et al. 2006) or by altering the in vivo activity of regulatory enzymes (Sandmeier et al. 2002; French et al. 2003). Such manipulations have affected cell biology on a large scale because of the centrality of translational capacity, enzymatic modification of chromatin structure, and the nucleolus (Perrin et al. 1998). Manipulation of rRNA transcription has been shown to alter gene expression at unlinked sites of the genome, reinforcing the view that the nucleolus is an important determinant in genome regulation (Maillet et al. 1996).
What has been absent in these studies is the ability to alter the rDNA as easily as the in trans-acting regulatory proteins. Molecular genetic analyses of repeated DNA have lagged behind the analysis of single-repeat sequences, in part because of the difficulty of altering repeated sequence in vivo. Genetic activities are often redundant, making mutation to recessive phenotypes difficult, and redundant homology does not allow precision during the use of gene targeting. Most past studies of the Drosophila rDNA have utilized alleles isolated from unrelated or distantly related sources, which may vary considerably (Lyckegaard and Clark 1989). Even chromosomes isolated from a common stock may differ twofold in rDNA content (Averbeck and Eickbush 2005). Although not mapped to the rDNA, chromosome polymorphisms can have considerable effects on gene activity (Spofford and Desalle 1991; Lemos et al. 2008).
We have developed a method to easily create an isogenic graded allelic series of rDNA copy number on the Y chromosome of Drosophila, which will circumvent some of these problems. Our approach uses one parental chromosome and derives and characterizes an allelic series within three generations. Further, we have developed a robust assay to quantify the extent of deletion within the rDNA. We have shown the efficacy and accuracy of these techniques through genetic and molecular confirmation of damage to the rDNA.
The utility of generating deletions within the rDNA is manifold, and we have demonstrated one by analyzing natural and induced magnification of the Y-linked rDNA array in males. Most studies have investigated magnification of the X-linked rDNA from weakly bobbed alleles to wild type in bobbed flies, which presumably put some pressure on the rDNA to magnify to supply rRNAs. The details of magnification are not understood in Drosophila, although it is clear that some chromosomes are capable of magnification while others are not (Procunier and Tartof 1978; Komma and Endow 1986, 1987). Spontaneous, large increases in rDNA array size, in both the germline and the soma, have supported the view that rDNA magnification occurs through unequal sister-chromatid exchange (Endow and Komma 1986), although reversion may occur through other means (Marcus et al. 1986; Terracol et al. 1990). We showed genetically that the reversion of bobbed maps to the Y chromosome and that the array grows in size concomitant with the reversion.
The qPCR technique that we developed to characterize the array length allowed us to monitor increases in array length in individuals as the rDNA underwent magnification. By combining the genetic measurements of translational capacity and the real-time quantitative polymerase chain reaction, we have defined the threshold of rDNA copy number necessary for organismal viability. In the absence of an X-linked array, deletions of the Y-linked rDNA (to ∼260 copies) cause a bobbed phenotype or, if more extreme (to ∼190), a lethal phenotype. We have established the threshold for the bobbed phenotype by using our allelic series in three different ways. First, deletions defined thresholds of lethal to bobbed and bobbed to wild type. Second, lethal-to-bobbed and bobbed-to-wild-type magnification reinforced those defined limits. Third, magnification by I-CreI expression further reinforced the severe bobbed and mildly bobbed phenotypes. Our findings show that an isogenic chromosome allelic series has clearly defined the limits of phenotypic expressivity, unlike previous studies that revealed broad ranges for these thresholds, further highlighting the utility of our approach.
Arrays allowed to magnify in flies that provide rDNA in trans exhibit a slow increase in rDNA array size that is most consistent with stochastic small increases outnumbering stochastic decreases expected for unequal sister-chromatid exchange between arrays. Thus, the Y chromosome rDNA arrays are similar to the X chromosome arrays: they are capable of magnification, even in the absence of a special inducing chromosome (Ritossa 1968; Hawley and Tartof 1985; Komma and Endow 1986). In previous studies of magnification, not every Y chromosome was able to magnify. In fact, a great number of rDNA alleles are stable (Lindsley and Zimm 1992). It is possible that a magnifying element similar to the one characterized on the Ybb0 chromosome existed on both the Y10A and Y10B chromosomes prior to our work, and deletion merely revealed its presence. It is also possible that induction of I-CreI induced our chromosomes to become magnifying chromosomes, possibly by epigenetic remodeling of the rDNA or by activation of resident R1 or R2 transposable elements. Since magnification acts through an unknown mechanism, we cannot state why our chromosomes magnify after reduction.
