Abstract
Eukaryotes harbor a highly conserved mitochondrial pathway for fatty acid synthesis (FAS), which is completely independent of the eukaryotic cytosolic FAS apparatus. The activities of the mitochondrial FAS system are catalyzed by soluble enzymes, and the pathway thus resembles its prokaryotic counterparts. Except for octanoic acid, which is the direct precursor for lipoic acid synthesis, other end products and functions of the mitochondrial FAS pathway are still largely enigmatic. In addition to low cellular levels of lipoic acid, disruption of genes encoding mitochondrial FAS enzymes in yeast results in a respiratory-deficient phenotype and small rudimentary mitochondria. Recently, two distinct links between mitochondrial FAS and RNA processing have been discovered in vertebrates and yeast, respectively. In vertebrates, the mitochondrial 3-hydroxyacyl-acyl carrier protein dehydratase and the RPP14 subunit of RNase P are encoded by the same bicistronic transcript in an evolutionarily conserved arrangement that is unusual for eukaryotes. In yeast, defects in mitochondrial FAS result in inefficient RNase P cleavage in the organelle. The intersection of mitochondrial FAS and RNA metabolism in both systems provides a novel mechanism for the coordination of intermediary metabolism in eukaryotic cells.
Mitochondrial research has been enjoying a renaissance during the last two decades because of major discoveries of previously unknown or overlooked processes such as mitochondrial fusion and fission, mechanisms and regulation of transcription and translation, iron-sulfur cluster biogenesis, structure and assembly of respiratory chain complexes, mitochondria-to-nucleus “retrograde” signaling, mechanisms of mitochondrial inheritance, programmed cell death, and the role of mitochondria in human disease. This review focuses on the process of fatty acid biosynthesis discovered relatively recently in mitochondria.
The de novo synthesis of fatty acids in eukaryotes can take place in at least two subcellular compartments: in the cytoplasm (FAS2 type I) and in mitochondria (FAS type II). Type II synthesis has its genesis in bacteria and is also found in plant chloroplasts (1). Why has the bacterial type FAS pathway been maintained in mitochondria, when the “classic” cytoplasmic pathway provides most of the cellular fatty acids? Surprisingly, respiratory competence in yeast is dependent on the ability of mitochondria to synthesize fatty acids.
Mitochondria in almost all eukaryotic organisms contain their own genome encoding a small number of primarily hydrophobic subunits of the respiratory chain complexes and ATP synthase (2). The genes are transcribed mainly in one or two long transcripts in many organisms from mammals to Schizosaccharomyces pombe (reviewed in Refs. 3 and 4) or as several multigenic transcripts such as in Saccharomyces cerevisiae (reviewed in Ref. 5). tRNA processing is essential for the expression of mitochondrial mRNAs because tRNAs are interspersed between the mRNAs in the genome-long vertebrate precursor RNA (3) and are also co-transcribed with many of the Saccharomyces protein-encoding mRNAs (5). RNase P is responsible for the endonucleolytic cleavages at the 5′-ends of the mature tRNAs (6), whereas a distinct tRNA endonuclease frees the 3′-ends (7). We have shown that fatty acid biosynthesis in mitochondria is linked to RNase P expression in vertebrates (8) and assembly and/or activity in Saccharomyces (9).
An intersection of two pathways frequently provides a point for controlling metabolic fluxes. In some systems, intersecting pathways are switched on or off in parallel, whereas in others, the pathways are regulated reciprocally. Our hypothesis is that the intersection of the mitochondrial FAS II pathway with RNA processing has been maintained throughout evolution as a means to regulate mitochondrial function relative to the nutritional state of the cell. The details of the intersection in yeast and vertebrates are distinct. The mitochondrial FAS pathway in yeast controls the maturation or activity of mitochondrial RNase P, which cleaves the 5′-leaders of mitochondrial precursor tRNAs (9). In humans and in other vertebrates as well, a nuclear bicistronic mRNA encodes both the RPP14 subunit of RNase P and 3-hydroxyacyl-ACP dehydratase (HTD2) in the FAS II pathway (8).
