Abstract
Staphylococcus aureus is known to activate mammalian immune cells through Toll-like receptor 2 (TLR2). We recently demonstrated that a lipoprotein fraction obtained from S. aureus by Triton X-114 phase partitioning is a potent activator of TLR2. In this study, we separated TLR2-activating lipoproteins expressed in S. aureus and characterized an N-terminal structure. The lipoprotein fraction of S. aureus was prepared by glass bead disruption followed by Triton X-114 phase partitioning. The TLR2-activating molecules were mainly detected in the mass range of 30–35 kDa. Seven lipoproteins were identified by the mass spectra of their tryptic digests. Among them, three lipoproteins were separated by preparative SDS-PAGE and proved to activate TLR2. After digestion with trypsin in the presence of sodium deoxycholate, the N terminus of the lipopeptide was isolated from lipoprotein SAOUHSC_02699 by normal phase high pressure liquid chromatography and characterized as an S-(diacyloxypropyl)cystein-containing peptide using tandem mass spectra. The synthetic lipopeptide counterpart also stimulated the cells via TLR2. These results showed that the diacylated lipoprotein from S. aureus acts as a TLR2 ligand in mammalian cells.
Bacterial infection is one of the major causes of death. Staphylococcus aureus, the most common Gram-positive pathogen, is a major source of mortality in medical facilities (1). The pathogen causes various infectious diseases, including sepsis, endocarditis, and pneumonia. During the infection, S. aureus activates cells and evokes serious inflammation in the host. TLR2 2 has been shown to play a crucial role in the host response to S. aureus (2). However, a detailed understanding of the molecular components that interact with TLR2 in S. aureus cells has not yet been obtained. One of the reported TLR2 ligands was peptidoglycan (PGN) (3), a cell wall component of most bacteria. However, Travassos et al. (4) recently showed that PGN from several bacteria that were highly purified by removal of lipoproteins or lipoteichoic acid (LTA) were not detected by TLR2. Moreover, the minimal active components of the PGN, muramyl dipeptide and desmuramyl dipeptide (γ-d-glutamyl diaminopimelic acid), were determined to be ligands of the intracellular innate immune receptor Nod2/Nod1 (5, 6), suggesting that PGN is not a ligand of TLR2. Another candidate of TLR2-activating ligands is LTA, a cell surface glycoconjugate of Gram-positive bacteria (3). Morath et al. (7) reported that LTA from S. aureus is a potent stimulator of cytokine release, whereas our group demonstrated that LTA from enterococci, also a major Gram-positive pathogen, has no cytokine-producing activity (8, 9). Furthermore, Han et al. (10) showed that LTA from pneumococci is 100-fold less potent than staphylococcal LTA. These observations suggested that LTA is not a common ligand of TLR2 in Gram-positive pathogens.
We also found that the enterococcal LTA fraction contains some contaminants other than LTA and that these components activate immune cells through TLR2 (8, 11). However, their structures were not identified at that time. TLR2 is known to be a predominant receptor for lipoproteins derived from various bacteria. We have previously shown that the lipoprotein-containing fraction from S. aureus stimulates activation of immune cells through TLR2 (12). Recently, Stoll et al. (13) constructed a lipoprotein diacylglycerol transferase deletion (Δlgt) mutant of S. aureus, which is unable to carry out lipid modification of prelipoproteins. It was demonstrated that the mutant completely lacks palmitate-labeled lipoproteins and that its cells and crude lysate induce much less proinflammatory cytokines than the wild type. Bubeck et al. (14) also reported that an S. aureus variant that lacks lipoproteins is able to escape immune recognition and cause lethal infections. Furthermore, we showed that the activities of an LTA fraction derived from a Δlgt mutant are largely decreased when compared with those from the same fraction of the wild type (15). These results suggest that the lipoproteins in S. aureus appear to be the TLR2 ligand for the immune system. In the present study, we separated lipoproteins from S. aureus and elucidated the TLR2-activating structure.
EXPERIMENTAL PROCEDURES
Bacterial Strain and Bacterial Components—S. aureus SA113, a restriction-deficient mutant of NCTC8325 (16), was cultured in brain heart infusion broth (Eiken, Tokyo, Japan) at 37 °C for 6 h with constant shaking in a culture bag (CB20-1, Fujimori Kogyo Co., Ltd., Tokyo, Japan) before being harvested by centrifugation (5,000 × g for 15 min at 4 °C). The membrane fraction was prepared by bead disruption of the cells as described (13). The lipoprotein fraction was obtained by Triton X-114 phase partitioning of the membrane fraction as described (17) and was designated as Sa-M-TX.
