Abstract
Ewing sarcoma family of tumors (ESFT) is an undifferentiated neoplasm of the bone and soft tissue. ESFT is characterized by a specific chromosomal translocation occurring between chromosome 22 and (in most cases) chromosome 11, which generates an aberrant transcription factor, EWS-FLI1. The function of EWS-FLI1 is essential for the maintenance of ESFT cell survival and tumorigenesis. The Hedgehog pathway is activated in several cancers. Oncogenic potential of the Hedgehog pathway is mediated by increasing the activity of the GLI family of transcription factors. Recent evidence suggests that EWS-FLI1 increases expression of GLI1 by an unknown mechanism. Our data from chromatin immunoprecipitation and promoter reporter studies indicated GLI1 as a direct transcriptional target of EWS-FLI1. Expression of EWS-FLI1 in non-ESFT cells increased GLI1 expression and GLI-dependent transcription. We also detected high levels of GLI1 protein in ESFT cell lines. Pharmacological inhibition of GLI1 protein function decreased proliferation and soft agar colony formation of ESFT cells. Our results establish GLI1 as a direct transcriptional target of EWS-FLI1 and suggest a potential role for GLI1 in ESFT tumorigenesis.
Ewing Sarcoma Family of Tumors (ESFT)2 affects patients between the ages of 3 and 40 with most cases occurring during the second decade of life. It is an undifferentiated small round cell tumor of the bone and soft tissue with an unknown cell of origin. Currently, the cure rate for patients with localized disease is only 70%, and is less than 30% for patients showing metastatic disease despite intensive multimodal treatment strategies (1). There is a need for more effective therapies to treat ESFT, especially in patients with metastases. ESFT is characterized by chromosomal translocations occurring between the genes for the TET (TAF15, EWS, and TLS) family member protein EWS and members of the ETS family of DNA-binding transcription factors. In ∼90% of the cases, the translocation occurs between chromosome 22 and chromosome 11 (2). This results in expression of a fusion protein EWS-FLI1, which acts as an aberrant transcription factor whose persistent expression is necessary to maintain the viability of ESFT cells (3–5). The ability of EWS-FLI1 to alter transcription of several target genes such as PTPL1, ID2, and TGFβ-RII is very important to its function in tumor formation and progression (6–9).
The Hedgehog (Hh) pathway is activated in several cancers such as basal cell carcinoma, medulloblastoma, rhabdomyosarcoma, and cancers of the pancreas, lung, colon, stomach, and prostate (10–18). The pathway is composed of three Hh ligands (Sonic, Indian, and Desert Hh) that all bind to the Patched1 receptor. In the absence of Hh ligand, Patched inhibits another transmembrane protein, Smoothened. When Patched is engaged by the Hh ligand, Smoothened is activated because of diminished inhibitory signal from Patched. The signal is then transduced to the important downstream effectors GLI1, GLI2, and GLI3, which act as transcription factors. GLI1 is the most potent isoform at inducing cellular transformation (19). Many of the mutations in human cancers result in overexpression of Hh ligand, inactivation of Patched1, or increased activation of Smoothened. However, in some tumors the pathway is activated by increasing the activity of GLI. Loss of inhibitory molecules SUFU/REN (20, 21), GLI gene amplifications (22–24), chromosomal translocations involving GLI (25), and increased GLI protein stability (26) are mechanisms that activate GLI function in human cancers.
Several recent publications suggest that EWS-FLI1 increases expression of GLI1 mRNA and/or protein in ESFT (27–29). In addition, several genes in the Hedgehog signaling pathway have been shown to be associated with metastasis in a microarray study of ESFT patient samples (30). Because EWS-FLI1 functions as a transcription factor, we hypothesized that GLI1 is a direct transcriptional target of EWS-FLI1 and may play a role in ESFT tumorigenesis. We present data that GLI1 is a direct transcriptional target of EWS-FLI1 and is important in ESFT tumorigenesis. Therefore, GLI1 could be a novel therapeutic target in ESFT.
EXPERIMENTAL PROCEDURES
Cell Lines and Hedgehog Pathway Drugs—COS7 cells were grown in DMEM with 10% fetal bovine serum (FBS). All of the ESFT cell lines were grown in RPMI with 10% FBS and 1% HEPES with the exception of SKES and A673. A673-inducible EWS-FLI1 shRNA cell line has been previously described (31). SKES cells were grown in McCoy's 5a medium with 15% FBS. A673 cells were grown in DMEM with 10% FBS and 1% sodium pyruvate. HepG2 cells were grown in DMEM with 10% FBS and 1% nonessential amino acids. Cyclopamine was purchased from Calbiochem. The GLI1 inhibitor NSC75503 was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute. All of the compounds were dissolved in DMSO.
Chromatin Immunoprecipitation (ChIP) Assay—The chromatin immunoprecipitation experiments were performed according to the manufacturer's protocol (Upstate Biotechnology, Inc., Lake Placid, NY) and as published before (6). Briefly, the cell lysates were cross-linked with 1% formaldehyde for 15 min. Samples immunoprecipitated with anti-FLI1 antibody, no antibody control, or the input were amplified with PCR (see supplemental Table S1 for primer sequences).
Luciferase Assays—GLI1 activity was assessed using a pGL3-Basic luciferase construct containing 8× GLI1-binding sites attached to a chicken lens crystalline promoter (pGL38xGLI) (American Type Culture Collection-Johns Hopkins Special Collections, deposited by P. A. Beachy, Manassas, VA) and a Renilla-TK mutant construct (Kindly provided by Dr. Stephen Byers, Georgetown University). COS7 cells were cotransfected with an EWS-FLI1 expression vector or empty vector control (CIneo) in addition to the pGL38xGLI luciferase and Renilla TK mutant constructs with FuGENE 6 (Roche Applied Science) according the manufacturer's protocol. In studies using drug treatment, the cells were treated with drug 24 h after transfection, and then luciferase activity was measured 48 h after transfection. GLI1 promoter activity was measured using the GLI1/pGL3 construct, which contains the full-length GLI1 promoter (kindly provided by Dr. Philip Iannaccone, Northwestern University). An NF-κB responsive construct that has 5× NF-κB-binding sites (Stratagene, La Jolla, CA) was used as a negative control. COS7 cells were transfected with either empty vector control or EWS-FLI1 construct in addition to the luciferase constructs with FuGENE 6 (Roche Applied Science) according the manufacturer's protocol. Luciferase activity was measured 24 h after transfection. The mutant GLI1 promoter was created by mutating all the possible FLI1 DNA-binding sites GGAA and TTCC to GCTA and TAGC. These mutations have been previously shown to abrogate EWS-FLI1 binding ability (32). All of the luciferase assays were performed using a dual luciferase assay kit according the manufacturer's protocol (Promega, Madison, WI).