Mitochondrial FAS Enzymes and Pathways
The cytosolic FAS type I pathway comprises one (a homodimer of α2-subunits in higher eukaryotes) or two (a heterododecamer of α6β6-subunits in fungi) multifunctional polypeptides, whereas the mitochondrial pathway comprises independent monofunctional polypeptides that carry out individual steps in FAS (Fig. 1). The first component identified in the mitochondrial pathway was isolated from Neurospora crassa. Labeling with [14C]pantothenic acid led to the identification of an ACP (10), and it was shown subsequently that the Neurospora protein is indeed involved in de novo synthesis of fatty acids in mitochondria (11). Other eukaryotes were also shown to contain a mitochondrial form of ACP (12). The initial identification of mitochondrial ACP as a structural component of the membrane-bound bovine mitochondrial respiratory Complex I was puzzling (13), but it was later shown that mammalian mitochondria also contain a soluble form of ACP (14).
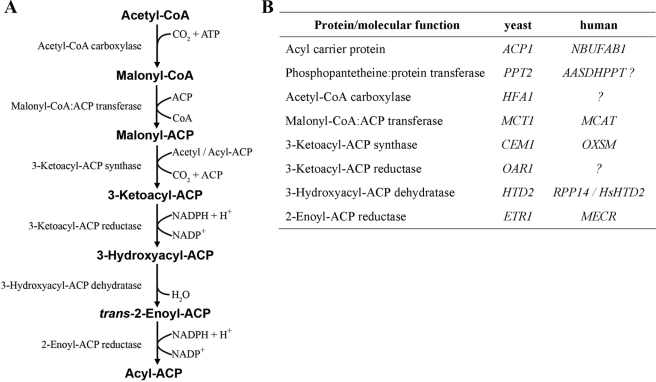
FIGURE 1.
Reactions and proteins of the mitochondrial FAS pathway. A, individual reactions of the FAS II pathway. As discussed in text, it remains to be experimentally demonstrated that generation of acyl-ACP exceeds a chain length of C8 in vivo and that longer fatty acids have a role in mitochondrial physiology. B, protein components of the mitochondrial FAS pathway and the corresponding genes in yeast and humans. All proteins of the mitochondrial FAS pathway required to synthesize saturated fatty acids have been identified and at least partially characterized in bakers' yeast. The enzymes responsible for carboxylation of acetyl-CoA and the 3-ketoacyl-ACP reduction reaction in humans have not yet been determined.
Since the initial discovery of mitochondrial ACP, the mitochondrial FAS pathway has been well characterized, particularly in S. cerevisiae. This model organism does not contain Complex I but expresses mitochondrial ACP (encoded by the ACP1 gene) and the entire remaining retinue of enzymes required for the synthesis of saturated fatty acids in mitochondria. The yeast malonyl-CoA:ACP transferase (Mct1) (15), phosphopantetheine transferase (Ppt2) (16), ketoacyl synthase (Cem1) (17), and ketoacyl reductase (Oar1) (15) are very similar to their prokaryotic counterparts, whereas the yeast HFA1 gene has been found to encode a mitochondrial acetyl-CoA carboxylase homologous to cytosolic Acc1 (18). The yeast proteins that catalyze the final two steps of the fatty acid elongation cycle, Htd2 and 2-enoyl-ACP reductase (Etr1), do not have clear sequence similarities to known prokaryotic FAS type II enzymes. Mutations in the yeast HTD2 gene were identified through a plasmid-loss, colony-sectoring screen in which the complementing Escherichia coli homolog was expressed on a plasmid (19). Our search for 2-enoyl-CoA reductase activity in yeast led to the identification of mitochondrial Etr1 in Candida tropicalis and its homolog in S. cerevisiae, Etr1/Mrf1′ (20). In contrast to an initial observation of nuclear localization for Etr1 (21), a series of subsequent experiments demonstrated that the bulk of the protein was localized to the mitochondrial matrix (20). Etr1 displayed enoyl thioester reductase activity and was necessary to support the growth of yeast cells on a non-fermentable carbon source (20).
