Our understanding of the role of non-B DNA structures in mutagenesis and human disease has been an exciting odyssey, but the road traveled has not always been linear or without challenges. Hence, this prologue will provide an overview and some guideposts for the route. Even in my postdoctoral years in the mid-1960s, I realized that DNA sequence was affecting its properties and probably its conformation. Because this concept was heretical at the time and because substantial further work was necessary to cement these concepts, the work moved slowly. I was a postdoctoral fellow with Professor H. Gobind Khorana (Fig. 1) at the Enzyme Institute of the University of Wisconsin-Madison from 1964 to 1966. During that time, the Khorana and Nirenberg teams solved the genetic code, and Gobind shared the Nobel Prize in 1968 with Marshall Nirenberg and Bob Holley (tRNA sequence) for these discoveries. My job as part of this project was to prepare and characterize a number of simple repeating DNA sequences that were subsequently transcribed by me and other workers to provide RNAs of defined repeating sequences. Using these RNAs, we determined the codon assignments; Gobind's brilliance and our hard work made the elucidation of the genetic code (i.e. the language between DNA, RNA, and proteins) straightforward (1).
FIGURE 1.
Professor H. Gobind Khorana.
As part of my postdoctoral studies, I utilized the newly developed Beckman Spinco Model E analytical ultracentrifuge to determine the buoyant densities and other properties of these DNA polymers of repeating sequences (2). From these determinations, there was no doubt that sequences with identical base compositions had very different properties, and further work in my Wisconsin laboratory (described below) during my Assistant and Associate Professorship days verified this conclusion.
My laboratory has always been involved in DNA structural problems. However, as opposed to the vast majority of other workers in the field, we were much less interested in small oligonucleotides that can be investigated by x-ray crystallographic or NMR methodologies but instead were interested in large molecules that were in aqueous solution. After all, this was the genuine interface with biology. However, a complication related to this unorthodox interest was that virtually all methodologies for determining structures and their biological functions had to be developed and were not carryovers from other fields (3).
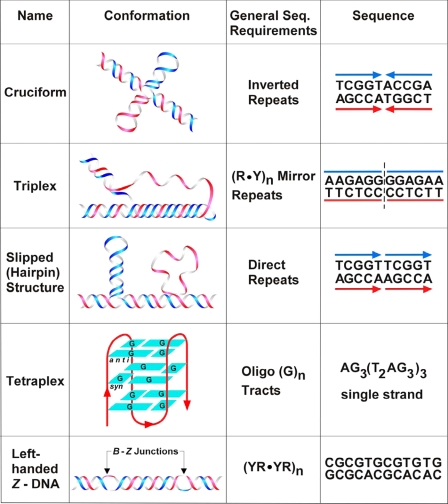
Most educators know the old adage that graduate students really do not understand the magnitude of the problems that they are encountering because, if they did, they would never undertake their thesis project. In some ways, I believe that my scientific career adheres to this principle. The 1970s were engaged in biophysical and biochemical investigations on restriction fragments and then recombinant molecules to evaluate the types of non-B DNA structures (described below) that were found at simple repeating sequences. However, a major change occurred in 1990 when I moved to Texas and learned of the involvement of simple repeating triplet sequences in hereditary neurological diseases such as myotonic dystrophy, fragile X syndrome, and Friedreich ataxia. The groundwork laid in Madison and at the University of Alabama at Birmingham (UAB) from the mid-1960s to 1990 was important for elucidating the molecular mechanisms involved in genetic instabilities that are responsible for the etiology of these diseases. Interestingly, the neurological and human genetics communities were quite receptive to the concept of non-B DNA structures (cruciforms, triplexes, slipped structure DNA, quadruplexes, and left-handed Z DNA) (Fig. 2), and thus, major advances were made in the past two decades in the relationship between non-B DNA structures and human diseases. Parenthetically, the term “non-B DNA structures” refers to all DNA conformations other than the orthodox right-handed Watson-Crick structure.
FIGURE 2.
Non-B DNA conformations involved in genomic disorders. Reprinted with permission from Ref. 56.
In the late 1990s and early 2000s, the convergence of bioinformatics, human genetics, molecular biology, and genomics provided powerful new tools for our investigations. In 2004, Albino Bacolla and colleagues discovered that breakpoints for gross deletions that cause a number of human diseases coincide with the presence of non-B DNA structures (described below). These and later studies have cemented the role of non-B DNA structures in human disease. Because of the strength of these techniques and the compelling nature of the results that were obtained, these concepts have been widely accepted and extended by the biological and medical communities.
In summary, my teams and I worked for 39 years (1965–2004) to determine the types of non-B DNA structures that exist and to attempt to broach their biological functions. However, my theories and predictions were substantially ahead of the science and methodologies that could be used to rigorously demonstrate these genetic functions. Machiavelli (1513 A.D.) stated, “It must be remembered that there is nothing more difficult to plan, more doubtful of success, nor more dangerous to manage, than the creation of a new system. For the initiator has enmity of all who would profit by preservation of the old institution and merely lukewarm defenders in those who would gain by the new ones.” I certainly agree with Machiavelli but might add that when one turns out to be correct in the end (even if it takes four decades), it is exceedingly rewarding.
The Wisconsin Years (1964–1981): DNA Is Polymorphic
My early educational years at Ohio Wesleyan University (1956–1960) and my Ph.D. with Professor Klaus Hofmann at the University of Pittsburgh (1960–1964) on the synthesis of the first analogs of ACTH and melanocyte-stimulating hormone have been reviewed previously (4).
During my postdoctoral fellowship, I learned that Gobind Khorana (Fig. 1) was truly a genius in the field of polynucleotide synthesis. His investigations on DNA and RNA as well as chemical methodologies for the synthesis of oligonucleotides at the University of British Columbia in the 1950s and later at the Enzyme Institute in Madison were unique; virtually no other labs were engaged in these types of chemical studies. Indeed, the early 1960s were rudimentary days in this field with the chemical synthesis of ATP, DPN (NAD), and then di- and trinucleotides. When I joined his lab in September of 1964, the term genetic code had not even been coined yet, according to my memory. However, when I left his laboratory 24 months later, the entire code was finished, and he shared the Nobel Prize in 1968 for these discoveries. His brilliance in pioneering the field of polynucleotide synthesis has been of tremendous benefit to the fields of nucleic acids, molecular biology, gene cloning, and genomics and is largely taken for granted at this time.
After my postdoctoral years, I moved to my Assistant Professorship in the Department of Biochemistry at the University of Wisconsin-Madison. Gobind suggested that I take the fourteen DNA polymers of defined repeating nucleotide sequences that I had prepared and characterized and utilize them in any way that I wished. I realized that this family of simple and well defined DNA sequences was a gold mine for evaluating the role of DNA sequence in determining DNA properties and structure. Thus, my students and I from 1966 to ∼1972 investigated a large number of behaviors, including helix-coil transitions, buoyant density analyses, actinomycin D binding studies, netropsin binding, circular dichroism, triplex formation, viscosity, x-ray diffraction, lac repressor binding studies, in vitro DNA synthesis and RNA synthesis (transcription), interferon binding, etc. (3). By all of these methodologies, it was clear that DNA was polymorphic; different sequences gave rise to different properties and probably structures. Thus, the concept (3) that DNA was not an inert repository of genetic information but instead had conformational features along its chain that provided important genetic signals was born. However, it must be remembered that these notions were at least 7–10 years ahead of DNA sequencing and that genomics was at least 25 years into the future. The power of this strategy was having the DNA molecules of known repeating nucleotide sequences.