FSL-1, a synthetic diacylated lipopeptide, was purchased from EMC microcollections (Tübingen, Germany). Escherichia coli O55:B5 lipopolysaccharide (Sigma-Aldrich) was further purified by sodium deoxycholate reextraction as described (18). SAOUHSC_02699 di-O-palmitoyl (Pam2) lipopeptide 10-mer (Pam2CGNNSSKDKDG) and Pam2CSK4 were synthesized in our laboratory. The synthetic details will be described elsewhere.
Separations of Proteins—Analytical SDS-PAGE was performed by the Tris-glycine method (19) using a mini PAGE chamber AE-6530 and an AE-8450 power supply (Atto Corp., Tokyo, Japan) with a 12.5% gel. Proteinous materials were visualized by silver or Coomassie Brilliant Blue staining, and visualization of acidic materials, such as LTA, was performed by Alcian blue staining.
Proteins eluting between 25 and 40 kDa in the analytical SDS-PAGE were separated using a preparative electrophoresis apparatus, AE-6750 (Atto Corp.), according to the manufacturer's instructions. The eluates were analyzed by SDS-PAGE and subjected to acetone precipitation to remove any contaminating SDS. The precipitates were dissolved in 20 mm octylglucoside. The concentration of lipoprotein was estimated by SDS-PAGE with silver staining and adjusted to 10 μg/ml.
Monocyte Western Blotting—Monocyte Western blotting was carried out by the method described previously (20). Briefly, stimuli were separated by SDS-PAGE, and the resolved stimuli in the gel were transblotted to a nitrocellulose membrane by the method of Towbin et al. (21) using an AE-6677 semidry blotting apparatus (Atto Corp.). The membrane of each lane was cut into 4-mm strips, and each strip was separately dissolved in dimethyl sulfoxide. The solution was poured into phosphate-buffered saline to precipitate stimuli-coated particles, which consist of stimuli and nitrocellulose. After washing three times with phosphate-buffered saline, the particle suspension in assay medium was applied to a luciferase assay using the Ba/mTLR2 cells described below.
Mass Spectrometry—Mass spectra were obtained by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) with an Axima QIT TOF mass spectrometer (Shimadzu, Kyoto, Japan) equipped with a nitrogen 337 nm laser. 2,5-Dihydroxybenzoic acid was used as the matrix at a concentration of 10 mg/ml in aqueous 50% acetonitrile containing 0.1% trifluoroacetic acid. Peptides were analyzed in positive and reflectron mode. Tandem mass (MS/MS) spectra were obtained using argon as a collision gas.
Identification of Lipoproteins and Lipopeptides—The proteins that were separated by analytical SDS-PAGE were digested with trypsin as described previously (22). The peptides were analyzed by MALDI-TOF-MS and identified by a data base (the National Center for Biotechnology Information nonredundant protein data base) search using the MASCOT software from Matrix Science (23). The proteins were designated by the locus tags for the genome of S. aureus NCTC8325, which is the parent strain of SA113 (16).
To isolate the N-terminal lipopeptide, the lipoprotein fraction was subjected to phase transfer surfactant-aided trypsin digestion (24). The tryptic digest was subjected to HPLC using a normal phase Daisopak SP-120-5-SIL-P column (250 × 4.6 mm, DAISO, Co., Ltd., Osaka, Japan). The digests were eluted using a gradient program (solvent A, chloroform, methanol, water = 65/20/3, v/v/v; solvent B, chloroform, methanol, water = 65/40/3, v/v/v) at a flow rate of 1.0 ml/min and were fractionated into 1-ml portions. The N-terminal lipopeptide was detected using a luciferase assay. The purified lipopeptide was characterized by MALDI-TOF-MS.
Luciferase Assay—Ba/F3 cells that stably expressed p55IgκLuc, an NF-κB/DNA binding activity-dependent luciferase reporter construct (Ba/κB), murine TLR2 and the p55IgκLuc reporter construct (Ba/mTLR2), and murine TLR4/MD-2 and the p55IgκLuc reporter construct (Ba/mTLR4/mMD-2) were kindly provided by Prof. K. Miyake (Institute of Medical Science, University of Tokyo, Japan). The NF-κB-dependent luciferase activity in these cells was determined as described previously (12).
