Abstract
Array comparative genomic hybridization (aCGH) has been used primarily to detect copy-number variants between two genomes. Here we report using aCGH to detect single nucleotide mutations on oligonucleotide microarrays with overlapping 50-mer probes. This technique represents a powerful method for rapidly detecting novel homozygous single nucleotide mutations in any organism with a sequenced reference genome.
A major roadblock in genetic research lies in the molecular identification of mutations responsible for an observed phenotype. Traditional positional cloning techniques are laborious, time-consuming, and sometimes impractical for mapping mutations to regions smaller than a few mega-base pairs, particularly in regions with low recombination frequencies such as the centers of Caenorhabditis elegans chromosomes (Barnes et al. 1995). Sequencing such a large region still remains impractical for most laboratories, and as a result many mutations remain uncharacterized. Recently, array comparative genomic hybridization (aCGH) has been used to detect single nucleotide variation in the 12.5-Mb yeast genome using short 25-mer probes (Gresham et al. 2006). Here we demonstrate the use of 50-mer probes to detect single nucleotide mutations in the 100-Mb C. elegans genome.
aCGH has been used to detect many types of genome diversity in a variety of organisms (Gresham et al. 2008). We have been using aCGH with exon-centric tiling arrays of 50-mer oligonucleotide probes to screen for deletions in the C. elegans genome following mutagenesis with trimethylpsoralen (TMP) and ultraviolet (UV) irradiation (Maydan et al. 2007). In one set of experiments utilizing a microarray with probes targeting primarily exons on C. elegans chromosome II, we screened individuals homozygous for a mutagenized chromosome II. In these experiments we identified three statistically significant putative mutations (P-values ranged from 2.7 × 10−5 to 1.8 × 10−14 according to one-sample t-tests). These putative mutations affected just a few adjacent overlapping probes and produced modest signals comparable to those normally observed for heterozygous deletions. We hypothesized that very small homozygous mutations (much shorter than the length of a probe) could produce signals of this magnitude. The mutations would have to be very small to target only a few overlapping probes and permit some hybridization of complementary sequence to the array. Mutations of this size would not have produced statistically significant signals on our whole-genome tiling arrays because each mutation would affect only one or two probes.
Our hypothesis was confirmed when PCR and DNA sequencing identified single nucleotide mutations in all three mutants. The strain VC10078 carries gk802, an A → T transversion allele of syd-1 at II: 7586645 (see Figure 1), causing a nonconservative amino acid substitution [I(887) → K]; VC10079 contains allele gk803, an A → G transition at nucleotide II: 10825740, which results in a synonymous base-pair substitution in mix-1 at the third position of a codon for leucine (CUA → CUG); and VC10077 carries gk801, an allele with two closely linked mutations in Y46E12BL.2: a G → A transition at II: 15240024, causing a conservative amino acid substitution [V(714) → I], and an A → G transition at II: 15240052, resulting in a nonconservative amino acid substitution [Y(723) → C].
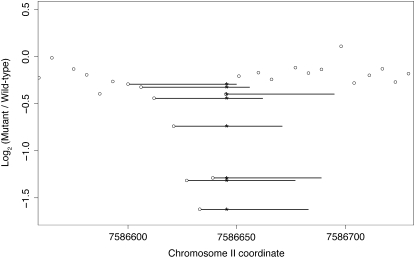
Figure 1.—
Novel detection of an A → T transversion in syd-1. Normalized log2 ratios of fluorescence intensities (mutant/wild type) are plotted as open circles at the first base of each 50-mer probe. The length of each probe targeted by the SNP is illustrated by a horizontal bar, and the position of the SNP is indicated by an asterisk. Multiple adjacent overlapping probes targeted the point mutation, so its effect on hybridization was assayed several times. Aberrant fluorescence ratios at probes targeting the SNP stand out from nearby probes targeting the wild-type sequence. Nematodes were grown on NGM agar plates spread with a lawn of Escherichia coli strain OP50 or χ1666. A mixed-stage population of VC1415 {unc-4(e120)/mIn1[mIs14 dpy-10(e128)] II} was subjected to mutagenesis with TMP at 10 μg/ml for 1 hr followed by UV irradiation for 90 sec at 340 μW/cm2 and then placed on food at 20°. Both unc-4 and dpy-10 mutations are recessive, and the mIn1 inversion suppresses recombination along the middle of chromosome II from lin-31 to rol-1 (Edgley and Riddle 2001); the mIs14 element confers a semidominant GFP signal confined to the pharyngeal muscle. After 48 hr, 30 gravid wild-type GFP+ P0 adults were singly picked onto 60-mm petri plates and allowed to self. Seven wild-type GFP+ F1 progeny were singly picked from each parent for a total of 210 clones, from which 100 were selected that segregated viable fertile Unc-4 F2 progeny. Single gravid Unc-4 progeny were picked from each of these plates and used to establish 100 new populations homozygous for unc-4 and any newly induced mutations within the genetic interval balanced by mIn1. Nematode populations were grown to