Abstract
We apply here comparative genome hybridization as a novel tool to identify the molecular lesion in two Caenorhabditis elegans mutant strains that affect a neuronal cell fate decision. The phenotype of the mutant strains resembles those of the loss-of-function alleles of the cog-1 homeobox gene, an inducer of the fate of the gustatory neuron ASER. We find that both lesions map to the cis-regulatory control region of cog-1 and affect a phylogenetically conserved binding site for the C2H2 zinc-finger transcription factor CHE-1, a previously known regulator of cog-1 expression in ASER. Identification of this CHE-1-binding site as a critical regulator of cog-1 expression in the ASER in vivo represents one of the rare demonstrations of the in vivo relevance of an experimentally determined or predicted transcription-factor-binding site. Aside from the mutationally defined CHE-1-binding site, cog-1 contains a second, functional CHE-1-binding site, which in isolation is sufficient to drive reporter gene expression in the ASER but in an in vivo context is apparently insufficient for promoting appropriate ASER expression. The cis-regulatory control regions of other ASE-expressed genes also contain ASE motifs that can promote ASE neuron expression when isolated from their genomic context, but appear to depend on multiple ASE motifs in their normal genomic context. The multiplicity of cis-regulatory elements may ensure the robustness of gene expression.
GENE regulatory information is hardwired into genomic DNA in the form of cis-regulatory control regions that are recognized by specific trans-acting factors (Davidson 2001; Hobert 2008a). To understand developmental processes, it is of paramount importance to decode such regulatory information. A variety of different approaches, including reporter gene assays, chromatin immunoprecipitation, and bioinformatic approaches, have identified a large number of cis-regulatory control modules embedded in the genome of metazoan organisms (Davidson 2001). However, in the vast majority of cases the importance of defined transcription-factor-binding sites has not been verified by the strict genetic criteria of assessing the phenotypic consequence of a mutation in a cis-regulatory element in its normal chromosomal and organismal context. In addition to the tedious reverse engineering of cis-regulatory mutations in metazoans, classic forward genetic mutant screens are a potential source of mutations that disrupt cis-regulatory elements. Even though such screens have been amply conducted in the nematode Caenorhabditis elegans, few cis-regulatory point mutations that disrupt defined transcription-factor-binding sites and result in an experimentally verified gene expression defect have been described in C. elegans (Conradt and Horvitz 1999; Sarin et al. 2007). Apparent reasons for the paucity of mutational validation of regulatory regions are the following: first, reverse engineering of mutations in the genomes is difficult; second, transcription-factor-binding sites tend to be quite degenerate, making their disruption by a single point mutation through a standard, nondirected chemical mutagenesis protocol a relatively rare event; and third, if nondirected chemical mutagenesis is employed, the resulting point mutations are hard to localize because cis-regulatory elements can localize at a great distance from the locus whose expression is controlled by the cis-regulatory element. This “needle-in-a-haystack” problem means that mutant alleles of a given locus that do not alter protein-coding regions are often not pursued further.
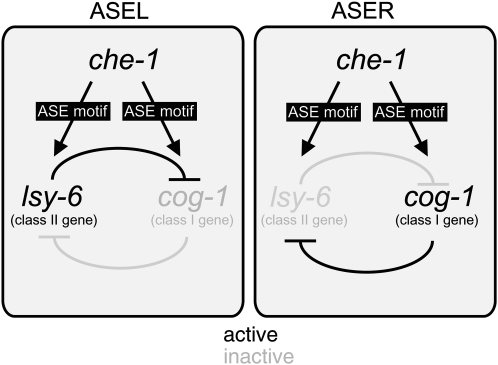
We describe in this article cis-regulatory alleles of the homeobox gene cog-1. The cog-1 gene, the C. elegans ortholog of vertebrate GTX/Nkx6.1 (Palmer et al. 2002), is involved in a specific neuronal cell fate decision in the nervous system of C. elegans (Chang et al. 2003). In wild-type animals, the bilaterally symmetric pair of ASE sensory neurons is specified by the zinc-finger transcription factor CHE-1 (Chang et al. 2003; Uchida et al. 2003). CHE-1 controls the expression of genes that are expressed in the left and right ASE neurons, including a specific subset of regulatory genes that are required to make ASEL and ASER express a distinct set of putative chemoreceptor genes encoded by the gcy gene family (Chang et al. 2003; Etchberger et al. 2007) (Figure 1). These regulatory che-1 target genes fall into two classes, class I and class II genes. Class I genes promote ASER fate (Figure 1). Hence, mutations in these genes, termed class I laterally symmetric (lsy) mutants result in a 2 ASEL phenotype. Class II genes promote ASEL fate and, hence, class II lsy mutants display a 2 ASER phenotype (Figure 1). Class I and class II genes inhibit each other's expression in a double-negative feedback loop (Johnston et al. 2005; Hobert 2006) (Figure 1). cog-1 is a class I regulatory gene that is expressed in ASER where it is required to induce ASER fate (Chang et al. 2003). As inferred by 18 alleles that affect the protein-coding region of cog-1, loss of cog-1 results in a loss of ASER fate and aberrant execution of ASEL fate in ASER (Chang et al. 2003; Sarin et al. 2007). cog-1 expression in the ASE neurons genetically depends on the zinc-finger transcription factor che-1 (Chang et al. 2003). cog-1 expression is restricted to ASER by the action of the microRNA (miRNA) lsy-6, a class II regulatory gene, which downregulates cog-1 expression in ASEL (Johnston and Hobert 2003).
