Abstract
IRSp53 is an essential intermediate between the activation of Rac and Cdc42 GTPases and the formation of cellular protrusions; it affects cell shape by coupling membrane-deforming activity with the actin cytoskeleton. IRSp53 is highly expressed in neurons where it is also an abundant component of the postsynaptic density (PSD). Here we analyze the physiological function of this protein in the mouse brain by generating IRSp53-deficient mice. Neurons in the hippocampus of young and adult knock-out (KO) mice do not exhibit morphological abnormalities in vivo. Conversely, primary cultured neurons derived from IRSp53 KO mice display retarded dendritic development in vitro. On a molecular level, Eps8 cooperates with IRSp53 to enhance actin bundling and interacts with IRSp53 in developing neurons. However, postsynaptic Shank proteins which are expressed at high levels in mature neurons compete with Eps8 to block actin bundling. In electrophysiological experiments the removal of IRSp53 increases synaptic plasticity as measured by augmented long term potentiation and paired-pulse facilitation. A primarily postsynaptic role of IRSp53 is underscored by the decreased size of the PSDs, which display increased levels of N-methyl-d-aspartate receptor subunits in IRSp53 KO animals. Our data suggest that the incorporation of IRSp53 into the PSD enables the protein to limit the number of postsynaptic glutamate receptors and thereby affect synaptic plasticity rather than dendritic morphology. Consistent with altered synaptic plasticity, IRSp53-deficient mice exhibit cognitive deficits in the contextual fear-conditioning paradigm.
Rho GTPases such as Cdc42, Rac, and Rho control key events in neuronal cell biology, including the generation of neuronal polarity and morphology, establishment of dendritic spines, the generation of postsynaptic specializations and synaptic plasticity (1, 2). Specificity in these processes is thought to arise through control of different downstream targets which are recognized and activated by the active, GTP-bound forms of Rho family members. The insulin receptor substrate of 53 kDa (IRSp53)3 is an essential mediator between activated Rac or Cdc42 and the formation of lamellipodia or filopodia, respectively. GTPase binding to IRSp53 enables interactions of its SH3 domain with downstream effectors WAVE2, Mena, Eps8, or N-WASP, all of which are known regulators of actin dynamics (3–6). In addition, the N-terminal IRSp53/missing in metastasis homology domain of IRSp53 assists in generating cellular protrusions by bundling actin filaments (5, 7, 8) and promoting membrane curvature (9, 10). Expression of IRSp53 is particularly high in the brain, and consequently IRSp53 contributes to the formation of dendritic spines in the cultured hippocampal neuron model (11).
Via the SH3 domain and a C-terminal PDZ binding motif, IRSp53 also bridges postsynaptic shank and PSD-95 family members (11–14). A significant enrichment in the postsynaptic density (PSD) of excitatory synapses suggests that Rac/Cdc42 signaling via IRSp53 may exert an as yet undefined role in postsynaptic signal transduction.
Here we have investigated the physiological role of IRSp53 by analyzing IRSp53-deficient mice. Our data indicate that the cellular and physiological functions of IRSp53 depend on the effector proteins present. Changes in neuronal morphology due to IRSp53 deficiency that were determined in primary developing cultured neurons in vitro are not observed in brains of developing and adult mice in vivo. The interaction of IRSp53 with the postsynaptic shank proteins abrogates the effects of IRSp53 on the actin cytoskeleton while revealing a synaptic function, which is evidenced by the enhanced synaptic plasticity in IRSp53 KO mice as well as their inability to learn in the hippocampus-dependent contextual fear-conditioning task.
EXPERIMENTAL PROCEDURES
Generation of IRSp53-deficient Mice—Embryonic stem cell clone XG-757 (Baygenomics; San Francisco, CA) was found by BLAST analysis to carry a transgenic insertion (“exon trap”) within the mouse IRSp53 gene. After injection of XG-757 cells into blastocysts and implantation, numerous chimeric animals were obtained. These gave rise to heterozygous animals carrying the transgene insertion after cross-breeding with C57BL/6 mice. Genotyping was performed using a forward primer in exon 3, one reverse primer in intron 3 for the WT allele, and one reverse primer in the exon trap insertion for the mutant allele. Animals used for most experiments in this study were in a 129P2/C57BL/6 hybrid background, with the exception of CA1 long term potentiation (LTP) and the behavioral experiments, which were performed after back-crossing for seven generations into the C57BL/6 strain.