Because the DNA polymers of repeating nucleotide sequences were synthesized with DNA polymerases, it was important to understand the mechanism of their formation and to utilize different types of polymerases for their preparation. Thus, my laboratory had several side excursions into the related fields of DNA polymerase, ligase, exonuclease, and nucleoside diphosphokinase mechanisms as well as reverse transcription.
Roger Wartell and I (3) stated in 1974, “The observation that the overall conformation of a DNA can be dictated by the sequence of its constituent nucleotides is pregnant with implications.” This was certainly an understatement! Likewise, I remarked in 1969 (5), “Thus, the two DNAs may have sufficiently different configurations, dictated by their base sequences, to determine this difference in their (actinomycin binding) behavior.” I could not have been more on target in the interpretation of these experiments; however, the rigorous demonstration of their physiological and/or pathological roles required waiting for genomics, human genetics, molecular genetics, and the molecular basis of genomic disorders, in ∼2004, to verify these predictions.
A Nature News and Views article (quoted in Ref. 6) in 1979 declared, “...the idea of DNA as an inert repository of genetic information seems really to have breathed its last.” Whereas I was delighted to see this analysis of our work, it required the next 25 years to nail down the DNA conformational features in solution and to extend this work to biology, where an obligate role for the non-B DNA structures was established.
In the 1970s, six critical experiments provided insightful new information on the polymorphic nature of DNA (7). First, triplexes were characterized between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences as well as the inhibition of transcription by the three-stranded structures (8). This work was in concert with the earlier studies of T. Miles, G. Felsenfeld, A. Rich, M. Gellert, D. Davies, P. Sigler, and R. Langridge (9) and the contributions by the group of M. Chamberlin (10). Second, left-handed DNA was described in a DNA polymer (11, 12). Two years later, F. Pohl and T. Jovin (13) reported physical studies on a related DNA polymer and suggested the possibility of a left-handed DNA conformation for one of the isomeric conformers. Nine years after the first proposal of a left-handed DNA structure, A. Rich and associates (14) reported the x-ray structure in a single crystal of a hexanucleotide containing strictly alternating C and G residues in a left-handed Z helix. This observation was quickly confirmed by R. Dickerson and associates (15) on an identical DNA sequence but with four repeating units. Hence, the groundwork was laid for an exciting era of investigations on left-handed DNA (reviewed in Refs. 16 and 17). Third, it was realized by Bob Blakesley in my lab that single-stranded phage DNAs (φX174 and M13) were not truly single-stranded but contained regions of double strandedness as recognized by the ability of certain restriction endonucleases to specifically cleave at their canonical sites (18, 19). This discovery was confirmed by N. Zinder and colleagues (20), but unfortunately, they wrongly proposed that the “half-sites” were cleaved in their single-stranded form rather than the folded-back duplex sites; Blakesley et al. (19) rigorously proved that the cleavage site was a duplex. Fourth, bent DNA was discovered (21–23) in a polynucleotide polymer consisting of a DNA·RNA hybrid joined to a DNA·DNA duplex. This discovery was followed after a four-year time period by the important findings of P. Englund, D. Crothers, J. Griffith, and others that certain kinetoplast DNA fragments were highly bent (elaborated below). Fifth, Nikos Panayotatos and I (24) discovered cruciforms in supercoiled DNAs (Fig. 2). D. Lilley (25) rapidly confirmed our discovery of cruciforms and inverted repeat sequences as recognizable structural features in supercoiled DNAs. Sixth, the widespread use of high pressure liquid chromatography fractionation of DNA restriction fragments (reviewed in Ref. 7) as well as gene cloning (reviewed in Ref. 6) enabled us to focus our attention specifically on bacterial promoters, T7 phage late promoters, terminators, the Escherichia coli lactose operator and promoter, the left-ward operator-promoter of bacteriophage λ, the origin of replication of bacteriophage T7, and the polyoma virus origin of replication.
The University of Wisconsin-Madison was an exciting institution for maturing into a productive educator and scientist. The Department of Biochemistry and related departments had extremely high standards with a number of very distinguished senior faculty, including Henry Lardy, Bob Burris, Harry Steenbock, Julian Davies, Bill Reznikoff, Chris Raetz, Julius Adler, Howard Temin, Helmut Bienert, David Green, Jack Gorski, Hatch Echols, Harlyn Halvorson, Gobind Khorana, Chuck Kurland, Oliver Smithies, Josh Lederberg, Matsayasu Nomura, Bob Bock, Karl Paul Link, Jim Crow, and Moe Cleland. The quality of the graduate students at Wisconsin was superb; how could I fail in this environment? Furthermore, a senior and highly respected faculty member, Harlyn Halvorson, had approached the administration of the University of Wisconsin in ∼1960 to explain the developing area of molecular biology (26). Luckily, the Wisconsin administration agreed, and in approximately a seven-year time period, eight or more related biological and chemical departments hired at least 40 Assistant Professors. This extremely competitive and energetic group of faculty created a marvelous, vibrant environment for the growth of a junior faculty member. Also, my sabbatical experience (1976–1977) with Dr. Walter Eckhart in the Tumor Virology Group of the Salk Institute (La Jolla, CA) was beneficial in giving me perspectives on DNA tumor viruses and eucaryotic genetics.
University of Alabama at Birmingham (1981–1990)
In 1980, I was asked to let my name be considered for the Chairmanship of the Department of Biochemistry at UAB, Schools of Medicine and Dentistry. By this time, I had been broached by at least 25 other institutions to consider departmental chairmanships, and none seemed very interesting; I suspected that UAB would also fall into this category. However, I did visit the institution and was moderately impressed, but the ensuing months revealed the extent to which the administration of the University was creative, energetic, and truly interested in building basic sciences in Birmingham. They promised to construct a new nine-story building if I would move to Birmingham and would provide a number of faculty positions. In 1982, the building was completed and occupied by my associates and me after my wife, Dotty, and I moved to Birmingham in 1981. By this time, our son and daughter, Kevin and Cindy, had left home for their college pursuits at the University of Wisconsin-Madison and Vanderbilt University, respectively. My ten years at UAB were great. The faculty was interesting, productive, and adroit at grantsmanship and was working on important research problems. The administration was lively and creative and made the entire experience completely enjoyable. UAB fully lived up to its promises, including a large number of faculty hired in the Department and in related disciplines.
My mentor, Gobind Khorana, has repeatedly stated that it is beneficial for a scientist to move approximately every ten years: “You only have chances at three careers because it takes approximately ten years to set up a laboratory, build a program, etc.” I cannot agree more because we are all profoundly influenced by our faculty colleagues. Thus, if you pick the right institutions and if the mix of research programs is beneficial, the results can be fantastic. I was one of the faculty at the University of Wisconsin-Madison who people thought would “never move” because I held a number of distinguished faculty positions and leadership roles with training grants and educational programs, etc. I viewed my move to UAB with some trepidation because I was passionate about continuing my research endeavors and was not fully confident that I could maintain my research program along with its funding in the new environment. However, it turned out that this was no problem at UAB, but instead my program took off intellectually, in part because of the environment and in part because of the external factors in the international research community in other labs developing stronger interests in non-B DNA structures.
Left-handed DNA—Shortly after my arrival at UAB, the left-handed DNA field (11–13) was substantially reinvigorated by the almost simultaneous demonstration by the x-ray crystallography groups of Rich and Dickerson that self-complementary tetranucleotide or hexanucleotide sequences with repeating C and G bases had a left-handed helix. This rediscovery of left-handed DNA (reviewed above) was extremely welcome because it 1) verified our past predictions of the role of sequence in DNA properties and structures, 2) offered a dramatic DNA structural change for future investigations, and 3) provided a tractable strategy for evaluating the role of left-handed helices in plasmids and chromosomes that had been replicated within cells. All of these predictions were realized.