Figure 1.—
Overview of the system. ASEL/R fate is controlled by a bistable feedback loop, which contains the ASEL fate inducer lsy-6, a miRNA, and the ASER fate inducer cog-1, a homeobox gene (Johnston et al. 2005). Even though both genes are asymmetrically expressed in either ASEL (lsy-6) or ASER (cog-1), both genes contain ASE motifs in their cis-regulatory regions, which are activated by the zinc-finger transcription factor CHE-1 (Etchberger et al. 2007). Cis-regulatory mutations were isolated from genetic screens in the ASE motif of lsy-6 (Sarin et al. 2007) and cog-1 (this article).
Our previous screens for ASE fate mutants independently isolated two recessive mutant alleles, ot119 and ot201, which display the same phenotype as recessive, loss-of-function cog-1 alleles; that is, the ASER neuron fails to appropriately express ASER fate markers and ectopically expresses ASEL fate (Sarin et al. 2007). Several lines of evidence suggested that these two alleles are cog-1 alleles: first, through SNP mapping the alleles were found to map in the same genetic interval as cog-1 (Sarin et al. 2007); second, they fail to complement the class I Lsy phenotype of a canonical cog-1 allele (Table 1); and third, the mutant phenotype can be rescued by an ∼41-kb genomic region (fosmid WRM067cF11) that contains the cog-1 gene and several neighboring genes (Sarin et al. 2007). However, sequencing of the cog-1 coding sequences, 5′- and 3′-UTRs, and all introns revealed no molecular lesion in animals harboring the ot119 or ot201 allele. In contrast, all 18 recessive cog-1 alleles that we have retrieved affect either protein-coding regions or splice junctions (Sarin et al. 2007). Therefore, it remained unclear if and how the ot119 and ot201 alleles affect cog-1 function.
TABLE 1.
ot119 and ot201 specifically affect cog-1 function in the ASER neuron but not in other cell types
| Canonical cog-1 mutant phenotypes
|
||||
|---|---|---|---|---|
| Genotype | % animals with ASER defect (class I Lsy phenotype = ectopic lim-6∷gfp expression) | % animals with VulB2 defect (loss of ceh-2∷gfp expression) | % animals with Egl defecta | % animals with Pvl defectb |
| Wild type | 0 (n > 100) | 0 (n = 58) | 0 (n = 30) | 0 (n = 30) |
| ot119 | 63 (n = 36)c | 0 (n = 24) | 0 (n = 30) | 0 (n = 30) |
| ot201 | 89 (n = 54)c | 0 (n = 27) | 0 (n = 30) | 0 (n = 30) |
| sy607d | 100 (n = 35)c | 90 (n = 20)e | 100 (n = 30)f | 87 (n = 30)f |
| ot119/sy607 | 90 (n = 20) | 11 (n = 19) | 0 (n = 30) | 0 (n = 30) |
| ot201/sy607 | 35 (n = 24) | 3 (n = 32) | 0 (n = 30) | 0 (n = 30) |
Markers used are otIs114 (lim-6∷gfp) (Chang et al. 2003) and syIs54 (ceh-2∷gfp) (Inoue et al. 2005).
Defined as animals with reduced brood size, including the bag-of-worms phenotype.
Pvl, protruding vulva phenotype.
This phenotype has already been described in Sarin et al. (2007); animals have been newly scored with similar results.
sy607 is a strong loss-of-function or null allele of cog-1 that affects its coding region (Palmer et al. 2002).
Similar to results reported by Inoue et al. (2005).