Cytokine Assay—Eight-week-old male BALB/c mice were obtained from Kyudo (Kumamoto, Japan). The animals received humane care in accordance with our institutional guidelines and the legal requirements of Japan. Stimulation of thioglycolate-elicited peripheral exudate cells and Histopaque-separated human peritoneal blood mononuclear cells (PBMC) from a healthy donor (M. F.) and a cytokine assay for secreted murine or human TNF-α were performed as described (15).
RESULTS
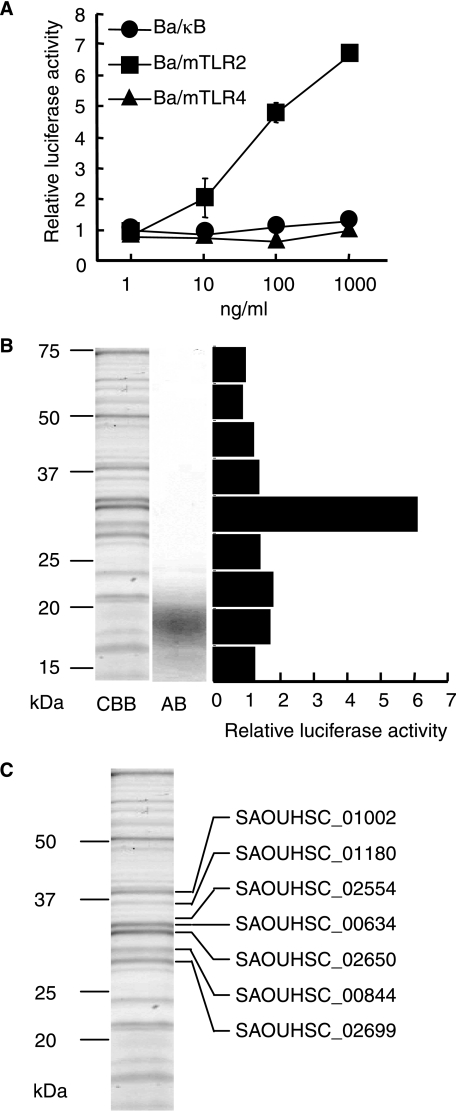
Identification of Lipoproteins in S. aureus—The lipoprotein fraction was obtained from S. aureus cells using glass bead disruption followed by Triton X-114 phase partitioning. Four mg of the lipoprotein fraction (Sa-M-TX) were obtained from 10 liters of bacterial cell culture. Sa-M-TX activated murine TLR2 expressing cells (Ba/mTLR2) at 10 ng/ml but not murine TLR4 and MD-2 expressing Ba/mTLR4/mMD-2 cells or the negative control Ba/κB cells (Fig. 1A). The SDS-PAGE profile of Sa-M-TX showed that it contained Coomassie Brilliant Blue (CBB)-positive proteins and Alcian blue (AB)-positive components (Fig. 1B). The active molecules in Sa-M-TX were analyzed by monocyte Western blotting. TLR2-mediated NF-κB activation was strongly detected in a molecular mass range of 30–35 kDa (Fig. 1B). Thus, we subjected the Sa-M-TX proteins with molecular masses of around 30–35 kDa to in-gel tryptic digestion. At least seven lipoproteins, SAOUHSC_00634, -00844, -01002, -01180, -02554, -02650, and -02699, were identified by a combination of peptide mass fingerprinting and MS/MS spectra (Fig. 1C).
FIGURE 1.
Identification of lipoproteins in S. aureus. A, the NF-κB activation induced by Sa-M-TX in Ba/κB, Ba/mTLR2, or Ba/mTLR4/mMD-2 cells for 4 h was measured with a luciferase assay. The results are shown as relative luciferase activity, which was determined as the ratio of stimulated to nonstimulated activity. The data represent the mean ± S.E. obtained from three independent experiments. B, SDS-PAGE profiles of Sa-M-TX and separation of TLR2-activating components in Sa-M-TX. The gels were separated with 12.5% gel and visualized by Coomassie Brilliant Blue (CBB) or Alcian blue (AB). NF-κB activation was detected by monocyte Western blotting using a luciferase assay in Ba/mTLR2 cells. The results are shown as relative luciferase activity, which was determined as the ratio of stimulated to nonstimulated activity. C, identification of lipoproteins in Sa-M-TX. Each protein were separated with 12.5% gel and identified by in-gel tryptic digestion followed by mass analysis.
