Abstract
Monofunctional and bifunctional classes of Rel proteins catalyze pyrophosphoryl transfer from ATP to 3′-OH of GTP/GDP to synthesize (p)ppGpp, which is essential for normal microbial physiology and survival. Bifunctional proteins additionally catalyze the hydrolysis of (p)ppGpp. We have earlier demonstrated that although both catalyze identical the (p)ppGpp synthesis reaction, they exhibit a differential response to Mg2+ due to a unique charge reversal in the synthesis domain; an RXKD motif in the synthesis domain of bifunctional protein is substituted by an EXDD motif in that of the monofunctional proteins. Here, we show that these motifs also determine substrate specificities (GTP/GDP), cooperativity, and regulation of catalytic activities at the N-terminal region through the C-terminal region. Most importantly, a mutant bifunctional Rel carrying an EXDD instigates a novel catalytic reaction, resulting in the synthesis of pGpp by an independent hydrolysis of the 5′Pα-O-Pβ bond of GTP/GDP or (p)ppGpp. Further experiments with RelA from Escherichia coli wherein EXDD is naturally present also revealed the presence of pGpp, albeit at low levels. This work brings out the biological significance of RXKD/EXDD motif conservation in Rel proteins and reveals an additional catalytic activity for the monofunctional proteins, prompting an extensive investigation for the possible existence and role of pGpp in the biological system.
The adaptability to changing environments determines the survival of an organism. Microorganisms utilize a hyperphosphorylated guanine nucleotide guanosine 5′-(tri- or di)phosphate,3′-diphosphate ((p)ppGpp),6 also known as the “alarmone” to cope up with unfavorable environmental conditions. (p)ppGpp, through a phenomenon termed the “stringent response,” accomplishes this by a rapid shutdown of active transcription, translation, and up-regulation of protein degradation and amino acid synthesis (1). The major effect of stringent response is so far attributed to the interactions of ppGpp to RNA polymerase, σ factors, and DksA (2, 3). Apart from its effect on transcription and translation, (p)ppGpp is implicated in the regulation of a wide variety of physiological processes including sporulation, antibiotic production, nucleotide and fatty acid metabolism, surface organelle production, and more importantly, in the virulence of pathogenic organisms (4).
(p)ppGpp is synthesized and hydrolyzed by two distinct domains in the N-terminal region of the Rel family of proteins (5–7). They are further grouped into a Rel/SpoT bifunctional class (that synthesize and hydrolyze) and a RelA monofunctional class (that can only synthesize) (8). Although both domains are present in all Rel proteins, monofunctional proteins lack hydrolysis activity due to the absence of a conserved HDXXED motif in the hydrolysis domain (9). Although the synthesis and hydrolysis activities are confined to the N-terminal ∼385 amino acids, the C-terminal region (∼385–750) ensures their regulation (10, 11). In Streptococcus equisimilis (RelS. eq), a bifunctional protein, the C-terminal region renders a negative regulatory effect on the synthesis activity and facilitates hydrolysis; deletion of the C-terminal region enhances synthesis activity by ∼12-fold while significantly reducing the hydrolysis activity (∼150-fold) (11). A direct interaction of C-terminal region with the synthesis domain has also been reported (10); however, the key players remain elusive.
During stress, Rel proteins catalyze the transfer of a pyrophosphate from ATP to the 3′-OH of GTP or GDP to synthesize pppGpp or ppGpp, respectively, collectively termed as (p)ppGpp (7, 12, 13). As favorable conditions are restored, the stringent response can be reversed by the hydrolysis of (p)ppGpp to GTP/GDP and pyrophosphate (PPi) by the bifunctional Rel proteins. Although both GTP and GDP can act as pyrophosphate acceptors for (p)ppGpp synthesis, the Rel proteins seem to display differential preference for either GDP or GTP; Rel from RelS. eq prefers GTP over GDP (11), and Rel from Mycobacterium tuberculosis (RelM. tb) (14) and RelA from Escherichia coli (RelAE. coli) (15) are reported to utilize both GTP and GDP with equal efficiency. However, as the intracellular concentration of GDP is very low, it is believed that GTP would be the principal pyrophosphate acceptor (1), and this notion is substantiated by the presence of phosphatases like GppA that hydrolyze pppGpp to ppGpp (16, 17). On the other hand, in E. coli ppGpp has been shown to be the most potent molecule to elicit stringent response (1, 4), which would imply a preference for GDP. In line with this, a closer analysis revealed that differences in (p)ppGpp synthesis by RelAE. coli and RelM. tb, reported in our earlier work, are not in consensus with the aforesaid equal preference for GTP and GDP.
Earlier, using the N-terminal regions of Rel proteins, we had shown that they differ in utilizing Mg2+ for (p)ppGpp synthesis and had attributed this difference to a charge reversal in the synthesis domain where an RXKD in the bifunctional protein is substituted to an EXDD in monofunctional proteins (8). Here, using the full-length proteins, we further explore the significance of the distinct conservation of these motifs in Rel proteins. In contrast to the earlier reports, we found that monofunctional RelAE. coli utilizes GDP and bifunctional RelM. tb utilizes GTP as the principal pyrophosphate acceptor, and this specificity is determined by the EXDD and RXKD motifs, respectively. We further find that the presence of an RXKD motif also leads to cooperative nucleotide binding, whereas EXDD does not. Interestingly, in bifunctional protein the substitution RXKD → EXDD (in RelM. tb) led to a drastic reduction in (p)ppGpp synthesis (where → indicates the interchange of the motifs). In contrast, a similar reversal in monofunctional proteins (i.e. EXDD → RXKD in RelAE. coli) resulted in enhanced synthesis. Analogous effects were not found when the N-terminal regions of the proteins were employed (8), implying a critical role for these motifs in determining the regulation of catalytic activities through their interaction with the C-terminal region. The most important finding, however, is that RXKD → EXDD substitution in the bifunctional RelM. tb resulted in the synthesis of a novel molecule that we identify as pGpp. Inspired by this observation, we probed the ability of RelAE. coli, which naturally carries an EXDD motif, to synthesize pGpp. The presence of pGpp in this reaction, albeit at low levels, opens the avenue to explore the significance of pGpp versus (p)ppGpp in microbial physiology.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification—rel from M. tuberculosis was amplified using forward (5′-TTAGAATTCCATATGACCGCCCAACGCAGCACCACC-3′) and reverse (5′-GGATCCAAGCTTTTACCAGTCGAGCAGCTGACGCATCCA-3′) primers from genomic DNA. Amplicon was digested with NdeI and HindIII and cloned into corresponding sites in pET-28 expression vector (Novagen). E. coli relA gene was amplified from its genomic DNA using primers (5′-GTTGCGGCATATGGTTGCGGTAAGAAGTGC-3′) and (5′-AATAAGCTTTAAGCTGCGTACTTCGTCGAG-3′) and similarly cloned into pET-28.