Similar to results reported by Palmer et al. (2002).
ot119 and ot201 are cis-regulatory alleles of the cog-1 locus:
Rather than manually sequencing the entire ∼40-kb fosmid that rescues the ot119 and ot201 phenotype, we utilized an alternative technique, comparative genome hybridization (CGH). CGH serves to detect sequence variations between two differentially labeled DNA samples that are hybridized to a microarray (Kallioniemi et al. 1992). To achieve high resolution, the microarray can be designed to contain densely spaced oligonucleotides (oligonucleotide array comparative genome hybridization, or aCGH). aCGH has been used successfully to detect chemically induced variations between different C. elegans genomes as well as natural variations in gene number between different C. elegans isolates (Jones et al. 2007; Maydan et al. 2007). For example, using an array that probed for protein-coding exons, the technique has been used to identify gene deletions and to map chromosomal deficiencies (Jones et al. 2007; Maydan et al. 2007). In an accompanying article in this issue, Maydan et al. (2009) describe that this method can be extended to identify single nucleotide alterations. We use CGH as a cost-effective alternative method to manual DNA sequencing, whose implementation is made easy through the ability to outsource the microarray synthesis and hybridization to NimbleGen and the use of software described in Maydan et al. (2009).
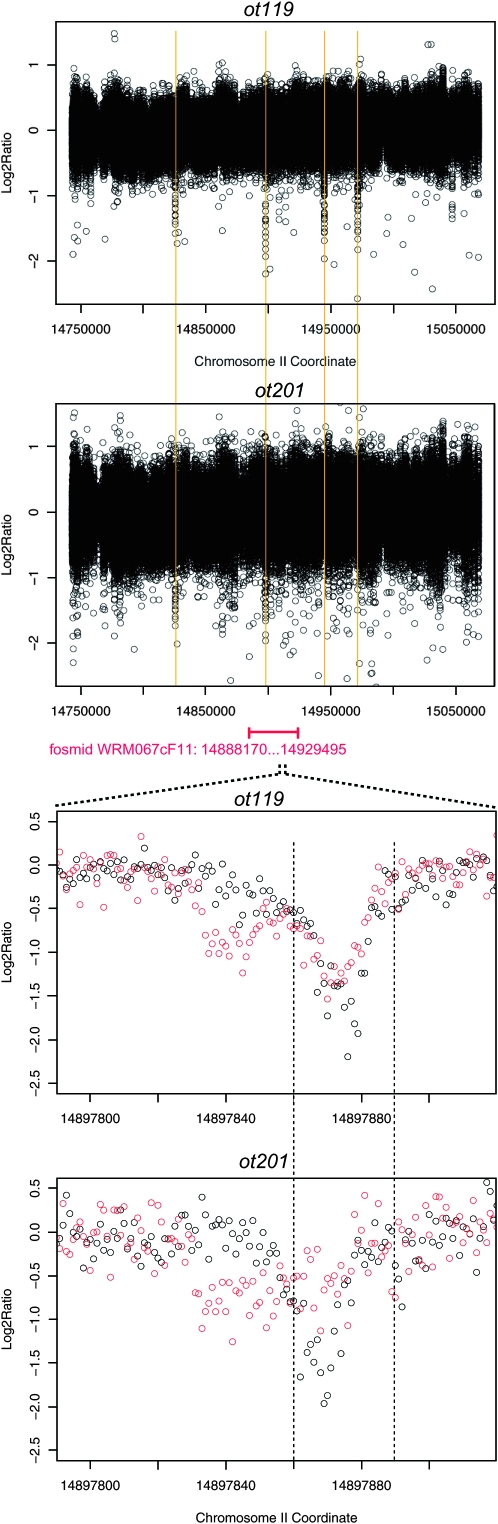
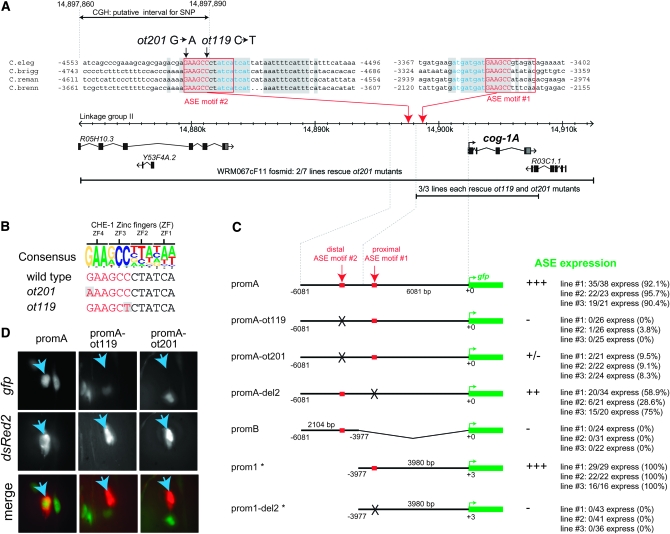
Using an automated oligonucleotide design program (see accompanying article by Maydan et al. 2009), we designed an oligonucleotide array containing 379,690 50-mer oligos to identify by aCGH the molecular lesions in the independently isolated ot119 and ot201 alleles. These oligos entirely tile the region between coordinates 14,743,042 and 15,068,429 on chromosome II on the plus and minus strand, with an oligo spacing of one base. This ∼352-kb region encompasses the ∼41-kb genomic interval (14,888,170–14,929,495) in the fosmid WRM067cF11 that rescues the ot119 and ot201 mutant phenotypes. DNA isolated from ot119 and ot201 and a wild-type reference were differentially labeled and hybridized to the array (as described in more detail in Maydan et al. 2009). Given the similar genetic behavior of ot119 and ot201, we focused on variants that are present at roughly the same location in both data sets and, as a first pass, focused on the genomic region covered by the fosmid that rescues the ot119 and ot201 defects (Figure 2). One set of candidate variants fulfills these criteria (Figure 2; bottom panels). We manually sequenced this region using standard Sanger sequencing and identified two closely clustered mutations in ot119 and ot201 animals (Figure 3A). An alignment of this genomic region from four related nematode species reveals that both mutations lie within a 17-bp sequence window that is 100% conserved in all four species (shading in Figure 3A). This region harbors a good match to the so-called ASE motif (Figure 3C), a predicted binding site for the CHE-1 zinc-finger transcription factor (Etchberger et al. 2007). CHE-1 is genetically required for expression of cog-1 in the ASE neurons (Chang et al. 2003). Invariant core sequences of the ASE motif that are predicted to bind to zinc fingers 3 and 4 of CHE-1, respectively (Etchberger et al. 2007), are affected in ot119 and ot201.
