Abstract
Pregnane X receptor (PXR) is a ligand-dependent transcription factor, regulating gene expression of enzymes and transporters involved in xenobiotic/drug metabolism. Here, we report that protein arginine methyltransferase 1 (PRMT1) is required for the transcriptional activity of PXR. PRMT1 regulates expression of numerous genes, including nuclear receptor-regulated transcription, through methylating histone and non-histone proteins. Co-immunoprecipitation and histone methyltransferase assays revealed that PRMT1 is a major histone methyltransferase associated with PXR. The PXR ligand-binding domain is responsible for PXR-PRMT1 interaction as determined by mammalian two-hybrid and glutathione S-transferase (GST) pull-down assays. The chromatin immunoprecipitation (ChIP) assay showed that PRMT1 was recruited to the regulatory region of the PXR target gene cytochrome P450 3A4 (CYP3A4), with a concomitant methylation of arginine 3 of histone H4, in response to the PXR agonist rifampicin. In mammalian cells, small interfering RNA (siRNA) knockdown and gene deletion of PRMT1 greatly diminished the transcriptional activity of PXR, suggesting an indispensable role of PRMT1 in PXR-regulated gene expression. Interestingly, PXR appears to have a reciprocal effect on the PRMT1 functions by regulating its cellular compartmentalization as well as its substrate specificity. Taken together, these results demonstrated mutual interactions and functional interplays between PXR and PRMT1, and this interaction may be important for the epigenetics of PXR-regulated gene expression.
Pregnane X receptor (PXR)2 is an orphan nuclear receptor that regulates metabolism and disposition of various xenobiotics and endobiotics (1). These physiological functions of PXR are achieved through coordinating transcriptional regulation of Phase I and Phase II drug-metabolizing enzymes as well as the “Phase III” transporters (2). The structural flexibility in the ligand binding pocket enables PXR to function as a xenobiotic receptor through interacting with a wide range of structurally diverse compounds (3). Xeno- and endobiotics that activate PXR include a variety of prescription and nonprescription drugs, herbal medicines, environmental toxicants, and bile acids (4). The transcriptional activity of PXR is not only regulated by ligands, but also by various signal transduction pathways including NF-κB-regulated inflammatory pathway (5). We have shown that the transcriptional activity of PXR is negatively regulated by NF-κB (5), which may play a role in the pathogenesis of inflammatory bowl diseases and colon cancer (6).
Post-translational modifications on the N termini of histones have been shown to play critical roles in gene regulation including the regulation of transcriptional activity by nuclear receptors. These modifications include phosphorylation, acetylation, methylation, and ubiquitination (7). It is believed that the combination of modifications of the chromatin-associated histone and non-histone proteins, and the interplays between these modifications create a marking system (“histone code”), which is part of the epigenetic mechanisms for gene regulation (8).
The protein arginine methyltransferases (PRMTs) including PRMT1, PRMT2, and PRMT4 (CARM1) were shown to be nuclear receptor coactivators (9–12). These enzymes regulate gene expression through methylating histone and non-histone proteins, and the methylation marks are important for the nuclear/steroid receptor-mediated transcriptional activity. PRMT1 is a major arginine methyltransferase which methylates H4R3 and H2AR3. Recent evidence suggests that histone modification by PRMT1 set the stage for subsequent histone modifications (13), and there is an intricate interplay between H4R3 methylation and other histone modifications. For example, arginine methylation (H4R3) by PRMT1 facilitates H4 acetylation but H4 acetylation inhibits methylation of H4R3 (14). These observations suggest that histone modifications during transcription proceed in a unidirectional sequence and to complete a transcription cycle, methylated H4R3 has to be demethylated, followed by acetylation and then deacetylation or replacement (15). Understanding the role of these histone modification enzymes in nuclear receptor-regulated gene expression will help us understand the epigenetic mechanism of gene regulation and provide important basis for drug/therapeutic designs to effectively intervene in pathological processes such as tumorigenesis and inflammatory responses.
In this study, we identified PRMT1 to be a major HMT associated with PXR, and we demonstrated that PRMT1 is a required histone methyltransferase for PXR transcriptional activity. PRMT1 regulates PXR transcriptional activity by direct association with PXR in a ligand-dependent manner, and PXR agonist rifampicin caused recruitment of PRMT1 to the regulatory region of PXR target gene CYP3A4. Knockdown of PRMT1 through siRNA or gene deletion inhibited PXR transcriptional activity. Furthermore, we found that PXR plays a critical role in regulating PRMT1 cellular compartmentalization and substrate preference.
EXPERIMENTAL PROCEDURES
Materials—DMSO, rifampicin, PCN, anti-FLAG M2 antibody, and anti-FLAG M2-agarose affinity beads were from Sigma. Core histones were from Roche (Indianapolis, IN). S-Adenosyl-l-[methyl-3H]methionine ([3H]SAM) and [35S]-methionine were from PerkinElmer (Waltham, MA). Recombinant histone H4, acetylated H4 N-terminal peptides, recombinant PRMT1, as well as anti-acetyl-(pan)H4, anti-H4(Me2)R3, and anti-PRMT1 antibodies were purchased from Upstate (Millipore, Billerica, MA). Goat and mouse anti-PXR antibodies, and isotype IgGs were from Santa Cruz Biotechnology (Santa Cruz, CA). Nitrocellulose and polyvinylidene difluoride membranes were from Bio-Rad.