Protein expression was carried out in E. coli. BL21 pLys cells from 2 liters of growth media were resuspended in 50 mm Tris-HCl (pH 8), 300 mm NaCl, 1 mm protease inhibitor mixture (Sigma), 1 mm dithiothreitol, 1% Triton X-100 and 10% glycerol. Cells were lysed by sonication. The supernatant was collected by centrifugation at 35,000 relative centrifugal force for 30 min followed by loading onto a nickel-nitrilotriacetic acid column (Amersham Biosciences). The column was washed with 10 column volumes of wash buffer containing 20 mm Tris-HCl (pH 8.0), 500 mm NaCl, 1 mm dithiothreitol, and 50 mm imidazole. The protein was eluted using a gradient of 20 column volumes (0–0.5 m) of imidazole, and the protein eluted at 40% of the gradient. The purified protein was then subjected to gel-filtration chromatography (Superdex 200, Amersham Biosciences). The buffer was exchanged with 50 mm HEPES (pH 8), 150 mm NaCl, and 1 mm dithiothreitol. The monomer fraction of the protein was collected, which had a final concentration of 0.3–0.5 mg/ml and was stored at –80 °C after snap-freezing in liquid N2. The total yield was 0.5 mg.
(p)ppGpp Synthesis Assay—pppGpp/ppGpp synthesis assays were carried out as described earlier (8). A 5-μl reaction volume contained 50 mm HEPES (pH 8.0), 100 mm NaCl, 1 mm dithiothreitol, 10 mm MgCl2 for RXKD-containing proteins, 50 mm for EXDD-containing proteins (8), 5 mm GTP/GDP, 5 mm ATP, 1 μCi of [γ-32P]ATP, and 5 μm WT or MT-Rel proteins at 37 °C for 10–30 min. ATP and GTP/GDP were varied from 0 to 20 mm (with corresponding variations in Mg2+ concentrations) for cooperative ATP and GTP/GDP binding studies. The reactions were stopped by adding 1 μl of 6 m formic acid. The mixture was then centrifuged at 13,000 rpm for 10 min. 5 μl of the sample was spotted on the polyethyleneimine-coated TLC (Merck), resolved in 1.5 m KH2PO4 (pH 3.4) buffer, and subjected to autoradiography to detect the formation of (p)ppGpp. For quantitation, the spots corresponding to (p)ppGpp were cored out from polyethyleneimine-coated TLC, and the counts were determined.
To determine the kinetic constants for GTP/GDP, reactions were carried out as described above with varying concentrations of GTP/GDP (0–15 mm), 15 mm ATP for 10 min at 37 °C. For the RXKD-containing proteins, Mg2+ concentration was equal to that of the total nucleotides and for EXDD-containing proteins, 5-fold higher concentrations were used. 1 μCi of [γ-32P]ATP/reaction was used as tracer for determining concentrations of the product. Km and Vmax values were determined by nonlinear regression analysis carried out with GraphPad prism software.
pGpp Synthesis Assay—The efficiency of MT-RelM. tb to synthesize pGpp when GTP and GMP are provided as substrates was compared in Fig. 4C. For these, synthesis reactions were carried out as above using 5 mm GTP/GMP as the substrate along with 5 mm ATP and 1 μCi of [γ-32P]ATP and spotted on a polyethyleneimine-TLC. For quantitation, the spots corresponding to pGpp were cored out from polyethyleneimine-TLC, and the counts were determined.
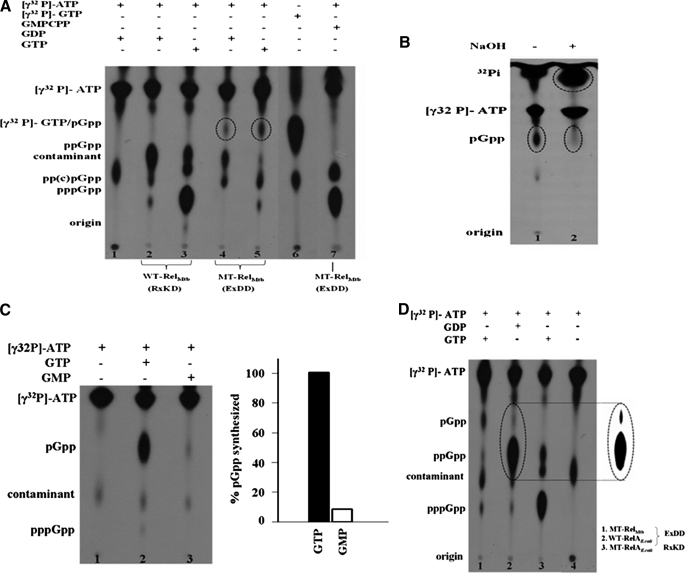
FIGURE 4.
A, the EXDD substitution in RelM. tb results in the synthesis of pGpp due to the formation of a new catalytic center. (p)ppGpp synthesis assays were carried out using [γ-32P]ATP, GDP, GTP, and GMPCPP with WT-RelM. tb and MT-RelM. tb, as indicated above the autoradiogram. No protein was used for reactions represented by lanes 1 and 6. Lane 1 contains [γ-32P]ATP as a negative control, whereas lane 6 contains [γ-32P]GTP alone to assess the mobility of the new spot. B, the new product, pGpp, is intrinsically unstable under alkaline condition. Lanes 1 and 2 denote the stability of pGpp (circled) in the absence and presence of NaOH (0.3 n), respectively. An apparent increase in the amount of 32Pi released is also indicated by a circle in lane 2. C, MT-RelM. tb utilizes GMP inefficiently for pGpp synthesis. Lane 1 contains [γ-32P]ATP as a negative control, and lanes 2 and 3 show pGpp formation using equal amounts (5 mm) of GTP and GMP, respectively. pGpp synthesized was quantitated and is shown on the right. The highest amount of pGpp synthesized (with GTP as substrate) was taken to be 100%. D, RelAE. coli that naturally contains an EXDD motif shows the presence of pGpp. (p)ppGpp synthesis assays were carried out with MT-RelM. tb and WT-RelAE. coli using [γ-32P]ATP, GTP, and GDP as indicated. The control reaction in lane 4 contains only [γ-32P]ATP and no protein. Contaminant present in the radioactive samples is also indicated. For better clarity, the contrast for the region containing pGpp and ppGpp, in lane 2, was enhanced, as shown in the inset.