Figure 2.—
aCGH primary data. For each individual 50-mer probe, the normalized log2 (sample fluorescence intensity/reference fluorescence intensity) is plotted at a chromosomal coordinate corresponding to the end of the oligonucleotide with the smallest coordinate, i.e., the 5′-end for probes on the plus strand and the 3′-end for probes on the minus strand. (Top two panels) The log2 ratio for the whole region represented on the microarray but only for probes following the plus strand template. The vertical yellow lines correspond to candidate SNPs in ot119. (Bottom two panels) A small interval around the most promising candidates. The black and red circles correspond to probes designed to follow, respectively, the plus and minus strand template. The shift between the position of the minima for the plus and minus strand oligonucleotides is expected and is due to fact that on the NimbleGen platform a SNP induces a larger perturbation effect on the hybridization process when it is located close to the protruding end freely floating in the solution. The vertical dashed lines are provided to guide the eye and are reproduced in Figure 3A. More details can be found in the supplemental Materials and Methods.
Figure 3.—
Location of ot119 and ot201 and their effect on reporter gene expression. (A) Genomic region containing the cog-1 locus. Coordinates refer to base pairs on linkage group II. See Figure 2 for explanation of the stippled interval. Conserved ASE motifs are highlighted and numbers next to the sequence indicate positions relative to the ATG start codon of the longer cog-1 isoform. The nucleotides in blue are putative transcription-factor-binding sites linked to the ASE motifs; they occur in opposite orientation and differ in relative location to each ASE motif. Shaded boxes indicate 100% conservation between all species. Black lines indicate DNA injected into the respective mutant strain to test for rescue of the ASE mutant phenotype. (B) Alignment of the cog-1 ASE motif and its mutated versions in ot109 and ot201 animals to the ASE consensus motif. “ZF” indicate the zinc fingers of CHE-1 with which it contacts its cognate binding sequence (Etchberger et al. 2007). (C) Reporter constructs. “ASE expression” indicates expression in at least one (ASER) or two ASE cells expressing gfp; note that the apparent left/right asymmetric expression of this reporter gene is brought about by transcriptional autoregulation of the translationally controlled COG-1 protein (Johnston et al. 2005). Expression was scored in young adults in a otIs151 transgene background to allow identification of the ASE neurons. In the one case in which an intermediate level of penetrance was observed (promA-del2), the brightness of the gfp signal seemed to vary in those animals where expression is observed in ASE, compared to the wild-type construct where little of such variance was observed. More details on constructs can be found in the supplemental Materials and Methods. Constructs with an * have been described in Etchberger et al. (2007) and are shown for comparison only. (D) gfp images of three animals, each expressing the indicated cog-1 reporter gene fusion and a chromosomally integrated transgene, ceh-36∷dsRed2 (otIs151), used to label ASER. Images of the green and red channel of the same animal in the same position are merged in the last set of panels. Blue arrows indicate ASER.