In the 1980s, my laboratory demonstrated the following: negative supercoiling causes the B-to-Z transition; the B-Z junction is specifically cleaved by S1 nuclease and the BAL31 nuclease; supercoiling is a sensitive indicator of the B-to-Z transition; B DNA and Z DNA can coexist in close proximity; Z DNA perturbs the conformation of neighboring “B DNA”; a variety of left-handed conformations are caused by different environmental conditions; (T-G)n·(C-A)n adopts a left-handed structure; plasmids containing alternating CG tracts have unusual biological properties; the conditions that cause the structural transformations were evaluated; B-Z DNA junctions contain few, if any, non-paired bases at physiological superhelical densities; sequence perturbations within the alternating purine-pyrimidine tracts are permitted for Z DNA formation; thermodynamic parameters for the B-to-Z transition were established; and the in vivo existence of left-handed DNA was demonstrated using a genetic/biochemical assay (27). This may be my most creative paper. The capacity of sequences to adopt either cruciforms or left-handed Z DNA was determined; the B-Z DNA equilibrium in vivo was perturbed by biological processes; Z DNA was stabilized in vivo by localized supercoiling due to transcription; flanking AT-rich tracts cause a structural distortion in the Z DNA tract in plasmids; and left-handed Z DNA was used to study the in vivo supercoil density in the E. coli chromosome. All of this work has been reviewed (28–31).
A major emphasis for all of this work was to attempt to establish a biological function for left-handed Z DNA; the discovery of the in vivo existence of left-handed DNA in bacterial cells was demonstrated using a genetic/biochemical assay (27). However, a number of efforts from my lab as well as others on a worldwide basis were relatively unproductive until ∼2000 when it was realized (described below) that left-handed DNA serves as a recognition signal for double-strand break formation, which is involved in genetic translocations responsible for a family of hereditary neurological diseases. A large number (at least 26) of talented research teams, in addition to the scientists acknowledged above, have contributed to the field of left-handed DNA. Also, in the 1980s, we conducted investigations on bent DNA, cruciforms, intramolecular triplexes in plasmids, and anisomorphic DNA.
Bent DNA—We continued our investigations (described above) on bent (curved) DNA (21–23) by collaboratively investigating the highly bent fragment of Crithidia fasciculata kinetoplast DNA (32). The Crothers and Englund labs discovered the bent helical structure three years after the initial description of DNA bending due to the anomalous gel mobility of restriction fragments from kinetoplast DNAs (33). In a sequence of 200 bases, the bent region contains 18 runs of four to six A residues, with 16 of these runs in the same strand. In some parts of this sequence, the A runs are regularly spaced with a periodicity of ∼10 bp. This spacing is nearly in-phase with the twists of the DNA helix. We proposed that because of their periodic spacing, the small bends associated with each A run add up to produce substantial curvature in this molecule (32). In other investigations with Jack Griffith and Paul Englund (34), we developed a strategy for evaluating the effect of sequence-directed bends or other polymorphisms in the context of a super-twisted DNA circle by electron microscopy. At present, DNA bending (intrinsic curvature) is widely believed to be a critical component of the binding of certain proteins to DNA target sites (35, 36).
Cruciforms—The early work on cruciforms was highly controversial because a small but influential group of workers in the field believed that structures of this type could not exist as genuine physical entities within DNA. However, the discovery (24, 25) of cruciforms revealed that a cruciform was a recognizable special feature in a supercoiled DNA molecule. This pair of discoveries was significant because it served as a harbinger for a family of other non-B DNA structures to be characterized as genuine physical entities that could have biological significance. Subsequently, Charles Singleton and I (37) revealed the relationship between the superhelical density of a plasmid and cruciform formation. Approximately 22 kcal/mol are required to generate the cruciform in pVH51. In addition, the effects of cations, temperature, and stem length on the supercoil-induced transition from the linear form to the cruciform state were investigated (38). Other studies (39–42) investigated the roles of DNA sequences in recombinant plasmids that contain perfectly alternating purine-pyrimidine base pairs that could adopt either left-handed Z DNA structures or cruciforms; in general, the left-handed DNA conformations were the more stable structures under a variety of conditions. D. Lilley (42) has contributed substantially to our understanding of the structures of DNA helical junctions in cruciforms.
These discoveries laid the groundwork for the realization that cruciforms are important chromosomal structures that signal the breakpoints involved in recombinational repair that are integral to a large number of translocations, copy number variations, etc., responsible for a myriad of human diseases (see below). Indeed, I noted with interest that an entire Federation of American Societies for Experimental Biology (FASEB) Summer Research Conference named “Structurally Ambiguous DNA” was held on 7/6–11/2008 on this topic. Thus, cruciforms may be the hottest DNA structural topic in the field of genetic translocations that are involved in human diseases. Unfortunately, some geneticists use the term palindromes to refer to the inverted repeat sequences that form cruciforms; the term palindrome (meaning to run backwards) is incorrectly used in this context.
Intramolecular Triplexes in Plasmids—As described above, the history of DNA triplexes dates back to the 1950s with investigations on DNA and RNA polymers. However, in the mid-1980s, we became very interested in returning to our old love of DNA triplexes (8) because a variety of studies from different laboratories had demonstrated that oligopurine·oligopyrimidine tracts in large restriction fragments and in plasmids demonstrated quite unusual properties. Of course, we suspected that these properties were due to the formation of triplexes at the sites. However, the proof of intramolecular triplexes in recombinant plasmids was a very substantial challenge because we were investigating the types of structures formed by a relatively small sequence (<70 bp) on the background of a plasmid composed of 4000–6000 bp. Several models were proposed by other laboratories, which I felt were likely to be incorrect (43). In addition, M. Frank-Kamenetskii and associates had performed biophysical measurements in recombinant plasmids and had favored the idea of triplexes (which they called H DNA) (reviewed in Refs. 30, 43, and 44).
Upon returning to the area of triplexes at oligopurine·oligopyrimidine tracts in plasmids, we investigated the influence of DNA sequence on the formation of unusual properties (43). However, I challenged Jeff Hanvey to unequivocally prove that an intramolecular triplex was formed within a recombinant plasmid. This was no small task because a 30-bp insert comprised only ∼0.5% of the base pairs in the recombinant molecule. However, Hanvey performed an ingenious experiment (45) that proved beyond any reasonable doubt that a folded-back triplex structure was adopted at the 30-bp repeating GAA·TTC repeat (which we now know is involved in the etiology of Friedreich ataxia). Hanvey performed judicious site-directed mutagenesis studies and demonstrated that a mutation at one position in the insert caused a biochemical perturbation at the other end because of the folded-back structure in the intramolecular triplex. This experiment ended any debate in the field about the validity of intramolecular triplexes in plasmids and clearly supported the view of Frank-Kamenetskii and colleagues.
The late 1980s were exceedingly good years because my associates and I had a wonderful time exploring a number of facets of triplexes in recombinant plasmids, including the following: the effect of loop size on triplex formation and stability; the effect of base composition and non-central interruptions on formation and stability; the effect of length, supercoiling, and pH; the effects of metal ions that cause isomerization of certain triplexes; the demonstration that GC-rich flanking tracts decrease the kinetics of formation; the demonstration that central, non-purine·pyrimidine sequences in oligo(G·C) tracts and metal ions influence the formation of triplex isomers; and the formation of an intramolecular triplex from non-mirror repeated sequences containing different types of triplexes.
