Abstract
The cardiac IKs potassium channel is a macromolecular complex consisting of α-(KCNQ1) and β-subunits (KCNE1) and the A kinase-anchoring protein (AKAP) Yotiao (AKAP-9), which recruits protein kinase A) and protein phosphatase 1 to the channel. Here, we have tested the hypothesis that specific cAMP phosphodiesterase (PDE) isoforms of the PDE4D family that are expressed in the heart are also part of the IKs signaling complex and contribute to its regulation by cAMP. PDE4D isoforms co-immunoprecipitated with IKs channels in hearts of mice expressing the IKs channel. In myocytes isolated from these mice, IKs was increased by pharmacological PDE inhibition. PDE4D3, but not PDE4D5, co-immunoprecipitated with the IKs channel only in Chinese hamster ovary cells co-expressing AKAP-9, and PDE4D3, but not PDE4D5, co-immunoprecipitated with AKAP-9. Functional experiments in Chinese hamster ovary cells expressing AKAP-9 and either PDE4D3 or PDE4D5 isoforms revealed modulation of the IKs response to cAMP by PDE4D3 but not PDE4D5. We conclude that PDE4D3, like protein kinase A and protein phosphatase 1, is recruited to the IKs channel via AKAP-9 and contributes to its critical regulation by cAMP.
The high specificity of cAMP cellular effects in the heart results from distinct localization of hormonal receptors (1) and from the compartmentalization of key molecular elements of cAMP signaling in subcellular microdomains. This compartmentalization is achieved via protein kinase A-anchoring proteins (AKAPs),2 adaptor proteins that have been shown to recruit multiple regulatory elements to target proteins. AKAPs have been shown to interact with protein kinase A (PKA), phosphatases, and adenylate cyclases and also with 14-3-3 proteins and other proteins involved in cAMP signaling (2, 3). Over the past few years, an increasing number of reports have shown that phosphodiesterases (PDEs) also interact with AKAPs (4). PDEs constitute the sole route for degrading cyclic nucleotides in cells. In mammalian tissues, 11 PDE families (PDE1 to PDE11) have been identified. They mainly differ by (a) the sequence of their N terminus; (b) their specificity for cAMP (versus cGMP); (c) their expression pattern; and (d) their protein-protein interactions (5, 6). In the mammalian heart, the temporal and spatial dynamics of cAMP gradients are predominantly controlled by members of the PDE3 and PDE4 subfamilies, with a prevailing role of PDE4 enzymes, which are cAMP-specific PDEs (3, 7, 8).
The PDE4 family is composed of four subfamilies, corresponding to four genes (PDE4A, PDE4B, PDE4C, and PDE4D), which all comprise several isoforms that are distinguished from each other by their unique N-terminal sequence (3, 9). This isoform-specific N terminus plays a key role in the compartmentalization of the different PDE4s because it determines their targeting either to selective intracellular membranes or to specific signaling complexes such as AKAPs (3). Interestingly, until now, one major PDE4D isoform was shown to interact with AKAPs; PDE4D3 associates with AKAP-18 (10), AKAP-250 (11, 12), AKAP-450 (12, 13), and mAKAP (14). Given the predisposition of PDE4D3 to associate with AKAPs, it is possible that this PDE interacts with other AKAPs that have yet to be identified. One candidate is AKAP-9, an adaptor protein that binds to IKs channels in the heart and that enables the distinct regulation of IKs channel activity by cAMP (15).
IKs potassium channels are major regulators of cardiac electrical activity, particularly during sympathetic stimulation (16, 17). These channels, which are composed of four KCNQ1 α-subunits assembled with KCNE1 β-subunits (18, 19), form a macromolecular complex with key proteins involved in cAMP signaling; AKAP-9 recruits PKA and protein phosphatase 1 to the KCNQ1 C terminus and regulates the PKA phosphorylated state of a single serine residue (Ser-27) located in the KCNQ1 N terminus (15). To be translated into maximal functional regulation of channel activity, the presence of KCNE1 subunits is required (20), and both KCNQ1 and the constitutively bound AKAP-9 must be PKA phosphorylated (15, 21–23).
Here, we report that in the mammalian heart, the cAMP-dependent activity of IKs channels is not only regulated by cAMP-specific PDE4 activity but that this is achieved through a spatially constrained regulatory module where the PDE4D3 isoform is sequestered with IKs channels through the action of the multifunctional PKA anchor protein, AKAP-9. This allows the functional response of IKs channels to cAMP to be modulated specifically by AKAP-9-sequestered PDE4D3. We conclude from this work that PDE4D3 is an integral part of the IKs macromolecular complex in the heart and contributes to its regulation by the cAMP-dependent signaling cascade.
EXPERIMENTAL PROCEDURES
Transgenic Mice and Isolation of Cardiac Ventricular Myocytes—Adult transgenic mice expressing hKCNE1-hKCNQ1 fusion protein in the heart have been previously described (15). Mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). Hearts were then removed, and ventricular myocytes were isolated using the methodology of Mitra and Morad (24) following protocols previously described (25) and approved by the Institutional Animal Care and Use Committee at Columbia University. Ventricular myocytes were used in electrophysiology experiments within 6 h following isolation.