Cells—HepG2 and CV-1 cells were cultured in Dulbecco's modified Eagle's medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (Sigma) and 1× antibiotic and antimycotic (Invitrogen, Carlsbad, CA). PXR-HepG2 and PXR-HT29 stable transfectants were created as described in Ref. 5. Wild-type and PRMT1-null ES cells were obtained from Mark Bedford (MD Anderson, Houston, TX) and cultured according to Ref. 17.
Plasmids—Plasmids expressing GST-fused PXR fragments have been created in our laboratory. DNA sequences coding different PXR fragments were PCR-amplified and subcloned into pGEX-5X-3 expression vector (Amersham Biosciences). pACT, pBIND, and pG5-luc were purchased from Promega (Madison, WI) for the mammalian two-hybrid assay. pBIND-PXR (Gal4-PXR) and pACT-PRMT1 (VP16-PRMT1) were constructed by inserting PCR-amplified human PXR DNA sequence into pBIND vector and PCR-amplified PRMT1 DNA sequence into pACT vector following the manufacturer's recommendation (Promega).
Co-immunoprecipitation—PXR-HepG2 cells were washed with PBS and homogenized in the Co-IP lysis buffer (20 mm Hepes, pH 7.4, 125 mm NaCl, 1% Triton X-100, 10 mm EGTA, 2 mm Na3VO4, 50 mm NaF, 20 mm ZnCl2, 10 mm sodium pyrophosphate, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride). 1× complete protease inhibitor mixture (Sigma) was added before use. Mouse liver tissues were homogenized in the same (above) lysis buffer. After centrifugation (12,000 × g in a microcentrifuge at 4 °C for 15 min), supernatant fractions were collected and incubated with antibodies and GammaBind Plus-Sepharose beads (Amersham Biosciences) for 2 h at 4 °C on a rotary shaker. Corresponding isotype IgG was used as a negative control. The beads were washed three times, and the precipitated protein complexes were analyzed with HMT assay or Western blot.
Histone Methyltransferase Assay and Peptide Sequencing Analysis—The PRMT1 (Upstate) HMT assay was based on the manufacturer's recommendation. In brief, 2 μg of core histones, 2 μg of H4, or 0.4 μg of H4 N-terminal peptides were incubated with the immunoprecipitated HMT complexes or recombinant PRMT1 at 30 °C for 90 min, in 1× HMT buffer (50 mm Tris, pH 9.0, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol) with S-adenosyl-l-[methyl-3H]methionine ([3H]SAM) as the methyl donor. The reaction mix was separated in 16% SDS-PAGE, and the separated proteins were transferred to the polyvinylidene difluoride membrane for autoradiography and staining with Ponceau BS red dye (Sigma). The radioactive proteins identified by autoradiography were excised from the membrane for N-terminal sequencing by Edman degradation. The radioactivity in the Edman degradation fractions corresponding to amino acid residues was determined by liquid scintillation counting.
Western Blot—Proteins were analyzed by SDS-PAGE and then transferred to a nitrocellulose membrane. After over 4 h of blocking in 5% milk with TBST buffer (20 mm Tris-HCL, pH 7.6, 137 mm NaCl, 0.5% Tween 20), the blot was incubated with appropriate primary antibodies at 37 °C overnight. After washing with TBST buffer for 30 min, the membrane was then subjected to 1:2000 corresponding alkaline phosphatase-conjugated secondary antibodies for 2 h. After another wash with TBST for 30 min, the membrane was exposed to Nitro Blue tetrazolium/BCIP as the substrate (Promega).
Transient Transfection and Luciferase Assay—Cells were seeded in 12-well plates. When growth reached 50% confluence, cells were transfected with plasmid DNA for 12 h using Lipofectamine (Invitrogen). The transfected cells were treated with chemicals or vehicle for an additional 48 h. The luciferase assay was performed using a luciferase assay system kit, according to the manufacturer's recommendation (Promega).
Mammalian Two-hybrid Assay—The mammalian two-hybrid assay was performed using Checkmate Mammalian Two Hybrid System (Promega). CV-1 cells were seeded in 12-well plates and transient transfected with pBIND-PXR, pACT-PRMT1, and pG5-luc. 12 h after transfection, cells were treated with rifampicin (10 μm, 48 h), and luciferase activity was determined with Polarstar optima luminometer (BMG Laboratory).
GST Pull-down Assay—The GST pull-down assay was performed as described (18). Briefly, [35S]methionine-labeled full-length PRMT1 protein was generated with a TnT-coupled Reticulocyte Lysate System (Promega) using the SP6 promoter-driven cDNA plasmid as the template. PCR-generated PXR cDNA fragments were inserted in-frame into pGEX-5X-3 (Amersham Biosciences). The plasmids were expressed in Escherichia coli (BL21), and fusion polypeptides were purified with glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instruction. Twenty micrograms of each fusion polypeptide (estimated by comparison with bovine serum albumin in an SDS-PAGE gel with Coomassie Blue staining) was incubated with 20 μl of radiolabeled PRMT1 in a total volume of 200 μl of binding reaction buffer (20 mm Hepes pH 7.9, 1% Triton X-100, 20 mm dithiothreitol, 0.5% bovine serum albumin, and 100 mm KCl) for 3 h at 4 °C. After incubation, beads were washed three times with the same buffer without bovine serum albumin. The bound proteins were eluted by boiling in the SDS-PAGE sample buffer and resolved by 12% SDS-PAGE gel electrophoresis. The signals were detected by autoradiography. The input control was 2 μl of the radioactive PRMT1.