Furthermore, we investigated the capacity of different types of naturally occurring sequences to adopt triplexes, including the following: the murine immunoglobulin-α switch region, the site of unequal sister chromatid exchange, the origin of replication near the dhfr locus in Chinese hamster ovary cells, the regulatory region of human papilloma virus type 11, and the human γ-globin 5′-flanking region in the fetal hemoglobin gene. Other investigations revealed that intermolecular triplexes were effective inhibitors of the EcoRI methylase and restriction endonuclease and demonstrated an intermolecular bitriplex that was named nodule DNA and the alteration of a triplex structure by a single-stranded DNA-binding protein (reviewed in Refs. 16, 44, and 46). Like cruciforms, triplexes are also frequently found at double-strand break sites in chromosomes, which are responsible for a large number of hereditary diseases (see below).
Anisomorphic DNA—Anisomorphic DNA (47) is an intriguing structure but remains to be fully characterized. The site of segment inversion of herpes simplex virus type 1 contains a series of tandem repeats with a purine bias on one strand and high T + C content (DR2 repeats) capable of adopting a non-B DNA structure under a variety of conditions. Plasmids containing eight or more contiguous copies of the DR2 repeat undergo a series of supercoil-driven conformational transitions that result in different extents of relaxation. We proposed (48) that anisomorphic DNA shows a buckling of the pyrimidine-rich strand of the DR2 sequences at a deformation due to an unequal rise per residue or a different angle of rotation or certain types of slipped pairing between the complementary strands. These studies have been reviewed (16, 46). The biological function of anisomorphic DNA is unclear.
Houston, Texas (1990 to Present)
Life was proceeding beautifully in Birmingham in 1989–1990; science was excellent; my wife, Dotty, and I were comfortable in our spacious home; the grandchildren were beginning to arrive; and life was good when Texas A&M University (TAMU) broached me repeatedly, declaring that a new research institute was being built in Houston that they boasted is the largest medical center in the United States. Upon visiting the Institute, I realized that they were not joking, and promises were made to me to completely fund the building, faculty, postdoctoral fellows, and graduate students along with support staff. I agreed to serve as the Founding Director of the Institute of Biosciences and Technology (IBT) in Houston on a permanent basis and for 24 months to simultaneously function as the Chairman of the Department of Biochemistry and Biophysics in College Station, which is located 90 miles from Houston. The IBT building was under construction in Houston but was not finalized until late 1991. Thus, we lived in both College Station and Houston for a two-year period; at this time, we designed and constructed a new home less than one mile from the Houston IBT building. In 1990–1994, I finalized the construction and occupancy of the eleven-story IBT building; hired a number of faculty; built the Center programs; recruited support staff to initiate the new research institute; did capital development for construction and program development; and developed positive interrelationships with the Baylor College of Medicine, the University of Texas Health Science Center, the M. D. Anderson Cancer Center, Rice University, and the University of Houston. The Institute was developing beautifully by 1994. However, in my fourth year of this peripatetic and frantic activity, TAMU underwent dramatic and repetitive administrative changes, and the IBT faculty and I were informed by Dr. Ray M. Bowen, the then President of TAMU, that they could not (would not) uphold their written commitments to me and the IBT faculty. Of course, this was disastrous news for the long-term prognosis of IBT. This declaration elicited my resignation as Director of the Institute, and I have enjoyed a full-time research career since 1994 because of this debacle; further comments are provided below under “University Administration.”
Many positive features were realized on my move to Houston, including the location of IBT in the heart of the Texas Medical Center and my association with extremely fine colleagues, including Jim Lupski, Karen Vasquez, John Wilson, Tetsuo Ashizawa, Lian Gao, Dick Brennan, Bill Brinkley, Tom Caskey, David Nelson, Richard Gibbs, Art Beaudet, Huda Zoghbi, and others at the Baylor College of Medicine and the other Houston medical and educational institutions. 1990–1991 was a time of great excitement in the field of hereditary neurological diseases because several laboratories almost simultaneously discovered the presence of triplet repeat sequences in the myotonic dystrophy gene as well as the fragile X syndrome gene (49). I distinctly remember hearing a lecture at Baylor by Tom Caskey in which he described David Nelson's discovery of the presence of very long tracts of CGG·CCG repeats in the 5′-untranslated region of the fragile X gene. Clearly, the inference was that the long tracts interfered in some way with transcription or translation of the genes, and I was immediately reminded of my work in the mid-1960s with Khorana on DNA polymers of simple repeating sequences; I knew from my work at Wisconsin that these molecules in the polymer form had unorthodox properties and probably conformations. Thus, it was intriguing to evaluate their behaviors in recombinant plasmids and chromosomes. Therefore, my move to Houston turned out to be a scientific bonanza in terms of DNA structural investigations and their relationship to human genetic diseases.
Hence, my laboratory took another turn in the road to investigate the conformational properties of simple repeating sequences related to hereditary neurological diseases. Likewise, the mechanisms of the genetic instabilities that cause these diseases were fascinating, and we had most of the tools for model systems to investigate the roles of DNA replication, repair, and recombination in these critical processes. Thus, my move to Houston has been extremely good from a scientific standpoint.
Molecular Mechanisms of Genetic Instabilities in Hereditary Neurological Diseases—Tremendous progress has been realized over the past 18 years in our understanding of the molecular mechanisms responsible for the expansions and deletions (genetic instabilities) of repeating tri- and tetranucleotide sequences associated with a number (∼20) of hereditary neurological diseases (49). The diseases are myotonic dystrophy types 1 and 2, fragile X syndrome, and Friedreich ataxia (FRDA), which are related to the massive expansions of repeat sequences (CTG·CAG, CCTG·CAGG, CGG·CCG, and GAA·TTC, respectively). These instabilities occur by replication, recombination, and repair processes, probably acting in concert, due to slippage of the DNA complementary strands relative to each other. These diseases and their mechanisms of instabilities have been thoroughly reviewed (49–52). The biophysical properties of the folded-back repeating sequence strands play a critical role in these instabilities (49). Non-B DNA structural elements (hairpins and slipped structures) and DNA unwinding elements (tetraplexes, triplexes, and sticky DNA) (Fig. 2) are important components of the instability mechanisms (reviewed in Refs. 49 and 50). The replication mechanisms are influenced by pausing of the replication fork, orientation of the repeat strands, location of the repeat sequences relative to replication origins, and the flap endonuclease (52). Methyl-directed mismatch repair, nucleotide excision repair, and repair of damage caused by mutagens are also important. Genetic recombination and double-strand break investigations in model systems (51, 52) have provided important information on the expansion mechanisms. Other important contributors to our understanding of the molecular mechanisms of these genetic instabilities include the labs of Drs. Usdin, Mirkin, Sinden, Wilson, McMurray, Griffith, and Kunkel.