Morphological Analysis of Mouse Brains—Mouse brains were subjected to Golgi staining, sectioning, and embedding according to published procedures (15, 16). 150-μm coronal sections were used for all analyses. For spine analysis, sections were viewed using an Axiovert 135 microscope equipped with 100× objective, Hamamatsu camera, and OpenLab software. Images of adjacent focal planes were stacked and analyzed using ImageJ. Length, width, and the number of spines per dendrite length were logged into an Excel spreadsheet using a modified macro from the ImageJ software. Dendrite branching in young animals was analyzed after importing stacks of microscopic images into ImageJ and tracing neurons through different layers of the stack. Because of their strongly increased complexity, analysis of dendrites in the CA1 region of adult hippocampi required the use of a Neurolucida setup at the Institute for Biology, Free University of Berlin (kindly provided by Prof. Dietmar Kuhl, Berlin).
Neuronal Primary Culture—Hippocampal neurons were prepared from P0 animals (mice) or E19 animals (rats) and cultivated and transfected as described (17) using Neurobasal media supplemented with B-27 (Invitrogen). Neurons were fixed after 14 days in culture with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS, and stained as described (17). Mouse anti-MAP2 (1:2000; Sigma) was used as the primary antibody followed by Cy3-labeled secondary antibody. Immunostaining was visualized using a Zeiss Axiovert 135 microscope equipped with a Hamamatsu camera or a confocal microscope (Zeiss LSM 510). Openlab 2.2.5 software (Improvision) was used for image analysis.
Dendritic branching was quantified as described (17). The number of branch points per dendrite was quantified and logged into an Excel spreadsheet. Statistical significance was in each case determined by Student's t test.
Biochemical Analyses—Fusion proteins (full-length His6-tagged IRSp53; GST-Cdc42; GST-IRSp53-SH3; GST-Eps8-N-PPP, residues 201–250; GST-shank3-PPP, residues 877–938; GST-shank1-PPP, residues 858–970) were generated using standard methods.
For actin bundling, monomeric G-actin was polymerized at room temperature in F-actin buffer (5 mm Tris-HCl, pH 7.8, 0.2 mm ATP, 1 mm dithiothreitol, 0.1 mm CaCl2, 1 mm MgCl2, and 100 mm KCl). F-actin was mixed with varying concentrations of recombinant proteins and incubated at room temperature for 30 min as described (5). Actin was then labeled with rhodamine-phalloidin, and 0.1% 1,4-diazabicyclo(2.2.2)octane and 0.1% methyl cellulose were added to the mixture. Samples were mounted between a slide and a coverslip coated with poly-lysine and imaged by fluorescence microscopy. The number of actin bundles in at least 10 randomly chosen microscopic fields was counted blindly by at least two investigators. PSDs were prepared as described (12) from forebrains of 2-week- and 2-month-old mice. For immunoprecipitation analysis, hippocampi were prepared from rats at different developmental stages. After lysis in immunoprecipitation buffer (50 mm Tris/HCl, pH 7.5, 120 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA, plus protease inhibitors pepstatin, leupeptin, and trasylol), samples were cleared by centrifugation and subjected to immunoprecipitation using immobilized anti-IRSp53. Material insoluble in immunoprecipitations buffer was solubilized in 1% SDS and again cleared by centrifugation. Samples were analyzed by Western blotting.
Electron Microscopy—Anesthetized animals were perfusion fixed with 2.5% glutaraldehyde in 0.1 m phosphate buffer with 1% sucrose, pH 7.3. Brains were post-fixed with 1% osmium tetroxide for 4 h. Small hippocampal tissue pieces were dehydrated through an ethanol series, stained with 2% uranyl acetate, and embedded in epoxy resin (Epon 812, Fluka, Germany). Ultrathin sections (70–80 nm) were cut with a diamond knife and collected on 300-mesh grids. Sections were stained with lead citrate and examined at a voltage of 80 kV using an EM10 transmission electron microscope (Carl Zeiss, Germany). Images were scanned at 600 dots/inch; PSDs were identified as dense membrane-associated structures in apposition to nerve terminals. For each animal at least 20 sections were analyzed. All PSDs from each section were scored for length, width, and area using ImageJ in combination with Excel. For all morphological analyses, the person evaluating the microscopic images was blind to the experimental conditions.