With the exception of the hfa1Δ mutant, which is completely respiratory-deficient only at higher temperatures, yeast strains with lesions in the mitochondrial FAS pathway exhibit several severe mitochondrial dysfunction phenotypes when grown at 30 °C. All of the strains are respiratory-deficient, exhibit a loss of mitochondrial cytochromes, and have low levels of lipoic acid. In contrast to the small mitochondria seen in deletion mutants, overexpression of Etr1 or Htd2 results in swelling of the mitochondrial compartment (19, 20).
Subsequent to the yeast work, several human FAS type II homologs were isolated and characterized (Fig. 1), and most have been shown to complement the yeast mutant phenotypes (8, 22, 23). In the cases that have been reported, the mammalian genes show strong expression in tissues with a high rate of respiration.
Lipoic Acid and Beyond
Neither the range of fatty acids produced by the mitochondrial FAS II pathway nor their roles in cellular metabolism have been determined. In fungi, there is good experimental support for the hypothesis that the pathway is the sole source of the octanoic acid precursor required for the production of the lipoic acid cofactor essential for PDH, α-ketoglutarate dehydrogenase, and glycine cleavage system function (9, 24–26). Witkowski et al. (26) demonstrated carbon flow from a two-carbon precursor to lipoic acid to the apoH protein (lipoic acid acceptor) of the glycine cleavage system when bovine heart mitochondrial soluble extract was incubated with labeled acetyl-ACP and malonyl-CoA. Lipoic acid is regarded as a nutritional requirement for mammals (27), and mammalian mitochondrial enzymes required for the attachment of the free acid to target proteins have been characterized (28, 29). Thus, it was puzzling when a cDNA encoding a mitochondrial lipoic acid synthase was identified in a mammalian heart library (30). Subsequently, it was shown that homozygotic inactivation of the lipoic acid synthase gene in mouse causes embryonic lethality, and embryo survival cannot be rescued by supplementing the diet of pregnant mothers with lipoic acid (31). Thus, critical developmental processes require that lipoic acid be synthesized in vivo in mammalian mitochondria.
In addition to octanoic acid, there is evidence accumulating that the FAS II pathway also synthesizes longer fatty acids. In all cases, enzymes of the mitochondrial FAS pathway (KAS/CEM1, HTD2, and MECR/ETR1) show broad substrate specificity with regard to chain length. The active sites accept substrates with short chains (C2) all the way up to C14–16 fatty acid derivatives (22, 32, 33). Interestingly, the recently published crystal structure of human MECR/ETR1 revealed a ligand-binding pocket deep enough to accommodate acyl groups up to 16 carbons in chain length (33). These observations of active-site pocket size suggest that the mitochondrial FAS pathway may produce fatty acids longer than octanoic acid. Curiously, the human CEM1 enzyme shows biphasic catalytic efficiencies, peaking with the use of C6 and C10 substrates, as does ETR1 with C8 and C12–14 substrates (22, 33). These kinetic properties may facilitate the generation and accumulation of the octanoyl-ACP required for lipoic acid production, and some ACP molecules may have the acyl chains extended.
Data obtained from work with isolated Neurospora mitochondria indicate that the longest fatty acids produced by the mitochondrial FAS pathway in this fungus are myristic (C14) and hydroxymyristic acids (11). The Trypanosoma brucei mitochondrial FAS pathway synthesizes C16 palmitoic acid (34). Where do these longer fatty acids end up? Interestingly, disruption of the T. brucei mitochondrial FAS pathway by the introduction of small interfering RNA against ACP resulted in cellular phospholipid composition changes (35). Some of the phenotypes of yeast mitochondrial FAS-deficient mutants such as loss of cytochromes and mitochondrial swelling upon overexpression of Etr1 or Htd2 cannot be explained simply by the loss or overproduction of octanoic acid/lipoic acid alone, and therefore, a physiological function for longer fatty acids produced by the mitochondrial FAS pathway or derivatives thereof must be postulated.