Sticky DNA and FRDA—In 1996 at the Baylor College of Medicine, Dr. Massimo Pandolfo discovered that FRDA was caused by an intronic GAA·TTC repeat expansion (reviewed in Refs. 49 and 53). I was amazed when Massimo informed me that the triplet repeat sequence was my old friend (GAA·TTC) with all purines in one strand and pyrimidines in the complementary strand, which we knew, from approximately a dozen of our papers in the 1980s, loved to form triplexes. Hence, studies on this DNA and its genetic and pathological manifestations have been a wonderful and productive experience. FRDA, the most common inherited ataxia, is caused by the transcriptional silencing of the FXN gene, which codes for the 210-amino acid frataxin, a mitochondrial protein involved in iron-sulfur cluster biosynthesis. The expansion of the GAA·TTC tract in intron 1 to as many as 1700 repeats elicits the transcriptional silencing by the formation of non-B DNA structures (triplexes or sticky DNA), the formation of a persistent DNA·RNA hybrid, or heterochromatin formation. The sticky DNA adopted by the long repeat sequence also elicits profound mutagenic, genetic instability, and recombination behaviors (reviewed in Ref. 53). Furthermore, it has been possible to develop early stage therapeutics involving polyamides or histone deacetylase inhibitors (53).
The transitioning of my laboratory in 1990 from a pure interest on DNA structures and biochemical processes to DNA structures with pathological and medical implications has been rewarding. First, any doubts that were held in the biochemical community about the role of non-B DNA structures prior to 1990 were eliminated due to their involvement in human hereditary neurological diseases by the human genetics and medical communities. In fact, all workers in the field have readily accepted the concept that slipped DNA structures are critical for the expansion and deletion mechanisms (50, 52). Also, the applicability of DNA structural biology to human genomic diseases has breathed new life into this area of investigation.
DNA Structural Properties of Repeating Tri- and Tetranucleotide Repeats—We continued our DNA structural investigations in the 1990s and 2000s on repeating tri- and tetranucleotide repeats in recombinant plasmids and, to a lesser extent, in restriction fragments. Interesting observations included the following: certain restriction fragments containing triplet repeat sequences exhibit anomalously rapid gel electrophoretic mobilities, probably due to their slipped DNA structures; all ten triplet repeat sequences were cloned and investigated for their ability to form underwound DNA conformations; repeating TTA·TAA sequences form an underwound DNA structure; the nucleosome assembly capacity of repeating triplet sequences is anomalous in both cases examined; restriction fragments containing CTG·CAG and CGG·CCG repeats are more flexible and highly writhed than random B DNA and therefore may act as sinks for the accumulation of supercoil density; and nascent DNAs form hairpins during DNA synthesis primer alignment in vitro for the myotonic dystrophy type 1 and fragile X syndrome repeat sequences. All of these investigations have been reviewed (50, 52).
Genomic Rearrangements, DNA Structure, and Human Genomic Disorders—Life in the Wells lab changed dramatically again in the early 2000s when my long-time colleague Dr. Albino Bacolla discovered that breakpoints of gross deletions coincide with non-B DNA conformations (54). This discovery was profound because it amalgamated the topics of DNA structure with human genetics along with bioinformatics and medicine. Breakpoints had been recognized for years to be important in the etiology of various translocations, but this discovery introduced the concept of non-B DNA conformations as signals for the specific breakpoints. This and subsequent work have revealed that slipped structures, cruciforms, triplexes, tetraplexes, and perhaps other non-B DNA structures, including left-handed Z DNA (Fig. 2), are formed in chromosomes and elicit far-reaching genetic consequences via recombination/repair. Repeating sequences, in their non-B conformations, cause gross genomic rearrangements (translocations, deletions, insertions, inversions, and duplications). These rearrangements are the genetic basis for numerous human genomic diseases, including polycystic kidney disease, adrenoleukodystrophy, follicular lymphomas, and spermatogenic failure (55). At least 70 diseases fall into this category (56–58).
The overarching concept is that chromosomal DNA normally exists in an orthodox right-handed B form for most of the time. However, certain non-B DNA conformations (Fig. 2) are formed at specific loci. These non-B conformations are in equilibrium with the right-handed B form. When the sequences are in a non-B conformation, at small direct or inverted repeat homologies, DNA repair systems recognize the altered base pairs at DNA structures that are at or near the breakpoints that trigger recombination/repair events and give rise to double-strand breaks. These double-strand breaks may reside either on the same chromosome or on two distinct chromosomes. After DNA joining and healing, the mutagenic events include gross deletions, translocations, inversions, insertions, and duplications. Through numerous steps in pathology, these genetic events ultimately give rise to genomic disorders (58).
An extremely compelling component of these experiments is the DNA sequence analyses, to the base pair, of the sites of double-strand breakage and rejoining as mediated by the cellular recombination/repair systems; close examination of the sequences of the healed breakpoints reveal a portion (half) of a cruciform (or a triplex) where the cleavage site occurred at the non-paired loops. Thus, the remnants of the non-B DNA structures that actually cause the double-strand break remain as telltale traces of the non-B DNA structure that was once present.
Bioinformatics analyses (59) demonstrate that long homopurine·homopyrimidine sequences that are characteristic of genes expressed in brain and the pseudoautosomal region exist in chromosomes. Thus, these sequences have interesting biological functions.
To tease apart the contributions of DNA sequence versus the non-B DNA conformations adopted by the sequences, Wojciechowska et al. (60) developed an ingenious family of experiments that rigorously demonstrated that the non-B DNA conformations are the culprit regarding mutagenesis, not the sequences per se. Long repeating tracts of CTG·CAG, CCTG·CAGG, and GAA·TTC were studied in E. coli and in three types of mammalian fibroblast-like cells by genetic and biochemical studies, including the in vivo modulation of global negative supercoil density using topoisomerase mutants in E. coli; the in vivo cleavage of hairpin loops that are an obligate consequence of slipped strand structures, cruciforms, and intramolecular triplexes; inactivation of the SbcC protein; and genetic instability studies with plasmids containing long repeating sequence inserts that do or do not adopt non-B structures in vitro. This important work revealed that the non-B DNA conformations are critical for the mutagenesis mechanisms, not the sequence per se in its orthodox right-handed structure.
Folded DNA Structures and Abundance of Simple Repeats in Chromosomes—During the course of our genomic investigations, A. Bacolla et al. (61) recognized that the abundance of certain repeating tetranucleotide sequences was extremely variable in the human genome, ranging from no copies at all to ∼15,000/genome. Upon examination of the sequence features of the repeating tetranucleotides that were absent, he predicted that the sequences that were most likely to fold into hairpin loop arrangements were the sequences that were absent. Biophysical studies on 82 synthetic single-stranded oligonucleotides revealed an inverse correlation between the stability of folded-back hairpin and quadruplex structures and the sequence representation for repeats ≥30 bp in length in nine vertebrate genomes (61). We concluded that DNA structure, i.e. hairpins, quadruplexes, and base stacking, determines the number and length distribution of microsatellite repeats in vertebrate genomes over evolutionary time and may have also potentiated repeat length polymorphisms (61). Hence, an elegant and simple biophysical explanation may be responsible for the abundance of certain types of sequences over evolutionary time.
In summary, DNA structure is a remarkably powerful factor that dictates evolution. Who would have dreamed about the power of DNA sequences and conformations in the 1960s from work on repeating DNA polymers!
University Administration
As stated above, I accepted the Chairmanship of the Department of Biochemistry in the Schools of Medicine and Dentistry at UAB in 1981, where I served until 1990. UAB was undergoing major restructuring and improvements. Thus, this was an exciting time. Furthermore, I accepted the Chairmanship of the Department of Biochemistry and Biophysics in the College of Agriculture and Life Sciences at TAMU in 1990 for a 24-month period while simultaneously serving as the Founding Director of IBT in Houston with satellite research programs in College Station. As part of these administrative responsibilities, I did the following: hired more than 50 faculty and helped them build their educational and research programs (a most rewarding experience), built three new research and education buildings, restructured graduate and undergraduate programs, developed new programs of research and administration, and conducted capital development. Simultaneously, I maintained my active research program. The UAB faculty numbered ∼49 individuals, whereas the TAMU faculty was ∼35 in number.