Immunohistochemistry—For pre-embedding immuno light microscopy and electron microscopy, animals were perfused with 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 m phosphate buffer, pH 7.4. Brains were cut into 30- and 150-μm-thick Vibratome sections, respectively, and immersed in 2.3 mol/liter sucrose in PBS overnight at 4 °C for cryoprotection. Thereafter, they were subjected to two freeze-thaw cycles in liquid nitrogen to aid penetration of immunoreagents, incubated with 10% horse serum containing 0.2% bovine serum albumin (BSA) (blocker) for 15 min, and left overnight at 4 °C in IRSp53 antibody (1:25) in PBS containing 1% PS and 0.2% BSA (Carrier). After washing in PBS, sections were incubated in biotinylated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA) diluted 1:1000 in Carrier for 90 min. After rinsing, they were incubated with ABC (Vector Laboratories) diluted 1:100 in PBS for 90 min. The sections were washed in PBS and reacted in diaminobenzidine-H202 solution (Sigma) for 10 min. The 30-μm sections were mounted on glass coverslips for light microscopic observation, and the 100-μm-thick sections were further fixed with 1% osmium tetroxide (w/v), dehydrated in an ascending series of ethanol, and embedded in Epon (Carl Roth GmbH & Co. KG, Karlsruhe, Germany). Ultrathin sections were examined with a Zeiss EM 902.
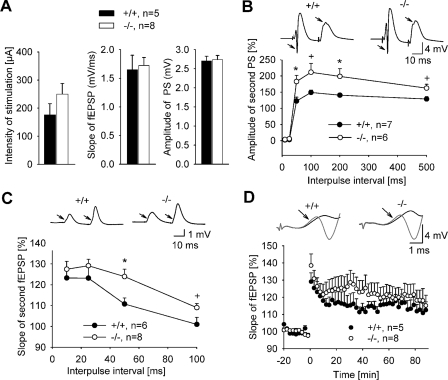
Electrophysiological Recordings—For in vivo recordings in the dentate gyrus, mice were anesthetized with urethane (Sigma, 1.5 g/kg body weight), and tracheotomies were performed as described (18). The recording electrode was placed 2 mm posterior and 1.5 mm lateral to bregma. The stimulation electrode was inserted under an angle of 70 degrees 0.5 mm anterior and 1.5 mm lateral to lambda. An additional hole was drilled on the surface of the contralateral frontal cortex for the ground electrode made of chlorided silver wire. A stimulating electrode was made of two 125-μm Teflon-insulated stainless steel wires, whereas a recording electrode was constructed of 75-μm Teflon-insulated tungsten wire. Electrodes were advanced until large positive field excitatory postsynaptic potentials (fEPSPs) with superimposed negative population spikes were evoked. The z-coordinate was adjusted to maximize the field responses to a biphasic current pulse of 100 μs and 200–400 μA elicited using an isolated pulse stimulator (Model 2100; A-M Systems, Everett, WA). The stimulation strength for recordings before and after induction of LTP was selected to provide a population spike of 2–3 mV. The responses were collected every 30 s, and averages of five consecutive sweeps were used for analysis. LTP was induced by 6 trains (inter-train interval of 20 s) of 6 bursts (inter-burst interval of 200 ms) of 6 pulses applied at 400 Hz. The stimulation intensity used for induction of LTP was two times higher than the one used for base-line recordings. Only experiments without rundown of fEPSPs (>90% of base line) were included in the analysis. The recorded signals were amplified ×100 and filtered (0.1–5 kHz bandpass) using a differential AC amplifier (Model 1700; A-M Systems) and digitized using a Pico ADC board (18). The signals were acquired and measured using the LTP Program (19). For measurements of the slope of fEPSPs, only the initial part (∼0.5 ms) of the responses was used, as shown by arrows in Fig. 4D.
FIGURE 4.