Immunoblotting—Whole cell lysates from cells grown to near confluency were subjected to SDS-PAGE and then transferred to a Immobilon-P membrane (Millipore, Billerica, MA). The membranes were then subject to blocking in 5% nonfat dry milk in 1× TTBS (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Tween 20) for 1 h. Dilutions for primary were anti-GLI1(L42B10) at 1:1000 (Cell Signaling, Boston, MA), anti-FLI1 (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:2000, and anti-actin-horseradish peroxidase (C-11, Santa Cruz Biotechnology) at 1:3000 or β-tubulin (MP Biomedicals, Solon, OH) at 1:5000. Primary antibodies were added to the membrane in 5% nonfat dry milk in 1× TTBS for 2 h. The membrane was then washed three times in 1× TTBS, and horseradish peroxidase-linked anti-rabbit or anti-mouse secondary antibody (GE Healthcare) in 5% nonfat dry milk was added for 1 h. The blots were than washed three times in 1× TTBS and then developed using Millipore Immobilon Western chemiluminescent horseradish peroxidase substrate per the manufacturer's instructions (Millipore Corporation, Billerica, MA). Chemiluminescence was detected using a Fujifilm LAS-3000 imaging system. Densitometry values were obtained using Multigauge software (FUJIFILM Corp).
Reverse Transcription (RT)-PCR—Total RNA was extracted from cell lines by TRIzol (Invitrogen) and reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. PCR was performed as previously described (33).
siRNA Experiments—Control scrambled siRNA was purchased from Invitrogen. The sequence is as follows 5′-AAAGTCATCGTGACTACGACG-3′. ON-TARGETplus SMART-pool GLI1 siRNA was purchased from Dharmacon (Dharmacon, Inc., Chicago, IL) that contains four distinct siRNA species targeting different sequences of the GLI1 transcript. TC-32 and TC-71 cells were electroporated in Opti-MEM medium (Invitrogen) using a Cell-Porator (Invitrogen). For TC-32 cells, 4 million cells were electroporated with 500 nm siRNA at 350 V and plated in triplicate at 1,000 cells/well in a 96-well plate. Viable cells were quantified using 10 μl of WST-1 reagent (Roche Applied Science) according to the manufacturer's protocol after 5 days. The rest of the cells were plated in a 6-well dish and were lysed after 5 days for subsequent immunoblotting. For TC-71 cells, 4 million cells were electroporated with 250 nm siRNA at 300 V and plated in triplicate at 5,000 cells/well in a 96-well plate. Viable cells were quantified using 10 μl of WST-1 reagent (Roche Applied Science) according to the manufacturer's protocol after 3 days. The rest of the cells were plated in a 6-well dish and were lysed after 3 days for subsequent immunoblotting. OD values were converted to cell numbers by doing a WST cell titration experiment for both TC-32 and TC-71 and then performing a linear regression using Prism Graphpad 4.0 for Macintosh.
Cellular Proliferation and Soft Agar Assays—Cellular proliferation was assessed by triplicate plating at a density of 1000 cells/well in a 96-well plate. Cyclopamine or 75503 at varying concentrations or vehicle alone (1% DMSO) were added in media containing 5% FBS to cells 4 h after plating once the cells had attached. Fresh medium containing drug or vehicle was added every 2 days. Viable cells were quantified using 10μl of WST-1 reagent (Roche Applied Science) according to the manufacturer's protocol after 72 h. For soft agar assays, 6% SeaPlaqueGTG agarose (Lonza, Rockland, ME) was prepared in phosphate-buffered saline as a stock solution and kept in 65 °C water bath. Bottom agar at 0.6% concentration was added as 1 ml to each well of a 12-well cell culture plate and left to solidify at room temperature. Five thousand of TC32 cells were resuspended in 1 ml of top agar (0.4%) and plated as triplicate. The plate was kept at room temperature for 10–15 min until the top agar solidified and placed in a 37 °C cell culture incubator for 2–3 weeks. Colonies were stained with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for 2 h at 37 °C. Images of colonies were taken by Fuji LAS-3000 imaging system, and colony size and counting analysis was carried out by using Multigauge Colony Count Software. The experiments were repeated at least twice.
RESULTS
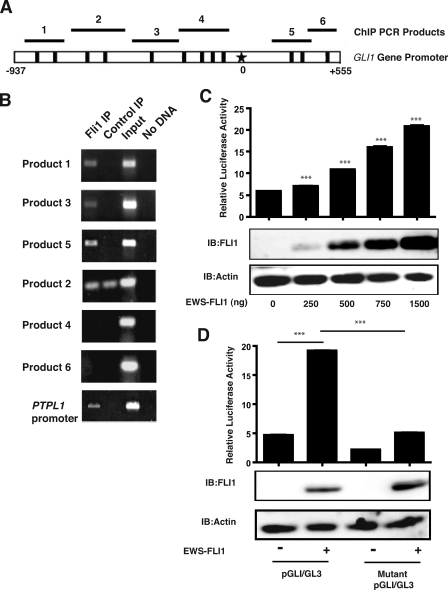
GLI1 Is a Direct Transcriptional Target of EWS-FLI1—Modulation of EWS-FLI1 expression was associated with increased GLI1 mRNA levels in cDNA array experiments (29). An observed increase in GLI1 mRNA can be either a direct or indirect effect of EWS-FLI1 expression. Therefore, we evaluated the possibility of GLI1 promoter being a direct target of EWS-FLI1. The GLI1 promoter has previously been mapped and characterized (34, 35). Analysis of the GLI1 promoter sequence identified 12 potential EWS-FLI1-binding sites, which are characterized by GGAA (Fig. 1A) (36). To show a direct interaction between EWS-FLI1 and the GLI1 promoter in ESFT cells, we performed ChIP experiments (Fig. 1B). We used the FLI1 antibody for immunoprecipitation of EWS-FLI1 because FLI1 is not expressed in ESFT cells (6). We designed six primer pairs to cover the entire length of GLI1 promoter (Fig. 1A). ChIP assay revealed interaction between EWS-FLI1 protein and multiple segments of the GLI1 promoter. Primers pairs 1, 3, and 5 gave positive ChIP results, whereas primers 4 and 6 were negative (Fig. 1B). Primer pair 2 gave same intensity bands in both the FLI1 IP and the control IP. Therefore, it was considered a negative result because of nonspecific product formation. PTPL1, a known direct transcriptional target of EWS-FLI1 (6), was used as a positive control. Control immunoprecipitations were performed in the absence of primary antibody, and negative controls for the PCR were performed in the absence of DNA. These results suggest that EWS-FLI1 is binding directly to elements of the GLI1 promoter in ESFT cells. The following experiments were designed to study the functional outcome of EWS-FLI1 protein and the GLI1 promoter interaction.