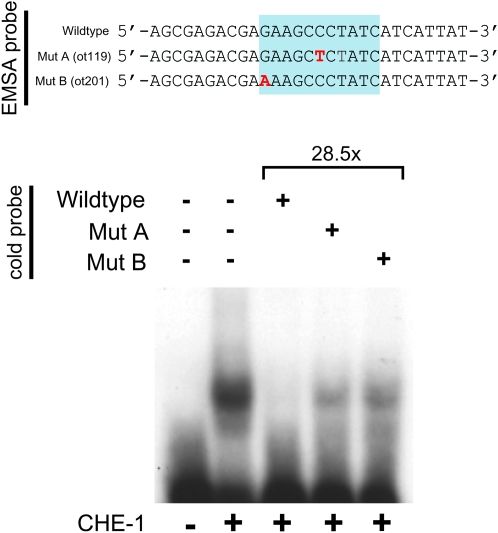
We first corroborated that the ASE motif affected in ot119 and ot201 mutants is indeed a binding site for CHE-1 in vitro using electrophoretic mobility shift assay with bacterially produced CHE-1 protein. We find that CHE-1 indeed binds this ASE motif in vitro (Figure 4). Moreover, both ot119 and ot201 mutations significantly reduce CHE-1 binding to the ASE motif in vitro (Figure 4), a notion consistent with the invariant nature of the bases affected by ot119 and ot201. To test whether the ASE motif is also required for cog-1 expression in vivo, we generated a series of gfp reporter constructs that monitor cis-regulatory control elements in the cog-1 locus. A fusion of 6 kb of sequences upstream of the cog-1 start codon to gfp shows expression in the sites previously reported to express cog-1, namely vulval cells and head neurons, including ASER (Palmer et al. 2002; Chang et al. 2003). Introducing the ot119 and ot201 mutations into this reporter gene construct results in a loss of gfp expression in the ASER neurons (Figure 3, C and D). This effect is restricted to ASER, consistent with the ot119 and ot201 alleles affecting the binding of the ASE-neuron-specific transcription factor CHE-1. Also consistent with ot119 and ot201 affecting only cog-1 expression in ASE, ot119 and ot201 mutant animals display none of the pleiotropies associated with a complete loss of cog-1 gene function. That is, ot119 and ot201 animals do not display egg-laying defects or obvious defects in vulval morphology (i.e., no Pvl or Cog phenotype) and do not affect expression of the vulval VulB2 cell fate marker ceh-2∷gfp, which is lost in canonical cog-1 mutant strains (Table 1). Moreover, ot119 and ot201 complement the Egl and Pvl phenotype of the severe cog-1 allele sy607 but do not complement the ASE (Lsy) phenotype of sy607 (Table 1). We conclude that ot119 and ot201 specifically affect the che-1-induced expression of the ASER inducer cog-1, resulting in a loss of ASER fate.
Figure 4.—
CHE-1 binds to the ASE motif of the wild-type cog-1 locus but not to the mutated ASE motif found in ot119 or ot201 animals. Gel shifts were done with bacterially purified CHE-1 protein as previously described (Etchberger et al. 2007). Binding to the mutated versions was assessed by cold-competition assays, using an ∼30× excess of cold wild-type or mutated probe.
ot119 and ot201 reveal an unanticipated feature in the regulation of the cog-1 locus. Upon the initial identification and description of the ASE motif, present in a large battery of ASE-expressed genes, we noted an ASE motif upstream of cog-1 (ASE motif 1 in Figure 3A), which we found to be both required and sufficient to drive expression of a cog-1 reporter gene in ASE (Figure 3C) (Etchberger et al. 2007). However, the ot119 and ot201 alleles identify another previously unstudied and more distally located ASE motif (ASE motif 2 in Figure 3A) that apparently is critical for in vivo expression of cog-1. The importance of the distal ASE motif 2 is counterintuitive for two reasons. First, as mentioned above, a 4-kb proximal regulatory element that contains the proximal ASE motif 1, but not motif 2, is sufficient to drive reporter gene expression in ASE (Figure 3C, prom1) (Etchberger et al. 2007). Second, a genomic piece that contains the cog-1 locus and the 4-kb proximal regulatory element that contains ASE motif 1, but not ASE motif 2, is able to rescue the mutant phenotype of ot119 and ot201 animals, in which motif 2 is mutated (black line in Figure 3A). Third, in contrast to the 4-kb region containing ASE motif 1 (prom1), a 2-kb genomic region containing the distal ASE motif 2, identified through the ot119 and ot201 alleles, is not sufficient to drive reporter gene expression (promB in Figure 3C). However, the importance of the distal ASE motif 2 becomes obvious in the context of the above-mentioned reporter in which 6 kb upstream sequences of the cog-1 locus are fused to gfp (promA in Figure 3C). If mutated in this context, reporter gene expression is completely lost. That is, in the 6-kb promoter context, the unaffected proximal ASE motif 1 is not sufficient to support enough visible reporter gene expression (promA-ot119 and promA-ot201 in Figure 3C). Mutating the proximal ASE motif 1 in the context of the 6-kb promoter region also affects reporter expression in ASE, but to a much lesser extent than mutating the distal motif 2 (promA-del2 in Figure 3C). The overall sequence context therefore appears to have an important impact on ASE motif function in a manner that we do not currently understand. However, if we keep the sequence context parameter constant and compare the relevance of both ASE motifs in the context of the 6-kb promoter fragment, we can nevertheless conclude from our mutational analysis that both ASE motifs contribute to cog-1 expression, albeit to notably different extents.