pppGpp Hydrolysis Assay—pppGpp hydrolysis assays were carried out in 5-μl reaction volumes containing 50 mm HEPES (pH 8.0), 100 mm NaCl, 1 mm dithiothreitol, 10 mm MnCl2, 5 mm pppGpp32, and 5 μm WT or MT-RelM. tb proteins at 37 °C for 30 min. The reaction was stopped by adding 1 μl of 6 m formic acid. The mixture was then centrifuged at 13,000 rpm for 10 min. 5 μl of the sample was spotted on the polyethyleneimine-coated TLC (Merck), resolved in 1.5 m KH2PO4 (pH 3.4) buffer, and subjected to autoradiography to detect the release of PPi. For quantitation, the spots corresponding to PPi were cored out from polyethyleneimine-coated TLC, and the counts were determined.
Intrinsic Chemical Stability of the New Product—MT-RelM. tb was employed in a (p)ppGpp synthesis assay, and the stability of the new product (see “Results”) formed, pGpp, was tested for its identity from its isoform, ppGp. After the synthesis reaction for 30 min, the enzyme was heat-inactivated, and 1 μl of 2 n NaOH was added to 5 μl of the reaction. It was spotted on a polyethyleneimine-coated TLC (as above) together with a negative control devoid of NaOH.
Site-directed Mutagenesis—Site-directed mutagenesis was carried out by overlapping PCR method as described earlier (8). The following forward and reverse primers were used, with desired change in the codon sequence. In RelM. tb Arg-348 and Lys-350 were mutated to Glu and Asp, respectively (RelM. tb R348E/K350D forward primer, 5′-ATGGCGGGTGAGTTCGACGACTACATCGC-3′; RelM. tb R348E/K350D reverse primer 5′-GCGATGTAGTCGTCGAACTCACCCGCCATC-3′). In RelAE. coli Glu-306 and Asp-308 were mutated to Arg and Lys, respectively (RelAE. coliE306R/D308K forward primer, 5′-CGCCACCTGCCCGGGCGGTTTAAGGATTACGTCGC-3′; RelAE. coli E306R/D308K, reverse primer, 5′-GCGACGTAATCCTTAAACCGCCCGGGCAGGTGGCG-3′). In each case, the fragments were amplified (Pfu DNA polymerase) using the forward primer of the gene and the reverse primer containing the mutation and the reverse primer of the gene and the forward primer with the mutation. Amplified fragments were gel-purified. Equal quantities of both fragments were used as template for the full-length amplification of the gene with mutation using the initial primers. The PCR-amplified mutant gene was digested with NdeI and HindIII and cloned into corresponding sites in pET-28 vector. The mutations were confirmed by sequencing.
Intrinsic Tryptophan Fluorescence—Tryptophan fluorescence studies were carried out using LS 55 Fluorescence Spectrometer (PerkinElmer Life Sciences) at room temperature. The protein was monitored with an excitation wavelength of 280 nm (slit width of 2.5 nm) and emission wavelength of 300–500 nm (slit width of 5 nm).
RESULTS
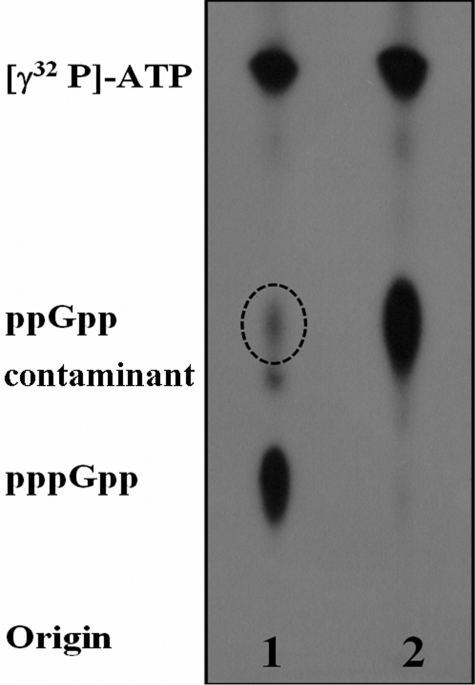
RelAE. coli Utilizes GDP as the Primary Pyrophosphate Acceptor—In our earlier work, we reported a differential effect of Mg2+ on (p)ppGpp synthesis by the N-terminal fragments of monofunctional and bifunctional Rel proteins (8). When synthesis reactions were carried out using the N-terminal regions of RelAE. coli and mutant RelM. tb (RXKD → EXDD) with ATP and GTP as substrates, we were surprised to observe an additional spot in the autoradiograms apart from pppGpp (8). Here, the same assay was performed using full-length RelAE. coli (WT-RelAE. coli). Like the N-terminal fragments, it also showed an additional spot in the autoradiograms (Fig. 1, dotted circle in lane 1) apart from the anticipated pppGpp (pentaphosphate) (lane 1, Fig. 1). Based on its position in polyethyleneimine-coated TLC, the additional spot appeared to be the tetraphosphate, ppGpp. To confirm this GTP was replaced by GDP, which resulted in the formation of ppGpp (tetraphosphate) (lane 2, Fig. 1). The position of the additional spot indeed corresponds to the ppGpp produced in lane 2. However, ppGpp synthesis in lane 1 was intriguing, as no GDP was provided in the reaction except a possible contaminant in the GTP pool that might arise from the intrinsic hydrolysis of GTP. The formation of ppGpp utilizing even the trace amount of contaminant GDP would indicate a very high affinity of RelAE. coli for GDP. This rationale would be contrary to the earlier reports that suggested equal affinity of WT-RelAE. coli for GTP and GDP (15). Interestingly, the additional spot could also be detected with the N-terminal fragment of mutant RelM. tb that had an EXDD motif (8) like WT-RelAE. coli. Taken together, these results indicated EXDD-containing proteins to have a higher affinity for GDP than GTP. On the other hand, Rel from S. equisimilis (RelS. eq) was reported to have a higher affinity for GTP over GDP (11), and RelM. tb was reported to utilize GTP and GDP with equal efficiency (14). Interestingly, both RelM. tb (14) and RelS. eq (11) are bifunctional proteins with an RXKD motif. To clarify the role of these motifs, if any, in dictating the specificities for GTP and GDP, the following experiments were conducted.
FIGURE 1.