starvation on three 60-mm petri plates, harvested by washing, centrifugation, and aspiration of the supernatant, and frozen at −80° in 2.5 vol of worm lysis buffer [50 mm KCl; 10 mm Tris–HCl, pH 8.3; 2.5 mm MgCl2; 0.45% NP-40 (Igepal); 0.45% Tween-20; 0.01% gelatin; 300 μg/ml Proteinase K]. Crude lysates were prepared from frozen samples by incubation at 65° for 2 hr. Genomic DNA was prepared from the lysates as described previously by Maydan et al. (2007). The filters used to select the 50-mer oligonucleotides for the exon-centric chromosome II chip have been described by Maydan et al. (2007). DNA sample handling, labeling with Cy3 (mutants and CB4856) or Cy5 [wild-type N2 (VC196) reference], hybridization, and imaging were performed by NimbleGen (Selzer et al. 2005). Extraction of fluorescence intensities and data normalization were performed as previously described (Maydan et al. 2007). Many experiments were performed using the same chromosome II array design, which allowed an approximate determination (by simply averaging) and subsequent subtraction of local bias in the log2 ratio signal for individual experiments. The signature of a SNP in the log2 ratio signal is similar to that of a deletion in CGH except that the log2 ratio shows only a modest reduction for the affected probes, and of course only a few probes are affected.
Dense tiling with oligonucleotides is necessary to obtain sufficient statistical power to detect single nucleotide alterations. In a previous study (Flibotte et al. 2009) we have shown that a window of ∼20 bases contains a strong log2 ratio signal (see Figure 1 in Flibotte et al. 2009), and since we require about four probes to target the mutated site, this allows a maximum probe spacing of ∼5 bases. The plot in Flibotte's figure also shows that it would be useful to target both strands and use the small shift in the peak position on opposite strands to help distinguish single nucleotide polymorphisms (SNPs) from artifacts. Utilizing these probe spacing guidelines, we conducted an additional 13 aCGH experiments comparing homozygous mutants to their parental strains, using 50-mer oligonucleotide microarrays probing regions from 0.65 to 2.60 Mb in length that are known to include unidentified mutations based on prior mapping experiments. The probe spacing, i.e., the distance between the 5′-ends of consecutive probes, on these arrays ranged from 1 to 5 bp, and all known repeats were excluded from the array designs. Unlike our previous exon-centric arrays, no other constraints were applied to the oligonucleotides. Note that, while probes for both strands are desirable, we were not able to include them for the majority of the 13 experiments, because the interval to be tested was too large to allow probes for both strands. All microarrays were manufactured by Roche NimbleGen with oligonucleotides synthesized at random positions on the arrays. Mutant strains were generated by standard ethyl methanesulfonate (EMS) mutagenesis, which yields approximately one single nucleotide mutation every 100–400 kb (Anderson 1995; Cuppen et al. 2007), and then were serially backcrossed with their parental strains.
From these experiments we selected 58 candidate single nucleotide mutations on the basis of a visual inspection of the data and identification using a segmentation algorithm (Maydan et al. 2007) or a sliding-window technique. We then performed PCR and DNA sequencing to gauge the accuracy of our mutation predictions. For each candidate mutation, we calculated a SNP score by averaging the log2 fluorescence ratios (mutant/wild-type) in a small window containing probes putatively affected by the mutation and renormalizing by subtracting from that the average log2 ratio in the immediate flanking regions. This renormalization is necessary to account for local bias, which varies both among and within experiments and makes the detection of SNPs more difficult since artifacts associated with a strong local bias in the log2 ratio could easily be confused with the signature expected for a SNP. Unlike previous observations that mutations near the centers of 25-mer probes are most inhibitory to efficient hybridization (Sharp et al. 2007), we observed that mutations located away from the glass slide and freely floating in the solution closer to the 5′-ends of our 50-mer probes produced a larger perturbation to the hybridization process, with a maximum perturbation at 7 bases in from the 5′-end (probably due to steric effects; again see Figure 1 in Flibotte et al. 2009). The location of the window used to calculate the score reflects this observation. This sensitivity to mutations at the 5′-end of NimbleGen probes has also been observed by Wei et al. (2008). The sequencing results (summarized in Figure 2A) confirmed the presence of a single nucleotide mutation in 16 of the candidates for an overall success rate or specificity of 28%. All mutations were either C-to-T or G-to-A transitions, as expected from EMS mutagenesis. The locations of the mutations were usually predicted to within <10 bp of their true positions and to within 1 bp in one case.