Normal synaptic transmission but altered synaptic plasticity in IRSp53-deficient mice in vivo. A, electrophysiological recordings in the dentate gyrus in vivo. Data represent the mean ± S.E. of three parameters characterizing basal synaptic transmission; that is, intensity of stimulation used to evoke population spikes of 2–3 mV (left), slope of fEPSPs (middle) and amplitude of population spikes (PS, right) collected at this stimulation strength. B, paired-pulse modulation of population spike (arrows) was elicited by supramaximal stimulation of the angular bundle. Data shown represent the mean ± S.E. of the ratio (expressed in %) between amplitudes of population spikes elicited by the second and first pulses. There is an increase in paired-pulse facilitation in KO mice (*, p < 0.05; +, p < 0.1; t test). C, paired-pulse facilitation of fEPSPs elicited by subthreshold double-pulse stimulation of the angular bundle with inter-pulse intervals of 10–100 ms. The data represent the mean ± S.E. of the ratio between slopes of fEPSPs elicited by the second and first pulses. There is an increase in paired-pulse facilitation in IRSp53 KO mice (*, p < 0.05; +, p < 0.1; t test). D, theta-burst stimulation (applied at time = 0) elicited a robust LTP in both WT and KO mice. Examples of responses collected before and after the induction of LTP are shown above the LTP profiles. Arrows point to the initial phase of fEPSPs where the measurements of slope were made. Data represent the mean ± S.E. of slope of fEPSPs expressed in % regarding the base line (time from -20 to 0).
Acute hippocampal slices were prepared from adult (5–6-month-old) mice. IRSp53 KO animals and age-matched WT animals from the same line were used. In brief, mice were anesthetized and decapitated, and the brains were quickly removed and placed into ice-cold constantly aerated (95% O2, 5%CO2) artificial cerebrospinal fluid (125 mm NaCl, 2.5 mm KCl, 2 mm MgCl2, 26 mm NaHCO3, 1.25 mm NaH2PO4, 2 CaCl2, 25 glucose) for 3 min. Hippocampi were isolated and cut into 400-μm-thick transversal slices with a Vibratome (VT 1000S; Leica, Nussloch, Germany), and sections were kept submerged in a storage chamber in aerated artificial cerebrospinal fluid. Before electrical recording slices were left in the storage chamber for ≥1 h at room temperature so that neuronal activity could recover to base line. For the extracellular recordings, slices were transferred to a recording chamber and kept submerged at 32 °C while constantly being perfused (1.5 ml/min) with aerated recording-artificial cerebrospinal fluid (125 mm NaCl, 2.5 mm KCl, 1 mm MgCl2, 26 mm NaHCO3, 1.25 mm NaH2PO4, 2 mm CaCl2, 25 mm glucose). Schaffer collaterals in area CA3 were stimulated by monopolar tungsten electrodes (9–10 megaohms), and fEPSPs were recorded by a glass electrode (4–15 megaohms) filled with 3 m NaCl and placed in the stratum radiatum of CA1. Synaptic field potentials were elicited with a frequency of 0.1 Hz, and LTP was induced with a theta burst stimulation of 10 bursts with four pulses each (100 Hz, 100-μs pulse duration, 200-ms inter-burst interval) which was repeated 3 times with an interval of 10 s. Data were collected, and slopes of the fEPSPs were calculated with a LabView (National Instruments, Munich, Germany)-based program. Paired-pulse facilitation was calculated as the second fEPSP slope divided by the first fEPSP slope a as percentage for inter-stimulus interval of 10, 20, 40, 80, 160 ms.
Behavioral Analysis—Adult male mice (10 for each genotype) were studied in the open field (to analyze locomotion, exploration, and anxiety), accelerated Rotarod (motor coordination) and contextual fear conditioning (learning and memory) tests, as described (20). The open field test was performed in a box (50 × 50 cm and 40 cm high) illuminated with white light (100 lux). Mice were started from one corner of the box, and their behavior was analyzed for 15 min. Distance moved, mean velocity, and time spent in the center (an imaginary 25 × 25-cm square in the middle of the arena) were analyzed with the software EthoVision (Noldus, Wageningen, The Netherlands).
In the accelerated Rotarod test mice had to walk on a turning, corrugated rod (3.2 cm in diameter) (Acceler Rotarod for mice Jones & Roberts, TSE Systems, Bad Homburg, Germany). Mice underwent 2 familiarization trials followed by 3 trials (the intertrial interval was about 45 min). The familiarization trials were performed at low (4 rpm) constant speed for a maximum duration of 3 min. Trials 1–3 were performed with the accelerating rod, starting with 4 rpm up to 40 rpm within 4 min, with a maximum duration of 5 min. On the following day, a fourth trial on the accelerating rod was carried out. The performance was evaluated by scoring the latency to fall.