In addition to lipoic acid, myristoyl-ACP was generated in the bovine heart mitochondrial extracts mentioned above (26). Although a number of transferases using acyl-ACPs instead of CoA esters as acyl group donors and glycerol-containing lipids as acceptors are found in plastids and prokaryotes (36), to the best of our knowledge, this type of transferase has not been identified in mammals. Therefore, perhaps with the exception of the 3-hydroxymyristyl-ACP found in Complex I of bovine heart mitochondria (37), the destiny of other long chain acyl groups synthesized on ACP in protists, parasites, and mammals remains to be determined.
Fatty Acid/Lipoic Acid Synthesis Is Required for tRNA Processing in Yeast
Some 15 years ago, there was an initial hint of a role for mitochondrial lipids in mitochondrial RNA metabolism. It was shown that mitochondrial tRNA processing in S. cerevisiae was perturbed in a strain that had a mutation in LIP5, which encodes lipoic acid synthase (38). This enzyme inserts two sulfurs into octanoic acid to form lipoic acid (39). Mitochondrial tRNA processing requires RNase P, a ribonucleoprotein complex that processes the 5′-ends of tRNAs. In turn, the assembly of RNase P requires processing of a mitochondrial precursor RNA containing the RPM1 RNA subunit of RNase P and tRNAPro. A crucial early step in processing is the RNase P cleavage at the 5′-end of the tRNA, which releases RPM1 RNA for further 5′- and 3′-trimming steps. Fully processed RPM1 RNA assembles with the Rpm2 protein to form the active holoenzyme (40).
A recent screen in S. cerevisiae for mutants defective in mitochondrial RNA processing focused on identifying genes encoding previously uncharacterized enzymes involved in the 5′- and 3′-trimming of multigenic precursor RNAs (9). A strain with a deletion in the HTD2 gene, which encodes the dehydratase in the mitochondrial FAS pathway, was deficient in processing of the RPM1 precursor RNA, specifically at the RNase P cleavage site, which releases tRNAPro from the RPM1-tRNAPro precursor RNA (Fig. 2).
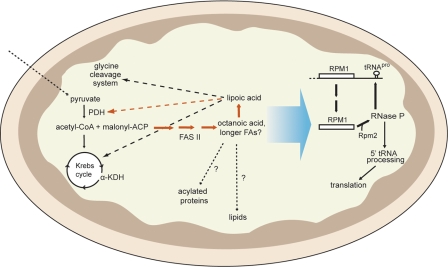
FIGURE 2.
Metabolic fluxes and functional links associated with the mitochondrial FAS type II pathway in yeast. Solid arrows represent metabolic fluxes, and dashed arrows indicate functional links. The large arrow indicates that an unidentified product of fatty acid or lipoic acid synthesis or a derivative thereof affects the efficiency of RNase P processing of the RPM1-tRNAPro precursor RNA. Alternative sources of acetyl-CoA other than that generated by PDH (pyruvate bypass, amino acid breakdown, or transfer of extramitochondrially produced acetyl units) have been omitted for clarity. FAs, fatty acids; α-KDH, α-ketoglutarate dehydrogenase.
How does Htd2, a FAS enzyme, influence RNA processing? Several cases of mitochondrial proteins that have two independent functions have been reported. For example, the Neurospora tyrosyl-tRNA synthetase is required also for the excision of Group I introns in mitochondrial precursor RNAs (41). Different surfaces of the enzyme bind to tRNA and to Group I introns, respectively (42). In another case, the Saccharomyces mitochondrial RNA polymerase has a separate domain that is required for protein synthesis (43). To investigate whether Htd2 is also a dual-function protein, we surveyed the deletion strains affecting FAS II enzymes and found that they were all deficient in RPM1 RNA maturation. Thus, accumulation of the RPM1-tRNAPro precursor RNA was not the result of a defect in one protein directly involved in RNA processing but was due to loss of a product of the complete mitochondrial FAS pathway.