Small Interference RNA—Two small interfering RNA-expressing plasmids were constructed by cloning the sequences targeting PRMT1 at coding region sequences 756–773 (19) (siPRMT1–28) and 353–371(siPRMT1–11) into pSilencer 5.1 plasmids according to the manual (Ambion). The targeting plasmids were created by inserting 5′-GATCCGATCCACTGGTGGGAGAACTTCAAGAGAGTTCTCCCACCAGTGGATTTTTTTGGAAAAGCT-3′ (siPRMT1–28) and 5′-GATCCGCTCCATGTTTCATAACCGGTTCAAGAGACCGGTTATGAAACATGGAGTTTTTTGGAAAAGCT-3′ (siPRMT1–11). The siRNA plasmids and the scramble siRNA control were co-transfected with PXR-directed reporter plasmid pGL3-3A4-Luc (5) into PXR-HepG2 cells. The transfected cells were treated with rifampicin (10 μm, 48 h). Luciferase activity and PRMT1 protein expression were determined with luminometry and Western blotting, respectively.
Chromatin Immunoprecipitation (ChIP)—ChIP assay was performed according to the manufacturer's protocol from Upstate, using the ChIP assay kit with modifications. Briefly, PXR-HepG2 cells were treated with rifampicin (10 μm, 2 h) and DMSO (vehicle control). Cells were cross-linked with 1% formaldehyde for 15 min at room temperature, and then the reaction was stopped by incubating in glycine with a final concentration of 0.125 m for 5 min. Cells were washed three times with cold PBS and harvested by scraping with cell scraper. Then the cells were lysed in the SDS lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris-HCl, pH 8.1) on ice for 10 min. The samples were sonicated into DNA fragments of 0.2–1 kb (checked by agarose gel electrophoresis/ethidium bromide staining) and microcentrifuged at maximal speed for 10 min at 4 °C. The supernatant was precleared by rotating with 60 μl of Salmon Sperm DNA/protein-agarose slurry for 30 min at 4 °C and then aliquoted after centrifugation. 20 μl was saved as input and 200 μl (equal to one-fifth the amount of cells from one 100% confluent 15-cm dish) was used for each antibody. Each 200-μl supernatant was diluted with 800 μl of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, and 167 mm NaCl) and incubated with the specific antibody (1 μg/sample) at 4 °C overnight. A mock precipitation without antibody was used as negative control. The next day, 60 μl of salmon sperm DNA/protein-agarose slurry was added to each sample and incubated at 4 °C for another 2–4 h. The beads were then washed for 3–5 min with 1 ml of each buffers listed: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 500 mm NaCl), and LiCl wash buffer (0.25 m LiCl, 1% IGE-PAL-CA630, 1% deoxycholic acid (sodium salt), 1 mm EDTA, 10 mm Tris-HCl, pH 8.1). After all washes, pellets were suspended by vertex with 150 μl of freshly prepared elution buffer (0.1 m NaHCO3, 1% SDS) for 15 min, and then supernatant was collected. This elution progress was repeated once again, and in total 300-μl elutes were collected. The one-tenth input was diluted with dilution buffer to a total volume of 300 μl. Elutes and diluted inputs were incubated in 0.3 m NaCl at 65 °C for 4 h to reverse formaldehyde cross-linking. Then 10 μl of 0.5 m EDTA, 20 μl of 1 m Tris-HCl, pH 6.5, and 20 μg of proteinase K were added to the sample and incubated at 45 °C for 1 h. DNA was extracted with phenol/chloroform and then incubated with 10 μg of glycogen in 75% ethanol at -20 °C overnight. After precipitation by centrifuging at 12,000 × g for 30 min at 4 °C, the recovered DNA pellets were dissolved in 30 μl of distilled water. The DNA target in the sample was determined by real-time quantitative PCR in triplicates with a 1-μl sample. Three independent experiments were performed. Amplifications were performed in the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) with SYBR Green Master Mix (Applied Biosystems). The PCR primers used were: forward primer, 5′-GTCCCAATTAAAGGTCATAAAGC-3′ and reverse primer, 5′-CTTGAACCGACATGATTTCAAG-3′.
Statistical Analysis—Statistical evaluations were conducted using two-tailed t test with triplicates for each treatment. A p value of less than 0.01 was considered to be statistically significant. Data are the means ± S.D. of three independent results.
Immunofluorescence Microscopy—Cells were seeded in chamber slides and kept in standard cell culture condition. For microscopy, the cells were washed with PBS and then fixed with freshly prepared 4% paraformaldehyde in PBS at 4 °C for 10 min. After three washes, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. After another three washes, cells were blocked with 10% donkey serum in PBS/Tween (0.1% Tween20 in PBS) at room temperature for 3 h. Primary antibodies (mouse anti-PXR antibody with a dilution of 1:100, rabbit anti-PRMT1 antibody with a dilution of 1:500) in the blocking buffer were incubated with cells at 4 °C overnight. The corresponding isotype IgG was used as negative control. After washing with PBS/Tween for three times, cells were incubated with anti-mouse red-fluorescent Alexa Fluor 568 or anti-rabbit green-fluorescent Alexa Fluor 488 dyes (Invitrogen) in the PBS/Tween for another 2 h at room temperature. Cells were washed for three times and DAPI (Vector Laboratory, Burlingame, CA) was added. The results were analyzed by fluorescence microscopy (Olympus IX71) equipped with Olympus DP70 digital camera.