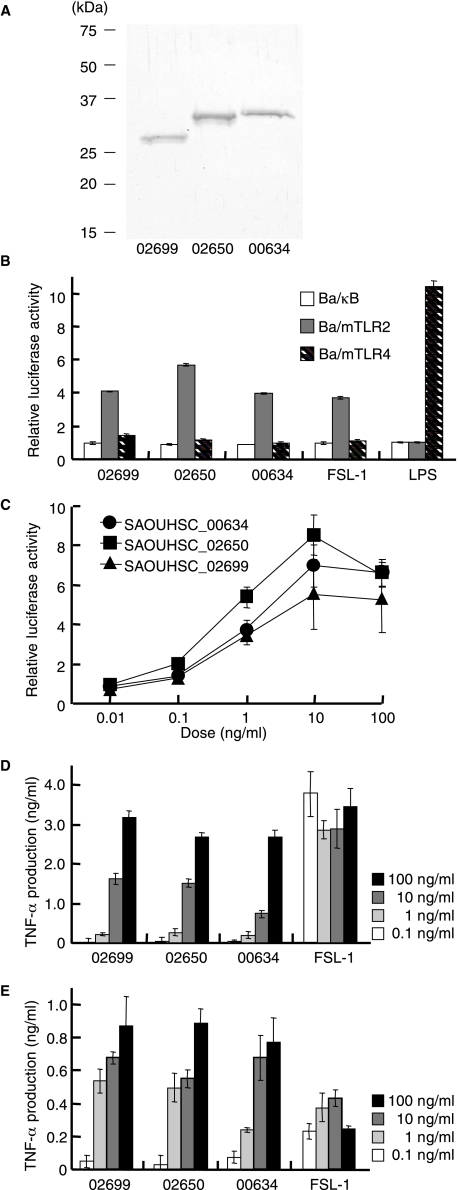
Lipoprotein Immunological Activities—The lipoproteins were separated by preparative SDS-PAGE using a 12.5% gel. Among seven identified lipoproteins, we could separate three lipoproteins, SAOUHSC_02699, -02650, and -00634 (Fig. 2A). About 1 μg of each lipoprotein were obtained from 250 μg of Sa-M-TX. The TLR2-dependent activities of the lipoproteins were detected by NF-κB activation in TLR-expressing cells. All lipoproteins induced NF-κB activation in Ba/mTLR2 cells but not in Ba/mTLR4/mMD-2 or Ba/κB cells (Fig. 2B). The activities of the lipoproteins were observed at 0.1 ng/ml (Fig. 2C) and were about 100-fold higher than that of Sa-M-TX, which was shown in Fig. 1A. They also stimulated human PBMC and murine peritoneal exudate cells to induce TNF-α dose dependently (Fig. 2, D and E). The other four lipoproteins could not be analyzed because of the low content in Sa-M-TX.
FIGURE 2.
Immunological activities of lipoproteins, SAOUHSC_02699, -02650, and -00634 separated by preparative SDS-PAGE using 12.5% gel. A, SDS-PAGE profile of lipoproteins. They were separated in 12.5% gel and visualized by silver staining. B, the NF-κB activation induced by 1 ng/ml of the lipoprotein, 0.1 ng/ml FSL-1, and 10 ng/ml lipopolysaccharide (LPS) in Ba/κB, Ba/mTLR2, or Ba/mTLR4/mMD-2 cells. C, dose-dependent NF-κB activation induced by lipoproteins in Ba/mTLR2 cells. The cells were incubated with stimuli for 4 h. NF-κB activation was measured with a luciferase assay. The results are shown as relative luciferase activity, which was determined as the ratio of stimulated to nonstimulated activity. The data represent the mean ± S.E. obtained from three independent experiments. D, TNF-α production induced by the lipoprotein in human PBMC. E, TNF-α production induced by the lipoprotein in murine peritoneal exudate cells. The levels of TNF-α in the culture supernatants of the cells incubated with the indicated concentration of the stimuli for 4 h were measured by enzyme-linked immunosorbent assay. The data represent the mean ± S.E. obtained from three independent experiments.