FIGURE 1.
EWS-FLI1 binds to and activates the GLI1 promoter. A, a map of the full-length GLI1 promoter is shown. The black boxes represent the 12 GGAA sites, which are possible EWS-FLI1-binding sites. The star represents the transcription start site. The black bars with numbers show the approximate positions of the PCR products shown in ChIP experiments. B, chromatin immunoprecipitation was performed by cross-linking the DNA of TC-71 cells followed by enrichment of DNA-protein complexes with anti-FLI1 antibody. Six different primer pairs were designed to cover all 12 possible GGAA binding sites on both strands in the GLI1 promoter and then used to amplify DNA by PCR. The PTPL1 primers served as a positive control for positive EWS-FLI1 binding. Control IP was no antibody. Input represents the cross-linked DNA prior to immunoprecipitation. C, COS7 cells were cotransfected with 0, 250, 500, 750, 1000, and 1500 ng of EWS-FLI1 and the GLI1 promoter luciferase construct (GLI1/pGL3). Bars represent the means of the relative luciferase activity, which is the calculated by dividing the luciferase activity by the Renilla activity used as a transfection control. The error bars are the standard deviations. (***, p < 0.001, using a two-tailed Student's t test). Transfection assays were performed in triplicate. D, COS7 cells were cotransfected with and without EWS-FLI1 and the wild type GLI1 promoter luciferase construct or a mutant GLI1 promoter construct that has the FLI1 DNA-binding sites mutated. The bars represent the means of the relative luciferase activity, which is calculated by dividing the luciferase activity by the Renilla activity used as a transfection control. The error bars are the standard deviations (***, p < 0.001 using a two-tailed Student's t test). Transfection assays were performed in triplicate and were repeated twice. IB, immunoblotting.
GLI1 Gene Promoter Is Activated by EWS-FLI1—We examined whether EWS-FLI1 activates transcription from the GLI1 promoter. COS7 cells were cotransfected with EWS-FLI1 in a CIneo expression vector and the full-length GLI1 gene promoter in a pGL3 basic luciferase vector (GLI1/pGL3). We showed that when EWS-FLI1 protein is expressed, it significantly increased activity of the GLI1 promoter. The experiment has been repeated a total of seven times with the average fold increase of 2.5 (data not shown). The activity of the GLI1 promoter is also increased in a dose-dependent manner compared to the empty vector control (Fig. 1C) with the highest expression of EWS-FLI1 giving a 3.5-fold increase in promoter activity. Transfection of EWS-FLI1 did not increase an NF-KB-responsive promoter (data no shown), which was used as a negative control to rule out the possibility that EWS-FLI1 can nonspecifically activate any reporter construct.
We then mutated all the possible FLI1 DNA-binding sites as shown in Fig. 1A to see whether we could abrogate the ability of EWS-FLI1 to increase activity of the GLI1 promoter. When we expressed both the WT and mutant GLI1 promoter luciferase constructs in COS7 cells cotransfected with EWS-FLI1, we saw a 4-fold decrease in activity that reduced the activity back to the WT control.
EWS-FLI1 Enhances GLI1 Protein Expression and Activity in COS7 Cells—Following the observation of an increase in GLI1 promoter activity upon EWS-FLI1 expression, we then looked at the effects of expressing EWS-FLI1 on GLI1 protein expression and the activity of GLI1 as a transcription factor. We transfected COS7 cells with EWS-FLI1 and examined endogenous GLI1 protein expression by Western blot. We observed a 1.4-fold increase in GLI1 protein expression when COS7 cells expressed EWS-FLI1 protein (Fig. 2A). COS7 cells were also cotransfected with EWS-FLI1 and a luciferase reporter construct (pGL38xGLI) that contains a GLI1-responsive promoter to examine the effect of EWS-FLI1 on GLI1 transcriptional activity (Fig. 2B). The pGL38xGLI construct contains eight GLI DNA-binding sites attached to the chicken lens crystalline promoter followed by the luciferase gene. The difference between the construct used in Fig. 1 and pGL38xGLI used in Fig. 2 is that one is measuring activity of the GLI1 gene promoter (GLI1/pGL3) stimulated by EWS-FLI1 protein, and the other is measuring activity of a GLI1-responsive promoter (pGL38xGLI) stimulated by GLI1 protein. Expression of EWS-FLI1 in COS7 cells significantly increased pGL38xGLI reporter activity (Fig. 2B). These data further suggest that EWS-FLI1 induces GLI1 protein expression to a functionally significant level, which provides enhanced GLI1 transcriptional activity.
FIGURE 2.
EWS-FLI1 expression in COS7 cells leads to an increase in endogenous GLI1 protein expression and transcriptional activity. A, COS7 cells were transfected with EWS-FLI1 or an empty vector control. Whole cell lysates were subjected to SDS-PAGE and subsequent immunoblotting with GLI1, FLI1, or actin antibody. FLI1 immunoblotting confirmed the expression of EWS-FLI1 and actin was used as a loading control. The values below the top panel are the densitometry values and are given as fold increase over the empty vector control. B, COS7 cells were cotransfected with and without EWS-FLI1 and a pGL38xGli responsive luciferase construct. Twenty-four hours after transfection, the cells were treated with the cyclopamine or 75503 GLI inhibitor at a concentration of 30 μm in 1% DMSO for 24 h. The columns represent the means of the relative luciferase activity, which is calculated by dividing the luciferase activity by the Renilla activity used as a transfection control. The error bars are the standard deviations (*, p < 0.05; ***, p < 0.001 using a two-tailed Student's t test). Transfection assays were performed in triplicate and repeated three times. One representative experiment is shown in this figure. IB, immunoblotting.