Cell Culture and Transfection—Chinese hamster ovary (CHO) cells (American Type Cell Culture) cultured in Ham's F12 medium and incubated at 37 °C (5% CO2) were used for biochemistry experiments (immunoprecipitations and Western blots). For patch clamp experiments, we generated a CHO cell line that stably expresses AKAP-9 by using the Flp-In system (Invitrogen). This cell line was maintained in a medium containing hygromycin B (500 μg/ml, Invitrogen). For both cell lines, transient transfection was carried out by using Lipofectamine with Lipofectamine Plus reagents (Invitrogen). Depending on experiments, the following DNAs were transfected: human KCNQ1, human KCNE1, the fusion protein hKCNE1-hKCNQ1 (i.e. IKs channels complex) (15), human AKAP-9, human VSV-epitope-tagged PDE4D3, and human VSV-epitope-tagged PDE4D5 (26). Control experiments with wild type PDE4D isoform (i.e. without VSV tag) were carried out to confirm that the VSV tag did not affect the protein-protein interactions probed (data not shown). For electrophysiological experiments, CD8 DNA was transfected along with DNAs of interest, and Dynabeads M-450 anti-CD8 beads were used to identify transfected cells (Dynal Biotech ASA, Oslo, Norway). These cells were plated onto 35-mm plastic Petri dishes after 24 h and used for patch clamp recordings 48 h after transfection.
Immunoprecipitation and Western Blot Experiments—CHO cells were lysed 48 h after transfection in lysis buffer containing: 150 mm NaCl, 10 mm Tris, 1 mm EDTA, 1% Triton X-100, pH 7.4, and Complete protease inhibitor mixture (Roche Diagnostics) at 4 °C for 1 h. Lysates were then centrifuged (15,000 × g) for 25 min, and supernatants were collected and used for either Western blot or immunoprecipitation experiments. Mice hearts were homogenized in phosphate-buffered saline and Complete protease inhibitor mixture at 4 °C with a Tissue-miser homogenizer (Fisher Scientific). Immunoprecipitation experiments were carried out by using either CHO cell lysates or heart homogenates with a modified radioimmune precipitation assay buffer containing (in mm): 150 NaCl, 50 Tris, 1 EDTA, 0.25% Triton X-100 (pH 7.4). A commercial KCNQ1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used to immunoprecipitate and detect KCNQ1 and the fusion protein hKCNE1-hKCNQ1, whereas a custommade AKAP-9 antibody (22) was used to immunoprecipitate and detect AKAP-9. Protein G-Sepharose beads were used to immobilize the immunocomplex, and the immunoprecipitates were washed extensively by using radioimmune precipitation assay buffer. Immunoprecipitates, lysates, and homogenates were size-fractionated on 7.5% SDS-PAGE. The presence of PDE4D family enzymes was performed using an antibody that only reacts with PDE4 isoforms from the PDE4D subfamily (26). This antibody was raised in sheep against a glutathione S-transferase fusion protein containing the C-terminal 40 amino acids of PDE4D, a sequence common to all of the known, active PDE4D isoforms (but different from PDE4A, PDE4B, and PDE4C isoforms). This construct has previously been used to raise PDE4D-specific antisera that have been verified in previous publications (26–28), and the antiserum here was identically PDE4D-specific, not detecting representative PDE4A, PDE4B, and PDE4C isoforms (data not shown). In Western blot experiments using mouse heart, recombinant PDE4D1, PDE4D3, and PDE4D5 expressed in COS1 cells were used as molecular weight markers (“standards”).
Single Cell Electrophysiology—Cells were plated in culture dishes, which were then placed on the stage of an inverted microscope (Nikon Diaphot 200, Nikon Instruments Inc., Melville, NY). IKs was recorded at room temperature with the whole-cell configuration of the patch clamp technique (series resistance was 2–3 megaohms) using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA). Voltage clamp protocols have been described previously (29) and consisted of 2-s depolarizing steps to +60 mV (from a holding potential of -75 mV) followed by 2-s repolarizing pulses to -40 mV during which IKs tail current was measured (stimulation frequency was 0.06 Hz). External solutions contained the following (in mm): 132 NaCl, 4.8 KCl, 2 CaCl2, 1.2 MgCl2, 10 HEPES, 5 glucose (pH = 7.4). In experiments with ventricular myocytes, E-4031 (5 μm, Wako, Osaka, Japan) and nisoldipine (1 μm, gift from Bayer AG, Leverkusen, Germany) were used to block IKr and ICa, respectively, and Na+ channels were inactivated by preceding IKs activation pulses with prepulses to -40 mV for 20 ms. Internal solution was (in mm): 110 potassium aspartate, 5 ATP-K2, 11 EGTA, 10 HEPES, 5.5 CaCl2 (free intracellular calcium was 100 nm), 1 MgCl2 (pH = 7.3). In experiments with ventricular myocytes, 100 μm IBMX (Sigma-Aldrich) and 1 μm rolipram (Sigma-Aldrich) were applied in external solutions to inhibit all phosphodiesterases and PDE4s, respectively (30, 31). In some experiments with CHO cells, 200 μm cAMP (Sigma-Aldrich) and 0.2 μm okadaic acid (OA; Calbiochem) were added to the internal solution to stimulate IKs current.