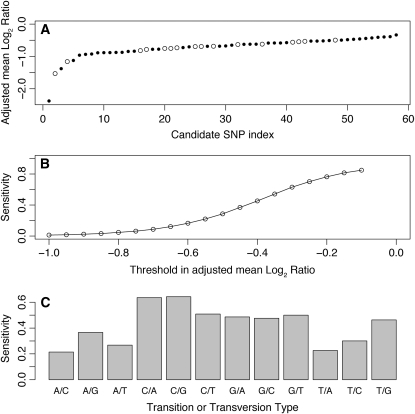
Figure 2.—
Estimation of the sensitivity and specificity of the current SNP detection technique. The SNP score, or adjusted mean log2 ratio, corresponds to the average log2 ratio of the probes where the mutation is located in a window 13 bases wide near the 5′-end of the oligonucleotide, which is away from the slide and therefore freely floating in the solution, and is renormalized by subtracting the mean of the log2 ratio in the immediate left and right 50-base-wide flanking regions for oligonucleotides not overlapping the mutation. (A) The SNP score is shown for the candidate SNPs that we have sequenced with the candidates ordered according to their score. The open and solid circles represent the candidates confirmed and not confirmed by sequencing, respectively. For example, a score < −0.45 would include all 16 confirmed cases and 36 nonconfirmed candidates, corresponding to a specificity of 31%. Microarrays were designed by tiling the target regions with equally spaced overlapping 50-mer oligonucleotides without any filtering except for the elimination of the repeats listed in WormBase. The earlier arrays were designed using the WormBase data freeze version WS170 while the most recent designs used version WS180. A single 380,000-oligonucleotide array was designed for each region of interest except for one experiment in which two arrays have been used to cover a genomic region 4.9 Mb wide. Mutation candidates were selected by analyzing the CGH data by visual inspection and use of a segmentation algorithm (Maydan et al. 2007) or a sliding-window technique. PCR was used to amplify products of a few hundred base pairs surrounding the candidate regions. DNA sequencing of these products precisely identified each mutation. (B) The detection sensitivity for the SNPs in the CB4856 (Hawaiian) experiments is shown as a function of the threshold in the SNP score. Using a threshold of −0.45 as before would correspond to a sensitivity of 37%. (C) The sensitivity is shown separately for each transition and transversion type when using the same threshold of −0.45. The natural isolate CB4856 and all mutant strains were prepared from isogenic cultures of worms. Nematode populations were grown to starvation on three 60-mm petri plates. DNA preparation, CGH, and other array data analyses were performed as described in Figure 1. From all the CB4856 SNPs present in WormBase data freeze WS170, we selected 2639 that were far enough from all the known mutations in that strain to minimize the presence of mutations in the immediate flanking regions of the selected SNPs. Once again the only filter used in the design process was to eliminate the known repeats. Each SNP was represented on the array by a maximum of 150 50-mer oligonucleotides spaced 1 bp apart, ≤50 oligonucleotides affected by the mutation, and ≤50 oligonucleotides for each immediate left and right flanking region. For each SNP, the set of probes alternated between the sequence from the plus and minus strand templates; thus, for a given strand, the minimum spacing between probes was equal to 2 bases. For this experiment, we performed dye-flip hybridizations to evaluate the Cy3/Cy5 bias. In that experiment, each SNP log2 ratio profile was measured four times, with two separate hybridizations and on both strands each time. When calculating the SNP detection sensitivity, each of the four profiles was considered as a separate measurement since each profile is associated with an oligonucleotide spacing of 2 bp, which is more representative of the SNP detection experiments that we used to evaluate the specificity of the technique. We could have averaged those four profiles to reduce the standard deviation before calculating the sensitivity, but this would not have allowed a direct and meaningful comparison with the data from our SNP detection experiments.
To estimate the sensitivity of our single nucleotide mutation detection technique, we performed aCGH experiments to test our ability to detect 2639 known SNPs in the CB4856 strain isolated in Hawaii (see Figure 2 legend for details of the array design). Examples of all possible transitions and transversions were detected. The SNP detection sensitivity is shown in Figure 2B for various thresholds in the SNP score described above. At the reasonable threshold of −0.45, the specificity (the percentage of predicted SNPs that are real) would be 31% with a sensitivity (the percentage of real SNPs that are successfully detected) of 37%. In other words, with the current SNP detection technique we could expect to detect roughly one of every three SNPs present in the targeted region and have to sequence roughly three candidates to detect a real SNP. As expected, the SNP detection sensitivity of the current technique depends on the type of transition or transversion being investigated, and, as can be seen in Figure 2C, the sensitivity reaches ∼50% for the most commonly induced EMS-generated mutations (C to T and G to A).