To rule out that the observed increase in the GLI1-responsive promoter (pGL38xGLI) may be due to direct transcriptional activity by EWS-FLI1, we used pharmacological inhibitors of GLI1 protein (Fig. 2B). We used NSC75503, which has been previously characterized as a specific inhibitor of GLI1 transcriptional activity (37). We also used cyclopamine, which is a Smoothened antagonist (38). Cyclopamine was expected to have little to no effect on GLI1 activation by EWS-FLI1, because EWS-FLI1 acts downstream of Smoothened to activate GLI1. When COS7 cells were transfected with EWS-FLI1 and treated with the GLI1 inhibitor 75503, we observed a significant decrease in GLI1 transcriptional activity as measured by the pGL38xGLI reporter construct (Fig. 2B). Cyclopamine decreased GLI1 activity as well but to a much lesser extent (Fig. 2B). These results suggested that the EWS-FLI1-induced increase in GLI1 responsive promoter (pGL38xGLI) activity is mediated by an increase in expression of activated GLI1 protein.
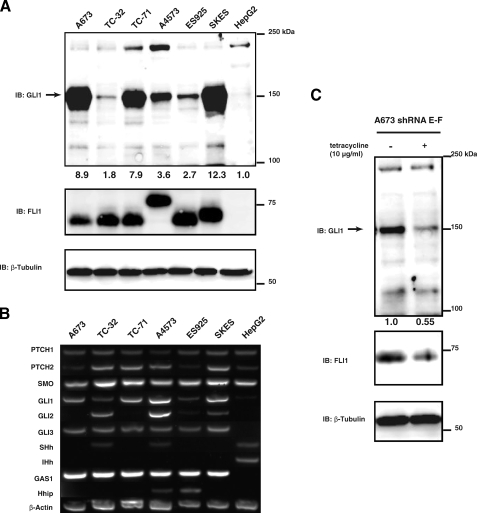
GLI1 Is Expressed in ESFT Cell Lines—Because the GLI1 promoter is activated by EWS-FLI1, we explored whether GLI1 protein is expressed in ESFT cell lines. We surveyed GLI1 protein expression in a panel of six ESFT cell lines that express EWS-FLI1. HepG2, a hepatocellular carcinoma cell line, was used as a negative control because it expresses low levels of GLI1 protein. All six ESFT cell lines examined had considerably higher GLI1 expression compared with HepG2 cells as determined by densitometric analysis of Western blots. On average, ESFT cells had a 6.2-fold higher level of GLI1 protein expression when compared with HepG2 cells (Fig. 3A). We also performed RT-PCR analysis for Hedgehog pathway components on ESFT and HepG2 cells (Fig. 3B). All ESFT cells lines express GLI1 as well as the GLI target GAS1. A4573 and ES925 also express the GLI target Hhip. HepG2 cells do not express any GLI genes or GAS1. β-Actin was used as a positive control. We therefore conclude that GLI1 is expressed in a majority of ESFT cell lines, which suggests that EWS-FLI1-induced GLI1 expression is a biologically relevant observation.
FIGURE 3.
GLI1 is expressed in ESFT cell lines, and knockdown of EWS-FLI1 decreases GLI1 expression in ESFT cell line A673. A, whole cell lysates from six different ESFT cell lines and HepG2 were subjected to SDS-PAGE. GLI1 expression was determined by immunoblotting with a GLI1 antibody. The arrow indicates the GLI1 band at 150 kDa. All of the ESFT cell lines express EWS-FLI1 as shown by immunoblotting with a Fli1 antibody. The size difference is due to the type of gene fusion that occurs. β-Tubulin was used a loading control. The numbers under the top panel are densitometry values. They are given as fold increase over the HepG2 cell line after normalizing each band to its corresponding loading control band value. B, total RNA from ESFT and HepG2 cell lines was isolated and analyzed by RT-PCR for Hedgehog pathway components. All of the ESFT cells lines express GLI1 as well as the GLI target GAS1. HepG2 does not express any GLI genes or GAS1. Of the ESFT cells only TC-32 and A4573 expresses Hh ligand. β-Actin was used as a positive control. Desert Hedgehog was also examined and was negative for all cell lines (data not shown). C, A673 cells stably transfected with a tetracycline inducible shRNA for EWS-FLI1 were plated and treated with 0 or 10 μg/ml of tetracycline for 96 h. After 96 h the cells were lysed, and whole cell lysates were subjected to SDS-PAGE and subsequent immunoblotting with GLI1, FLI1, or β-tubulin antibody. FLI1 immunoblotting confirmed the knockdown of EWS-FLI1 in tetracycline treated cells and actin was used as a loading control. IB, immunoblotting.
Knockdown of EWS-FLI1 Decreases GLI1 Protein Levels—A673 cells stably transfected with shRNA for EWS-FLI1 were treated with tetracycline to induce shRNA expression. A673 cells were used because they are the only ESFT cell line that can tolerate EWS-FLI1 knockdown. After 96 h EWS-FLI1 expression was decreased upon tetracycline treatment in the shRNA cells (Fig. 3C). When EWS-FLI1 expression was decreased this caused a subsequent 45% decrease in GLI1 levels as determined by densitometric analysis. These data further suggest that EWS-FLI1 alters GLI1 protein expression in ESFT cells.
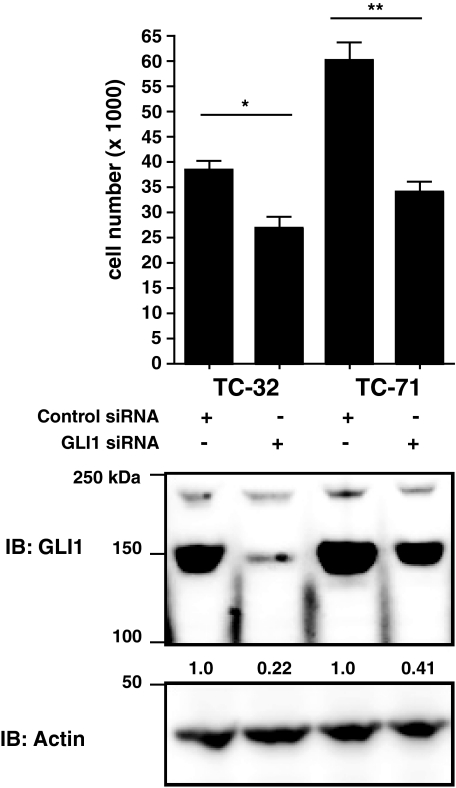
Knockdown of GLI1 in ESFT Cells Decreases Proliferation—TC-71 and TC-32 cells were electroporated with control scrambled or GLI1 siRNA. GLI1 siRNA reduced GLI1 protein in TC-32 cells by 78% and in TC-71 cells by 59% (Fig. 4). After 3 days, TC-71 cells expressing GLI1 siRNA had a 43% decrease in proliferation compared with the control (Fig. 4). The result was statistically significant. After 5 days, TC-32 cells expressing GLI1 siRNA had a 30% decrease in proliferation compared with the control (Fig. 4). This result was also statistically significant. These experiments were repeated four times, and GLI1 protein expression was reduced on average 50% with a standard deviation of 25.7% for TC-32 cells and 55% with a standard deviation of 10.2% for TC-71 cells. Proliferation on average was inhibited 31% with a standard deviation of 14.90% for TC-32 cells and a 36% decrease in proliferation with a standard deviation of 10.0% for TC-71 cells.