RESULTS
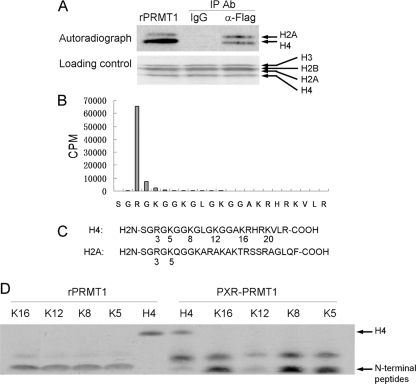
Association of PRMT1 with PXR in HepG2 Cells—Histone methyltransferases have been demonstrated to be transcriptional regulators for nuclear receptors. To analyze PXR-associated histone methyltransferase(s) (HMT), we created a cell line (PXR-HepG2), by stable transfection of FLAG-tagged human PXR into HepG2 cells, which lack PXR (5). The HMT(s) associated with rifampicin-activated PXR were detected by co-immunoprecipitation followed by histone methyltransferase assay with core histones as the substrates and radiolabeled S-adenosyl-l-[methyl-3H]methionine ([H3]SAM) as the methyl donor. Methylated histones were detected by autoradiography following SDS-PAGE. As shown in Fig. 1A, the methyltransferase activity was associated with FLAG-tagged PXR in the precipitated complex and the precipitated HMT(s) methylated both histones H4 and H2A.
FIGURE 1.
Histone methyltransferase activity is associated with PXR. A, PXR-HepG2 cells were treated with rifampicin (10 μm, 2 h) and used to perform co-immunoprecipitation/HMT assay with anti-FLAG antibody. The precipitates and recombinant PRMT1 were subjected to HMT assay, respectively, with core histones as the substrates and [3H]SAM as the methyl donor. Methylated histones were analyzed by autoradiography. B, methylated H4 in A was subjected to N-terminal sequencing analysis. The radioactivity associated with Edman degradation fractions was determined by liquid scintillation counting. C, illustration of N-terminal sequences of histone H2A and H4 with the common “SGRGK” motif. D, substrate specificity comparison of the PXR-assocated HMT and recombinant PRMT1. Same molar pre-acetylated H4 N-terminal peptides (0.4 μg, 2 kDa) and recombinant H4 (2 μg, 11 kDa) (Upstate) were used as the substrates and [3H]SAM was used as the methyl donor. The pre-acetylated H4 peptides are 20-amino acid N-terminal peptides with K5, K8, K12, or K16 individually acetylated. Methylation was detected by autoradiography.
To identify the amino acid residue(s) methylated by PXR-associated HMT(s), the methylated H4 was analyzed by N-terminal sequencing. As shown in Fig. 1B, arginine 3 (H4R3) was the only methylated residue among the 23 N-terminal amino acids analyzed. PRMT1 has been demonstrated to be the predominant enzyme responsible for this site-specific methylation (20). H2A shares the same N-terminal “SGRGK” sequence motif with H4, therefore it was also methylated by PRMT1 (Fig. 1A, left lane). PRMT5 is another enzyme that has been shown to methylate H4R3 in this SGRGK motif (21). However, PRMT5 is also known to methylate histone H3 (22). Under our experimental conditions, we found that major HMT associated with PXR-methylated H2A and H4, but not H3.
It has been reported that acetylation of H4 inhibits methylation of H4R3 by recombinant PRMT1 (14). The pre-acetylation on H4K5, K8, K12, or K16 inhibits the methylation at similar level (14). However, in our experiments, the pre-acetylation on K12 significantly inhibits the methylation of H4R3 by the PXR-associated complex while pre-acetylation on the other lysines was less effective in the inhibition (Fig. 1D). The recombinant PRMT1 showed the same substrate methylation preference regardless the acetylation status of the test peptide. These results suggested that the substrate specificity of PRMT1 can be regulated when it is in association with PXR.
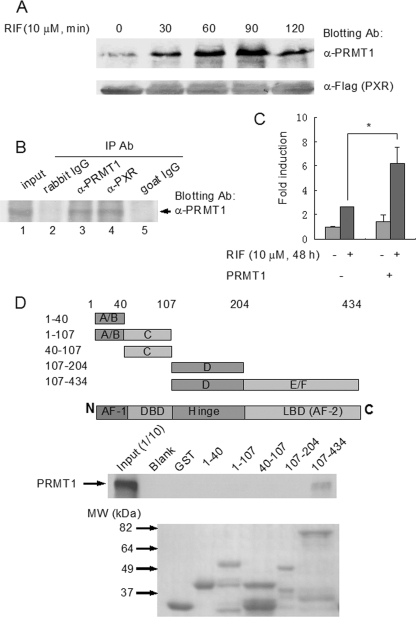
Ligand-dependent Physical and Functional Interaction between PRMT1 and PXR—To analyze the effects of PXR ligand on the interaction between the receptor and PRMT1, we performed co-immunoprecipitation assay with the protein extracts of PXR-HepG2 cells treated with the PXR ligand rifampicin. The complexes precipitated with anti-FLAG antibody were eluted with FLAG tag and analyzed by Western blotting. PRMT1 was found to associate with PXR in a ligand-dependent manner (Fig. 2A).
FIGURE 2.