The optimal probe length for single nucleotide mutation detection by CGH is unclear and likely depends on the hybridization conditions. Single nucleotide mutations should have a greater impact on hybridization to shorter oligonucleotides, but longer oligonucleotides allow a greater number of overlapping probes to target a given single nucleotide mutation, and arrays with longer oligonucleotides tend to have better standard deviations in log2 ratios (Sharp et al. 2007). Further experiments are needed to determine the optimal probe length to achieve the greatest sensitivity and specificity as a function of the size of the targeted region; such an optimal length will probably vary with the complexity of the genome being studied.
Although this technique is particularly well suited to detecting SNPs generated by EMS mutagenesis, some single nucleotide mutations may not be detectable by aCGH even with higher probe densities than we have used here. We suspected that some of the Hawaiian SNPs that we failed to detect might have been missed because they were found in regions with significant homology to other regions of the genome. In these cases, multiple regions of the genome could have hybridized to our probes, making it difficult for the effect of a SNP on the log2 ratios to be detectable. However, filtering the oligonucleotide properties according to our best practices and standard microarray design recommendations (Flibotte and Moerman 2008) failed to improve the SNP detection sensitivity, which makes this possibility unlikely. It is also possible that SNPs are more difficult to detect with aCGH when present in the background of the Hawaiian genome because this genome has significant structural variation relative to the N2 reference genome (Maydan et al. 2007); consequently, for a more typical SNP detection experiment the sensitivity of the technique might be slightly better than what we have reported here. However, limiting the analysis to SNPs that are located far away from other known polymorphisms did not improve the SNP detection sensitivity, which makes this possible source of interference also unlikely. Finally, we have not yet attempted to detect heterozygous single nucleotide mutations using this technique, but this would be nearly impossible to accomplish with current microarrays.
The ability of aCGH to detect homozygous single nucleotide mutations in addition to deletions and duplications makes it possible to quickly and affordably identify mutations mapped by traditional positional cloning approaches. A clear example of the feasibility of this technique is demonstrated in an accompanying article in this issue, O'Meara et al. (2009), where two single base lesions were mapped to the promoter of the gene cog-1 using aCGH. We recommend a maximum probe spacing of ≤5 bp to have a reasonable chance at successful SNP detection with this technique, This probe spacing corresponds to ∼2 Mbp of genomic sequence on a microarray with 380,000 probes, the oligo capacity of the chips that we used in this study. We prefer to apply this SNP detection technique only in situations where the mutation is mapped to a maximum of a 1-Mbp region, as this provides denser coverage of the mutation site and allows us to target both strands. Targeting both strands should result in fewer false positives. Further reducing the size of the candidate region should improve the likelihood of successful base-change detection as more probes target any specific base. If any sequences in the mapped region can be excluded (such as noncoding DNA, repeat elements, or genes that can be ruled out as candidate genes), the probe density can be further increased in specific regions of interest. It is of course possible to use more than one microarray to probe the candidate region if the region is too large to achieve the desired probe density on a single array. Also, when the search region is small enough to allow very high density tiling, one can take advantage of the fact that the effect of a SNP on hybridization is dependent on its position in the probe by including probes that target both strands and then by pursuing primarily candidates showing a small shift between the plus and minus strand log2 ratio profiles.
To make the current SNP detection technique more accessible, we have mounted a web application to design oligonucleotide microarrays. The application can be found at http://hokkaido.bcgsc.ca/SNPdetection/. Downstream analysis tools to calculate and normalize the log2 ratios are also available on the same web site. Given the criteria set by the user, such as the probe target region and strand(s), the oligonucleotides are selected in a way to evenly distribute the probes across the selected region. The placement of these probes are selected to avoid repeat regions, noncoding regions (optional), and specific probe sequences that cannot be synthesized due to the cycle number constraint in NimbleGen's manufacturing process. Once the criteria have been selected the file is sent to the user in a format ready for submission to NimbleGen. We recommend that users start with the constraints that we describe in this article. Currently, the probe selection application has been set to support the C. elegans and Drosophila melanogaster genomes, but genomes from other species will be added upon request.