FIGURE 4.
Knockdown of GLI1 by siRNA decreases proliferation of ESFT cells. TC-32 and TC-71 cells were electroporated with control scrambled or GLI1 siRNA. TC-32, were plated in triplicate at a density of 1,000 cells/well in a 96-well plate. TC-71 cells were plated at a density of 5,000 cells/well in a 96-well plate. After 3 (TC-71) or 5 (TC-32) days cell proliferation was measured by a WST assay. Proliferation is shown as the number of viable cells/well. The experiment was done in triplicate and repeated four times. Shown is a representative experiment. The error bars are the standard deviations (**, p < 0.01; ***, p < 0.001 using a two-tailed Student's t test). Whole cell lysates from TC-32 and TC-71 cells treated with control or GLl1 siRNA reagents were subjected to SDS-PAGE. GLI1 expression was determined by immunoblotting with a GLI1 antibody. Actin was used a loading control. The numbers under the top panel are densitometry values. They are given as fold increase over the control cell line after normalizing each band to its corresponding loading control band value. IB, immunoblotting.
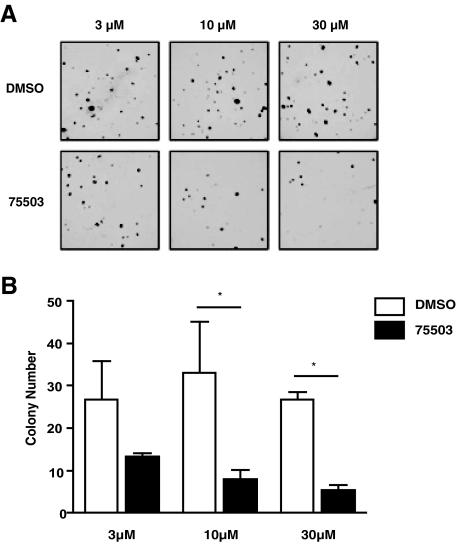
Targeting GLI1 with a Small Molecule Inhibitor Decreases ESFT Proliferation and Soft Agar Colony Formation—We next examined whether GLI1 plays a role in EWS-FLI1 tumorigenesis. The ESFT cell lines TC-32 and TC-71 were treated with the GLI1 inhibitor 75503 at 10 and 30 μm in 1% DMSO. HepG2 cells were used as a negative control to show that the observed effect was not due to general cell toxicity. After 72 h, the 75503 compound at 10 μm had 43% inhibition of cellular proliferation in TC-71 cells and 31% inhibition in TC-32 cells. There was no growth inhibition in the negative control, HepG2 cells. At a concentration of 30 μm, the 75503 compound had 60% inhibition in TC-71 cells, 84% inhibition in TC-32 cells, and 23% inhibition in HepG2 cells (Fig. 5A). We also treated TC-32 and TC-71 cells with cyclopamine at 10 and 30 μm and compared it with the 75503 compound (Fig. 5B). Cyclopamine was expected to have a smaller effect in inhibiting proliferation of ESFT cells because the EWS-FLI1 acts downstream of Smoothened to activate GLI1. We were able to show that at both 10 and 30 μm, the 75503 compound inhibited proliferation to a greater extent in TC-71 and TC-32 cells compared with cyclopamine. At 10 μm the 75503 compound had 43% inhibition in TC-71 and 31% inhibition in TC-32 cells, both of which were significantly better than cyclopamine. At 30 μm the 75503 compound had 58% inhibition in TC-71 and 84% inhibition in TC-32 cells, both of which were again significantly better than cyclopamine. We then compared the effects of cyclopamine on proliferation in ESFT cells to DAOY cells, a medullablastoma cell line. This experiment demonstrated that we are able inhibit proliferation in a cyclopamine-sensitive cell line but not ESFT cells. At 10 μm TC-71 cells had 1.9-fold greater proliferation, and TC-32 cells had 2.0-fold greater proliferation when compared DAOY cells. At 30 μm TC-71 cells had 2.6-fold greater proliferation, and TC-32 cells had 3.4-fold greater proliferation when compared with DAOY cells (Fig. 5C). To show that the decrease in proliferation was due to the effect of the inhibitor on GLI1 transcription activity in ESFT cells, we transfected TC-32 cells with the pGL38xGLI1 reporter construct. Forty-eight hours after transfection, 10 and 30 μm of cyclopamine or 75503 was added (Fig. 5D). Luciferase activity was measured 24 h after drug addition. Cyclopamine did not inhibit reporter activity of the pGL38xGLI construct in TC-32, whereas the 75503 compound significantly inhibited reporter activity at both 10 and 30 μm. The GLI1 inhibitor 75503 was also able to inhibit soft agar colony formation in TC-32 cells (Fig. 6). The 75503 compound significantly inhibited colony formation at 10 and 30 μm. These results indicate that GLI1 may play a role in cell proliferation and anchorage-independent growth of ESFT tumor cells.
FIGURE 5.