(p)ppGpp synthesis reactions carried out using EXDD motif-containing proteins shows the formation of ppGpp (dotted circle) from the contaminant GDP in the GTP pool. The assays were carried out using WT-RelAE. coli with GTP and [γ-32P]ATP (lane 1) and with GDP and [γ-32P]ATP (lane 2). A contaminant present in the radioactive samples is circled.
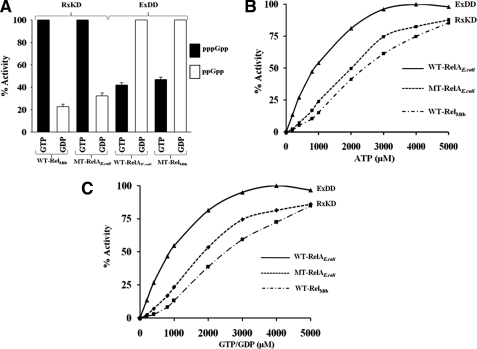
EXDD and RXKD Motifs Determine Substrate Specificity and Nucleotide Binding—To examine the specificities for GTP/GDP and the role of EXDD/RXKD motifs in governing this specificity, we used four Rel proteins; two wild types (WT-RelAE. coli with an EXDD and WT-RelM. tb with an RXKD) and their mutants where the motifs were interchanged (MT-RelAE. coli with EXDD → RXKD and MT-RelM. tb with RXKD → EXDD). Each protein was subjected to an independent synthesis reaction using ATP and either GTP or GDP as substrates. The highest amount of product synthesized (pppGpp when GTP is the substrate and ppGpp when GDP is the substrate) by each protein was taken as 100% as shown in Fig. 2A. Here, RXKD-containing proteins (WT-RelM. tb and MT-RelAE. coli) show a distinct preference for GTP over GDP, whereas the EXDD containing proteins (WT-RelAE. coli and MT-RelM. tb) conversely prefer GDP over GTP. The preference for GTP/GDP by these motifs is further substantiated by the kinetic constants, as shown in Table 1. GDP is preferred over GTP by WT-RelAE. coli (EXDD) with a lower Km and a higher Vmax, reflected in ∼15-fold higher catalytic efficiency (Vmax/Km) for GDP over GTP. On the other hand, RXKD-containing WT-RelM. tb and MT-RelAE. coli show a preference for GTP over GDP, shown by their lower Km and higher Vmax for GTP. These values are not in agreement with the kinetic constants reported earlier (5, 15) for the wild type proteins RelAE. coli and RelM. tb, as they were determined in the presence of cofactors (mRNA, tRNA, and/or ribosome) or denaturants (methanol or detergents) that were absent in our study. Nevertheless, these results demonstrate the importance of the motifs RXKD/EXDD in governing substrate specificities in Rel proteins.
FIGURE 2.
A, effect of RXKD and EXDD motifs in determining specificities for GTP and GDP. Independent (p)ppGpp synthesis assays were carried out as in Fig. 1 using the proteins and either GTP or GDP along with [γ-32P]ATP, as indicated, and % activity is shown. The highest amount of product synthesized (pppGpp when GTP as substrate and ppGpp when GDP as the substrate) by each protein was taken as 100%. B and C, RXKD motif renders cooperative ATP (B) and GTP (C) binding. % (p)ppGpp synthesized by different Rel proteins using GDP for EXDD- and GTP for RXKD-containing proteins is plotted against varying ATP (B). Similarly, in C, GTP or GDP was varied for RXKD- and EXDD-containing proteins, respectively.
TABLE 1.
The kinetic constants for GTP and GDP in RXKD and EXDD containing Rel proteins
All the values were determined in the absence of cofactors such as mRNA, tRNA, and/or ribosomes or denaturants such as methanol and detergents, unlike the earlier studies (5, 15). Values obtained from three independent experiments were used for calculating the S.D. MT-RelM. tb (EXDD) was not used here, as (p)ppGpp synthesis was reduced by 90% (see Fig. 3A).
| Protein (motif) | KmGTP | KmGDP | Vmax GTP | Vmax GDP | Vmax/Km (GTP) | Vmax/Km (GDP) |
|---|---|---|---|---|---|---|
| μm | μm | μm pppGpp formed/min | μm ppGpp formed/min | |||
| WT-RelAE. coli (EXDD) | 3703 ± 379 | 532 ± 2.5 | 60 ± 1.7 | 142 ± 0.65 | 162 | 2667 |
| MT-RelAE. coli (RXKD) | 1156 ± 89 | 2201 ± 58.5 | 249.13 ± 7.55 | 49.98 ± 1.5 | 2155 | 227 |
| WT-RelM. tb (RXKD) | 1579 ± 70.9 | 2315 ± 250 | 164.7 ± 0.47 | 62.5 ± 3.5 | 1043 | 270 |
It appears that the monofunctional RelAE. coli prefers GDP over GTP, and the bifunctional RelM. tb prefers GTP over GDP, like RelS. eq. As co-operative ATP binding was reported for RelS. eq (11), we examined the same for RelM. tb and RelAE. coli to investigate the effect of the motifs RXKD/EXDD, if any. The aforesaid (p)ppGpp synthesis assays were carried out to examine cooperativity in nucleotide binding. Percent (p)ppGpp synthesized (% activity) was plotted against increasing ATP (Fig. 2B) and GTP (Fig. 2C) concentrations for WT-RelAE. coli, MT-RelAE. coli, and WT-RelM. tb. WT-RelM. tb, like RelS. eq (11), having an RXKD motif, also exhibited a cooperative ATP binding as suggested by the sigmoidal nature of the curve (Fig. 2B). Similarly, cooperative GTP binding also was observed for WT-RelM. tb (Fig. 2C). WT-RelAE. coli, with an EXDD motif, on the other hand, displayed a hyperbolic behavior indicating the absence of cooperativity for both ATP and GTP binding. However, the hyperbolic behavior was reversed to sigmoidal behavior when MT-RelAE. coli, having an RXKD motif, was employed (Fig. 2, B and C). A reversal to hyperbolic nature in MT-RelM. tb, with an EXDD motif, could not be shown, as the activity of the protein was compromised significantly (see below). These experiments indicate that RXKD motif, but not EXDD, renders cooperative nucleotide binding.