How is the FAS type II biosynthetic pathway tied to tRNA processing through the biogenesis or activity of RNase P? In yeast, a product of the mitochondrial FAS pathway is required for either 1) specific maturation of RPM1 RNA and subsequent assembly of the RNase P holoenzyme or 2) enhancement of RNase P activity. A simple explanation would be that the Rpm2 protein component of RNase P undergoes a hitherto undiscovered lipoic acylation, but we have shown that this is not the case (9). Alternatively, a fatty acid or lipoic acid could associate non-covalently with the RNase P enzyme or with the substrate, or another protein with fatty acid/lipoic acid association could chaperone RNase P assembly or activity. Our hypothesis that a mitochondrial FAS pathway product plays a direct role in the maturation of RNase P is reinforced by the finding that these pathways are linked also in vertebrates. Although the details of this connection are different for yeast and vertebrates, the outcome may be the same. The connections in both phyla hint at a mechanism for the regulation of mitochondrial gene expression in response to cellular metabolism.
Genetic Linkage between a Mitochondrial FAS Enzyme and an RNase P Subunit in Vertebrates
Human mitochondrial FAS II components such as malonyl-CoA:ACP transferase (44), β-ketoacyl synthase (OXSM) (22), and ETR1 (23) were identified based on their similarity to corresponding bacterial and yeast proteins, but human homologs of fungal Htd2 or prokaryotic fabA-or fabZ-type dehydratases were not found by sequence matching (19). Instead, cDNAs encoding the human dehydratase were identified by a functional cloning approach in which the respiratory-deficient htd2-1 yeast mutant strain was transformed with human cerebellum and kidney libraries, and transformants were selected for their ability to grow on medium containing a non-fermentable carbon source (8). The isolation of human HTD2 allowed the identification of a highly similar mitochondrial homolog in T. brucei, which had evaded characterization before the identity of the human enzyme was established (32). In addition, the recently characterized Mycobacterium tuberculosis 3-hydroxyacyl-ACP dehydratase is homologous to human HTD2 (45). Interestingly, all three proteins resemble the phaJ-type dehydratases involved in polyhydroxyalkanoate synthesis rather than the paradigmal E. coli fabA or fabZ dehydratases.
Surprising, however, was the finding that the plasmids rescuing the htd2-1 yeast mutation contained human RPP14 cDNAs, which encode one subunit of the mammalian mitochondrial RNase P complex. Closer analysis of the cDNAs revealed an additional 3′-open reading frame encoding mitochondrial HTD2. The HTD2 gene was shown to be responsible for rescuing the yeast mutation. The RPP14-HTD2 bicistronic mRNA encodes two proteins with seemingly widely disparate roles and is expressed most abundantly in heart and liver, human tissues with robust mitochondrial function.
The emergence of this unusual bicistronic arrangement in bony fish implies that the mRNA structure has been preserved for 400 million years. Almost all eukaryotic mRNAs are monocistronic because translation is initiated by the small ribosomal subunit, which scans from the cap along the 5′-untranslated region in search of the first AUG start codon (46). How translation of the HTD2 dehydratase coding sequence is initiated 121 nucleotides 3′ of the RPP14 stop codon is a topic for future investigation. Regardless of the mechanism of translation initiation for the downstream reading frame, the fact that the physical association has been maintained over evolutionary time suggests that this arrangement is not spurious but allows co-transcriptional regulation and is advantageous in the coordination of mitochondrial gene expression.
Regulatory Loops
We propose that a positive feedback loop exists in yeast to regulate mitochondrial function in response to pyruvate availability in glucose-grown cells (Fig. 2). FAS produces octanoic acid, which is the substrate for lipoic acid synthesis. Lipoic acid is attached to the E2 subunit of PDH and is required for the conversion of pyruvate to acetyl-CoA, which feeds into the FAS pathway. Thus, when the pyruvate supply is low, less acetyl-CoA would be available for the FAS pathway, resulting in lower levels of lipoic acid and PDH activity. A second positive feedback loop exists in the yeast mitochondrial RNase P maturation process, in which processing of the RNase P RNA precursor, RPM1-tRNAPro, requires the action of the RNase P enzyme itself (Fig. 2). Our data show that these two regulatory loops are linked; disruption of the FAS pathway results in lower levels of processed RPM1-tRNAPro precursor RNA and all other mitochondrial tRNAs (9). Although not substantiated experimentally, this feedback loop may have a direct effect on the synthesis of mitochondrially encoded components of the respiratory chain complexes.