In the contextual fear conditioning test mice had to learn the association between the unconditioned (electric footshock) and conditioned (context) stimuli. Mice were conditioned in the context, a chamber (23.5 × 23.5 cm and 19.5 cm high) with Plexiglas walls and ceiling and a stainless grid floor from which an electric shock could be elicited. The chamber was illuminated by white light (10 lux). Mice were placed in the center of the cage and received three electric footshocks (0.25 mA, 1 s) at 120, 160, and 200 s. At 240 s the recoding ended, and the mouse was immediately returned into its home cage. Twenty-four hours after conditioning, mice were placed again in the context for 3 min. The conditioned response was analyzed by quantifying the percentage of time spent freezing (defined as absence of body movements for at least 1 s). Freezing behavior was automatically analyzed using a modified version of the system Mouse-E-Motion (Infra-e-motion, Hamburg, Germany). All behavioral data were analyzed by two-way analysis of variance for repeated measurements having a genotype as between group factor and time interval (for the open field test) and trial (for the accelerated Rotarod and contextual fear conditioning tests) as within group factor. When appropriate, Newman-Keuls post-hoc analyses were performed.
RESULTS
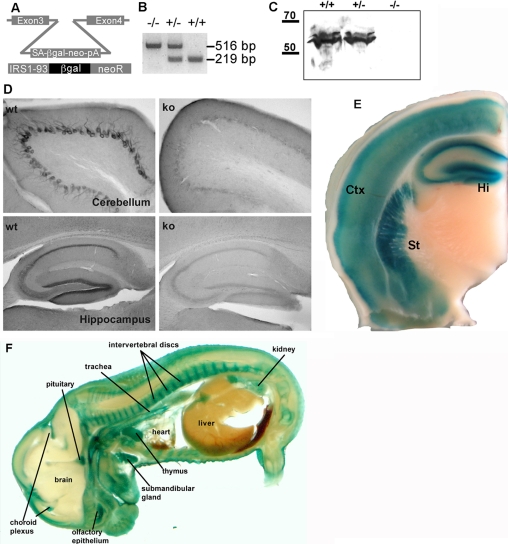
Generation of IRSp53-deficient Mice—We generated IRSp53-deficient animals to determine the functional relevance of the protein in the nervous system. For this we used an ES cell line which harbored an insertion of an exon trapping construct (coding for a neomycin phosphoribosyl transferase/β-galactosidase fusion protein) within intron 3 of the mouse IRSp53 gene (Fig. 1, A and B). Injection into blastocysts yielded offspring with efficient germ line transmission, leading to mice heterozygous and eventually homozygous for the mutant IRSp53 gene. Two independent lines of mice were obtained. The exon trapping cassette leads to a IRSp53/neo/β-galactosidase fusion protein which contains only 73 (of 521) amino acids of IRSp53, leading to a complete functional loss of the protein. Accordingly, IRSp53 immunoreactivity is lost from brains of homozygous mutant mice in immunohistochemical as well as Western blot analyses using a C terminus-directed antibody (Fig. 1, C and D). Genotyping of offspring from heterozygous crosses showed that IRSp53 KO mice were born at a reduced mendelian ratio (6.6% and 7.2% in the 2 lines of mice we generated with respect to the expected 25%). Survival rates improved to 12% for one of the lines after back-crossing into a C57Bl6 background. Surviving KO animals appeared healthy and displayed no gross abnormalities.
FIGURE 1.
Generation of IRSp53-deficient mice. A, structure of the exon trap vector integration site in intron 3 of the IRSp53 gene observed in the embryonic stem cell line XG-757. The integrated DNA fragment contains a splice acceptor (SA), coding sequences forβ-galactosidase (βGal), and neomycin phosphoribosyl transferase (neo) as well as a poyladenylation signal (pA). The predicted fusion protein expressed in these cells contains the first 73 amino acids of IRSp53 followed by βGal and neoR proteins. B, PCR genotyping of the exon trap insertion site; KO allele, 516 bp; WT allele, 219 bp. C, Western blot analysis of brain lysates from WT and heterozygous and KO mice using anti-IRSp53. The blots were deliberately overloaded and overexposed to rule out the presence of trace amounts of the protein which might arise due to alternative or aberrant splicing. D, immunostaining of the cerebellum and hippocampus from WT and IRSp53 KO mice using a C-terminal anti-IRSp53 antibody. In both brain areas staining of cell bodies and intervening layers is reduced to background levels in KO mice. E, a brain section of a heterozygous mouse was stained forβ-galactosidase activity to map IRSp53 expression. Note the strong staining in cortex (Ctx), striatum (St), and hippocampus (Hi). F, section of a (+/-) mouse embryo stained for β-galactosidase activity. Several tissues with particular high expression levels are indicated; note the almost complete absence of staining in brain and spinal chord.