Data Analysis—Patch clamp data, shown as mean ± S.E., were acquired using pCLAMP 8.0 (Axon Instruments) and analyzed with Origin 7.0 (OriginLab, Northampton, MA) and Clampfit 8.2 (Axon Instruments). Statistical data analysis was assessed with Student's t test for simple comparisons; differences at p < 0.05 were considered to be significant. For quantification of immunoprecipitation experiments, gels were loaded with similar amounts of cell extracts, and all films were developed for the same exposure time. Western blots were then scanned, and the signal intensity of immunoreactive bands was quantified using ImageJ software. Following a quantification method that we previously reported (23), the amount of PDE4D recovered in the immunoprecipitation pellet was corrected for input error and normalized to allow comparisons. In each case, averaged data from three experiments are shown and expressed as mean ± S.E. Statistical data analysis was assessed with ANOVA and Bonferroni's test; differences of p < 0.05 were considered to be significant.
RESULTS
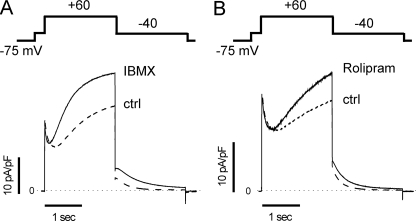
Phosphodiesterases Regulate IKs Channels in Murine Ventricular Myocytes—To determine whether PDE regulate cAMP-dependent activity of IKs channels in the heart, we used a murine model that we have previously characterized in which IKs, channels expressed in a targeted manner in the heart, were shown to respond to cAMP (15). Voltage clamp experiments were carried out on ventricular myocytes freshly isolated from mouse heart, and IKs channels were recorded with a stimulation protocol consisting of a series of 2-s depolarizing steps to +60 mV, from a holding potential of -75 mV followed by 2-s repolarizing steps to -40 mV, during which tail current amplitude (Itail) was measured. Recordings were first carried out in control conditions and then in the presence of 100 μm IBMX, a non-selective PDE inhibitor (30), applied in the bath solution. As shown in Fig. 1A, IBMX significantly increased IKs from 4.21 ± 0.86 pA/pF to 7.57 ± 1.61 pA/pF (n = 3, p < 0.05). In the mammalian heart, the temporal and spatial dynamics of cAMP gradients are mainly controlled by PDE4s (7). We then repeated our experiments using rolipram, a PDE4-selective inhibitor (9, 32) (Fig. 1B). At 1 μm, a concentration corresponding to the IC50 for PDE4 (30, 31), rolipram increased IKs from 4.44 ± 0.78 pA/pF to 6.21 ± 0.86 pA/pF (n = 3, p < 0.05). These experiments suggest that in ventricular myocytes isolated from our IKs mouse, phosphodiesterases in general and PDE4s in particular regulate IKs channels.
FIGURE 1.
Phosphodiesterases regulate IKs activity in cardiac myocytes. IKs was recorded in ventricular myocytes isolated from an IKs transgenic mouse (“Experimental Procedures”) using the voltage clamp protocol illustrated schematically above the current traces (stimulation frequency was 0.06 Hz). A, current traces averaged from three cells obtained in control (ctrl) conditions (dashed line) and in the presence of 100 μm IBMX (solid line). B, same as A but with 1 μm rolipram (solid line) instead of IBMX. In A and B, the horizontal dotted line indicates zero current.
PDE4D Is Associated with IKs Channels in the Heart—Recent studies have reported that members of the PDE4D subfamily physically interact, in a direct way or via an adaptor protein, with cardiac target proteins such as β-arrestin (33, 34), β-adrenergic receptors (33, 35), and ryanodine receptors (14, 36, 37). We next investigated whether PDE4D species are endogenously expressed in mouse heart and whether they interact with IKs channels. As shown in Fig. 2, using an antibody specifically directed against PDE4 isoforms of the PDE4D subfamily (see “Experimental Procedures”), we indeed detected immunoreactive PDE4D species in mouse heart, and their migration on SDS-PAGE was, for the most part, at the level of PDE4D3 standard, whereas a faint band was observed at the level of PDE4D5 standard. IKs channels were immunoprecipitated with KCNQ1 antibody, and we detected the presence of PDE4Ds in the immunoprecipitate (Fig. 2, left lane in the right panel and supplemental Fig. S1). These experiments show that in mouse heart, PDE4D isoforms are associated with IKs channels.
FIGURE 2.
PDE4Ds associate with IKs channels in mouse heart. Major endogenous PDE4D isoforms expressed in mouse heart lysate were detected in a Western blot (IB) experiment using PDE4D antibody (right panel). The migration profile was compared with PDE4D standards consisting of three recombinant PDE4D isoforms expressed in COS cells: PDE4D1 (lowest band), PDE4D3 (middle band), and PDE4D5 (upper band). Immunoprecipitation (IP) of IKs channels was carried out by incubating the IKs mouse heart homogenate with KCNQ1 antibody (+Ab) or with normal IgG (-Ab). A Western blot was then carried out with PDE4D antibody (right panels) and with KCNQ1 antibody (left panel). Data shown are representative of three experiments performed.
PDE4Ds Do Not Interact with IKs Channels in the Absence of AKAP-9—To determine more specifically the molecular determinants involved in PDE4D interactions with IKs channels and to see whether AKAP-9 is required for these interactions or not, we used a heterologous system where we can express channels alone or in the presence of AKAP-9. We first carried out immunoprecipitation experiments in CHO cells expressing KCNQ1, the IKs channel α-subunits, with either the PDE4D3 or the PDE4D5 long isoforms, which are both found in heart (34). In the illustrated experiment, we failed to observe noticeable amounts of either of these two PDE4D isoforms in each KCNQ1 immunoprecipitate (Fig. 3). Similarly, in cells expressing IKs channels (hKCNE1-hKCNQ1 fusion protein (38)), we did not observe noticeable amounts of either of these PDE4D isoforms in each immunoprecipitate, indicating that these two PDE4D species do not interact directly with the α-subunit, KCNQ1 (Fig. 3), or with the β-subunit, KCNE1 (Fig. 4). Summary data for experiments illustrated in Figs. 3 and 4 are provided in supplemental Fig. S3. Our experiments suggest that PDE4D interactions with IKs channels in myocytes is indirect, being mediated by an adaptor protein that is not endogenously expressed in CHO cells.