Treatment of ESFT cells with a small molecule inhibitor of GLI1 decreases proliferation. A, TC-32, TC-71, and HepG2 cells were plated in triplicate at a density of 1,000 cells/well in a 96-well plate. The cells were treated with 75503 at 10 and 30 μm in 1% DMSO. After 72 h cell proliferation was measured by a WST assay. Proliferation is shown as the percentage of growth over 1% DMSO-treated cells. The experiment was done in triplicate and repeated three times. The error bars are the standard deviations (**, p < 0.01; ***, p < 0.001 using a two-tailed Student's t test). B, TC-32 and TC-71 cells were plated in triplicate at a density of 1,000 cells/well in a 96-well plate. The cells were treated with cyclopamine and 75503 at 10 and 30 μm in 1% DMSO. After 72 h cell proliferation was measured by a WST assay. Proliferation is shown as the percentage of growth over 1% DMSO-treated cells. The experiment was done in triplicate and repeated three times. The error bars are the standard deviations (*, p < 0.05; **, p < 0.01; ***, p < 0.001 using a two-tailed Student's t test). Shown is a representative experiment. C, TC-32, TC-71, and DAOY cells were plated in triplicate at a density of 1,000 cells/well in a 96-well plate. The cells were treated with cyclopamine at 10 and 30 μm in 1% DMSO. After 72 h cell proliferation was measured by a WST assay. Proliferation is shown as the percentage of growth over 1% DMSO-treated cells. The experiment was done in triplicate. The error bars are the standard deviations (*, p < 0.05; **, p < 0.01; ***, p < 0.001 using a two-tailed Student's t test). D, TC-32 cells were transfected with the pGL38XGLI luciferase reporter construct and Renilla-TK construct. Forty-eight hours later, the cells were treated for 24 h with cyclopamine and 75503 at 10 and 30 μm. The bars represent the means of the relative luciferase activity, which is calculated by dividing the luciferase activity by the Renilla activity used as a transfection control. The error bars are the standard deviations (***, p < 0.001 using a two-tailed Student's t test). Transfection assays were performed in triplicate and were repeated twice. Shown is a representative experiment.
FIGURE 6.
Treatment of ESFT cells with a small molecule inhibitor of GLI1 decreases soft agar colony formation. A, TC-32 cells were plated in soft agar in triplicate at a density of 5,000 cells/well in a 12-well tissue culture plate. The cells were treated with DMSO or 75503 at 3, 10, and 30 μm, which was added on top every 2 days in 50 μl of volume. B, graph shown is the quantification of A. The error bars are the standard deviations (*, p < 0.05 using a one-way analysis of variance, Bonferroni's multiple comparison test).
DISCUSSION
The ability of EWS-FLI1 to cause a malignant phenotype is dependent in part on its ability to act as a transcription factor and alter gene expression. Many targets have been described as either directly or indirectly regulated by EWS-FLI1. Previous studies have suggested that EWS-FLI1 increases GLI1 expression. One study showed by microarray analysis that GLI1 gene expression was decreased in an ESFT cell line where EWS-FLI1 was knocked down by RNA interference (29). Another study showed that NIH3T3 cells transformed with EWS-FLI1 had increased expression of GLI1 and its transcriptional targets (27). However, neither study proposed a mechanism for how EWS-FLI1 increases GLI1 expression. Zwerner et al. (27) argued that increased expression of GLI1 protein could be an indirect effect of EWS-FLI1. Their work provided evidence that EWS-FLI1-induced expression of c-Myc may regulate GLI1 expression. In this study, we show that EWS-FLI1 directly binds to the GLI1 promoter and increases expression of active GLI1 protein. Our results do not rule out the potential role of c-Myc involvement in GLI1 gene expression. However, they strongly suggest that direct activation of GLI1 promoter by EWS-FLI1 is the primary mechanism behind increased GLI1 protein expression.
Inhibiting GLI1 with a small molecule inhibitor can decrease ESFT proliferation and anchorage-independent growth. These results were validated by knocking down GLI1 with siRNA and showing a subsequent decrease in proliferation. Zwerner et al. (27) also showed that an ESFT cell line, TC-32, had reduced anchorage independent growth when GLI1 expression was inhibited by shRNA. Our findings support their hypothesis that GLI1 is important for ESFT tumor phenotype. We showed that inhibition of GLI1 function reduced growth of TC-71 and TC-32 cell lines in culture and TC-32 growth in soft agar. In many of our experiments, we utilized a small molecule inhibitor of GLI1 as opposed to shRNA. Even though shRNA, siRNA, and antisense oligonucleotides are excellent experimental tools to reduce protein expression in vitro, their clinical applications have not been possible in any target in any tumor. Small molecules provide drugable properties that allow them to be optimized for better pharmakinetics and pharmacodynamics. A study of GLI1 gene amplification in childhood sarcomas examined eight ESFT cell lines and found that none of them had rearrangements or amplification of the GLI1 gene (24). These data support our findings that increased GLI1 expression in ESFT cells is due to EWS-FLI1 and not by other mechanisms such as gene amplification. Our work supports adding GLI1 to the list of genes that are directly regulated by EWS-FLI1 and contribute to ESFT tumorigenicity.
NSC75503 has been characterized as a specific inhibitor of GLI1 transcriptional activity (37). However, its specificity for GLI1 is not proven. It is likely that NSC75503 can also inhibit other GLI isoforms, GLI2 and GLI3. TC-32 cells express both GLI1 and GLI2 in contrast to TC-71 cells expressing only GLI1 (Fig. 3). Therefore, the differences observed between these two cell lines in respect to their NSC75503 sensitivity (Fig. 4), could potentially be due to expression levels of different GLI isoforms. Nevertheless, the difference in response to cyclopamine and NSC75503 suggests the activation of Hedgehog pathway at the GLI level. Hence, our findings support the hypothesis that EWS-FLI1-induced GLI1 expression is an important contributor to ESFT carcinogenesis.
GLI1 is often up-regulated in cancer by Hh pathway-dependent mechanisms such as overexpression of Hh ligand, inactivation of Patched1, or activation of Smoothened. However, recent evidence has also shown that Hedgehog-independent activation of GLI1 by Ras or transforming growth factor-β signaling pathways is important for tumor formation of many cancers such as pancreas, lung and colon (39–42). However, the effect of RAS and transforming growth factor-β signaling on GLI1 activation has not been shown to be a direct mechanism. We propose a novel mechanism of Hh-independent GLI1 activation whereby EWS-FLI1 directly activates GLI1 by binding to the GLI1 promoter, leading to an increase in expression of GLI1 protein. RT-PCR analysis of ESFT cell lines compared with control cell line showed higher expression levels of GLI1 and its target gene GAS1 (Fig. 3). Most ESFT cell lines showed no detectable hedgehog ligand expression or very low levels compared with control. Consistent with this finding, cyclopamine did not inhibit GLI1 transcriptional activity in ESFT cells (Fig. 4D). These findings further supported the hypothesis that the molecular mechanism of EWS-FLI1 activating GLI1 is independent of the upstream Hedgehog pathway.