On a practical level, we can also conclude that the sufficiency of a regulatory element to drive reporter gene constructs in a specific cell (as evidenced by the correct expression of the regulatory region that contains only proximal ASE motif 1) may not be an accurate reflection of the sufficiency of the regulatory element in vivo.
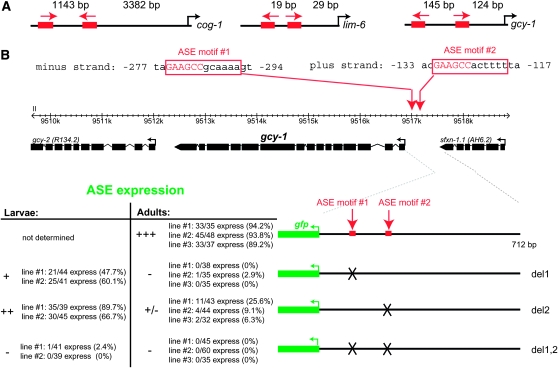
The gcy-1 locus also contains several functional ASE motifs:
Two cases in addition to cog-1 experimentally confirm the physiological relevance of duplicated ASE motifs. The cis-regulatory region of the LIM homeobox gene lim-6 contains two ASE motifs, and a mutation of either motif results in a loss of expression in ASE (Etchberger et al. 2007) (Figure 5A), similar to what we observe for cog-1 here. The cis-regulatory region of the gcy-1 locus, which encodes an ASER-expressed guanylyl cyclase (Ortiz et al. 2006), also contains two ASE motifs, and mutation of either leads to a loss of expression of the reporter in ASE (Figure 5, A and B). Each ASE motif when mutated alone has partial effects on ASE expression with the effect being more severe in adults than in larvae (Figure 5B). In contrast, mutating both motifs leads to a complete loss of ASE expression in both larval and adult stages. Moreover, the effects of ASE motif mutations are differential. Mutating ASE motif 1 appears to have stronger effects than mutating ASE motif 2, demonstrating that ASE motif 2 can function more independently from ASE motif 1 than vice versa (Figure 5B). This differential requirement is reminiscent of the differential requirement of ASE motifs in the cog-1 locus. We note that in all three cases mentioned here, there is no obvious pattern in the spacing between the two ASE motifs; spacing can vary from a few base pairs to >1000 bp (Figure 5A).
Figure 5.—
gcy-1 expression also depends on two ASE motifs. (A) Schematic of the location of the functional ASE motif in the cog-1 (Figure 3), lim-6 (Etchberger et al. 2007, 2009), and gcy-1 (B) loci. Spacings in between the 6-bp core (binding site for zinc fingers 3 and 4 of CHE-1) and translational start sites are shown. Arrows indicate the orientation of motifs. Note that we have previously documented a large number of cis-regulatory modules that drive expression in ASE and contain only a single ASE motif (e.g., gcy-5, gcy-7, lsy-6, etc.) and that completely isolated ASE motifs are sufficient to drive expression in the ASE neurons (Etchberger et al. 2007, 2009). (B) Reporter gene analysis of the gcy-1 locus. All constructs were generated by PCR fusion and scored in a otIs151 transgene background to allow identification of the ASE neurons. Mutations are complete deletions of the 12-bp site. The importance of individual ASE motifs is different, which is reminiscent of the cog-1 case, but not as drastic.
Multiplicity of ASE motifs is a common feature of ASE-expressed genes:
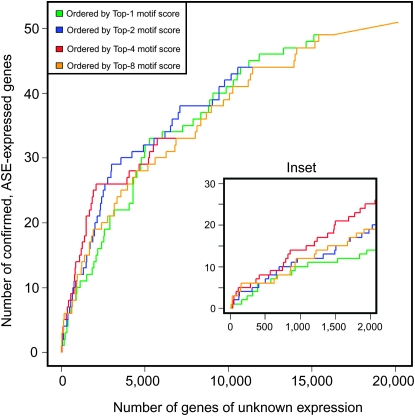
The presence of two ASE motifs in the examples discussed above prompted us to ask whether the occurrence of multiple ASE motifs is a common feature of ASE-expressed genes. We analyzed a data set of 52 genes that on the basis of reporter gene analysis are expressed in ASE (Etchberger et al. 2007) (supplemental Table 1). For the analysis we generated 10 separate orderings of the 20,183 C. elegans genes, ordering them respectively by the combined score of each gene's best N ASE motifs, where N varied from 1 to 10 (see supplemental methods). We then asked how well a given ordering isolated the 52 ASE-expressed genes at the top of the list. ASE-expressed gene enrichment toward the top of the list increases with the increasing number of motifs considered, reaching a peak at four motifs (Figure 6; including more than four motifs degrades the enrichment progressively). This indicates that the 52 ASE-expressed genes are indeed enriched in high-scoring matches to ASE motifs vs. the rest of the genome. Taken together, even though previous work has shown that single ASE motifs, isolated from their genomic context, are sufficient to drive gene expression in the ASE neurons (Etchberger et al. 2007, 2009), the presence of multiple ASE motifs appears to be a more reliable predictor of the expression of a gene in the ASE neuron than the presence of a single ASE motif.