I never felt compelled to conduct university administrative work but instead saw it as an integral part of the success of our research programs. Thus, it was important in that regard. I always felt that I did not go to graduate school to learn biochemistry to do university administration. In fact, university administration, for its own sake, was unrewarding.
Citizenship at the National Level
I always felt a desire to “give back” to the scientific community by participating in the organizational functions of scientific societies in addition to my research contributions. These scientific societies are of substantial benefit to the research and educational community. Thus, I served as President of the American Society for Biochemistry and Molecular Biology (ASBMB) (2000–2002). ASBMB has ∼12,000 members and is the home of the Journal of Biological Chemistry (JBC). During this time, a new journal (Molecular and Cellular Proteomics) was initiated with the excellent leadership of a number of talented ASBMB members (62). This was, without a doubt, the largest and most expensive initiative ever undertaken by ASBMB. Furthermore, a new newsletter (ASBMB Today) was brought into existence in 2002 to replace the former and more modest newsletter called ASBMB News. Both of these contributions have substantially strengthened our Society.
In addition, I served as an Associate Editor of JBC from 1978 to 1989 after serving for three years on the Editorial Board. This was one of the most rewarding experiences of my life. My interactions with other Associate Editors were lively and enlightening. The leadership of Dr. Herbert Tabor as the Editor of JBC is inspirational. The philosophy engendered in JBC by Herb Tabor should serve as a standard for all scientific journals; Herb believes that the author is important and has certain rights and should be favored in the case of disputes between referees, the Journal, and authors. His leadership is a major reason for the preeminent position of JBC in life sciences.
In 2003–2004, I served as the President of FASEB. FASEB (founded in 1912) is the largest and most prestigious consortium of biomedical research societies in the U. S. The combined membership is ∼86,000 individual scientists and scholars representing 24 biomedical research societies. ASBMB is one of the original societies within FASEB. While President, I initiated and organized a visit to the White House on November 20, 2003 with four Nobel Laureates to meet with Vice President Richard Cheney; Director of the Office of Management and Budget, Mr. Josh Bolton; Deputy Chief of Staff to President Bush, Ms. Harriet Miers; and Presidential Scientific Advisor, Dr. Jack Marburger. After our meeting with Vice President Cheney, he moved approximately $750 million into the National Institutes of Health (NIH) budget on the Thursday of the week of our visit. In addition, I organized a Coalition of American Scientific Society Presidents (CASSP) consisting of the Presidents of the American Physical Society, the American Mathematics Society, the American Chemical Society, and FASEB. The purpose of CASSP was to conduct joint advocacy for scientific research funding in the U. S.
I have always felt that it is important to be involved as a citizen in advocacy for enhanced funding for scientific and medical research. Accordingly, Mr. Peter Farnham (Director of Public Affairs of ASBMB) and I published a Letter to the Editor of Science (63) challenging the scientific community to become more involved in advocacy.
One person can make a difference. I know that from my initiatives in the White House (described above) and from my meetings with Senator Mark Hatfield (Republican, Oregon), who invited me to organize a consortium of prestigious biomedical scientists concerning the 1990 Excellence in Mathematics, Science, and Engineering Act, which was co-sponsored by Senators Hatfield and Kennedy.
The Academic Life
Although I became engaged in my research career almost by accident (4), this all-consuming intellectual challenge has been thoroughly fulfilling. The profound satisfaction of solving a complex biochemical puzzle has been a complete joy. Indeed, the realization that my hypotheses of the mid-1960s to early 1970s were “spot-on” correct as shown by our recent (2003–2008) studies engenders profound fulfillment. A 2008 Google search for “non-B DNA” revealed ∼25.1 million hits, and a PubMed search listed >590,000 references.
In addition, I have had the privilege of facilitating the education and training of a large number (>70) of postdoctoral fellows from the U. S. as well as 16 foreign countries; I contributed to the education of more than 45 Polish postdoctoral fellows as well as their wives, children, etc., who took technical positions, graduated from medical schools in the U. S., etc. This wonderful group of scientists added substantially to my research program over the years. Dr. Adam Jaworski (University of Lodz, Lodz, Poland) was an integral component in facilitating this cooperative program.
Fortunately, I have been funded continuously by NIH for 42 years and by the National Science Foundation (NSF) for 27 years. In fact, I was told that I received the first five-year NSF grant in the 1970s. In addition, I have enjoyed support from at least ten other organizations. The Robert A. Welch Foundation provided an Endowed Chair for the past 18 years; I cherish my affiliation with this Foundation and admire the extremely high standards of the Welch Foundation.
The NIH granting mechanisms within the U. S. are not straightforward. In fact, it was not always possible for me to be deliberate and up-front and declare what I was really working on, namely the role of DNA sequence in conformational properties and genetic processes. Instead, it was necessary for me to “sugarcoat” my proposals with other aims and goals. I have always found the NIH programs to be remarkably unimaginative and risk-averse. Might this have serious long-term consequences for science in the U. S.? Biotechnology industries have been of some interest over the years; I have participated actively with at least 50 chemical, pharmaceutical, biotechnology, legal, petrochemical, venture capital, and other companies and co-founded Alatech Associates, Inc. In general, I have found academic science and education to be far more fulfilling than biotechnology.
I was an Assistant Professor in 1966 at age 28 and Chairman of a department in 1980 at age 42, and my second Chairmanship and Directorship occurred in 1990 at age 52. Unfortunately, I believe that this maturation process has been substantially delayed for the next generation of scientists. This matter has been examined carefully by a national study group (www.amacad.org/), and appropriate action should be taken because it is counterproductive to attracting the best young minds into our science.
I am pleased to acknowledge the satisfaction and joy of mentoring 31 Ph.D. students and my technician, Jacquelynn E. Larson, who worked with me for 40 years and co-authored more than 50 excellent papers. The rough and tumble intellectual exchange was exciting and gave true meaning to the educational process. Everything good that has emanated out of my laboratory is due to my wonderful students and my 70+ postdoctoral fellows (photographs of my lab groups for more than 40 years can be found on my web site at www.ibt.tamhsc.edu/labs/cgr/. Science is a “people business.” People make science move.
Poetry composition is one of my avocations; I have written more than 100 poems. A sample entitled “Phase V” is on my web site at www.ibt.tamhsc.edu/labs/cgr/symposium.html.
While living this exciting and productive academic life, I have been blessed with my wonderful wife, Dotty, for the past 48 years. We have one son, Kevin, and one daughter, Cindy, along with six grandchildren. Life is perfect!
Acknowledgments
This work was supported, in whole or in part, by National Institutes of Health Grant ES11347. This work was also supported by the Friedreich's Ataxia Research Alliance, the Seek-a-Miracle Foundation, and the Robert A. Welch Foundation. I thank Dr. A. Bacolla, J. E. Larson, and D. S. Wells for comments on the manuscript.