Cyclopamine and a few other Smoothened antagonists are currently entering phase I trials (37). However, the growing evidence that in many cancers GLI1 activation occurs by a Hedgehog-independent mechanism, as we present in this study, suggests that the development of GLI inhibitors should be pursued. These novel agents could be used to treat cancers including ESFT, which have activation of the pathway at the level of GLI. Because cancers have multiple dysregulated pathways, it is unlikely that inhibition of GLI1 alone can provide a cure for ESFT. EWS-FLI1 activates other important genes contributing to cellular transformation. Therefore, the greatest benefit from specific GLI1 inhibitors will most likely come from combinatorial therapies.
Supplementary Material
Acknowledgments
We thank Dr. Philip Iannaccone for the GLI1 gene promoter luciferase construct, Dr. Olivier Delattre for the A673 inducible EWS-FLI1 shRNA cell line, and Dr. Steve Byers for the modified Renilla construct.
This work was supported, in whole or in part, by National Institutes of Health Grants CA88004 (to J. T.) and CA10841 (to A. Ü.). This work was also supported by a grant from the Children's Cancer Foundation of Baltimore, MD, and the Intramural Research Program of the NCI, National Institutes of Health.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: ESFT, Ewing sarcoma family of tumors; Hh, Hedgehog; DMSO, dimethyl sulfoxide; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; ChIP, chromatin immunoprecipitation; RT, reverse transcription; siRNA, small interfering RNA; shRNA, small hairpin RNA.
References
- 1.Grier, H. E., Krailo, M. D., Tarbell, N. J., Link, M. P., Fryer, C. J., Pritchard, D. J., Gebhardt, M. C., Dickman, P. S., Perlman, E. J., Meyers, P. A., Donaldson, S. S., Moore, S., Rausen, A. R., Vietti, T. J., and Miser, J. S. (2003) N. Engl. J. Med. 348 694–701 [DOI] [PubMed] [Google Scholar]
- 2.Sandberg, A. A., and Bridge, J. A. (2000) Cancer Genet Cytogenet. 123 1–26 [DOI] [PubMed] [Google Scholar]
- 3.Tanaka, K., Iwakuma, T., Harimaya, K., Sato, H., and Iwamoto, Y. (1997) J. Clin. Investig. 99 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouchida, M., Ohno, T., Fujimura, Y., Rao, V. N., and Reddy, E. S. (1995) Oncogene 11 1049–1054 [PubMed] [Google Scholar]
- 5.Chansky, H. A., Barahmand-Pour, F., Mei, Q., Kahn-Farooqi, W., Zielinska-Kwiatkowska, A., Blackburn, M., Chansky, K., Conrad, E. U., III, Bruckner, J. D., Greenlee, T. K., and Yang, L. (2004) J. Orthop. Res. 22 910–917 [DOI] [PubMed] [Google Scholar]
- 6.Abaan, O. D., Levenson, A., Khan, O., Furth, P. A., Uren, A., and Toretsky, J. A. (2005) Oncogene 24 2715–2722 [DOI] [PubMed] [Google Scholar]
- 7.Nishimori, H., Sasaki, Y., Yoshida, K., Irifune, H., Zembutsu, H., Tanaka, T., Aoyama, T., Hosaka, T., Kawaguchi, S., Wada, T., Hata, J., Toguchida, J., Nakamura, Y., and Tokino, T. (2002) Oncogene 21 8302–8309 [DOI] [PubMed] [Google Scholar]
- 8.Hahm, K. B., Cho, K., Lee, C., Im, Y. H., Chang, J., Choi, S. G., Sorensen, P. H., Thiele, C. J., and Kim, S. J. (1999) Nat. Genet. 23 222–227 [DOI] [PubMed] [Google Scholar]
- 9.Fukuma, M., Okita, H., Hata, J., and Umezawa, A. (2003) Oncogene 22 1–9 [DOI] [PubMed] [Google Scholar]
- 10.Tostar, U., Malm, C. J., Meis-Kindblom, J. M., Kindblom, L. G., Toftgard, R., and Unden, A. B. (2006) J. Pathol. 208 17–25 [DOI] [PubMed] [Google Scholar]
- 11.Hahn, H., Wicking, C., Zaphiropoulous, P. G., Gailani, M. R., Shanley, S., Chidambaram, A., Vorechovsky, I., Holmberg, E., Unden, A. B., Gillies, S., Negus, K., Smyth, I., Pressman, C., Leffell, D. J., Gerrard, B., Goldstein, A. M., Dean, M., Toftgard, R., Chenevix-Trench, G., Wainwright, B., and Bale, A. E. (1996) Cell 85 841–851 [DOI] [PubMed] [Google Scholar]
- 12.Thayer, S. P., di Magliano, M. P., Heiser, P. W., Nielsen, C. M., Roberts, D. J., Lauwers, G. Y., Qi, Y. P., Gysin, S., Fernandez-del Castillo, C., Yajnik, V., Antoniu, B., McMahon, M., Warshaw, A. L., and Hebrok, M. (2003) Nature 425 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheng, T., Li, C., Zhang, X., Chi, S., He, N., Chen, K., McCormick, F., Gatalica, Z., and Xie, J. (2004) Mol. Cancer 3 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reifenberger, J., Wolter, M., Weber, R. G., Megahed, M., Ruzicka, T., Lichter, P., and Reifenberger, G. (1998) Cancer Res. 58 1798–1803 [PubMed] [Google Scholar]
- 15.Pietsch, T., Waha, A., Koch, A., Kraus, J., Albrecht, S., Tonn, J., Sorensen, N., Berthold, F., Henk, B., Schmandt, N., Wolf, H. K., von Deimling, A., Wainwright, B., Chenevix-Trench, G., Wiestler, O. D., and Wicking, C. (1997) Cancer Res. 57 2085–2088 [PubMed] [Google Scholar]
- 16.Chi, S., Huang, S., Li, C., Zhang, X., He, N., Bhutani, M. S., Jones, D., Castro, C. Y., Logrono, R., Haque, A., Zwischenberger, J., Tyring, S. K., Zhang, H., and Xie, J. (2006) Cancer Lett. 244 53–60 [DOI] [PubMed] [Google Scholar]