Figure 6.—
Correlating ASE motif number and expression in ASE. The power of a gene's upstream ASE motifs to predict ASE expression depends on the number and score of each ASE motif. We define a gene's top-N motif score (which estimates the probability that all N motifs are functional; see materials and methods) as the product of the top-N upstream ASE motif scores for a given gene. Meanwhile we split the genes into a positive set, consisting of the 52 ASE-expressed genes [as confirmed by reporter–GFP construct experiments (Etchberger et al. 2007)] and a negative set the remainder of the genome. Then we sort the entire list of genes according to each top-N motif score, for N = 1, 2, 4, and 8 and measure how well the score criterion places the ASE-expressed genes near the top. To visualize this sorting, we generated “receiver operator characteristic” (ROC) curves. Intuitively, a ROC curve can be understood as follows: Starting at point (0, 0) at the top of the list (gene with best score), the graph moves up (y-axis) one gene if the next gene on the list is a positive and to the right (x-axis) if the gene is a negative, and so on until the last (20,183rd) gene is encountered [point (20131, 52)] at top right corner of graph. For example, point (2000, 25) on the red curve denotes that 25 ASE-expressed genes are found in the top 2025 genes (10% of the genome) in the list sorted using the top-4 motif score. Point (2000, 14) on the green curve, by contrast, shows that only 14 of the 52 ASE-expressed genes (27%) are recovered in the same-size list when sorted by the top-1 motif score (inset). Therefore, the top-4 motif score, which assumes four functional motifs and scores accordingly, is about twice as effective at identifying ASE-expressed genes than the top-1 motif score. This is statistical evidence that, on average, multiple ASE motifs are functional in ASE expression. Additionally, the underlying data from which these graphs are derived (supplemental Methods) may be interpreted as a probability estimate for ASE expression of all C. elegans genes and used to choose candidate ASE-expressed genes for further testing.
Conclusions:
Using mapping technology newly applied to de novo C. elegans mutant identification, we have identified here cis-regulatory mutations that affect single neuron-specific expression of the Nkx6-type homeobox gene cog-1, resulting in the aberrant execution of a neuronal cell fate decision. The relative rarity of cis-regulatory mutations, associated with a difficulty in reliably pinpointing such mutations, leaves the physiological relevance of the vast amount of cis-regulatory elements defined by reporter analysis, in vitro approaches, or in silico predictions essentially untested. We have confirmed here the importance of a previously defined regulatory “terminal selector motif,” the ASE motif. Terminal selector motifs are present in many terminal differentiation gene batteries that define the differentiated feature of a given neuron type and are activated by terminal selector genes, such as CHE-1 (Hobert 2008b).
The initial identification and analysis of the ASE motif presented us with a specific conundrum (Etchberger et al. 2007). On the one hand, we found that isolated ASE motifs are sufficient to drive reporter gene expression in ASE (Etchberger et al. 2007, 2009); moreover, larger genomic regulatory fragments, such as the 4-kb regulatory element that drives cog-1 expression in ASE or in many other regulatory elements that produce expression in ASE, contain only single recognizable ASE motifs (Etchberger et al. 2007). On the other hand, however, as expected from the small size of the ASE motif, the motif is very abundant in the genome and many genes that contain a good match with the ASE motif are not expressed in ASE (Etchberger et al. 2007). The data presented here explain at least parts of this conundrum. Our identification and validation of multiple ASE motifs in ASE-expressed genes show that, in their normal genomic context, genes appear to have a tendency to require multiple ASE motifs to be expressed in ASE—as deduced by a combination of bioinformatic analysis and experimental validation described here. It is important to emphasize that even though endogenous gene loci may display such requirements, as revealed here by the cis-regulatory cog-1 alleles, such requirements are not necessarily observed in reporter gene analysis, as revealed by the sufficiency of a single ASE motif, the proximal ASE motif 1, in the cog-1 locus. That is, even though many previously ASE-expressed cis-regulatory elements rely on single ASE motifs to function and even though an ASE motif can work in complete isolation (Etchberger et al. 2007, 2009), many ASE-expressed genes may in fact depend on multiple ASE motifs for expression in the ASE neurons in their normal genomic context.