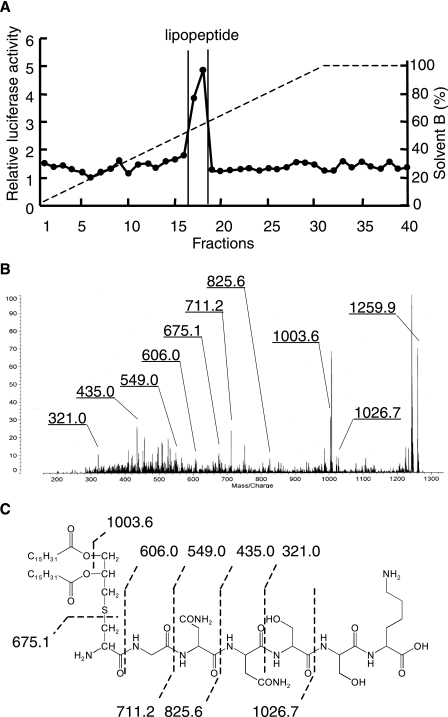
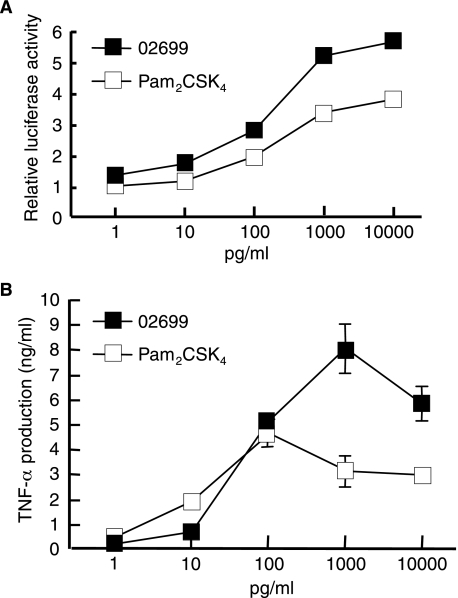
The N-terminal Structure of S. aureus Lipoprotein—Because the minimal active structure of bacterial lipoproteins is reported to be N-terminal acylated S-(2,3-dihydroxypropyl)cysteine-containing lipopeptide (25), the N-terminal structure of S. aureus lipoprotein was investigated. We attempted to separate N-terminal lipopeptides from three separated lipoproteins by conventional tryptic digestion followed by reverse or normal phase HPLC separation. Although active components were eluted, no lipopeptides were detected by mass spectrometry. This may have been caused by insufficient digestion of the N-terminal moiety of the lipopeptides, probably because of poor solubility or micelle formation. Therefore, we used phase transfer surfactant-aided trypsin digestion (24) to improve the efficiency of hydrophobic lipoprotein digestion. The lipoprotein SAOUHSC_02699 was thus able to be digested, and the digests were subjected to normal phase HPLC (Fig. 3A). A TLR2-activating component was found between fractions 17 and 18. The MALDI-TOF-MS spectra of the component showed a pseudomolecular ion [M+H]+ at m/z 1259.9. In the MS/MS spectra of the precursor ion at m/z 1259.9 (Fig. 3, B and C), C-terminal y ions were observed at m/z 606.0, 549.0, 435.0, and 321.0, which agreed with the N-terminal sequence of SAOUHSC_02699, GNNSSK. The ions at m/z 1026.7, 825.6, and 711.2 correspond to b ions of an S-(dipalmitoyloxypropyl)-cysteine-containing peptide. The ion at m/z 675.1 represents dehydroalanyl GNNSSK, which is formed by β-elimination of the 2,3-dipalmitoyloxypropylthio group. These results prove that the N-terminal structure of SAOUHSC_02699 is a diacyltype lipoprotein. The synthetic counterpart of its N-terminal lipopeptide 10-mer stimulated TLR2-dependent NF-κB activation in Ba/mTLR2 cells and TNF-α induction in PBMC (Fig. 4). The lipoprotein SAOUHSC_00634 and -02650 were also subjected to the phase transfer surfactant-aided trypsin digestion. Although active components were eluted from HPLC, no spectra corresponding to lipopeptide were obtained, suggesting that further improvement is required for complete digestion of some lipoproteins.
FIGURE 3.