References
- 1.Khorana, H. G., Buchi, H., Ghosh, H., Gupta, N., Jacob, T. M., Kossel, H., Morgan, A. R., Narang, S. A., Ohtuska, E., and Wells, R. D. (1966) Polynucleotide synthesis and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 31 39–49 [DOI] [PubMed] [Google Scholar]
- 2.Wells, R. D., and Blair, J. E. (1967) Studies on polynucleotides. LXXI. Sedimentation and buoyant density studies of some DNA-like polymers with repeating nucleotide sequences. J. Mol. Biol. 27 273–288 [DOI] [PubMed] [Google Scholar]
- 3.Wells, R. D., and Wartell, R. M. (1974) in Biochemistry Series One: Biochemistry of Nucleic Acids (Burton, K., ed) Vol. 6, pp. 41–64, Butterworth Publishers, UK [Google Scholar]
- 4.Wells, R. D. (2002) How I became a biochemist. IUBMB Life 53 137–138 [DOI] [PubMed] [Google Scholar]
- 5.Wells, R. D. (1969) Actinomycin binding to DNA: Inability of a DNA containing guanine to bind actinomycin D. Science 165 75–76 [DOI] [PubMed] [Google Scholar]
- 6.Wells, R. D., Goodman, T. C., Hillen, W., Horn, G. T., Klein, R. D., Larson, J. E., Muller, U. R., Neuendorf, S. K., Panayotatos, N., and Stirdivant, S. M. (1980) DNA structure and gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 24 167–267 [DOI] [PubMed] [Google Scholar]
- 7.Wells, R. D., Blakesley, R. W., Burd, J. F., Chan, H. W., Dodgson, J. B., Hardies, S. C., Horn, G. T., Jensen, K. F., Larson, J., Nes, I. F., Selsing, E., and Wartell, R. M. (1977) The role of DNA structure in genetic regulation. Crit. Rev. Biochem. 4 305–340 [DOI] [PubMed] [Google Scholar]
- 8.Morgan, A. R., and Wells, R. D. (1968) Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J. Mol. Biol. 37 63–80 [DOI] [PubMed] [Google Scholar]
- 9.Felsenfeld, G., and Miles, H. T. (1967) The physical and chemical properties of nucleic acids. Annu. Rev. Biochem. 36 407–408 [DOI] [PubMed] [Google Scholar]
- 10.Chamberlin, M. J. (1965) Comparative properties of DNA, RNA, and hybrid homopolymer pairs. Fed. Proc. 24 1446–1457 [PubMed] [Google Scholar]
- 11.Mitsui, Y., Langridge, R., Shortle, B. E., Cantor, C. R., Grant, R. C., Kodama, M., and Wells, R. D. (1970) Physical and enzymatic studies on poly d(I-C)·poly d(I-C), an unusual double-helical DNA. Nature 228 1166–1169 [DOI] [PubMed] [Google Scholar]
- 12.Grant, R. C., Kodama, M., and Wells, R. D. (1972) Enzymatic and physical studies on (dI-dC)n·(dI-dC)n and (dG-dC)n·(dG-dC)n. Biochemistry 11 805–815 [DOI] [PubMed] [Google Scholar]
- 13.Pohl, F., and Jovin, T. M. (1972) Salt-induced cooperative conformational change in a synthetic DNA: Equilibrium and kinetic studies with poly(dG-dC). J. Mol. Biol. 67 375–396 [DOI] [PubMed] [Google Scholar]
- 14.Wang, A. H., Quigley, G. J., Kolpak, F. J., Crawford, J. L., van Boom, J. H., van der Marel, G., and Rich, A. (1979) Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282 680–686 [DOI] [PubMed] [Google Scholar]
- 15.Drew, H., Takano, T., Tanaka, S., Itakura, K., and Dickerson, R. E. (1980) High-salt d(CpGpCpG), a left-handed Z-DNA double helix. Nature 286 567–573 [DOI] [PubMed] [Google Scholar]
- 16.Wells, R. D., Amirhaeri, S., Blaho, J. A., Collier, D. A., Dohrman, A., Griffin, J. A., Hanvey, J. C., Hsieh, W.-T., Jaworski, A., Larson, J. E., McLean, M. J., Rahmouni, A., Rajagopalan, M., Shimizu, M., Wohlrab, F., and Zacharias, W. (1990) in The Organization of the Bacterial Chromosome (Drlica, K., and Riley, M., eds) pp. 187–194, American Society of Microbiology, Washington, D. C.
- 17.Hebert, A., and Rich, A. (1996) The biology of Left-handed Z-DNA. J. Biol. Chem. 271 11595–11598 [DOI] [PubMed] [Google Scholar]
- 18.Blakesley, R. W., and Wells, R. D. (1975) “Single-stranded” DNA from φX174 and M13 is cleaved by certain restriction endonucleases. Nature 257 421–422 [DOI] [PubMed] [Google Scholar]
- 19.Blakesley, R. W., Dodgson, J. B., Nes, I. F., and Wells, R. D. (1977) Duplex regions in “single-stranded” φX174 DNA are cleaved by a restriction endonuclease from Haemophilus aegyptius. J. Biol. Chem. 252 7300–7306 [PubMed] [Google Scholar]
- 20.Horiuchi, K., and Zinder, N. D. (1975) Site-specific cleavage of single-stranded DNA by a Hemophilus restriction endonuclease. Proc. Natl. Acad. Sci. U. S. A. 72 2555–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selsing, E., Wells, R. D., Early, T. A., and Kearns, D. R. (1978) Two contiguous conformations in a nucleic acid duplex. Nature 275 249–250 [DOI] [PubMed] [Google Scholar]
- 22.Selsing, E., and Wells, R. D. (1979) Polynucleotide block polymers consisting of a DNA·RNA hybrid joined to a DNA·DNA duplex. J. Biol. Chem. 254 5410–5416 [PubMed] [Google Scholar]
- 23.Selsing, E., Wells, R. D., Alden, C. J., and Arnott, S. (1979) Bent DNA: visualization of a base-paired and stacked A-B conformational junction. J. Biol. Chem. 254 5417–5422 [PubMed] [Google Scholar]
- 24.Panayotatos, N., and Wells, R. D. (1981) Cruciform structures in supercoiled DNA. Nature 289 466–470 [DOI] [PubMed] [Google Scholar]
- 25.Lilley, D. M. J. (1980) The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 77 6468–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halvorson, H. O. (2007) Development of molecular biology at the University of Wisconsin, Madison. Biol. Cell 99 717–724 [DOI] [PubMed] [Google Scholar]
- 27.Jaworski, A., Hsieh, W.-T., Blaho, J. A., Larson, J. E., and Wells, R. D. (1987) Left-handed DNA in vivo. Science 238 773–777 [DOI] [PubMed] [Google Scholar]
- 28.Wells, R. D., Brennan, R., Chapman, K. A., Goodman, T. C., Phillip, Hart, A., Hillen, W., Kellogg, D. R., Kilpatrick, M. W., Klein, R. D., Klysik, J., Lambert, P., F., Larson, J. E., Miglietta, J. J., Neuendorf, S. K., O'Connor, T. R., Singleton, C. K., Stirdivant, S. M., Veneziale, C. M., Wartell, R. M., and Zacharias, W. (1982) Left-handed DNA helices, supercoiling, and the B-Z junction. Cold Spring Harbor Symp. Quant. Biol. 47 77–84 [DOI] [PubMed] [Google Scholar]
- 29.Blaho, J. A., and Wells, R. D. (1989) Left-handed Z-DNA and genetic recombination. Prog. Nucleic Acid Res. Mol. Biol. 37 107–126 [DOI] [PubMed] [Google Scholar]
- 30.Shimizu, M., Hanvey, J. C., and Wells, R. D. (1989) Intramolecular DNA triplexes in supercoiled plasmids. I. Effect of loop size on formation and stability. J. Biol. Chem. 264 5944–5949 [PubMed] [Google Scholar]
- 31.Wells, R. D. (1988) Unusual DNA Structures. J. Biol. Chem. 263 1095–1098 [PubMed] [Google Scholar]
- 32.Kitchin, P. A., Klein, V. A., Ryan, K. A., Gann, K. L., Raush, C. A., Kang, D. S., Wells, R. D., and Englund, P. T. (1986) A highly bent fragment of Crithidia fasciculata kinetoplast DNA. J. Biol. Chem. 261 11302–11309 [PubMed] [Google Scholar]