FIGURE 3.
PDE4D3 and PDE4D5 do not associate with KCNQ1 in the absence of AKAP-9. Immunoprecipitation (IP) experiments were carried out with CHO cells transfected with KCNQ1 and either of the recombinant PDE4Ds: PDE4D3 (left panel) or PDE4D5 (right panel). Cell lysates were incubated with KCNQ1 antibody (+) or with normal IgG (-), and KCNQ1 was immunoprecipitated with anti-KCNQ1. A Western blot (IB) was then carried out with PDE4D antibody (top panels) and with KCNQ1 antibody (bottom panels). * indicates nonspecific signal. Data shown are representative of three experiments performed.
FIGURE 4.
PDE4D3 and PDE4D5 do not associate with IKs channels in the absence of AKAP-9. Immunoprecipitation (IP) experiments were carried out with CHO cells transfected with hKCNE1-hKCNQ1 fusion protein and either of the recombinant PDE4Ds: PDE4D3 (left panel) or PDE4D5 (right panel). Cell lysates were incubated with KCNQ1 antibody (+) or with normal IgG (-), and the IKs channel was immunoprecipitated with anti-KCNQ1. A Western blot (IB) was then carried out with PDE4D antibody (top panels) and with KCNQ1 antibody (bottom panels). Data shown are representative of three experiments performed.
AKAP-9 Interacts Selectively with PDE4D3—We have previously shown that the adaptor protein AKAP-9 recruits PKA and protein phosphatase 1 to IKs channels (15). We thus investigated whether AKAP-9 also recruits PDE4D3 and/or PDE4D5 to IKs channels. CHO cells were transfected with the human AKAP-9, and either of the two PDE4Ds and AKAP-9 was then immunoprecipitated with AKAP-9 antibody (22). As illustrated in Fig. 5, AKAP-9 preferentially pulled down PDE4D3. In this regard, PDE4D3 and PDE4D5 differ exclusively at their isoform-specific N-terminal regions, which are encoded by distinct exons (26) and have been shown to exhibit distinct targeting functions, allowing PDE4D5 preferential interaction with β-arrestin and RACK1 (32, 39, 41, 42) and allowing for PDE4D3 interaction with mAKAP (14), for example. Our data thus indicate that residues within the unique 15-amino acid N-terminal region of PDE4D3 are critical for binding to AKAP-9.
FIGURE 5.
AKAP-9 selectively interacts with PDE4D3. A, immunoprecipitation (IP) experiments were carried out with CHO cells transfected with AKAP-9 and either of the recombinant PDE4Ds: PDE4D3 (left panel) or PDE4D5 (right panel). Cell lysates were incubated with AKAP-9 antibody (+) or with normal IgG (-), and AKAP-9 was immunoprecipitated with anti-AKAP-9. A Western blot (IB) was then carried out with PDE4D antibody (top panels) and with AKAP-9 antibody (bottom panels). B, quantification of the efficiency with which AKAP-9 pulls down PDE4D3 and PDE4D5. Data shown are representative of (A) or represent the means ± S.E. (B) of three experiments performed. *, p < 0.0001.
PDE4D3 Is Recruited to IKs Channels by AKAP-9—We next verified that AKAP-9 recruits PDE4D3 to IKs channels (Fig. 6). CHO cells were transfected with KCNQ1 and AKAP-9 in the absence (Fig. 6A, left panel) or presence (Fig. 6A, right panel) of recombinant PDE4D3. KCNQ1 was immunoprecipitated with KCNQ1 antibody and, as expected, a significant level of PDE4D3 was detected in the KCNQ1 immunoprecipitate only when AKAP-9 was present (Fig. 6B).
FIGURE 6.
PDE4D3 is recruited to IKs channels by AKAP-9. A, immunoprecipitation (IP) experiments were carried out with CHO cells transfected with KCNQ1 and recombinant PDE4D3 in the absence (left panel) or presence (right panel) of AKAP-9. Cell lysates were incubated with KCNQ1 antibody (+) or with normal IgG (-), and KCNQ1 was immunoprecipitated with anti-KCNQ1. A Western blot (IB) was then carried out with PDE4D antibody (top panels), KCNQ1 antibody (middle panels), and AKAP-9 antibody (bottom panels). B, quantification of the efficiency with which KCNQ1 pulls down PDE4D3 in the absence (-) and presence (+) of AKAP-9. Data shown are representative of (A) or represent the means ± S.E. (B) of three experiments performed. *, p < 0.001.