PRMT1 interacts with PXR in a ligand-dependent manner. A, PXR-HepG2 cells were treated with rifampicin (10 μm, 0, 30, 60, 90, 120 min) and subjected to co-immunoprecipitation with anti-FLAG antibody-coupled beads. The precipitates were eluted with 3× FLAG peptide and analyzed by Western blotting with PRMT1 antibody. Anti-FLAG antibody blotting was used to show the equal loading of the samples. B, liver tissue from a VP16-hPXR transgenic mouse was homogenized in the Co-IP lysis buffer and co-immunoprecipitated with goat anti-PXR (lane 4) and rabbit anti-PRMT1 antibodies (lane 3). Goat IgG (lane 5) and rabbit IgG (lane 2) were used as negative controls. 1:10 lysate was loaded as the input control (lane 1). Precipitates were analyzed by Western blotting with PRMT1 antibody. C, CV-1 cells were transfected with the bait plasmid, pBIND-PXR, and the reporter pG5-luc vector, with cotransfection of the prey plasmid pACT-PRMT1 or blank pACT plasmid. Six hours after transfection, cells were treated with rifampicin (10 μm) or vehicle for an additional 48 h. The interaction was characterized by luciferase activity. *, statistically significant difference (t test, p < 0.01). The data are the means ± S.D. of three independent results. D, mapping of the interactive domains of PXR with PRMT1 by GST pull-down assay. Various PXR fragments were fused with GST and the fusion peptides coupled with glutathione-Sepharose beads were incubated with radiolabeled PRMT1. The precipitated complexes were analyzed by autoradiography following SDS-PAGE (middle panel). Upper panel, illustration of PXR fragments. Lower panel, loading control of the GST-fused PXR fragments (Coomassie Blue staining).
To further analyze the interaction between PXR and PRMT1 in vivo, we performed co-immunoprecipitation assay with liver tissues from VP16-hPXR transgenic mice. In these mice, the mouse PXR has been replaced with human PXR, which has been fused with VP16 activation domain, resulting in constitutively active PXR in these animals (16). PRMT1 was found to specifically associate with PXR as determined by co-immunoprecipitation followed by Western blot analysis (Fig. 2B).
To further analyze the ligand-dependent PXR-PRMT1 interaction, we performed mammalian two-hybrid assay in CV-1 cells. Consistent with the ligand-dependent interaction in the co-immunoprecipitation assay (Fig. 1A), transient transfection of VP16-PRMT1 significantly enhanced the Gal4-PXR-driven luciferase expression upon PXR ligand rifampicin treatment (Fig. 2C).
To identify and characterize the interactive domains of PXR responsible for association with PRMT1, we performed GST pull-down assay using GST fusion peptides containing various domains of PXR (Fig. 2D). As shown in Fig. 2D, only the PXR fragment, which contains the hinge domain and the ligand binding domain, interacted with PRMT1. However, the hinge domain alone showed no interaction. Taken together, these results indicated that PRMT1 specifically associated with PXR ligand-binding domain, which is consistent with the ligand-dependent interaction manner. The PXR interaction appears to be PRMT1-specific. PRMT5 failed to interact with GST-PXR fusion peptides in the pull-down assay (data not shown).
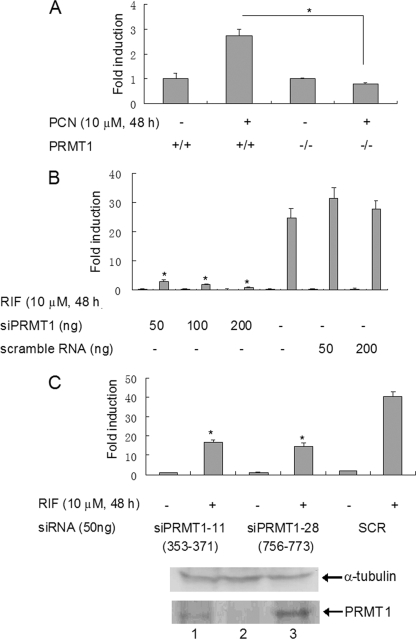
Requirement of PRMT1 for the Transcriptional Activity of PXR—The PRMT1-null mutation is embryonic lethal in homozygous mice. However, mouse embryonic stem cells survived without PRMT1 (23). We utilized these PRMT1 knock out mouse embryonic stem cells (ES) to analyze the role of PRMT1 in regulating PXR transcriptional activity in vivo. Gal4-mPXR (mouse PXR) and Gal4-responsive tk-UAS-luciferase reporter plasmids were co-transfected into the PRMT1-deficient and wild-type mouse ES cells. In the wild-type ES cells, mouse PXR agonist pregnenolone-16-α-carbonitrile (PCN) induced the PXR-driven luciferase reporter gene, whereas in the PRMT1(-/-) ES cells, PCN was not effective in the induction (Fig. 3A).
FIGURE 3.
PRMT1 is required for PXR transcriptional activity. A, PXR activity in PRMT1(-/-) ES cells. Gal4-driven luciferase reporter gene and Gal4-mPXR were transiently transfected into mouse PRMT1-null ES cells or wild-type ES cells. The transfected cells were treated with the receptor agonist PCN (10 μm, 24 h). Luciferase activity was determined by a luminometer. *, statistically significant difference (t test, p < 0.01). The data are the means ± S.D. of three independent results. B and C, the effect of siRNA knockdown of PRMT1 on PXR transcriptional activity. PXR-HepG2 cells were transfected with CYP3A4-luciferase. Two siRNAs targeting different sequences of PRMT1 (756–773 and 353–371) were used to knockdown PRMT1. Scrambled siRNA was used as the control (B and upper panel of C). The total PRMT1 protein expression was analyzed by Western blotting with PRMT1 antibody. Western blot with α-tubulin antibody was shown for loading control (C, lower panel). Lane 1, siPRMT1–11; lane 2, siPRMT1–28; lane 3, control. The reporter gene expression was measured by luciferase assay. *, statistically significant difference (t test, p < 0.01). The data are the means ± S.D. of three independent results.