Analysis of the brains of adult heterozygous animals by staining for β-galactosidase activity (which arises due to the expression of the IRSp53/neo/β-galactosidase fusion protein) revealed that cortex, striatum, and hippocampus exhibit high levels of β-galactosidase (and by inference IRSp53) expression (Fig. 1E), in agreement with a previous in situ hybridization analysis (21). Notably, in mouse embryos (shown here for day E15) the central nervous system is almost completely devoid of IRSp53 expression, as analyzed by β-galactosidase staining (Fig. 1F), indicating that expression is switched on postnatally (also see below). Strong expression of IRSp53 in mouse embryos is, however, observed in many peripheral tissues, including thymus, submandibular gland, pituitary, choroid plexus, olfactory epithelium, and the kidney (Fig. 1F).
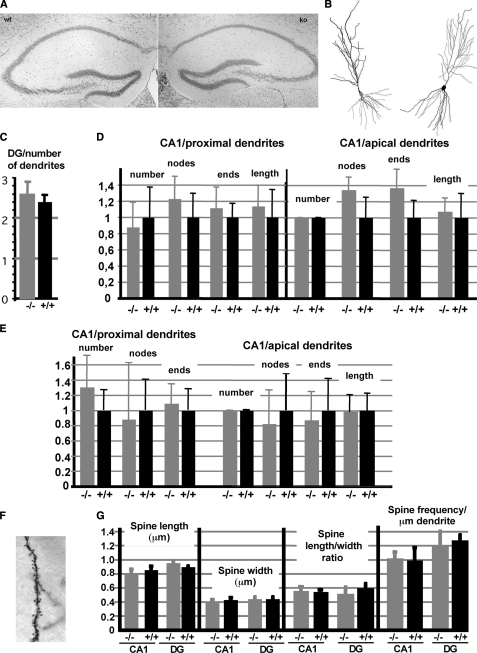
IRSp53 has been suggested to mediate the signaling from activated (GTP-bound) Rho GTPases leading to the generation of cellular protrusions (3, 5, 6). Because of the high expression of the protein in the hippocampus, we expected changes in dendritic or spine morphology in this brain region in KO mice. Therefore, we performed a thorough morphological analysis of hippocampi from WT and KO mice at the light microscopic level. The overall morphology appeared unchanged, as analyzed by Nissl staining (Fig. 2A). In addition, Golgi staining revealed that the numbers of primary dendrites and of dendritic branches were not changed in the hippocampus of IRSp53 KO mice (Fig. 2, B–D). To rule out a compensatory effect during development, we also analyzed dendritic branching in young animals (age 9 days); again no differences between WT and KO animals were observed in the CA1 area of the hippocampus (Fig. 2E). Earlier (i.e. prenatal) stages were not analyzed as IRSp53 is not expressed in the embryonic brain (see above, Fig. 1F). Because IRSp53 was previously shown to enhance the formation and maturation of dendritic spines in cultured neurons (11), we also analyzed spine morphology. When comparing Golgi-stained brain sections of WT and KO mice, we could not detect any significant changes in the length, width, frequency, and form (determined as length/width ratio) of dendritic spines in CA1 and dentate gyrus (DG) regions (Fig. 2, F and G). Thus, in the hippocampus, IRSp53 KO animals do not display any significant structural changes when examined at the light microscopic level either during early postnatal development or in the adult stage.
FIGURE 2.