PDE4D3 Regulates cAMP-dependent Activity of IKs Channels—Our biochemical experiments clearly show that PDE4D3 is an element within the IKs macromolecular complex. We then carried out functional experiments to determine whether PDE4D3 alters IKs channel activity. We used CHO cells expressing IKs channels (IKs fusion proteins) and AKAP-9, with or without expression of either PDE4D3 or PDE4D5. IKs channel activity, assessed with a voltage clamp protocol similar to the one used for ventricular myocytes experiments, was first recorded in cells dialyzed with control pipette solution. In these conditions, IKs tail current amplitude (Itail) measured 1 min after obtaining whole cell configuration was not significantly altered by the presence of either of the two PDE4Ds: 54.16 ± 12.64 pA/pF (n = 6) in the absence of recombinant PDE4Ds, 45.13 ± 12.15 pA/pF (n = 7) in cells transfected with PDE4D3 and 52.12 ± 6.53 pA/pF (n = 7) in cells transfected with PDE4D5. To compare the time course of the tail current between the different transfection conditions, we next normalized Itail recorded during 11 min to the initial Itail measured 1 min after obtaining the whole cell configuration. These results are shown in Fig. 7, D–F. After 11 min of recording, IKs showed little change over time whether cells were expressing the recombinant PDE4Ds or not; Itail decreased by 8.9 ± 10.5% (n = 6) in the absence of recombinant PDE4Ds, by 17.6 ± 4.7% (n = 6) in cells transfected with PDE4D3, and by 13.3 ± 4.5% (n = 6) in cells transfected with PDE4D5. These results suggest that the endogenous cAMP level in CHO cells may not be as high as in cardiac myocytes. We next repeated these experiments, but this time, we increased the cytosolic cAMP concentration by dialyzing cells with 200 μm cAMP and we used 0.2 μm OA to inhibit phosphatases and maximize IKs response to cAMP stimulation. In the absence of recombinant PDE4Ds, IKs was markedly increased by cAMP/OA (Fig. 7D). However, in cells transfected with PDE4D3, which we find binds to AKAP-9, cAMP/OA dialysis had little effect on IKs, suggesting that PDE4D3 prevented IKs channels from being fully stimulated by cAMP (Fig. 7E), after 11 min of recording, IKs was only increased by 3.6 ± 4.4% (n = 6, p < 0.01 versus control). In cells transfected with PDE4D5, which we find does not associate with AKAP-9, cAMP-induced increase in IKs was initially slow but eventually reached the same level as in cells without recombinant PDE4Ds after 11 min of recording (Fig. 7F): +65.77 ± 17.28%, n = 5, with PDE4D5 versus +63.4 ± 17.1%, n = 5, without PDE4Ds. Similar results were obtained when CHO cells were transfected with cDNA individually encoding KCNQ1 and KCNE1 subunits rather than IKs fusion proteins (data not shown). Altogether, these experiments show that PDE4D3 but not PDE4D5 regulates the cAMP-dependent activity of IKs channels.
FIGURE 7.
PDE4D3, but not PDE4D5, regulates cAMP-dependent activity of IKs channels. CHO cells stably expressing AKAP-9 were transfected with IKs channels (hKCNE1-hKCNQ1 fusion protein) alone (A and D, circles) or additionally with either PDE4D3 (B and E, triangles) or PDE4D5 (C and F, squares). Channel activity was elicited by the pulse protocol shown schematically in the figure applied once every 15 s. A–C, averaged current traces (n = 5–6) obtained after 11 min of recording for each transfection condition. Dashed lines indicate currents recorded with a control (ctrl) pipette solution, whereas solid lines represent currents recorded in cells dialyzed with 200 μm cAMP plus 0.2 μm OA. Dotted horizontal lines indicate zero current. D–F, time course of normalized tail current amplitude (Itail) for each transfection condition indicated in the corresponding panel above (D, PDE4Ds; E, + PDE4D3; F, + PDE4D5; n = 5–8) in cells dialyzed either with control pipette solution (open symbols) or with cAMP/OA (filled symbols). Dashed lines provide reference for invariant current over the 11-min recording period.
DISCUSSION
PDE4D3, but Not PDE4D5, Associates with AKAP-9 within IKs Macromolecular Complex—Our results indicate that PDE4Ds associate with the IKs macromolecular complex in murine cardiac myocytes. Using a biochemical approach, we were able to identify the predominant PDE4D variant (PDE4D3) that is part of the complex and, in addition, we have shown that PDE4D3 associates with AKAP-9 and is, in turn, recruited to KCNQ1, the α-subunit of the channel. AKAP-9 is a splice variant of AKAP-450 (39). PDE4D3 has previously been shown to interact with AKAP-450, but the binding site on the AKAP does not comprise the sequence of AKAP-9 (12, 13). These previous experiments were carried out using AKAP-450 fragments that, by disrupting the conformation of the full protein, could destabilize PDE4D3 interactions with other sites on the AKAP. The existence of several binding sites between a PDE4D isoform and its target protein has already been reported, namely for the binding of PDE4D3 to AKAP-18 (10) and for the binding of PDE4D5 to β-arrestin and also to RACK1 (40–42). Our findings suggest that PDE4D3 also has multiple binding sites on AKAP-450, at least one located within the sequence of AKAP-9 (this study) and at least one located downstream of this sequence (13).