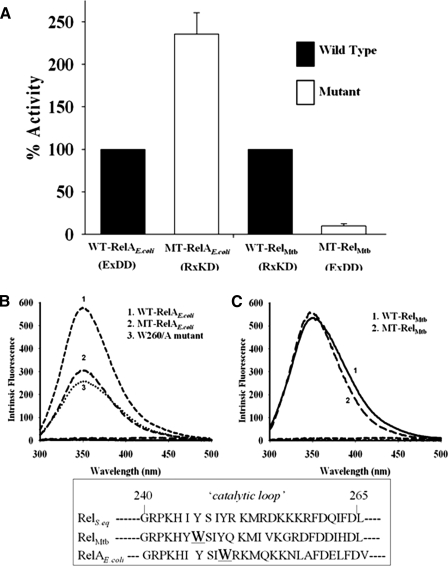
Effect of Interchanging EXDD and RXKD on (p)ppGpp Synthesis—To understand the effect of interchanging the motifs, the four proteins mentioned above were used to analyze their catalytic activities. (p)ppGpp synthesis and hydrolysis reactions were carried out as described under “Experimental Procedures.” Percent (p)ppGpp synthesized by these proteins utilizing GDP (for EXDD proteins) and GTP (for RXKD proteins) is shown in Fig. 3A. Intriguingly, MT-RelAE. coli (EXDD → RXKD) showed an enhanced synthesis (∼2.5-fold) compared with WT-RelAE. coli, whereas MT-RelM. tb (RXKD → EXDD) displayed a drastic reduction (∼90% of WT-RelM. tb) (Fig. 3A) without significantly affecting (p)ppGpp hydrolysis (data not shown). The contrasting behavior of these mutants led us to probe if any conformational change in the protein affected the synthesis activity. This was examined utilizing the intrinsic tryptophan fluorescence. A major difference in the fluorescence spectra was seen for WT-RelAE. coli versus MT-RelAE. coli (Fig. 3B). But on the contrary, RelM. tb proteins (WT-RelM. tb and MT-RelM. tb) did not exhibit any difference (Fig. 3C). As EXDD → RXKD in MT-RelAE. coli led to an enhanced (∼2.5-fold) synthesis and reduced fluorescence, we wanted to discern the candidate tryptophan that contributed significantly toward the reduced fluorescence. A sequence-based structural comparison with RelS. eq identified Trp-260 (see the inset, Fig. 3, B and C) as a potential residue based on its position in the catalytic loop. This loop is believed to play an important role in regulating the synthesis activity (8, 18). W260A mutant was, thus, prepared to examine its role, and as expected, the mutation resulted in a drastic reduction in intrinsic fluorescence (Fig. 3B), suggesting Trp-260 as the major contributor to the intrinsic fluorescence of WT-RelAE. coli. The implications of these observations are discussed under “Discussion.”
FIGURE 3.
A, interchanging RXKD and EXDD motifs in Rel proteins affects (p)ppGpp synthesis. (p)ppGpp synthesis was assayed for the indicated wild type and mutant proteins using either GTP or GDP along with [γ-32P]ATP. The activity of MT-RelAE. coli is plotted while considering WT-RelAE. coli activity to be 100%. The activities of WT-RelM. tb and MT-RelM. tb are also plotted similarly. B, intrinsic fluorescence exhibited by WT-RelAE. coli, MT-RelAE. coli, and W260A mutant of WT-RelAE. coli. C, intrinsic fluorescence exhibited by WT-RelM. tb and MT-RelM. tb. Emission spectra were recorded at λex of 280 nm. The inset shows a structure-based sequence comparison of the catalytic loop of RelS. eq, RelM. tb, and RelAE. coli, with the numbers corresponding to that of RelS. eq.
The Presence of EXDD Generates a New Catalytic Site—A reduction in the synthesis activity by MT-RelM. tb was unanticipated. However, we were surprised by the presence of a new spot (circled in lanes 4 and 5; Fig. 4A) in the autoradiograms of (p)ppGpp synthesis reactions catalyzed by MT-RelM. tb. The new spot was not seen when WT-RelM. tb (lanes 2 and 3) or the N-terminal domains (8) were used, and its migration was similar to that of [γ-32P]GTP (lane 6, Fig. 4A). Mobilities similar to GTP indicated that the new product might contain identical number of phosphates on the guanine nucleoside, suggesting pGpp, ppGp, or pppG as the possible molecules. Because [γ-32P]ATP was used in lanes 4 and 5, where the new molecule is formed, it must bear a radiolabeled pyrophosphate transferred from [γ-32P]ATP to the 3′-OH of the guanine nucleotide. This eliminates ppGp and pppG as candidates, and hence, the product should be 5′-pGpp-3′. This was further examined by intrinsic (in)stability of the new product under alkaline conditions. This is because the presence of β-phosphates at the 3′-OH is known to be alkali-labile (19). In Fig. 4B, the new product (circled in lane 1) disappears when the reaction was subjected to alkaline conditions (lane 2), indicating that the new product bears a radiolabeled 3′-β phosphate. Hence the new product would be 5′-pGpp-3′ and not its isoform, 5′-ppGp-3′, which is stable under alkaline conditions (19–21).
Interestingly, the formation of pGpp requires GMP, which was not supplied. We hypothesized that the GMP part of pGpp would have arisen either as a contaminant in GTP/GDP pool or as a result of an ester hydrolysis between Pα and Pβ of GTP/GDP, perhaps due to the formation of a novel catalytic center in MT-RelM. tb. However, Rel proteins were shown to utilize GMP very inefficiently (11, 22), and because GTP/GDP, the efficient substrates, were provided in excess, the possibility of pGpp production due to contaminant GMP might be negated. This was further substantiated by comparing the efficiency with which MT-RelM. tb utilizes GTP and GMP for pGpp synthesis, which is shown in lanes 2 and 3 of Fig. 4C, respectively. The amount of pGpp produced in these clearly depicts that GMP is a poor substrate for MT-RelM. tb as compared with GTP, ruling out the possibility of utilizing contaminant GMP from the GTP/GDP pool to synthesize pGpp.