Characterization of the N-terminal lipopeptide of SAOUHSC_02699. A, the elution profile of a tryptic digest of SAOUHSC_02699 using normal phase HPLC. The NF-κB activation of the fraction was determined using Ba/mTLR2 cells. B, the MS/MS spectrum of the lipopeptide. The precursor ion at m/z 1259.9 was decomposed using collision-induced dissociation (CID) mode. C, the structure of the lipopeptide. Interpretations of the fragment ions in the MS/MS spectra are indicated in the structure.
FIGURE 4.
Immunological activities of a synthetic lipopeptide of SAOUHSC_ 02699. A, NF-κB activation induced by the indicated doses of lipopeptide in Ba/mTLR2 cells for 4 h was measured with a luciferase assay. The results are shown as relative luciferase activity, which was determined as the ratio of stimulated to nonstimulated activity. The data represent the mean ± S.E. obtained from three independent experiments. B, TNF-α production induced by the lipoprotein in human PBMC. The levels of TNF-α in the culture supernatants of the cells incubated with the indicated concentration of the stimuli for 4 h were measured by enzyme-linked immunosorbent assay. The data represent the mean ± S.E. obtained from three independent experiments.
DISCUSSION
S. aureus is known to activate TLR2, but the principal molecule responsible for this activity has not been proven. In this study, we identified several lipoproteins including SAOUHSC_ 01002, -01180, -02554, -00634, -02650, -00844, and -02699 as candidates of TLR2-activating molecules (Fig. 1C). Some of the lipoproteins were identified as quinol oxidase (SAOUHSC_01002), and the ATP-binding cassette transporter (SAOUHSC_02699, SAOUHSC_00634) and the others were classified as hypothetical proteins. SAOUHSC_00634 has been previously reported as SitC, which acts as an iron-regulated ATP-binding cassette transporter in S. aureus and Staphylococcus epidermidis and is a major lipoprotein that is distributed throughout the cell wall (26). SAOUHSC_00634 and -01180 were also identified in the membrane fraction of S. aureus (13). Although some of lipoproteins reported (13) were not identified in our study, probably because of differences in the culture conditions, similar lipoproteins were extracted. Thus, we consider lipoproteins to be constitutively expressed in S. aureus.
SAOUHSC_02699, -02650, and -00634, which were separated by preparative SDS-PAGE (Fig. 2A), activate TLR2-expressing cells (Ba/mTLR2) but not murine TLR4- and MD-2-expressing Ba/mTLR4/mMD-2 cells or the negative control Ba/κB cells (Fig. 2B). They also exert strong TNF-α-inducing activity on murine and human immune cells (Fig. 2, D and E). These data indicate that the lipoproteins are the TLR2-activating ligands in S. aureus. Sa-M-TX contained LTA, which was visualized in the range of 15–23 kDa (Fig. 1B), whereas the activity of Sa-M-TX was observed in the range of 30–35 kDa (Fig. 1B). We have previously reported that the LTA fraction derived from an S. aureus lipoprotein knock-out mutant is 100-fold less potent than that of the wild-type (15). Although the LTA is still thought to be a TLR2-activating molecule (27), the principal molecules responsible for the TLR2 activity of Sa-M-TX are concluded to be lipoproteins.
Because the three lipoproteins separated were predominant constituents in Sa-M-TX and the sum of their activities is comparable with that of Sa-M-TX (Figs. 1A and 2B), they are responsible for most of the activity in Sa-M-TX. As for the TLR2-mediated recognition of whole S. aureus, it should include other lipoproteins, which depend on the condition of bacteria. Characterization of lipoproteins expressed in a pathogenic condition is important for the analysis of virulence factors by S. aureus in infectious diseases.