The specificity of interaction of PDE4D3 with AKAP-9 is likely to be due to the N terminus of this phosphodiesterase because it is the only part that distinguishes it from PDE4D5 isoform, which does not bind to AKAP-9. A similar isoform-specific association between PDE4D3 and mAKAP, an adaptor protein within the ryanodine receptor macromolecular complex, has been shown to depend on the unique N terminus sequence of PDE4D3 (14). Conversely, the unique 87-amino acid N-terminal region of PDE4D5 has been shown to target preferentially this particular isoform to the signaling scaffold proteins RACK1 (42, 43) and β-arrestin (27, 42). Interestingly, our functional experiments showed that PDE4D3 and PDE4D5 did not exert the same effects on cAMP-dependent activity of IKs channels. PDE4D5 only slowed down the cAMP-induced increase in IKs and, despite the PKA activation of the PDE activity (44), did not prevent the maximum level of IKs stimulation from being reached. However, PDE4D3 markedly reduced the IKs response to cAMP. The initial increase in IKs was weak and was followed by little change in current, which indicates the almost complete suppression of cAMP stimulation, most likely due to the PKA-induced increase in PDE4D3 activity (45). The differential regulation of IKs by the two PDE4D isoforms is unlikely to be due to a difference in enzymatic properties because both PDE4Ds share similar affinity for cAMP and because the maximal catalytic activity of PDE4D5 is more important than that of PDE4D3 (26). It is also unlikely to be due to a difference in protein expression level as both recombinant proteins were similarly overexpressed in CHO cells (see, for example, Fig. 3). Instead, the differential regulation of IKs by the two PDE4D isoforms is more likely to be due to a spatially localized component due to the selective recruitment of PDE4D3 to the IKs macromolecular complex by AKAP-9.
Role of PDE4s on cAMP-dependent Activity of IKs Channels in Cardiac Myocytes—Our functional experiments using IBMX, the non-selective PDE inhibitor, in ventricular myocytes, show that phosphodiesterases regulate the basal activity of IKs channels. This result is consistent with a previous study carried out with guinea pig sinoatrial myocytes (46). In the latter study, a part of the IBMX stimulatory effect was attributed to inhibition of PDE3s, the cGMP-inhibited PDEs. In the present study, using rolipram to inhibit the cAMP-specific PDE4 enzymes selectively, we show that PDE4s also contribute to the regulation of IKs activity in myocytes in the absence of β-adrenergic stimulation. The latter result contrasts with the regulation of voltage-dependent calcium channels (ICa), for which cAMP-dependent activity is only altered by PDE4s after β-adrenergic stimulation (47, 48). It is possible that the PDE4 regulating ICa has a lower affinity for cAMP than the one regulating IKs channels or that this PDE4 is recruited to the vicinity of calcium channels only after β-adrenergic receptor stimulation. Such a phenomenon has indeed been observed with β2-adrenergic receptors; PDE4D5 is recruited by β-arrestin to β2-adrenergic receptors only after their agonist stimulation (33, 34). Altogether, these findings further support the concept that different PDE isoforms are responsible for the fine regulation of the PKA phosphorylated state of target proteins not only during but also in the absence of β-adrenergic stimulation (3, 4, 30).
Implications for Heart Pathophysiology—The critical role played by cAMP regulation of IKs channels in human physiology and pathophysiology is illustrated by the following findings. Mutations in either KCNQ1 or KCNE1 IKs channel subunits that cause loss of channel function have been linked to inherited forms of the long QT syndrome, a disorder in which the repolarization phase of the cardiac AP is delayed, resulting in a prolongation of the QT interval on the electrocardiogram and polymorphic ventricular tachycardia, which can lead to sudden death (49). Elevated arrhythmia risk associated with exercise in long QT syndrome patients harboring mutations in either IKs subunit is likely due at least in part to disruption of β-adrenergic receptor-mediated regulation in these patients (50). On the other hand, inherited gain of function mutations in IKs channels have been associated with atrial fibrillation (51, 52). We have previously reported that it is possible to obtain a pharmacological gain of function of IKs channels with cAMP stimulation and that it can prolong the duration of atrial arrhythmia in mice (53). Our identification here of PDE4D3 as an element of the IKs macromolecular complex suggests that a similar gain of function might ensue if PDE4D3 were to become down-regulated in disease. Interestingly, such a phenomenon has been observed with the ryanodine receptor (RYR2) macromolecular complex in heart failure where the reduced interaction between PDE4D3 and RYR2 was associated with RYR2 hyperphosphorylation (37). Further work will be necessary to determine whether IKs hyperphosphorylation occurs in pathological conditions and whether this abnormal state is associated with a deficiency in PDE4D3 regulation in the IKs macromolecular complex.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01HL044365-16 from the NHLBI (to R. S. K.). This work was also supported by Medical Research Council (U. K.) Grant G0600765 (to M. D. H. and G. S. B.), European Union Grant LSHB-CT-2006-037189 (M. D. H.), and Fondation Leducq Grant 06CVD02 (M. D. H. and G. S. B.).
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures.
Footnotes
The abbreviations used are: AKAP, A kinase-anchoring protein; mAKAP, muscle AKAP; PKA, protein kinase A; PDE, phosphodiesterases; IKs, slowly activating potassium current; CHO, Chinese hamster ovary; IBMX, isobutylmethylxanthine; OA, okadaic acid; RYR2, ryanodine receptor 2; VSV, vesicular stomatitis virus; h, human; pF, picofarads.