Concluding Remarks and Future Directions
The conservation of mitochondrial FAS argues against the idea that the pathway is an evolutionary remnant of the ancestral endosymbiont. Rather, the view that this pathway plays a key role in cellular metabolism is supported by the observation that inactivation of any of the genes encoding mitochondrial FAS components in bakers' yeast results in compromised mitochondrial respiration. The simplest explanation for the respiratory deficiency of these mutants is a lack of cellular lipoic acid, which leads to subsequent inactivation of α-keto acid dehydrogenase complexes. However, the molecular link between the loss of lipoic acid and inefficient mitochondrial RNA processing in yeast has not been explained, and the elucidation of this connection will remain a major challenge for our future work. Intriguingly, diminished mitochondrial tRNA processing due to restricted mitochondrial RNase P activity may affect the rate of mitochondrial protein synthesis and explain the observed loss of functional respiratory complexes. Hence, the exploration at the molecular level of how a product of the FAS pathway affects the intramitochondrial maturation of the RPM1 RNA and tRNA processing in yeast should lead to a deeper understanding of how mitochondria coordinate mitochondrial metabolism with gene expression.
The link between the mitochondrial FAS II pathway and RNA processing is also reflected in the co-transcription of mammalian HTD2 with a subunit of RNase P. How vertebrate HTD2 is translated from the RPP14-HTD2 transcript is another unanswered question. This unusual bicistronic transcript may prove to be an interesting candidate for the study of regulatory mechanisms affecting translation initiation downstream of a stop codon in a higher eukaryote.
Of the remaining uncharacterized components of mitochondrial FAS in mammals, 3-ketoacyl-ACP reductase has resisted identification. Likewise, it is not clear which protein(s) are responsible for the generation of malonyl-CoA. Because all other components of the mitochondrial FAS pathway are localized in the mitochondrial matrix, 1) the malonyl group must be transported across the mitochondrial membranes from the cytosol to the mitochondria, or 2) malonyl-CoA must be synthesized in the mitochondrial matrix. It is noteworthy in this context that human mitochondrial malonyl-CoA:ACP transferase has been identified (44), and characterization of the human homolog of yeast Cem1, OXSM, has demonstrated the dependence of the condensing reaction on the presence of malonyl-ACP in the mitochondrial matrix (22).
Data available from reconstitution experiments show that the components of the mitochondrial FAS pathway can generate acyl groups up to a C16 chain length in vitro (26). This result is in agreement with the kinetic data obtained with purified mitochondrial FAS enzymes from various sources (22, 33). However, the ultimate function of the long chain acyl groups remains a mystery. One candidate for an example of long chain acylation is 3-hydroxymyristyl-ACP, a component of Complex I of mammalian mitochondria (37). Even though the phospholipid composition of mitochondria was shown to be affected in yeast strains with a disrupted mitochondrial FAS pathway (17), it is not known whether this is a direct effect due to decreased channeling of mitochondrially synthesized fatty acids into phospholipids or a pleiotropic effect due to a disturbed mitochondrial metabolism. Future labeling experiments designed to follow mitochondrially synthesized fatty acids and destination thereof may help to answer these important questions.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM34893 (to C. L. D.). This work was also supported by a grant from the Sigrid Juselius Foundation, the Academy of Finland, and Nordic Center of Excellence Programme in Food, Nutrition, and Health Project (070010) “MitoHealth” (to J. K. H.). This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: FAS, fatty acid synthesis; ACP, acyl carrier protein; PDH, pyruvate dehydrogenase.