Hypotheses that rDNA quantity is tied to aging, disease, or gene regulation exist but are difficult to test without some way to manipulate the rDNA array size (Spofford and Desalle 1991; Gotta et al. 1997; Palumbo et al. 1994; Johnson et al. 1999; Weber et al. 1999; Carmo-Fonseca et al. 2000; Martindill et al. 2007; Versa-Ostojć et al. 2008). Only by altering the initial size of the rDNA array, in an otherwise isogenic background, can the contribution of rDNA size to these pleiotropic phenomena be investigated. To advance beyond the stage where rDNA arrays are merely correlated to aging, cancer, or other diseases or cellular function, variables must be experimentally manipulated. Ideally, this would be done with a minimal perturbation to other factors. Such manipulations—facile, specific, and graded—are now possible with the rDNA arrays of Drosophila. If developing hypotheses linking genomewide gene regulation or aging and rDNA are correct, then we expect that deletions to the rDNA will profoundly affect these phenotypes. Our work has established a mode of generating an allelic series of rDNA deletions (or, possibly, expansions) on chromosomes of choice and has detailed a robust means for quantifying the rDNA array size. With these chromosomes, it will now be possible to study the role of the rDNA in nuclear biology and to address intriguing hypotheses connecting rDNA magnification, transcriptional regulation and developmental programs, inheritance of acquired characteristics, and complex diseases or cell states such as cancers or aging.
Acknowledgments
The authors thank James Erickson and Arne Lekven for reading the manuscript prior to submission. This work was funded by the National Institutes of Health grant GM076092 and partially by the March of Dimes Basil O'Conner Starter Scholars Award 05-1229.
References
- Ashburner, M., K. G. Golic and R. S. Hawley, 2005. Drosophila: A Laboratory Handbook, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Averbeck, K. T., and T. H. Eickbush, 2005. Monitoring the mode and tempo of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics 171 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, C. B., 1916. Non-disjunction as proof of the chromosome theory of heredity. Genetics 1 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., L. Mendes-Soares and I. Campos, 2000. To be or not to be in the nucleolus. Nat. Cell Biol. 2 107–112. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., F. M. Szumski and E. M. Lyckegaard, 1991. Population genetics of the Y chromosome of Drosophila melanogaster: rDNA variation and phenotypic correlates. Genet. Res. 58 7–13. [DOI] [PubMed] [Google Scholar]
- Cohen, S., K. Yacobi and D. Segal, 2003. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 13 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S., N. Agmon, K. Yacobi, M. Mislovati and D. Segal, 2005. Evidence for rolling circle replication of tandem genes in Drosophila. Nucleic Acids Res. 33 4519–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cicco, D. V., and D. M. Glover, 1983. Amplification of rDNA and type I sequences in Drosophila males deficient in rDNA. Cell 32 1217–1225. [DOI] [PubMed] [Google Scholar]
- Dobie, K. W., C. D. Kennedy, V. M. Velasco, T. L. McGrath, J. Weko et al., 2001. Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics 157 1623–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow, S. A., and D. J. Komma, 1986. One-step and stepwise magnification of a bobbed lethal chromosome in Drosophila melanogaster. Genetics 114 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, S. L., Y. N. Osheim, F. Cioci, M. Nomura and A. L. Beyer, 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersh, E. S., 1968. Mutants at the bobbed locus in Drosophila melanogaster: Relation to ribosomal RNA synthesis? Science 162 1139. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis et al., 1993. Type I repressors of P element mobility. Genetics 125 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G., and M. M. Golic, 1996. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G., M. M. Golic and S. Pimpinelli, 1998. Imprinted control of gene activity in Drosophila. Curr. Biol. 8 1273–1276. [DOI] [PubMed] [Google Scholar]
- Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy et al., 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, R. S., and K. D. Tartof, 1985. A two-stage model for the control of rDNA magnification. Genetics 109 691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, F. B., D. A. Sinclair and L. Guarente, 1999. Molecular biology of aging. Cell 96 291–302. [DOI] [PubMed] [Google Scholar]
- Komma, D. J., and S. A. Endow, 1986. Magnification of the ribosomal genes in female Drosophila melanogaster. Genetics 114 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komma, D. J., and S. A. Endow, 1987. Incomplete Y chromosomes promote magnification in male and female Drosophila. Proc. Natl. Acad. Sci. USA 84 2382–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komma, D. J., S. J. Glass and S. A. Endow, 1993. Constitutive magnification by the Ybb- chromosome of Drosophila melanogaster. Genet. Res. 62 205–212. [DOI] [PubMed] [Google Scholar]
- Lemos, B., L. O. Araripe and D. L. Hartl, 2008. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319 91–93. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Long, E., and I. B. Dawid, 1980. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49 727–764. [DOI] [PubMed] [Google Scholar]