References
- 1.Ostrom, R. S., and Insel, P. A. (2004) Br. J. Pharmacol. 143 235-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beene, D. L., and Scott, J. D. (2007) Curr. Opin. Cell Biol. 19 192-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houslay, M. D., Baillie, G. S., and Maurice, D. H. (2007) Circ. Res. 100 950-966 [DOI] [PubMed] [Google Scholar]
- 4.Baillie, G. S., Scott, J. D., and Houslay, M. D. (2005) FEBS Lett. 579 3264-3270 [DOI] [PubMed] [Google Scholar]
- 5.Lugnier, C. (2006) Pharmacol. Ther. 109 366-398 [DOI] [PubMed] [Google Scholar]
- 6.Conti, M., and Beavo, J. (2007) Annu. Rev. Biochem. 76 481-511 [DOI] [PubMed] [Google Scholar]
- 7.Mongillo, M., McSorley, T., Evellin, S., Sood, A., Lissandron, V., Terrin, A., Huston, E., Hannawacker, A., Lohse, M. J., Pozzan, T., Houslay, M. D., and Zaccolo, M. (2004) Circ. Res. 95 67-75 [DOI] [PubMed] [Google Scholar]
- 8.Rochais, F., Abi-Gerges, A., Horner, K., Lefebvre, F., Cooper, D. M., Conti, M., Fischmeister, R., and Vandecasteele, G. (2006) Circ. Res. 98 1081-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conti, M., Richter, W., Mehats, C., Livera, G., Park, J. Y., and Jin, C. (2003) J. Biol. Chem. 278 5493-5496 [DOI] [PubMed] [Google Scholar]
- 10.Stefan, E., Wiesner, B., Baillie, G. S., Mollajew, R., Henn, V., Lorenz, D., Furkert, J., Santamaria, K., Nedvetsky, P., Hundsrucker, C., Beyermann, M., Krause, E., Pohl, P., Gall, I., MacIntyre, A. N., Bachmann, S., Houslay, M. D., Rosenthal, W., and Klussmann, E. (2007) J. Am. Soc. Nephrol. 18 199-212 [DOI] [PubMed] [Google Scholar]
- 11.Willoughby, D., Wong, W., Schaack, J., Scott, J. D., and Cooper, D. M. (2006) EMBO J. 25 2051-2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCahill, A., McSorley, T., Huston, E., Hill, E. V., Lynch, M. J., Gall, I., Keryer, G., Lygren, B., Tasken, K., van Heeke, G., and Houslay, M. D. (2005) Cell. Signal. 17 1158-1173 [DOI] [PubMed] [Google Scholar]
- 13.Tasken, K. A., Collas, P., Kemmner, W. A., Witczak, O., Conti, M., and Tasken, K. (2001) J. Biol. Chem. 276 21999-22002 [DOI] [PubMed] [Google Scholar]
- 14.Dodge, K. L., Khouangsathiene, S., Kapiloff, M. S., Mouton, R., Hill, E. V., Houslay, M. D., Langeberg, L. K., and Scott, J. D. (2001) EMBO J. 20 1921-1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marx, S. O., Kurokawa, J., Reiken, S., Motoike, H., D'Armiento, J., Marks, A. R., and Kass, R. S. (2002) Science 295 496-499 [DOI] [PubMed] [Google Scholar]
- 16.Kass, R. S., and Wiegers, S. E. (1982) J. Physiol. (Lond.) 322 541-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh, K. B., and Kass, R. S. (1988) Science 242 67-69 [DOI] [PubMed] [Google Scholar]
- 18.Sanguinetti, M. C., Curran, M. E., Zou, A., Shen, J., Spector, P. S., Atkinson, D. L., and Keating, M. T. (1996) Nature 384 80-83 [DOI] [PubMed] [Google Scholar]
- 19.Barhanin, J., Lesage, F., Guillemare, E., Fink, M., Lazdunski, M., and Romey, G. (1996) Nature 384 78-80 [DOI] [PubMed] [Google Scholar]
- 20.Kurokawa, J., Chen, L., and Kass, R. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2122-2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurokawa, J., Motoike, H. K., Rao, J., and Kass, R. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 16374-16378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, L., Kurokawa, J., and Kass, R. S. (2005) J. Biol. Chem. 280 31347-31352 [DOI] [PubMed] [Google Scholar]
- 23.Chen, L., Marquardt, M. L., Tester, D. J., Sampson, K. J., Ackerman, M. J., and Kass, R. S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20990-20995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra, R., and Morad, M. (1985) Am. J. Physiol. 249 H1056-H1060 [DOI] [PubMed] [Google Scholar]
- 25.Dilly, K. W., Kurokawa, J., Terrenoire, C., Reiken, S., Lederer, W. J., Marks, A. R., and Kass, R. S. (2004) J. Biol. Chem. 279 40778-40787 [DOI] [PubMed] [Google Scholar]
- 26.Bolger, G. B., Erdogan, S., Jones, R. E., Loughney, K., Scotland, G., Hoffmann, R., Wilkinson, I., Farrell, C., and Houslay, M. D. (1997) Biochem. J. 328 539-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch, M. J., Baillie, G. S., Mohamed, A., Li, X., Maisonneuve, C., Klussmann, E., van Heeke, G., and Houslay, M. D. (2005) J. Biol. Chem. 280 33178-33189 [DOI] [PubMed] [Google Scholar]
- 28.Murdoch, H., Mackie, S., Collins, D. M., Hill, E. V., Bolger, G. B., Klussmann, E., Porteous, D. J., Millar, J. K., and Houslay, M. D. (2007) J. Neurosci. 27 9513-9524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terrenoire, C., Clancy, C. E., Cormier, J. W., Sampson, K. J., and Kass, R. S. (2005) Circ. Res. 96 e25-34 [DOI] [PubMed] [Google Scholar]