With the advent of whole-genome sequencing using new high-throughput sequencing machines (Hillier et al. 2008; Sarin et al. 2008) it might be asked whether SNP detection on microarrays is a reasonable technique for mutation detection. Deep sequencing is certainly a powerful method, but for now our method is easier to perform, as we have provided the web site for oligo design and data analysis. Mapping short reads and calling variants is still challenging using deep sequencing, but programs are coming online to make this much easier (see, for example, MAQ in Li et al. 2008). A CGH experiment can be done rapidly and involves less labor and, if desired, DNA labeling and hybridization can be outsourced to NimbleGen. This advantage will certainly be short lived as more and more sequencing machines become available and their use more transparent. Our CGH method is also less expensive, but this situation too will no doubt change in the future as deep sequencing becomes commonplace. At present it is difficult to compare the two methods for accuracy of mutation detection. We have measured a false-positive and false-negative rate for CGH in this article, but at present there is no comparable measure for deep sequencing. We suspect that with several short reads across an interval containing a mutation and with improvements in alignment programs such as MAQ that deep sequencing will become highly accurate. With either method one cannot avoid genetic mapping. For our SNP detection method one needs to do initial mapping to limit the mutation of interest to a small region of the genome. For deep sequencing one can sequence first, but one then has to determine which of several hundred changes in the genome is the causative change (Hillier et al. 2008 and our unpublished results). A more effective approach using deep sequencing is illustrated in Sarin et al. (2008) where the gene of interest was first mapped to a 4-Mb interval.
Acknowledgments
We thank Bin Shen, Owen Dadivas, and Sarah Neil for able technical assistance in PCR of candidate mutations and preparation of PCR products for sequencing. We thank Don Riddle, Harald Hutter, Michel Leroux, Nancy Hawkins, and Ralf Schnabel for graciously providing several C. elegans strains carrying mutations previously mapped to the intervals targeted by our arrays. The Hawaiian strain CB4856 was obtained from the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health National Center for Research Resources. This work was supported by grants from Genome Canada, Genome British Columbia, and the Michael Smith Research Foundation to D.G.M. and S.F.
References
- Anderson, P., 1995. Mutagenesis. Methods Cell Biol. 48 31–58. [PubMed] [Google Scholar]
- Barnes, T. M., Y. Kohara, A. Coulson and S. Hekimi, 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppen, E., E. Gort, E. Hazendonk, J. Mudde, J. van de Belt et al., 2007. Efficient target-selected mutagenesis in Caenorhabditis elegans: toward a knockout for every gene. Genome Res. 17 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley, M. L., and D. L. Riddle, 2001. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics 266 385–395. [DOI] [PubMed] [Google Scholar]
- Flibotte, S., and D. G. Moerman, 2008. Experimental analysis of oligonucleotide microarray design criteria to detect deletions by comparative genomic hybridization. BMC Genomics 9 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flibotte, S., M. L. Edgley, J. Maydan, J. Taylor, R. Zapf et al., 2009. Rapid high resolution single nucleotide polymorphism-comparative genome hybridization mapping in Caenorhabditis elegans. Genetics 181 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham, D., D. M. Ruderfer, S. C. Pratt, J. Schacherer, M. J. Dunham et al., 2006. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311 1932–1936. [DOI] [PubMed] [Google Scholar]
- Gresham, D., M. J. Dunham and D. Botstein, 2008. Comparing whole genomes using DNA microarrays. Nat. Rev. Genet. 9 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier, L. W., G. T. Marth, A. R. Quinlan, D. Dooling, G. Fewell et al., 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5 183–188. [DOI] [PubMed] [Google Scholar]
- Li, H., J. Ruan and R. Durbin, 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan, J. S., S. Flibotte, M. L. Edgley, J. Lau, R. R. Selzer et al., 2007. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 17 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara, M. M., H. Bigelow, S. Flibotte, J. F. Etchberger, D. G. Moerman et al., 2009. Cis-regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog-1 affect neuronal development. Genetics 181 1679–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin, S., S. Prabhu, M. M. O'Meara, I. Pe'er and O. Hobert, 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer, R. R., T. A. Richmond, N. J. Pofahl, R. D. Green, P. S. Eis et al., 2005. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 44 305–319. [DOI] [PubMed] [Google Scholar]
- Sharp, A. J., A. Itsara, Z. Cheng, C. Alkan, S. Schwartz et al., 2007. Optimal design of oligonucleotide microarrays for measurement of DNA copy-number. Hum. Mol. Genet. 16 2770–2779. [DOI] [PubMed] [Google Scholar]
- Wei, H., P. F. Kuan, S. Tian, C. Yang, J. Nie et al., 2008. A study of the relationships between oligonucleotide properties and hybridization signal intensities from NimbleGen microarray datasets. Nucleic Acids Res. 36 2926–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]