To test the effect of PRMT1 on PXR transactivation in human cells, we performed PRMT1 knockdown experiment in PXR-HepG2 cells with small interference RNA (siRNA). The DNA fragment, which encoded a 21-bp hairpin siRNA targeting at PRMT1 nucleotides 756–773 (19) was cloned into the pSilencer vector (Ambion). When this siRNA-expressing plasmid was transfected into the PXR-HepG2 cells, the PXR ligand-dependent activation of CYP3A4-luciferase reporter gene activity was dramatically inhibited (Fig. 3B). A similar result was obtained with another siRNA, which targets at PRMT1 nucleotides 353–371 (Fig. 3C, upper panel). The decreased PRMT1 protein expression in siRNA-transfected cells was confirmed by Western blotting analysis (Fig. 3C, lower panel).
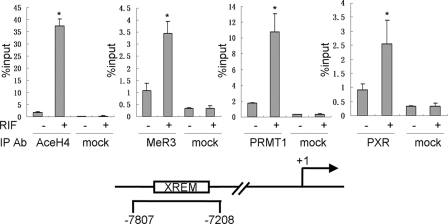
Recruitment of PRMT1 to the Regulatory Regions of PXR Target Gene CYP3A4—PXR was identified to be the major transcription factor regulating CYP3A4 through binding to the xenobiotic response enhancer module (XREM) about 8-kb upstream of the transcriptional starting site (24). The above results of physical and functional interactions between PXR and PRMT1 led us to hypothesize that PRMT1 is recruited to this regulatory region of CYP3A4 in response to PXR ligand stimulation. PXR-HepG2 cells were used in a ChIP assay to analyze the recruitment of PXR and PRMT1 as well as changes of histone modifications on the CYP3A4 regulatory region. Our results indicated that activation of PXR by rifampicin resulted in recruitment of PXR to the regulatory region of CYP3A4 as well as increasing of histone H4 acetylation which is indicative of transcriptional activation of the gene. Concomitantly, PRMT1 was also recruited to this CYP3A4 regulatory region in response to rifampicin treatment with increases in H4R3 methylation (Fig. 4).
FIGURE 4.
Recruitments of PRMT1, PXR, and changes of histone modifications in the CYP3A4 regulatory regions in response to PXR activation. PXR-HepG2 cells were treated with rifampicin (10 μm, 2 h). ChIP assay was performed to analyze the association of PXR, PRMT1, and changes of histone H4 acetylation and H4R3 methylation. Results were analyzed by quantitative real-time PCR. *, statistically significant difference (t test, p < 0.01). The data are the means ± S.D. of three independent results.
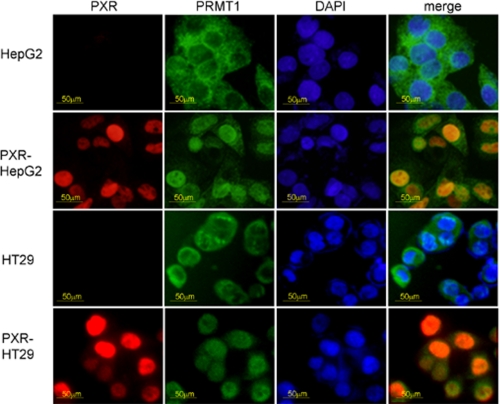
Regulation of PRMT1 Subcellular Localization by PXR—Results from the co-immunoprecipitation/HMT assay indicated that PXR regulates PRMT1 substrate specificity (Fig. 1D). Because PXR and PRMT1 interacted physically and functionally, it is possible that PXR may influence the cellular distribution of PRMT1. To test this possibility, we examined the subcellular localization of PRMT1 in cells with or without PXR. Interestingly, in human hepatoma cell line HepG2 which lacks PXR, PRMT1 was primarily localized in the cytoplasm region. However, in PXR-HepG2 cells where PXR is restored, PRMT1 was largely localized in the nucleus. The similar effects of PXR on PRMT1 were also observed in the human intestinal epithelial tumor cell line HT29, which also lacks PXR. In these cells, PRMT1 was mainly localized in the cytoplasm. Upon restoration of PXR expression by stable transfection, PRMT1 became localized in the nucleus (Fig. 5). These results suggest that PXR plays an important role in regulating the nuclear compartmentalization of PRMT1, which may affect the activity of PRMT1.
FIGURE 5.
PXR regulates PRMT1 subcellular localization as determined by immunofluorescence microscopy. Parental HepG2 and HT29 cells as well as the PXR stable transfectants PXR-HepG2 and PXR-HT29 were analyzed for PXR and PRMT1 subcellular localization by immunofluorescence microscopy.