Neuronal morphology is unchanged in IRSp53-deficient mice. A, architecture of the hippocampus of WT and KO mice visualized by Nissl staining. B, reconstruction of dendritic trees of pyramidal neurons from the CA1 region of WT (left) and KO (right) mice, as obtained by Golgi staining, followed by analysis with a Neurolucida system. C, number of primary dendrites emanating from granule cells in the dentate gyrus (10 neurons per animal, 4 animals per genotype recorded). D, quantification of CA1 pyramidal neuron morphology (eight neurons from four different animals per genotype). For both apical and proximal dendrites, the number of dendrites, number of branch points/nodes, and branch ends as well as total dendrite length were recorded. In all cases values were normalized to the average value obtained for WT animals. E, similar data were recorded from hippocampi from young (postnatal day 9) animals. 13 neurons per genotype were analyzed. F, section of a dendrite of a hippocampal neuron from a KO mouse visualized by Golgi staining. G, spine parameters of WT and KO mice obtained from Golgi stained sections as in E, spine length and width (in μm) were measured in dendrites originating from granule cells of the dentate gyrus (DG) and pyramidal neurons of the CA1 region of the hippocampus. Width/length ratios and spine densities were deduced from these measurements. Each data point represents measurements from four animals per genotype (±S.D.), with at least 10 dendritic segments per animal and a total of more than 300 spines analyzed per data point.
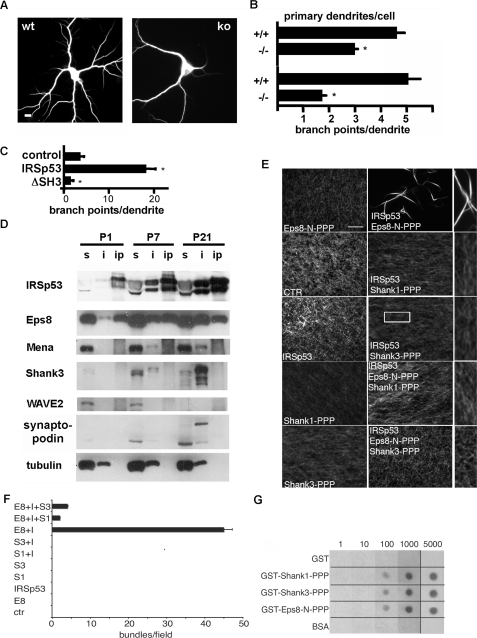
IRSp53-containing Complexes Differentially Regulate Actin Dynamics—Our findings contradict previous evidence implying IRSp53 as a key regulator of cellular morphology (3, 5, 11). As this evidence was based mostly on cultured cells in vitro, we also analyzed morphological parameters in cultured neurons derived from WT and KO mice. Under these conditions, we observed indeed a significant reduction in primary dendrites and dendritic branches (Fig. 3, A and B). These defects could be rescued by ectopic expression of the protein (supplemental Fig. S1). This is mirrored by the finding that the overexpression of IRSp53, but not of mutant lacking the SH3 domain (ΔSH3 in Fig. 3C) in cultured rat hippocampal neurons, increased dendritic branching. Collectively, these observations indicate that IRSp53 regulates morphogenesis of primary processes in neurons in vitro through interaction partners of its SH3 domain.
FIGURE 3.
Different IRSp53-based complexes control cellular morphology and actin dynamics. A, neurons prepared from hippocampi of WT and KO mice (14 days in vitro) were stained for the dendrite marker MAP2 (scale bar = 10 μm). B, quantification of effects of IRSp53 deletion on neuronal morphology, as determined from experiments shown in A. *, different from WT neurons; p < 0.001; n = 3. C, rat hippocampal neurons were transfected with enhanced green fluorescence protein alone (control) or in combination with IRSp53 or a IRSp53 ΔSH3 deletion mutant. Branching of dendrites was evaluated as in B but using enhanced green fluorescence protein fluorescence. *, different from control; p < 0.001; n = 3. D, hippocampi were prepared at different time points of postnatal developments. After solubilization in immunoprecipitation buffer, soluble (s) material was subjected to immunoprecipitation using immobilized anti-IRSp53 (ip). Insoluble material (i) was dissolved in 1% SDS. Samples were analyzed by Western blotting using the antibodies indicated. E and F, shank antagonizes actin bundling induced by the Eps8·IRSp53 complex. Fluorescence microscopy-based F-actin-bundling assay of IRSp53 in the presence or absence of various combinations of the proline-rich regions of Eps8 or Shank1 or -3. F-actin (1 μm) was incubated with the indicated concentrations of bovine serum albumin (BSA), used as a control (CTR), or the proline-rich region of Eps8 (Eps8-N-PPP, 0.5 μm) or Shank1 (Shank1-PPP, 2.0 μm) or Shank3 (Shank3-PPP, 2.0 μm) or IRSp53 (1.0 μm) alone (left) or in combination (center), as indicated. Actin filaments labeled with rhodamine-phalloidin were imaged using a fluorescence microscope. 2.5-Fold enlargements of portions of center panels are shown on the right, as indicated for the IRSp53/shank3-PPP experiment. The images are representative fields of at least four independent experiments. For quantification (F), at least 10 random microscopic fields for each assay were analyzed, and the number of actin bundles/field were counted. Data are the mean ± S.E. of triplicate experiments. All immunofluorescent images were acquired with 60× magnifying objective. Bar, 5 μm. G, equal amounts (10 pmol) of GST alone (GST) or GST-fused to the proline-rich region of Shank1 (GST-Shank1-PPP) or Shank3 (GST-Shank3-PPP) or Eps8 (GST-Eps8-N-PPP) or bovine serum albumin as a control were spotted on nitrocellulose and incubated with the indicated concentrations of purified His-tagged IRSp53, which was revealed by immunoblotting with anti-His6 antibody.