In the case of Gram-negative bacteria and mycoplasma, several lipoproteins have been identified as TLR2 ligands, and their N-terminal lipopeptides, which contain diacylated or triacylated S-(2,3-dihydroxypropyl)cystein, have been proven to be essential moieties for TLR2 activity using chemically synthesized compounds. Lipoproteins derived from Gram-negative bacteria, such as E. coli, Borrelia burgdorferi, Neisseria gonorrhoeae, and Porphyromonas gingivalis, have been proven to be triacylated lipoproteins (28–31). Mycoplasma, such as Mycoplasma fermentans and Mycoplasma salivarium, have been shown to possess diacylated lipoproteins (32, 33). However, it is not clear whether the lipoproteins in Gram-positive bacteria are diacylated or triacylated. In this study, we identified the N-terminal structure of S. aureus lipoprotein SAOUHSC_ 02699 as diacylated lipoproteins (Fig. 3). We also confirmed its activity using a synthetic counterpart (Fig. 4). In several bacteria, but not all, the N terminus of the diacylglyceryl-modified cysteine residue is fatty-acylated by a lipoprotein N-acyltransferase (lnt) (34). Stoll et al. (13) screened the published genome sequences of S. aureus strains for a gene encoding an lnt homolog and found no such protein. These data correspond to our data that the lipoproteins in S. aureus are diacylated.
Kurokawa et al. (35) recently reported that N terminus of S. aureus SitC lipoprotein is triacylated. They also suggested the existence of another type of N-acyltransferase distinct from lnt. Although the results did not agree with our analytical data, it is possible to consider that strain variation or cultural condition may affect the activity of the suggested enzyme. Further analysis must be necessary.
In conclusion, we identified TLR2-activating lipoproteins from S. aureus cells and characterized the N-terminal lipopeptide structure of a lipoprotein SAUOHSC_02699 as a diacylated one. Because these lipoproteins are considered to contribute to the virulence of S. aureus, further studies using protein expression or organic synthetic chemistry are now ongoing to clarify their immunobiological properties.
Acknowledgments
We thank Professor Kazuhisa Sugimura at Kagoshima University for measuring luciferase activity.
This work was supported by a research grant from The Uehara Memorial Foundation.
Footnotes
The abbreviations used are: TLR, Toll-like receptor; mTLR2, murine TLR2; LTA, lipoteichoic acid; MALDI-TOF-MS, matrix-assisted laser desorption ionization-time of flight mass spectrometry; MS/MS, tandem mass spectrometry; PBMC, peripheral blood mononuclear cell; PGN, peptidoglycan; HPLC, high pressure liquid chromatography; TNF, tumor necrosis factor.
References
- 1.Lowy, F. D. (1998) N. Engl. J. Med. 339 520-532 [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi, O., Hoshino, K., Kawai, T., Sanjo, H., Takada, H., Ogawa, T., Takeda, K., and Akira, S. (1999) Immunity 11 443-451 [DOI] [PubMed] [Google Scholar]
- 3.Schwandner, R., Dziarski, R., Wesche, H., Rothe, M., and Kirschning, C. J. (1999) J. Biol. Chem. 274 17406-17409 [DOI] [PubMed] [Google Scholar]
- 4.Travassos, L. H., Girardin, S. E., Philpott, D. J., Blanot, D., Nahori, M. A., Werts, C., and Boneca, I. G. (2004) EMBO Rep. 5 1000-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inohara, N., Ogura, Y., Fontalba, A., Gutierrez, O. Pons, F., Crespo, J., Fukase, K., Inamura, S., Kusumoto, S., Hashimoto, M., Foster, J. S., Moran, P. A., Fernandez-Luna, L. J., and Nuñez, G. (2003) J. Biol. Chem. 278 5509-5512 [DOI] [PubMed] [Google Scholar]
- 6.Chamaillard, M., Hashimoto, M., Horie, Y., Masumoto, J., Qiu, S., Saab, L., Ogura, Y., Kawasaki, A., Fukase, K., Kusumoto, S., Valvano, M. A., Foster, S. J., Mak, T. W., Nuñez, G., and Inohara, N. (2003) Nat. Immunol. 4 702-707 [DOI] [PubMed] [Google Scholar]