To verify the possibility that pGpp is formed due to a hydrolysis of the Pα-O-Pβ bond of GTP/GDP, GMPCPP, a GTP analogue with a carbon in place of an oxygen between Pα and Pβ was used to hinder the aforesaid ester hydrolysis (lane 7, Fig. 4A). Indeed, lane 7 shows the synthesis of pp(c)pGpp, but not pGpp, emphasizing the need for a hydrolysable oxygen between Pα and Pβ of GTP/GDP to synthesize pGpp. Hence, pGpp was synthesized due to the aforesaid novel catalytic center, which was further substantiated by the release of a pyrophosphate from [γ-32P]GTP (Fig. 5), as discussed below. It appears that introducing EXDD resulted in the formation of a new catalytic site to synthesize pGpp by either hydrolyzing GTP/GDP to GMP, which then accepts a pyrophosphate from ATP, or by hydrolyzing (p)ppGpp to pGpp. A critical role for the C-terminal region in catalyzing this reaction was brought out by the fact that only the full-length protein (carrying EXDD mutation) can synthesize pGpp. Therefore, the reaction catalyzed by full-length WT-RelAE. coli, possessing an EXDD, was also examined for the presence of pGpp. The autoradiogram in Fig. 4D, showing a spot at an identical position (lane 2), confirmed that it also synthesized pGpp, although the amounts are insignificant compared with the amount of (p)ppGpp. As anticipated, pGpp synthesis was abolished by MT-RelAE. coli, with EXDD → RXKD substitution (lane 3, Fig. 4D), reiterating the need of EXDD for this activity. Further experiments were designed to understand this novel catalytic subsite.
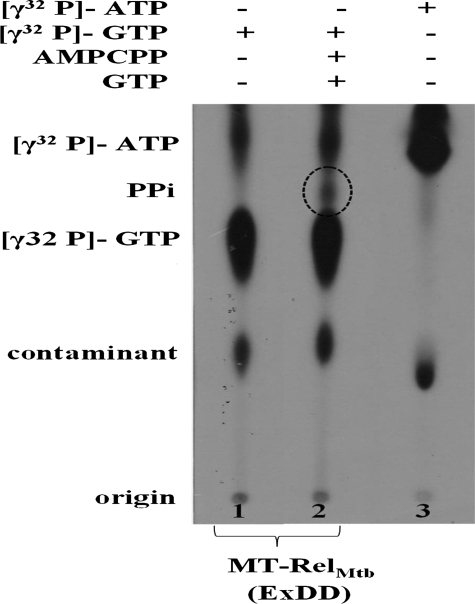
FIGURE 5.
The new catalytic center involves Pα-O-Pβ bond cleavage of GTP/GDP, (p)ppGpp, and is independent of the pyrophosphoryl transfer reaction from ATP. MT-RelM. tb was used in (p)ppGpp synthesis reactions with [γ-32P]GTP, GTP, and AMP-CPP, a non-hydrolysable ATP analog were used in lane 2. Lanes 1 and 3 contain only [γ-32P]GTP and only [γ-32P]ATP, respectively. The pyrophosphate (PPi) released due to Pα-O-Pβ bond cleavage is indicated in dotted circle.
The New Catalytic Subsite Functions Independent of the Pyrophosphoryl Transfer—Our results clearly demonstrate that EXDD motif along with the C-terminal region is capable of synthesizing pGpp, likely due to the hydrolysis of Pα-O-Pβ bond of GTP/GDP. However, to determine the (in)dependence of this reaction on the pyrophosphate transfer from ATP, a non-hydrolysable ATP analogue, AMP-CPP, was used to inhibit the same. In reactions where [γ-32P]GTP or [γ-32P]ATP alone was used as substrates, no additional spots were detected (lanes 1 and 3, Fig. 5). Interestingly, complementing [γ-32P]GTP with AMP-CPP led to the formation of labeled PPi (lane 2, Fig. 5), suggesting the hydrolysis of Pα-O-Pβ bond of GTP even in the absence of a pyrophosphate transfer from ATP. This demonstrated that the new catalytic subsite in the full-length protein, created by EXDD, functioned independent of pyrophosphate transfer from ATP but required ATP binding at the active site.
DISCUSSION
The Rel family of proteins is essential for microbial survival under stress by virtue of its ability to metabolize (p)ppGpp, the mediator of stringent response (1, 4, 6, 7, 13). That monofunctional and bifunctional Rel proteins, due to a unique charge reversal in the synthesis domain, differ in Mg2+ utilization to synthesize (p)ppGpp, was earlier reported by us (8). Driven by the observation that monofunctional proteins generate both pppGpp and ppGpp, although only GTP but not GDP was provided, together with ATP in the synthesis reaction, we set out to examine the effects rendered by the EXDD motif. Here, we uncover several other intriguing aspects of this seemingly simple charge reversal.
Here, for the first time we generalize substrate specificities for the Rel family of proteins based on the presence of a conserved motif in the nucleotide binding region of the synthesis domains. Proteins with an RXKD motif prefer GTP, and those with an EXDD prefer GDP as the principal pyrophosphate acceptor to synthesize (p)ppGpp. This preference is also indicated by the kinetic constants shown in Table 1. Although these values were determined in the absence of any cofactors or denaturants, unlike in the earlier reports (5, 15), we believe that RXKD and EXDD motifs continue to govern substrate specificity, although the absolute values of Km and Vmax would change in presence of the cofactors in vivo. So far, based on qualitative analysis, RelM. tb was inferred to utilize GTP and GDP with equal efficiency (14), but a quantitative study on RelS. eq showed a preference for GTP (11). Both being bifunctional RXKD-containing proteins, the motif-based generalization of specificity concurs with our finding that RelM. tb also prefers GTP (Fig. 2). That RelAE. coli with an EXDD motif utilizes GDP (Fig. 2) as the principal pyrophosphate acceptor is also in concurrence. However, it is not in agreement with the equal GTP/GDP efficiencies reported by Cochran and Byrne (15). Perhaps this discrepancy arises due to the methods used to prepare RelAE. coli; Cochran and Byrne (15) used the NH4Cl wash of 70 S ribosomes that also contained ribosome-associated GTPases (23), which would hydrolyze GTP used in the assays to GDP, leading to an apparent equal affinity. Also, the efficiency was calculated based on the total amount of both pppGpp and ppGpp synthesized. On the other hand, the procedures employed by us (see “Experimental Procedures”) ensured high purity of the protein. The implication of a high GDP affinity of RelAE. coli indicates its ability to synthesize ppGpp directly bypassing phosphatases GppA to hydrolyze pppGpp to ppGpp; ppGpp is known to be the major mediator of stringent response in E. coli (1, 4). In addition, this high GDP affinity may also be utilized by RelAE. coli to ensure a rapid supply of ppGpp necessary to maintain a high fidelity of amino acid biosynthesis (24, 25) and to elicit a prompt response to the dynamically changing environment. This is possible because active translation utilizes a large amount of GTP to ensure high fidelity (26, 27) and that may perhaps result in a higher local GDP concentration in the microenvironment around the ribosomes which could be exploited by RelAE. coli.