References
- 1.White, S. W., Zheng, J., Zhang, Y. M., and Rock, C.O. (2005) Annu. Rev. Biochem. 74 791–831 [DOI] [PubMed] [Google Scholar]
- 2.Burger, G., Gray, M. W., and Lang, B. F. (2003) Trends Genet. 19 709–716 [DOI] [PubMed] [Google Scholar]
- 3.Clayton, D. A. (1984) Annu. Rev. Biochem. 53 573–594 [DOI] [PubMed] [Google Scholar]
- 4.Schafer, B. (2005) Gene (Amst.) 354 80–85 [DOI] [PubMed] [Google Scholar]
- 5.Dieckmann, C. L., and Staples, R. R. (1994) Int. Rev. Cytol. 152 145–181 [DOI] [PubMed] [Google Scholar]
- 6.Hollingsworth, M. J., and Martin, N. C. (1987) Nucleic Acids Res. 15 8845–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. Y., and Martin, N. C. (1988) J. Biol. Chem. 263 13677–13682 [PubMed] [Google Scholar]
- 8.Autio, K. J., Kastaniotis, A. J., Pospiech, H., Miinalainen, I. J., Schonauer, M. S., Dieckmann, C. L., and Hiltunen, J. K. (2008) FASEB J. 22 569–578 [DOI] [PubMed] [Google Scholar]
- 9.Schonauer, M. S., Kastaniotis, A. J., Hiltunen, J. K., and Dieckmann, C. L. (2008) Mol. Cell. Biol. 28 6646–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody, S., and Mikolajczyk, S. (1988) Eur. J. Biochem. 173 353–359 [DOI] [PubMed] [Google Scholar]
- 11.Mikolajczyk, S., and Brody, S. (1990) Eur. J. Biochem. 187 431–437 [DOI] [PubMed] [Google Scholar]
- 12.Chuman, L., and Brody, S. (1989) Eur. J. Biochem. 184 643–649 [DOI] [PubMed] [Google Scholar]
- 13.Runswick, M. J., Fearnley, I. M., Skehel, J. M., and Walker, J. E. (1991) FEBS Lett. 286 121–124 [DOI] [PubMed] [Google Scholar]
- 14.Cronan, J. E., Fearnley, I. M., and Walker, J. E. (2005) FEBS Lett. 579 4892–4896 [DOI] [PubMed] [Google Scholar]
- 15.Schneider, R., Brors, B., Burger, F., Camrath, S., and Weiss, H. (1997) Curr. Genet. 32 384–388 [DOI] [PubMed] [Google Scholar]
- 16.Stuible, H. P., Meier, S., Wagner, C., Hannappel, E., and Schweizer, E. (1998) J. Biol. Chem. 273 22334–22339 [DOI] [PubMed] [Google Scholar]
- 17.Harington, A., Herbert, C. J., Tung, B., Getz, G. S., and Slonimski, P. P. (1993) Mol. Microbiol. 9 545–555 [DOI] [PubMed] [Google Scholar]
- 18.Hoja, U., Marthol, S., Hofmann, J., Stegner, S., Schulz, R., Meier, S., Greiner, E., and Schweizer, E. (2004) J. Biol. Chem. 279 21779–21786 [DOI] [PubMed] [Google Scholar]
- 19.Kastaniotis, A., Autio, K., Sormunen, R., and Hiltunen, J. (2004) Mol. Microbiol. 53 1407–1421 [DOI] [PubMed] [Google Scholar]
- 20.Torkko, J. M., Koivuranta, K. T., Miinalainen, I. J., Yagi, A. I., Schmitz, W., Kastaniotis, A. J., Airenne, T. T., Gurvitz, A., and Hiltunen, K. J. (2001) Mol. Cell. Biol. 21 6243–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazoe, M., Shirahige, K., Rashid, M. B., Kaneko, Y., Nakayama, T., Ogasawara, N., and Yoshikawa, H. (1994) J. Biol. Chem. 269 15244–15252 [PubMed] [Google Scholar]
- 22.Zhang, L., Joshi, A. K., Hofmann, J., Schweizer, E., and Smith, S. (2005) J. Biol. Chem. 280 12422–12429 [DOI] [PubMed] [Google Scholar]
- 23.Miinalainen, I., Chen, Z., Torkko, J., Pirila, P., Sormunen, R., Bergmann, U., Qin, Y., and Hiltunen, J. (2003) J. Biol. Chem. 278 20154–20161 [DOI] [PubMed] [Google Scholar]