- 33.Marini, J. C., Levene, S. D., Crothers, D. M., and Englund, P. T. (1982) Bent helical structure in kinetoplast DNA. Proc. Natl. Acad. Sci. U. S. A. 79 7664–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffith, J. D., Laundon, C. H., Rauch, C. A., Englund, P. T., Hsieh, W.-T., and Wells, R. D. (1988) Use of electron microscopy to examine sequence-directed DNA bending. J. Biomol. Struct. Dyn. 3 25–37 [Google Scholar]
- 35.Spurio, R., Falconi, M., Brandi, A., Pon, C. L., and Gualerzi, C. O. (1997) The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tissier, A., Kannouche, P., Mauffrey, P., Allemand, I., Frelat, G., Devoret, R., and Angulo, J. F. (1996) Molecular cloning and characterization of the mouse Kin17 gene coding for a Zn-finger protein that preferentially recognizes bent DNA. Genomics 38 238–242 [DOI] [PubMed] [Google Scholar]
- 37.Singleton, C. K., and Wells, R. D. (1982) Relationship between superhelical density and cruciform stability in plasmid pVH51. J. Biol. Chem. 257 6292–6295 [PubMed] [Google Scholar]
- 38.Singleton, C. K. (1983) Effects of salts, temperature, and stem length on supercoil induced formation of cruciforms. J. Biol. Chem. 258 7661–7668 [PubMed] [Google Scholar]
- 39.McLean, M. J., Larson, J. E., Wohlrab, F., and Wells, R. D. (1987) Reaction conditions affect the specificity of bromoacetaldehyde as a probe for DNA cruciforms and B-Z junctions. Nucleic Acids Res. 15 6917–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLean, M. J., and Wells, R. D. (1988) The role of DNA sequence in the formation of Z-DNA versus cruciforms in plasmids. J. Biol. Chem. 263 7370–7377 [PubMed] [Google Scholar]
- 41.Blaho, J. A., Larson, J. E., McLean, M. J., and Wells, R. D. (1988) Multiple DNA secondary structures in perfect inverted repeat inserts in plasmids. Right-handed B-DNA, cruciforms, and left-handed Z-DNA. J. Biol. Chem. 263 14446–14455 [PubMed] [Google Scholar]
- 42.Lilley, D. M. J. (2000) Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 33 109–159 [DOI] [PubMed] [Google Scholar]
- 43.Hanvey, J. C., Klysik, J., and Wells, R. D. (1988) Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine·oligopyrimidine inserts in plasmids. J. Biol. Chem. 263 7386–7396 [PubMed] [Google Scholar]
- 44.Wells, R. D., Collier, D. A., Hanvey, J. C., Shimizu, M., and Wohlrab, F. (1988) The chemistry and biology of unusual DNA structures adopted by oligopurine·oligopyrimidine sequences. FASEB J. 2 2939–2949 [PubMed] [Google Scholar]
- 45.Hanvey, J. C., Shimizu, M., and Wells, R. D. (1988) Intramolecular DNA triplexes in supercoiled plasmids. Proc. Natl. Acad. Sci. U. S. A. 85 6292–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells, R. D., Amirhaeri, S., Blaho, J. A., Collier, D. A., Dohrman, A., Griffin, J. A., Hanvey, J. C., Jaworski, A., Larson, J. E., Rahmouni, A., Rajagopalan, M., Shimizu, M., Wohlrab, F., and Zacharias, W. (1990) ICN-UCLA Symp. Mol. Cell. Biol. 127 79–91 [Google Scholar]
- 47.Wohlrab, F., and Wells, R. D. (1989) Slight changes in conditions influence the family of non-B DNA conformations of the herpes simplex virus type 1 DR2 repeats. J. Biol. Chem. 264 8207–8213 [PubMed] [Google Scholar]
- 48.Wohlrab, F., Chatturjee, S., and Wells, R. D. (1991) The herpes simplex virus I segment inversion site is specifically cleaved by a virus-Induced nuclear endonuclease. Proc. Natl. Acad. Sci. U. S. A. 88 6432–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells, R. D., and Ashizawa, T. (eds) (2006) Genetic Instabilities and Neurological Diseases, 2nd Ed., Elsevier Science Publishing Co., Inc., New York
- 50.Wells, R. D., Dere, R., Hebert, M., Napierala, M., and Son, L. S. (2005) Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 33 3785–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jakupciak, J. P., and Wells, R. D. (2000) Genetic instabilities of triplet repeat sequences by recombination. IUBMB Life 50 355–359 [DOI] [PubMed] [Google Scholar]
- 52.Bowater, R. P., and Wells, R. D. (2001) The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 66 159–202 [DOI] [PubMed] [Google Scholar]
- 53.Wells, R. D. (2008) DNA triplexes and Friedreich ataxia. FASEB J. 22 1625–1634 [DOI] [PubMed] [Google Scholar]
- 54.Bacolla, A., Jaworski, A., Larson, J. E., Jakupciak, J. P., Chuzhanova, N., Abeysinghe, S. S., O'Connell, C. D., Cooper, D. N., and Wells, R. D. (2004) Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. U. S. A. 101 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bacolla, A., Wojciechowska, M., Kosmider, B., Larson, J. E., and Wells, R. D. (2006) The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair 5 1161–1170 [DOI] [PubMed] [Google Scholar]
- 56.Wells, R. D. (2007) Non-B conformations, mutagenesis, and diseases. Trends Biochem. Sci. 32 271–278 [DOI] [PubMed] [Google Scholar]
- 57.Bacolla, A., and Wells, R. D. (2006) in Genomic Disorders: The Genomic Basis of Disease. Part II: Genomic Structure (Lupski, J. R., and Stankiewicz, P., eds) pp. 89–99, Humana Press, Totowa, NJ
- 58.Bacolla, A., and Wells, R. D. (2004) Non-B DNA conformations, genomic rearrangements, and genetic diseases. J. Biol. Chem. 279 47411–47414 [DOI] [PubMed] [Google Scholar]
- 59.Bacolla, A., Collins, J. R., Gold, B., Chuzhanova, N., Yi, M., Stephens, R. M., Stefanov, S., Olsh, A., Jakupciak, J. P., Dean, M., Lempicki, R. A., Cooper, D. N., and Wells, R. D. (2006) Long homopurine·homopyrimidine sequences are characteristic of genes expressed in brain and in the pseudoautosomal region. Nucleic Acids Res. 34 2663–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wojciechowska, M., Napierala, M., Larson, J. E., and Wells, R. D. (2006) Non-B DNA structures formed by long DNA repeats of DM1, DM2, and FRDA genes, not the sequences per se, promote mutagenesis in flanking DNA. J. Biol. Chem. 281 24531–24543 [DOI] [PubMed] [Google Scholar]
- 61.Bacolla, A., Larson, J. E., Collins, J. R., Li, J., Milosavljelic, A., Stenson, P. D., Cooper, D. N., and Wells, R. D. (2008) Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 18 1545–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wells, R. D. (2002) The birth of molecular and cellular proteomics. Mol. Cell. Proteomics 1 1–2 [Google Scholar]
- 63.Wells, R. D., and Farnham, P. (2006) Why aren't there more scientists advocating for funding? Science, 314 1081. [DOI] [PubMed] [Google Scholar]