DISCUSSION
PRMT1, which methylates arginine 3 of histone H4 (H4R3), is a major arginine methyltransferase in mammalian cells. Accumulating evidence indicates that PRMT1 plays a vital role in physiological and pathophysiological processes including development, nuclear receptor-regulated gene expression, and oncogenesis (12, 25–27). The mouse homozygous null mutant of PRMT1 is early embryonic lethal, attesting to the vital function of PRMT1 in the development and survival of the whole organism (23). In addition to modifying histones, PRMT1 has also been found to methylate non-histone proteins involved in DNA repair (28), DNA methylation (29), translational control, and maintenance of heterochromatic and euchromatic barrier (30), suggesting that PRMT1 regulates many aspects of gene expression. At the molecular level, intricate interplays between PRMT1 and other histone modification enzymes have been found. For example, methylation of H4R3 by PRMT1 promotes acetylation of histone H4, which leads to gene activation; however, acetylations of histone H4 inhibits H4R3 methylation (14), suggesting a unidirectional relay of histone marking processes in a transcription cycle (15). Based on our results, we propose that by direct interaction with PRMT1, nuclear receptors such as PXR initiate target gene transcription by recruiting PRMT1 to the regulatory region to accomplish the step of creating methyl marks on the chromatin.
In this study, we provide strong evidence indicating that PRMT1 is a major histone methyltransferase associated with PXR and plays an indispensable role in the transcriptional activity of PXR. We used an unbiased biochemical approach with FLAG-tagged PXR to precipitate the PXR-associated histone methyltransferases in HepG2 cells. In our Co-IP/methyltransferase assay with core histones H3, H2A, H2B, and H4 as substrates, H2A and H4 were methylated by the PXR-associated HMTs. We sequenced the N-terminal 23 amino acids of methylated H4 and found that H4R3 was the major methylated residue. The methylated H2A was most likely due to the common “SGRGK” motif shared by these peptides (Fig. 1C).
The results of Co-IP in both mouse liver tissue and PXR-transfected HepG2 cells indicated that PXR interacts with PRMT1. GST pull-down assay strongly suggested that the direct interaction between PXR and PRMT1 is through the PXR ligand binding domain. As indicated by the ChIP assay results, this direct interaction may play a role in recruitment of PRMT1 to the CYP3A4 regulatory region, where it promotes transcription through methylation and acetylation of chromatin as demonstrated here (Fig. 4). Another possibility is that by direct contact, PRMT1 methylates PXR and thus modifies its transactivity. For example, HNF-4α is methylated by PRMT1 and thus changes its activity of target gene regulation (25). We have tested this possibility by performing PRMT1 methyltransferase assay with GST-PXR as the substrate. In this assay, PRMT1 did not methylate the GST-PXR fusion peptide (data not shown).
The important role of PRMT1 in the transcriptional activity of PXR was further confirmed using two approaches: 1) knockdown of PRMT1 expression by PRMT1-specific siRNA drastically inhibited the PXR-regulated luciferase reporter gene activity, and 2) in PRMT1(-/-) cells PXR transcriptional activity was not detectable, suggesting an indispensable function of PRMT1 for the PXR-regulated gene expression.
Interestingly, our results indicate that PXR also regulates functions of PRMT1 in at least two aspects. 1) PXR regulates the PRMT1 substrate specificity. In comparison with the recombinant PRMT1, the PXR-associated PRMT1 demonstrated preference for certain pre-acetylated H4 peptides; whereas preacetylation of H4K12 is inhibitory to methylation of H4R3 by PRMT1 (Fig. 1D), acetylation of H4K5, H4K8, and H4K16 has no effect on the H4R3 methylation. 2) The presence of PXR has a significant effect on the cellular compartmentalization of PRMT1. In normal human hepatocytes that express PXR, PRMT1 is mostly localized in the nucleus (data not shown); however, in HepG2 cells that lack PXR, PRMT1 is localized mostly in the cytoplasm (Fig. 5). Stable transfection of PXR restores the PXR responses (5, 24), and PRMT1 is localized in the nucleus as demonstrated in this study (Fig. 5), suggesting PXR plays an important role in PRMT1 nuclear translocation. However, possibility cannot be completely eliminated that this phenomenon is unique to hepatocytes, and PXR overexpression causes PRMT1 nuclear translocation. To further analyze the role of PXR in PRMT1 nuclear translocation, we extend this observation to another cell line. We transfected PXR into colon epithelium cell line HT29, which lost endogenous PXR (Fig. 5). Similar to HepG2 cells, PRMT1 also translocated from cytoplasmic region into nucleus with PXR expression, lending addition support to the role of PXR in PRMT1 nuclear localization (Fig. 5).
Taken together, these results suggest that interaction between PXR and PRMT1 is reciprocal and not only PRMT1 regulates PXR transcriptional activity, but PXR also regulates the activity of PRMT1 through controlling its cellular compartmentalization in addition to substrate preferences. The effects of PXR on PRMT1 suggest that PXR has a rather general effect on the cellular processes that require PRMT1 and furthermore, function of PXR may go beyond the xenobiotic/drug metabolism to include many aspects of physiological/pathophysiological processes which require PRMT1.
This work was supported, in whole or in part, by NIEHS, National Institutes of Health Grants ES 09859 and ES 09106. This work was also supported by American Heart Association Grant 0355131Y. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PXR, pregnane X receptor; PRMT, protein arginine methyltransferase; CARM1, coactivator-associated arginine methyltransferase 1; CYP3A4, cytochrome P450 3A4; HMT, histone methyltransferase; Co-IP, co-immunoprecipitation; GST, glutathione S-transferase; XREM, xenobiotic response enhancer module; ChIP, chromatin immunoprecipitation; RIF, rifampicin; PCN, pregnenolone-16-α-carbonitrile; DMSO, dimethyl sulfoxide; PBS, phosphate-buffered saline.