Although significance to a high GDP affinity of RelAE. coli may be construed, the genesis of the differential GDP/GTP specificities of EXDD/RXKD motifs is intriguing. In the earlier work, based on the crystal structure of RelS. eq (18) and Mg2+-independent nucleotide binding in RXKD-containing proteins, we speculated that the phosphate groups of GTP could be coordinated by the positively charged Lys and Arg residues (8). Hence, the presence of γ-phosphate may explain the observed preference of RXKD for GTP. In the case of EXDD enzymes, we reasoned that the interactions provided by Lys and Arg of RXKD are compensated by interactions from an additional Mg2+, coordinated by the carboxyl groups of Glu and Asp of EXDD (8). They may bind guanine nucleotides by replacing the Mg2+ of GDP·Mg2+/GTP·Mg2+ complexes with the EXDD-coordinated Mg2+. A possible reasoning for the preference of these motifs for GTP/GDP may stem from their different modes of nucleotide binding and also from the difference in coordination of Mg2+ in GDP·Mg2+ and GTP·Mg2+ complexes. Mg2+ of GDP·Mg2+ coordinates Pα and Pβ, and that in GTP·Mg2+ would also involve Pγ, leading to a difference in the positioning of Mg2+ while binding to the protein. Such a difference in the position of Mg2+ is observed in the structures of ppGpp bound to RNA polymerase (2, 3) and GTP bound to Ras (28, 29). This differential positioning of Mg2+ may lead to differences in binding energy for motifs RXKD and EXDD to replace the GTP/GDP coordinated Mg2+ such that it is optimal for EXDD to bind GDP·Mg2+ and RXKD to bind GTP·Mg2+. Interestingly, K+ ion channels similarly exploit subtle differences in hydration energy to selectively transport K+ ions, but not Na+, although the latter is smaller in size (30). Although this speculation may explain the differential specificities in Rel proteins, evidently rigorous crystal structure analysis in the presence of GDP and GTP for both EXDD and RXKD-containing proteins will be needed to comprehensively understand this intriguing attribute.
Apart from substrate specificity, different modes of regulation in bifunctional and monofunctional Rel proteins may be inferred based on the finding that RXKD imparts cooperative ATP and GTP binding but not EXDD (Fig. 2, B and C). Perhaps a better regulation is achieved by RXKD for bifunctional proteins as they need to regulate two activities (i.e. synthesis and hydrolysis) at distinct domains. Delving further, we found that the C-terminal region exerts a negative regulatory effect on (p)ppGpp synthesis by means of interactions mediated through these motifs at the N-terminal region. RXKD → EXDD reversal in bifunctional RelM. tb led to a drastic reduction in (p)ppGpp synthesis, whereas EXDD → RXKD in RelAE. coli increased synthesis by 2.5-fold (Fig. 3A). However, when only the N-terminal regions of the same proteins were employed, a similar influence was not observed (8). Hence, the observed differences in synthesis are likely due to differences in interactions of the C-terminal region with the EXDD and RXKD motifs. An EXDD → RXKD interchange in MT-RelAE. coli apart from enhancing synthesis activity, exhibited a reduction in the intrinsic fluorescence (Fig. 3B). We attributed the reduced fluorescence in the full-length protein to a possible exposure of Trp-260 (in the catalytic loop) to an aqueous environment that was otherwise buried due to interactions by the C-terminal region. The presence of RXKD in the mutant would have altered the interactions between the synthesis domain and the C-terminal region and thereby enhanced synthesis. On the contrary, RXKD → EXDD in RelM. tb that displayed a severe reduction in synthesis did not show a difference in fluorescence, although here too the catalytic loop contained a tryptophan (see the inset, Fig. 3C). Unaltered fluorescence negates a major conformational change, and the reduction in (p)ppGpp synthesis would perhaps have arisen due to a strengthened interaction of EXDD with the C-terminal region in the mutant. Such strengthening may not influence the environment of the tryptophan in the catalytic loop but would be necessary to occlude water to facilitate the formation of a new catalytic site. Together with the observed increase in synthesis for RelS. eq (with RXKD) upon deleting the C-terminal region (11), we conclude that the C-terminal region regulates the activities of Rel proteins through interactions mediated by these motifs.
However, it would be interesting to compare the conformational changes seen in the full-length protein and the N-terminal half carrying only the catalytic domains (8). In the latter we had earlier proposed a “loop to helix” transition of the catalytic loop, present in the synthesis domain. Based on circular dichroism experiments, we had proposed that these structural changes occur in RXKD-containing Rel proteins (N-terminal part) with increasing Mg2+ (8). Here, although the observed change in intrinsic fluorescence is due to a Trp residue present in the same catalytic loop, the conformational changes appear to arise due to an interaction between the C-terminal region in the full-length protein and the EXDD motif, as discussed above.
Thus far our work reveals several interesting features of Rel proteins, attributed by the motifs RXKD and EXDD. However, an important finding is the additional catalytic reaction by EXDD-containing proteins to synthesize pGpp (Fig. 4). This new catalytic activity, involving the hydrolysis of the 5′ α-β ester bond of GTP/GDP/(p)ppGpp, occurs in same active site of the synthesis domain (Fig. 5). The key features of this additional reaction are as follows. 1) It requires the presence of an EXDD motif. 2) The reaction is independent of the pyrophosphate transfer reaction from ATP. 3) It requires both GTP/GDP and ATP bind to the pocket. 4) In addition, it also requires the C-terminal region, as the additional product, pGpp, was absent when the N-terminal regions alone were employed (8). The fact that pGpp synthesis was pronounced in RXKD → EXDD mutant of RelM. tb than in RelAE. coli, where EXDD is present naturally, made it possible to identify the existence of pGpp. A possible reason for the different amounts of pGpp produced may be provided based on the strength of interactions between the C-terminal region and the synthesis domain as described above; the stronger the interaction, the lower the (p)ppGpp synthesized and higher the pGpp synthesized. These conjectures raise several possibilities and warrant the structure determination of full-length Rel proteins.
In summary, our findings open new avenues to further explore the function of Rel proteins. It may appear that pGpp is a byproduct of (p)ppGpp synthesis by RelAE. coli. However one cannot rule out that depending on the biological context, RelAE. coli may fine-tune interactions between synthesis domain and C-terminal region to promote pGpp synthesis. Thus, Gram-negative bacteria may have an evolutionary advantage of having a monofunctional RelA in addition to SpoT, a bifunctional enzyme, to adjust to the dynamic environment more rapidly than the Gram positives, which need to cope with a single copy of bifunctional Rel. As RXKD → EXDD in bifunctional proteins introduced a third activity, i.e. pGpp synthesis, efficient regulation of three independent catalytic activities in a single polypeptide chain may not be viable and, hence, the absence of an EXDD in bifunctional proteins. This would be in line with the drastic reduction in (p)ppGpp synthesis upon RXKD → EXDD substitution in bifunctional RelM. tb. Similarly, monofunctional proteins would have overcome this constraint of regulating three activities by compromising the hydrolysis activity, ensured by the presence of SpoT in the organism and thereby facilitating the efficient acquisition of a new catalytic activity in the synthesis domain.