- 7.Morath, S., Geyer, A., and Hartung. T. (2001) J. Exp. Med. 193 393-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suda, Y., Tochio, H., Kawano, K., Takada, H., Yoshida, T., Kotani, S., and Kusumoto, S. (1995) FEMS Immunol. Med. Microbiol. 12 97-112 [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto, M., Yasuoka, J., Suda, Y., Takada, H., Yoshida, T., Kotani, S., and Kusumoto, S. (1997) J. Biochem. 121 779-786 [DOI] [PubMed] [Google Scholar]
- 10.Han, S. H., Kim, J. H., Martin, M., Michalek, S. M., Nahm, M. H. (2003) Infect. Immun. 71 5541-5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto, M., Imamura, Y., Morichika, T., Arimoto, K., Takeuchi, O., Takeda, K., Akira, S., Aoyama, K., Tamura, T., Kotani, S., Suda, Y., and Kusumoto, S. (2000) Biochem. Biophys. Res. Commun. 273 164-169 [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, M., Tawaratsumida, K., Kariya, H., Aoyama, A., Tamura, T., and Suda, Y. (2006) Int. Immunol. 18 355-362 [DOI] [PubMed] [Google Scholar]
- 13.Stoll, H., Dengjel, J., Nerz, C., and Götz, F. (2005) Infect. Immun. 73 2411-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bubeck. J. W., Williams, W. A., and Missiakas, D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 13831-13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto, M., Tawaratsumida, K., Kariya, H., Kiyohara, A., Suda, Y., Krikae, F., Kirikae, T., and Götz, F. (2006) J. Immunol. 177 3162-3169 [DOI] [PubMed] [Google Scholar]
- 16.Iordanescu, S., and Surdeanu, M. (1976) J. Gen. Microbiol. 96 277-281 [DOI] [PubMed] [Google Scholar]
- 17.Shibata, K., Hasebe, A., Sasaki, T., and Watanabe, T. (1997) FEMS Immunol. Med. Microbiol. 19 275-283 [DOI] [PubMed] [Google Scholar]
- 18.Hirschfeld, M., Ma, Y, Weis, J. H., Vogel, S. N., and Weis, J. J. (2000) J. Immunol. 165 618-622 [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 20.Wallis, R. S., Amir-Tahmasseb, M., and Ellner, J. J. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3348-3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, O. N., Wilm, M., Shevchenko, A., and Mann, M. (1999) Methods Mol. Biol. 112 513-530 [DOI] [PubMed] [Google Scholar]
- 23.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999) Electrophoresis 20 3551-3567 [DOI] [PubMed] [Google Scholar]
- 24.Masuda, T., Tomita, M., and Ishihama. Y. (2007) J. Proteome Res. 7 731-740 [DOI] [PubMed] [Google Scholar]
- 25.Hantke, K., and Braun, V. (1973) Eur. J. Biochem. 34 284-296 [DOI] [PubMed] [Google Scholar]
- 26.Cockayne, A., Hill, J. P., Powell, N. B., Bishop, K., Sims, C., and Williams, P. (1998) Infect. Immun. 66 3767-3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schröder, N. W., Morath, S., Alexander, C., Hamann, L., Hartung, T., Zähringer, U., Göbel, B. U., Webe, R., Jr., and Schumann, R. R. (2003) J. Biol. Chem. 278 15587-15594 [DOI] [PubMed] [Google Scholar]
- 28.Hirschfeld, M., Kirschning, C. J., Schwandner, R., Wesche, H., Weis, J. H., Wooten, R. M., and Weis, J. J. (1999) J. Immunol. 163 2382-2386 [PubMed] [Google Scholar]
- 29.Lee, H. K., Lee, J., and Tobias, S. P. (2002) J. Immunol. 16 8 4007-4012 [DOI] [PubMed] [Google Scholar]
- 30.Fisette, P. L., Ram, S., Andersen, M. J., Guo, W., and Ingalls, R. R. (2003) J. Biol. Chem. 278 46252-46260 [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto, M., Asai, Y., and Ogawa, T. (2004) Int. Immunol. 16 431-437 [DOI] [PubMed] [Google Scholar]
- 32.Mühlradt, P. F., Kiess, M., Meyer, H., Süssmuth, R., and Jung, G. (1997) J. Exp. Med. 185 1951-1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata K, Hasebe, A., Into, T., Yamada, M., and Watanabe, T. (2000) J. Immunol. 165 6538-6544 [DOI] [PubMed] [Google Scholar]
- 34.Sankaran, K., and Wu, H. C. (1994) J. Biol. Chem. 269 19701-19706 [PubMed] [Google Scholar]
- 35.Kurokawa, K., Lee, H., Roh, K-B., Asanuma, M., Kim, Y. S., Nakayama, H., Shiratsuchi, A., Choi, Y., Takeuchi, O., Kang, H. J., Dohmae, N., Nakanishi, Y., Akira, S., Sekimizu, K., and Lee, B. L. (January 12, 2009) J. Biol. Chem. 10.1074/jbc.M809618200 [DOI] [PMC free article] [PubMed]