The multiplicity of cog-1 alleles may be indicative of a principle that is mirrored in the recently described “shadow enhancers” in Drosophila (Hong et al. 2008). Chromatin immunoprecipitation data and reporter gene assays have shown that many Drosophila developmental control genes contain multiple enhancers that produce similar expression patterns. This multiplicity has been proposed to help ensure the precision of embryonic patterning (Hong et al. 2008). In light of our finding of the apparent sufficiency of individual regulatory motifs, contrasted by the joint requirement of multiple elements in vivo, it is conceivable that even though the defined Drosophila shadow enhancers work in isolation, they may be jointly required to drive correct levels of gene expression.
From a practical perspective, our findings provide a strong note of caution for interpreting both reporter gene analysis and rescue analysis. The importance of distally located cis-regulatory elements may be overlooked in transgenic approaches. Such distally located elements may provide robustness and tune the precise levels of gene expression, issues usually of less importance for multi-copy transgenic arrays in C. elegans. These notions underscore the importance of cis-regulatory alleles—and hence the value of extensive forward genetic screens (Sarin et al. 2007)—as they unambiguously demonstrate the relevance of regulatory information dissected by standard reporter analysis.
Acknowledgments
We thank Q. Chen for expert DNA injection, L. Cochella for generating one of the cog-1 reporter fusion constructs, the Caenorhabditis Genetics Center for providing strains, and members of the Hobert lab for comments on the manuscript. S.F. and D.G.M. acknowledge funding from Genome Canada, Genome British Columbia, and the Michael Smith Research Foundation. O.H. acknowledges funding by the National Institutes of Health (R01NS039996-05; R01NS050266-03). O.H. is an Investigator of the Howard Hughes Medical Institute.
References
- Chang, S., R. J. Johnston, Jr. and O. Hobert, 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt, B., and H. R. Horvitz, 1999. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98 317–327. [DOI] [PubMed] [Google Scholar]
- Davidson, E. H., 2001. Genomic Regulatory Systems. Academic Press, San Diego.
- Etchberger, J. F., A. Lorch, M. C. Sleumer, R. Zapf, S. J. Jones et al., 2007. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21 1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger, J. F., E. B. Flowers, R. Poole, E. Bashllari and O. Hobert, 2009. Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification. Development 136 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O., 2006. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb. Symp. Quant. Biol. 71 181–188. [DOI] [PubMed] [Google Scholar]
- Hobert, O., 2008. a Gene regulation by transcription factors and microRNAs. Science 319 1785–1786. [DOI] [PubMed] [Google Scholar]
- Hobert, O., 2008. b Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. USA 105 20067–20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, J. W., D. A. Hendrix and M. S. Levine, 2008. Shadow enhancers as a source of evolutionary novelty. Science 321 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., M. Wang, T. O. Ririe, J. S. Fernandes and P. W. Sternberg, 2005. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc. Natl. Acad. Sci. USA 102 4972–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, R. J., and O. Hobert, 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 845–849. [DOI] [PubMed] [Google Scholar]
- Johnston, R. J., Jr., S. Chang, J. F. Etchberger, C. O. Ortiz and O. Hobert, 2005. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA 102 12449–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. R., J. S. Maydan, S. Flibotte, D. G. Moerman and D. L. Baillie, 2007. Oligonucleotide array comparative genomic hybridization (oaCGH) based characterization of genetic deficiencies as an aid to gene mapping in Caenorhabditis elegans. BMC Genomics 8 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi, A., O. P. Kallioniemi, D. Sudar, D. Rutovitz, J. W. Gray et al., 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258 818–821. [DOI] [PubMed] [Google Scholar]
- Maydan, J. S., S. Flibotte, M. L. Edgley, J. Lau, R. R. Selzer et al., 2007. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 17 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan, J. S., H. M. Okada, S. Flibotte, M. L. Edgley and D. G. Moerman, 2009. De novo identification of single nucleotide mutations in Caenorhabditis elegans using array comparative genomic hybridization. Genetics 181 1673–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, C. O., J. F. Etchberger, S. L. Posy, C. Frokjaer-Jensen, S. Lockery et al., 2006. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, R. E., T. Inoue, D. R. Sherwood, L. I. Jiang and P. W. Sternberg, 2002. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev. Biol. 252 202–213. [DOI] [PubMed] [Google Scholar]
- Sarin, S., M. M. O'Meara, E. B. Flowers, C. Antonio, R. J. Poole et al., 2007. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176 2109–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, O., H. Nakano, M. Koga and Y. Ohshima, 2003. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130 1215–1224. [DOI] [PubMed] [Google Scholar]