References
- 1.Kliewer, S. A., and Willson, T. M. (2002) J. Lipid Res. 43 359-364 [PubMed] [Google Scholar]
- 2.Handschin, C., and Meyer, U. A. (2003) Pharmacol. Rev. 55 649-673 [DOI] [PubMed] [Google Scholar]
- 3.Watkins, R. E., Wisely, G. B., Moore, L. B., Collins, J. L., Lambert, M. H., Williams, S. P., Willson, T. M., Kliewer, S. A., and Redinbo, M. R. (2001) Science 292 2329-2333 [DOI] [PubMed] [Google Scholar]
- 4.Kliewer, S. A., Goodwin, B., and Willson, T. M. (2002) Endocr. Rev. 23 687-702 [DOI] [PubMed] [Google Scholar]
- 5.Gu, X., Ke, S., Liu, D., Sheng, T., Thomas, P. E., Rabson, A. B., Gallo, M. A., Xie, W., and Tian, Y. (2006) J. Biol. Chem. 281 17882-17889 [DOI] [PubMed] [Google Scholar]
- 6.Xie, W., and Tian, Y. (2006) Cell Metab. 4 177-178 [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides, T. (2007) Cell 128 693-705 [DOI] [PubMed] [Google Scholar]
- 8.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074-1080 [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., Ma, H., Hong, H., Koh, S. S., Huang, S. M., Schurter, B. T., Aswad, D. W., and Stallcup, M. R. (1999) Science 284 2174-2177 [DOI] [PubMed] [Google Scholar]
- 10.Koh, S. S., Chen, D., Lee, Y. H., and Stallcup, M. R. (2001) J. Biol. Chem. 276 1089-1098 [DOI] [PubMed] [Google Scholar]
- 11.Qi, C., Chang, J., Zhu, Y., Yeldandi, A. V., Rao, S. M., and Zhu, Y. J. (2002) J. Biol. Chem. 277 28624-28630 [DOI] [PubMed] [Google Scholar]
- 12.Rizzo, G., Renga, B., Antonelli, E., Passeri, D., Pellicciari, R., and Fiorucci, S. (2005) Mol. Pharmacol. 68 551-558 [DOI] [PubMed] [Google Scholar]
- 13.Huang, S., Litt, M., and Felsenfeld, G. (2005) Genes Dev. 19 1885-1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, H., Huang, Z. Q., Xia, L., Feng, Q., Erdjument-Bromage, H., Strahl, B. D., Briggs, S. D., Allis, C. D., Wong, J., Tempst, P., and Zhang, Y. (2001) Science 293 853-857 [DOI] [PubMed] [Google Scholar]
- 15.Tian, Y. Biochem. Pharmacol (2009) 77 670-680 [DOI] [PubMed] [Google Scholar]
- 16.Xie, W., Barwick, J. L., Downes, M., Blumberg, B., Simon, C. M., Nelson, M. C., Neuschwander-Tetri, B. A., Brunt, E. M., Guzelian, P. S., and Evans, R. M. (2000) Nature 406 435-439 [DOI] [PubMed] [Google Scholar]
- 17.Cote, J., Boisvert, F. M., Boulanger, M. C., Bedford, M. T., and Richard, S. (2003) Mol. Biol. Cell 14 274-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian, Y., Ke, S., Chen, M., and Sheng, T. (2003) J. Biol. Chem. 278 44041-44048 [DOI] [PubMed] [Google Scholar]
- 19.Robin-Lespinasse, Y., Sentis, S., Kolytcheff, C., Rostan, M. C., Corbo, L., and Le Romancer, M. (2007) J. Cell Sci. 120 638-647 [DOI] [PubMed] [Google Scholar]
- 20.Tang, J., Frankel, A., Cook, R. J., Kim, S., Paik, W. K., Williams, K. R., Clarke, S., and Herschman, H. R. (2000) J. Biol. Chem. 275 7723-7730 [DOI] [PubMed] [Google Scholar]
- 21.Nishioka, K., and Reinberg, D. (2003) Methods 31 49-58 [DOI] [PubMed] [Google Scholar]
- 22.Pal, S., Vishwanath, S. N., Erdjument-Bromage, H., Tempst, P., and Sif, S. (2004) Mol. Cell. Biol. 24 9630-9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawlak, M. R., Scherer, C. A., Chen, J., Roshon, M. J., and Ruley, H. E. (2000) Mol. Cell. Biol. 20 4859-4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodwin, B., Hodgson, E., and Liddle, C. (1999) Mol. Pharmacol. 56 1329-1339 [DOI] [PubMed] [Google Scholar]
- 25.Barrero, M. J., and Malik, S. (2006) Mol. Cell 24 233-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung, N., Chan, L. C., Thompson, A., Cleary, M. L., and So, C. W. (2007) Nat. Cell Biol. 9 1208-1215 [DOI] [PubMed] [Google Scholar]
- 27.Lim, Y., Kwon, Y. H., Won, N. H., Min, B. H., Park, I. S., Paik, W. K., and Kim, S. (2005) Biochim. Biophys. Acta 1723 240-247 [DOI] [PubMed] [Google Scholar]
- 28.Dery, U., Coulombe, Y., Rodrigue, A., Stasiak, A., Richard, S., and Masson, J. Y. (2008) Mol. Cell. Biol. 28 3058-3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan, C. P., and Nakielny, S. (2006) Mol. Cell. Biol. 26 7224-7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, S., Li, X., Yusufzai, T. M., Qiu, Y., and Felsenfeld, G. (2007) Mol. Cell. Biol. 27 7991-8002 [DOI] [PMC free article] [PubMed] [Google Scholar]