- Lyckegaard, E. M., and A. G. Clark, 1989. Ribosomal DNA and Stellate gene copy number variation on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 86 1944–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert, K. A., and K. G. Golic, 2005. Highly efficient sex chromosome interchanges produced by I-CreI expression in Drosophila. Genetics 171 1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert, K. A., W. J. Gong and K. G. Golic, 2008. Methods for homologous recombination in Drosophila. Methods Mol. Biol. 420 155–174. [DOI] [PubMed] [Google Scholar]
- Maillet, L., C. Boscheron, M. Gotta, S. Marcand, E. Gilson et al., 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10 1796–1811. [DOI] [PubMed] [Google Scholar]
- Marcus, C. H., A. E. Zitron, D. A. Wright and R. S. Hawley, 1986. Autosomal modifiers of the bobbed phenotype are a major component of the rDNA magnification paradox in Drosophila melanogaster. Genetics 113 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindill, D. M., C. A. Risebro, N. Smart, M. Franco-Viseras Mdel, C. O. Rosario et al., 2007. Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat. Cell Biol. 9 1131–1141. [DOI] [PubMed] [Google Scholar]
- Mayer, C., K. M. Schmitz, J. Li, I. Grummt and R. Santoro, 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 22 51–61. [DOI] [PubMed] [Google Scholar]
- McStay, B., and I. Grummt, 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24 131–157. [DOI] [PubMed] [Google Scholar]
- Palumbo, G., M. Berloco, L. Fanti, M. P. Bozzetti, S. Massari et al., 1994. Interaction systems between heterochromatin and euchromatin in Drosophila melanogaster. Genetica 94 267–274. [DOI] [PubMed] [Google Scholar]
- Perrin, L., O. Demakova, L. Fanti, S. Kallenbach, S. Saingery et al., 1998. Dynamics of the sub-nuclear distribution of Modulo and the regulation of position-effect variegation by nucleolus in Drosophila. J. Cell Sci. 111 2753–2761. [DOI] [PubMed] [Google Scholar]
- Procunier, J. D., and K. D. Tartof, 1978. A genetic locus having trans and contiguous cis functions that control the disproportionate replication of ribosomal RNA genes in Drosophila melanogaster. Genetics 88 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa, F. M., 1968. Unstable redundancy of genes for ribosomal RNA. Proc. Natl. Acad. Sci. USA 60 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa, F. M., and K. C. Atwood, 1996. Unequal proportions of DNA complementary to ribosomal RNA in males and females of Drosophila simulans. Proc. Natl. Acad. Sci. USA 56 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, L., 1981. Genetically induced mitotic exchange in the heterochromatin of Drosophila melanogaster. Genetics 99 443–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, L., 1996. Specificity of chromosome damage caused by the Rex element of Drosophila melanogaster. Genetics 144 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo et al., 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21 4959–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner, P., A. N. Brooks and T. M. Lowe, 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33 W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman, L. M., K. M. Stephens, J. H. Savage and R. J. Monnat, Jr., 1997. Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics 147 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen, A. W., and B. I. Kiefer, 1975. Regulation in rDNA-deficient Drosophila melanogaster. Cell 4 275–280. [DOI] [PubMed] [Google Scholar]
- Spofford, J. B., and R. DeSalle, 1991. Nucleolus organizer-suppressed position-effect variegation in Drosophila melanogaster. Genet. Res. 57 245–255. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., M. Ashburner and R. S. Hawley, 2000. Drosophila Protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Tartof, K. D., 1973. Regulation of ribosomal RNA gene multiplicity in Drosophila melanogaster. Genetics 73 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof, K. D., 1974. Unequal mitotic sister chromatin exchange as the mechanism of ribosomal RNA gene magnification. Proc. Natl. Acad. Sci. USA 71 1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracol, R., 1987. Differential magnification of rDNA gene types in bobbed mutants of Drosophila melanogaster. Mol. Gen. Genet. 208 168–176. [DOI] [PubMed] [Google Scholar]
- Terracol, R., and N. Prud'Homme, 1981. 26S and 18S rRNA synthesis in bobbed mutants of Drosophila melanogaster. Biochimie 63 451–455. [DOI] [PubMed] [Google Scholar]
- Terracol, R., and N. Prud'Homme, 1986. Differential elimination of rDNA genes in bobbed mutants of Drosophila melanogaster. Mol. Cell. Biol. 6 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracol, R., Iturbide, Y., and N. Prud'Homme, 1990. Partial reversion at the bobbed locus of Drosophila melanogaster. Biol. Cell 68 65–71. [DOI] [PubMed] [Google Scholar]
- Tautz, D., J. M. Hancock, D. A. Webb, C. Tautz and G. A. Dover, 1988. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol. Biol. Evol. 5 366–376. [DOI] [PubMed] [Google Scholar]
- Versa-Ostojć, D., T. Stanković, S. Stemberger-Papić, D. Vrdoljak-Mozetic, M. Manestar et al., 2008. Nuclear morphometry and AgNOR quantification: computerized image analysis on ovarian mucinous tumor imprints. Anal. Quant. Cytol. Histol. 3 160–168. [PubMed] [Google Scholar]
- Weber, J. D, L. J. Taylor, M. F. Roussel, C. J. Sherr and D. Bar-Sagi, 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1 20–26. [DOI] [PubMed] [Google Scholar]
- Wellauer, P. K., and I. B. Dawid, 1977. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell 10 193–212. [DOI] [PubMed] [Google Scholar]