- 30.Bethke, T., Meyer, W., Schmitz, W., Scholz, H., Stein, B., Thomas, K., and Wenzlaff, H. (1992) Br. J. Pharmacol. 107 127-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeves, M. L., Leigh, B. K., and England, P. J. (1987) Biochem. J. 241 535-541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houslay, M. D., and Adams, D. R. (2003) Biochem. J. 370 1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry, S. J., Baillie, G. S., Kohout, T. A., McPhee, I., Magiera, M. M., Ang, K. L., Miller, W. E., McLean, A. J., Conti, M., Houslay, M. D., and Lefkowitz, R. J. (2002) Science 298 834-836 [DOI] [PubMed] [Google Scholar]
- 34.Baillie, G. S., Sood, A., McPhee, I., Gall, I., Perry, S. J., Lefkowitz, R. J., and Houslay, M. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 940-945 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Richter, W., Day, P., Agrawal, R., Bruss, M. D., Granier, S., Wang, Y. L., Rasmussen, S. G., Horner, K., Wang, P., Lei, T., Patterson, A. J., Kobilka, B., and Conti, M. (2008) EMBO J. 27 384-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marx, S. O., Reiken, S., Hisamatsu, Y., Jayaraman, T., Burkhoff, D., Rosemblit, N., and Marks, A. R. (2000) Cell 101 365-376 [DOI] [PubMed] [Google Scholar]
- 37.Lehnart, S. E., Wehrens, X. H., Reiken, S., Warrier, S., Belevych, A. E., Harvey, R. D., Richter, W., Jin, S. L., Conti, M., and Marks, A. R. (2005) Cell 123 25-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, W., Xia, J., and Kass, R. S. (1998) J. Biol. Chem. 273 34069-34074 [DOI] [PubMed] [Google Scholar]
- 39.Witczak, O., Skalhegg, B. S., Keryer, G., Bornens, M., Tasken, K., Jahnsen, T., and Orstavik, S. (1999) EMBO J. 18 1858-1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger, G. B., McCahill, A., Huston, E., Cheung, Y. F., McSorley, T., Baillie, G. S., and Houslay, M. D. (2003) J. Biol. Chem. 278 49230-49238 [DOI] [PubMed] [Google Scholar]
- 41.Bolger, G. B., Baillie, G. S., Li, X., Lynch, M. J., Herzyk, P., Mohamed, A., Mitchell, L. H., McCahill, A., Hundsrucker, C., Klussmann, E., Adams, D. R., and Houslay, M. D. (2006) Biochem. J. 398 23-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baillie, G. S., Adams, D. R., Bhari, N., Houslay, T. M., Vadrevu, S., Meng, D., Li, X., Dunlop, A., Milligan, G., Bolger, G. B., Klussmann, E., and Houslay, M. D. (2007) Biochem. J. 404 71-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, K. J., Baillie, G. S., Hyde, E. I., Li, X., Houslay, T. M., McCahill, A., Dunlop, A. J., Bolger, G. B., Klussmann, E., Adams, D. R., and Houslay, M. D. (2007) Cell. Signal. 19 2612-2624 [DOI] [PubMed] [Google Scholar]
- 44.MacKenzie, S. J., Baillie, G. S., McPhee, I., MacKenzie, C., Seamons, R., McSorley, T., Millen, J., Beard, M. B., van Heeke, G., and Houslay, M. D. (2002) Br. J. Pharmacol. 136 421-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sette, C., and Conti, M. (1996) J. Biol. Chem. 271 16526-16534 [DOI] [PubMed] [Google Scholar]
- 46.Shimizu, K., Shintani, Y., Ding, W. G., Matsuura, H., and Bamba, T. (2002) Br. J. Pharmacol. 137 127-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kajimoto, K., Hagiwara, N., Kasanuki, H., and Hosoda, S. (1997) Br. J. Pharmacol. 121 1549-1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verde, I., Vandecasteele, G., Lezoualc'h, F., and Fischmeister, R. (1999) Br. J. Pharmacol. 127 65-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moss, A. J., and Kass, R. S. (2005) J. Clin. Investig. 115 2018-2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kass, R. S., Kurokawa, J., Marx, S. O., and Marks, A. R. (2003) Trends Cardiovasc. Med. 13 52-56 [DOI] [PubMed] [Google Scholar]
- 51.Chen, Y. H., Xu, S. J., Bendahhou, S., Wang, X. L., Wang, Y., Xu, W. Y., Jin, H. W., Sun, H., Su, X. Y., Zhuang, Q. N., Yang, Y. Q., Li, Y. B., Liu, Y., Xu, H. J., Li, X. F., Ma, N., Mou, C. P., Chen, Z., Barhanin, J., and Huang, W. (2003) Science 299 251-254 [DOI] [PubMed] [Google Scholar]
- 52.Hong, K., Piper, D. R., Diaz-Valdecantos, A., Brugada, J., Oliva, A., Burashnikov, E., Santos-de-Soto, J., Grueso-Montero, J., Diaz-Enfante, E., Brugada, P., Sachse, F., Sanguinetti, M. C., and Brugada, R. (2005) Cardiovasc. Res. 68 433-440 [DOI] [PubMed] [Google Scholar]
- 53.Sampson, K. J., Terrenoire, C., Cervantes, D. O., Kaba, R. A., Peters, N. S., and Kass, R. S. (2008) J. Physiol. (Lond.) 586 627-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.