- 24.Brody, S., Oh, C., Hoja, U., and Schweizer, E. (1997) FEBS Lett. 408 217–220 [DOI] [PubMed] [Google Scholar]
- 25.Jordan, S. W., and Cronan, J. E., Jr. (1997) J. Biol. Chem. 272 17903–17906 [DOI] [PubMed] [Google Scholar]
- 26.Witkowski, A., Joshi, A. K., and Smith, S. (2007) J. Biol. Chem. 282 14178–14185 [DOI] [PubMed] [Google Scholar]
- 27.Challem, J. J. (1999) Med. Hypotheses 52 417–422 [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara, K., Hosaka, H., Matsuda, M., Okamura-Ikeda, K., Motokawa, Y., Suzuki, M., Nakagawa, A., and Taniguchi, H. (2007) J. Mol. Biol. 371 222–234 [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara, K., Takeuchi, S., Okamura-Ikeda, K., and Motokawa, Y. (2001) J. Biol. Chem. 276 28819–28823 [DOI] [PubMed] [Google Scholar]
- 30.Morikawa, T., Yasuno, R., and Wada, H. (2001) FEBS Lett. 498 16–21 [DOI] [PubMed] [Google Scholar]
- 31.Yi, X., and Maeda, N. (2005) Mol. Cell. Biol. 25 8387–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Autio, K. J., Guler, J. L., Kastaniotis, A. J., Englund, P. T., and Hiltunen, J. K. (2008) FEBS Lett. 582 729–733 [DOI] [PubMed] [Google Scholar]
- 33.Chen, Z. J., Pudas, R., Sharma, S., Smart, O. S., Juffer, A. H., Hiltunen, J. K., Wierenga, R. K., and Haapalainen, A. M. (2008) J. Mol. Biol. 379 830–844 [DOI] [PubMed] [Google Scholar]
- 34.Stephens, J. L., Lee, S. H., Paul, K. S., and Englund, P. T. (2007) J. Biol. Chem. 282 4427–4436 [DOI] [PubMed] [Google Scholar]
- 35.Guler, J. L., Kriegova, E., Smith, T. K., Lukes, J., and Englund, P. T. (2008) Mol. Microbiol. 67 1125–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byers, D. M., and Gong, H. (2007) Biochem. Cell Biol. 85 649–662 [DOI] [PubMed] [Google Scholar]
- 37.Carroll, J., Fearnley, I. M., Shannon, R. J., Hirst, J., and Walker, J. E. (2003) Mol. Cell. Proteomics 2 117–126 [DOI] [PubMed] [Google Scholar]
- 38.Sulo, P., and Martin, N. C. (1993) J. Biol. Chem. 268 17634–17639 [PubMed] [Google Scholar]
- 39.Miller, J. R., Busby, R. W., Jordan, S. W., Cheek, J., Henshaw, T. F., Ashley, G. W., Broderick, J. B., Cronan, J. E., Jr., and Marletta, M. A. (2000) Biochemistry 39 15166–15178 [DOI] [PubMed] [Google Scholar]
- 40.Dang, Y. L., and Martin, N. C. (1993) J. Biol. Chem. 268 19791–19796 [PubMed] [Google Scholar]
- 41.Akins, R. A., and Lambowitz, A. M. (1987) Cell 50 331–345 [DOI] [PubMed] [Google Scholar]
- 42.Paukstelis, P. J., Chen, J. H., Chase, E., Lambowitz, A. M., and Golden, B. L. (2008) Nature 451 94–97 [DOI] [PubMed] [Google Scholar]
- 43.Rodeheffer, M. S., and Shadel, G. S. (2003) J. Biol. Chem. 278 18695–18701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, L., Joshi, A. K., and Smith, S. (2003) J. Biol. Chem. 278 40067–40074 [DOI] [PubMed] [Google Scholar]
- 45.Sacco, E., Covarrubias, A. S., O'Hare, H. M., Carroll, P., Eynard, N., Jones, T. A., Parish, T., Daffe, M., Backbro, K., and Quemard, A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozak, M. (2007) J. Cell. Biochem. 102 280–290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.