Acknowledgments
We sincerely thank the members of Prakash laboratory for input during the course of the study.
Author's Choice—Final version full access.
This work was supported by a Wellcome Trust, United Kingdom International Senior Research Fellowship (to B. P.) and generous support provided by the Department of Biotechnology, New Delhi (to B. P. and V. K. N.).
Footnotes
The abbreviations used are: (p)ppGpp, ppGpp (guanosine 5′-diphosphate,3′-diphosphate) and pppGpp (guanosine 5′-triphosphate,3′-diphosphate); pGpp (guanosine 5′-monophosphate,3′-diphosphate; AMP-CPP, adenosine 5′-(α,β-methylene)triphosphate; WT, wild type; MT, mutant.
References
- 1.Cashel, M., Gentry, D., Hernandez, V. J., and Vinella, D. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhardt, F. C., ed) Vol. 2, pp. 1458–496, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 2.Artsimovitch, I., Patlan, V., Sekine, S., Vassylyeva, M. N., Hosaka, T., Ochi, K., and Vassylyev, D. G. (2004) Cell 117 299–310 [DOI] [PubMed] [Google Scholar]
- 3.Perederina, A., Svetlov, V., Vassylyeva, M. N., Tahirov, T. H., Yokoyama, S., Artsimovitch, I., and Vassylyev, D. G. (2004) Cell 118 297–309 [DOI] [PubMed] [Google Scholar]
- 4.Potrykus, K., and Cashel, M. (2008) Annu. Rev. Microbiol. 62 35–51 [DOI] [PubMed] [Google Scholar]
- 5.Avarbock, A., Avarbock, D., Teh, J., Buckstein, M., Wang, Z., and Rubin, H. (2005) Biochemistry 44 9913–9923 [DOI] [PubMed] [Google Scholar]
- 6.Cashel, M., and Gallant, J. (1969) Nature 221 838–841 [DOI] [PubMed] [Google Scholar]
- 7.Haseltine, W. A., Block, R., Gilbert, W., and Weber, K. (1972) Nature 238 381–384 [DOI] [PubMed] [Google Scholar]
- 8.Sajish, M., Tiwari, D., Rananaware, D., Nandicoori, V. K., and Prakash, B. (2007) J. Biol. Chem. 282 34977–34983 [DOI] [PubMed] [Google Scholar]
- 9.Aravind, L., and Koonin, E. V. (1998) Trends Biochem. Sci. 23 469–472 [DOI] [PubMed] [Google Scholar]
- 10.Jain, V., Saleem-Batcha, R., and Chatterji, D. (2007) Biophys. Chem. 127 41–50 [DOI] [PubMed] [Google Scholar]
- 11.Mechold, U., Murphy, H., Brown, L., and Cashel, M. (2002) J. Bacteriol. 184 2878–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sy, J., and Lipmann, F. (1973) Proc. Natl. Acad. Sci. U. S. A. 70 306–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendrich, T. M., Blaha, G., Wilson, D. N., Marahiel, M. A., and Nierhaus, K. H. (2002) Mol. Cell. 10 779–788 [DOI] [PubMed] [Google Scholar]
- 14.Avarbock, D., Salem, J., Li, L., Wang, Z., and Rubin, H. (1999) Gene (Amst.) 233 261–269 [DOI] [PubMed] [Google Scholar]
- 15.Cochran, W. J., and Byrne, R. W. (1974) J. Biol. Chem. 249 353–360 [PubMed] [Google Scholar]
- 16.Keasling, J. D., Bertsch, L., and Kornberg, A. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7029–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda, A., Murphy, H., Cashel, M., and Kornberg, A. (1997) J. Biol. Chem. 272 21240–21243 [DOI] [PubMed] [Google Scholar]
- 18.Hogg, T., Mechold, U., Malke, H., Cashel, M., and Hilgenfeld, R. (2004) Cell 117 57–68 [DOI] [PubMed] [Google Scholar]
- 19.Cashel, M., and Kalbacher, B. (1970) J. Biol. Chem. 245 2309–2318 [PubMed] [Google Scholar]
- 20.Pao, C. C., and Gallant, J. (1979) J. Biol. Chem. 254 688–692 [PubMed] [Google Scholar]
- 21.Pao, C. C., Dennis, P. P., and Gallant, J. A. (1980) J. Biol. Chem. 255 1830–1833 [PubMed] [Google Scholar]
- 22.Sy, J. (1975) Biochemistry 14 970–973 [DOI] [PubMed] [Google Scholar]
- 23.Nishizuka, Y., and Lipmann, F. (1966) Proc. Natl. Acad. Sci. U. S. A. 55 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nègre, D., Cortay, J. C., Donini, P., and Cozzone, A. J. (1989) Biochemistry 28 1814–1819 [DOI] [PubMed] [Google Scholar]
- 25.Wagner, E. G. H., and Kurland, C. G. (1980) Mol. Gen. Genet. 180 139–145 [DOI] [PubMed] [Google Scholar]
- 26.Ogle, J. M, and Ramakrishnan, V. (2005) Annu. Rev. Biochem. 74 129–177 [DOI] [PubMed] [Google Scholar]
- 27.Rodnina, M. V., and Wintermeyer, W. (2001) Annu. Rev. Biochem. 70 415–435 [DOI] [PubMed] [Google Scholar]
- 28.Margarit, S. M., Sondermann, H., Hall, B. E., Nagar, B., Hoelz, A., Pirruccello, M., Bar-Sagi, D., and Kuriyan, J. (2003) Cell 112 685–695 [DOI] [PubMed] [Google Scholar]
- 29.Scheffzek, K., Ahmadian, M. R., Kabsch, W., Wiesmüller, L., Lautwein, A., Schmitz, F., and Wittinghofer, A. (1997) Science 277 333–338 [DOI] [PubMed] [Google Scholar]
- 30.Doyle, D. A., Cabral, J. M., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T., and MacKinnon, R. (1998) Science 280 69–77 [DOI] [PubMed] [Google Scholar]