- 17.Berman, D. M., Karhadkar, S. S., Maitra, A., Montes De Oca, R., Gerstenblith, M. R., Briggs, K., Parker, A. R., Shimada, Y., Eshleman, J. R., Watkins, D. N., and Beachy, P. A. (2003) Nature 425 846–851 [DOI] [PubMed] [Google Scholar]
- 18.Bian, Y. H., Huang, S. H., Yang, L., Ma, X. L., Xie, J. W., and Zhang, H. W. (2007) World J. Gastroenterol. 13 1659–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, H., Stephen, D., Joyner, A., and Curran, T. (2005) Oncogene 24 4026–4036 [DOI] [PubMed] [Google Scholar]
- 20.Taylor, M. D., Liu, L., Raffel, C., Hui, C. C., Mainprize, T. G., Zhang, X., Agatep, R., Chiappa, S., Gao, L., Lowrance, A., Hao, A., Goldstein, A. M., Stavrou, T., Scherer, S. W., Dura, W. T., Wainwright, B., Squire, J. A., Rutka, J. T., and Hogg, D. (2002) Nat. Genet. 31 306–310 [DOI] [PubMed] [Google Scholar]
- 21.Di Marcotullio, L., Ferretti, E., De Smaele, E., Argenti, B., Mincione, C., Zazzeroni, F., Gallo, R., Masuelli, L., Napolitano, M., Maroder, M., Modesti, A., Giangaspero, F., Screpanti, I., Alesse, E., and Gulino, A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10833–10838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinzler, K. W., Bigner, S. H., Bigner, D. D., Trent, J. M., Law, M. L., O'Brien, S. J., Wong, A. J., and Vogelstein, B. (1987) Science 236 70–73 [DOI] [PubMed] [Google Scholar]
- 23.Nessling, M., Richter, K., Schwaenen, C., Roerig, P., Wrobel, G., Wessendorf, S., Fritz, B., Bentz, M., Sinn, H. P., Radlwimmer, B., and Lichter, P. (2005) Cancer Res. 65 439–447 [PubMed] [Google Scholar]
- 24.Roberts, W. M., Douglass, E. C., Peiper, S. C., Houghton, P. J., and Look, A. T. (1989) Cancer Res. 49 5407–5413 [PubMed] [Google Scholar]
- 25.Dahlen, A., Fletcher, C. D., Mertens, F., Fletcher, J. A., Perez-Atayde, A. R., Hicks, M. J., Debiec-Rychter, M., Sciot, R., Wejde, J., Wedin, R., Mandahl, N., and Panagopoulos, I. (2004) Am. J. Pathol. 164 1645–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatia, N., Thiyagarajan, S., Elcheva, I., Saleem, M., Dlugosz, A., Mukhtar, H., and Spiegelman, V. S. (2006) J. Biol. Chem. 281 19320–19326 [DOI] [PubMed] [Google Scholar]
- 27.Zwerner, J. P., Joo, J., Warner, K. L., Christensen, L., Hu-Lieskovan, S., Triche, T. J., and May, W. A. (2008) Oncogene 27 3282–3291 [DOI] [PubMed] [Google Scholar]
- 28.Stegmaier, K., Wong, J. S., Ross, K. N., Chow, K. T., Peck, D., Wright, R. D., Lessnick, S. L., Kung, A. L., and Golub, T. R. (2007) PLoS Med. 4 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, R., Owen, L. A., Trem, D. J., Wong, J. S., Whangbo, J. S., Golub, T. R., and Lessnick, S. L. (2006) Cancer Cell 9 405–416 [DOI] [PubMed] [Google Scholar]
- 30.Schaefer, K. L., Eisenacher, M., Braun, Y., Brachwitz, K., Wai, D. H., Dirksen, U., Lanvers-Kaminsky, C., Juergens, H., Herrero, D., Stegmaier, S., Koscielniak, E., Eggert, A., Nathrath, M., Gosheger, G., Schneider, D. T., Bury, C., Diallo-Danebrock, R., Ottaviano, L., Gabbert, H. E., and Poremba, C. (2008) Eur. J. Cancer 44 699–709 [DOI] [PubMed] [Google Scholar]
- 31.Tirode, F., Laud-Duval, K., Prieur, A., Delorme, B., Charbord, P., and Delattre, O. (2007) Cancer Cell 11 421–429 [DOI] [PubMed] [Google Scholar]
- 32.Uren, A., Tcherkasskaya, O., and Toretsky, J. A. (2004) Biochemistry 43 13579–13589 [DOI] [PubMed] [Google Scholar]
- 33.Sacedon, R., Varas, A., Hernandez-Lopez, C., Gutierrez-deFrias, C., Crompton, T., Zapata, A. G., and Vicente, A. (2003) J. Histochem. Cytochem. 51 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, C. Z., Yang, J. T., Yoon, J. W., Villavicencio, E., Pfendler, K., Walter-house, D., and Iannaccone, P. (1998) Gene (Amst.) 209 1–11 [DOI] [PubMed] [Google Scholar]
- 35.Villavicencio, E. H., Yoon, J. W., Frank, D. J., Fuchtbauer, E. M., Walter-house, D. O., and Iannaccone, P. M. (2002) Genesis 32 247–258 [DOI] [PubMed] [Google Scholar]
- 36.Mao, X., Miesfeldt, S., Yang, H., Leiden, J. M., and Thompson, C. B. (1994) J. Biol. Chem. 269 18216–18222 [PubMed] [Google Scholar]
- 37.Lauth, M., Bergstrom, A., Shimokawa, T., and Toftgard, R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8455–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taipale, J., Chen, J. K., Cooper, M. K., Wang, B., Mann, R. K., Milenkovic, L., Scott, M. P., and Beachy, P. A. (2000) Nature 406 1005–1009 [DOI] [PubMed] [Google Scholar]
- 39.Stecca, B., Mas, C., Clement, V., Zbinden, M., Correa, R., Piguet, V., Beermann, F., and Ruiz, I. A. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji, Z., Mei, F. C., Xie, J., and Cheng, X. (2007) J. Biol. Chem. 282 14048–14055 [DOI] [PubMed] [Google Scholar]
- 41.Pasca di Magliano, M., Sekine, S., Ermilov, A., Ferris, J., Dlugosz, A. A., and Hebrok, M. (2006) Genes Dev. 20 3161–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dennler, S., Andre, J., Alexaki, I., Li, A., Magnaldo, T., ten Dijke, P., Wang, X. J., Verrecchia, F., and Mauviel, A. (2007) Cancer Res. 67 6981–6986 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.