We reasoned that the differences between in vivo and in vitro conditions may be due to the dynamic formation of different IRSp53-based complexes, whose activities may become limiting and, thus, functionally relevant only under controlled in vitro conditions but dispensable in vivo. To determine which of these complexes are relevant in the hippocampus (in particular, which interaction partners of the IRSp53 SH3 domain), we performed a biochemical analysis to identify the major IRSp53 binding proteins in the hippocampus during various developmental stages. Analysis of detergent soluble and insoluble fractions from hippocampi prepared at postnatal days 1–21 showed that IRSp53 expression is weak at postnatal day 1 (in agreement with the absence of expression before birth; see Fig. 1F) but increases significantly during postnatal development toward P21. IRSp53 is at all times equally present in the soluble and the insoluble fraction (Fig. 3D). Among the actin regulatory proteins tested, only Eps8, but not WAVE2 or Mena could be efficiently coimmunoprecipitated with IRSp53. Conversely, Shank proteins (shown here for Shank3) and a larger molecular weight form of synaptopodin (Synpo-long) were strongly enriched in the insoluble fraction (which includes the PSD). Thus, possible downstream effectors of IRSp53 may be very roughly divided into Eps8 in the soluble fraction and shank and synaptopodin proteins in the insoluble fraction. Overlay assays provided further indirect evidence that shank is the quantitatively most relevant target of Cdc42/IRSp53 signaling in the PSD (supplemental Fig. S2), suggesting an important role of this latter complex in mediating some of the synaptic alterations observed in KO mice.
Remarkably, the two major IRSp53/Eps8 and IRSp53/Shank complexes significantly differed in their ability to modulate actin cytoskeletal dynamics. Interaction of a proline-rich segment of Eps8 (Eps8-N-PPP motif) with the SH3 domain of IRSp53 is sufficient to activate actin bundling via the IRSp53 metastasis homology domain (Ref. 4 and Fig. 3, E and F). Conversely, neither Shank1 nor Shank3 PPP motifs, which bind IRSp53 with affinities comparable with those observed for Eps8 (Fig. 3G), were able to induce actin bundling in this type of assay (Fig. 3, E and F). Moreover, both Shank variants abrogated actin bundling induced by Eps8 when incubated together with the proline-rich fragment of Eps8. Thus, Shank antagonizes the actin bundling activity of IRSp53, which can be induced by Eps8. Consistently, coexpression of shank3 with IRSp53 blocked the increase in the number of primary dendrites observed when IRSp53 is expressed in cultured neurons (supplemental Fig. S3).
Altered Synaptic Plasticity in IRSp53-deficient Mice—As both IRSp53 and shank proteins are highly concentrated at synaptic sites (12), we analyzed hippocampal synaptic transmission in WT and KO mice at synapses formed by entorhinal cortex axons in the dentate gyrus in vivo and at Schaffer collateral synapses in hippocampal slices in vitro. For stimulation via the perforant path at the angular bundle, stimulation intensity for base-line recordings was adjusted to elicit population spikes with amplitudes of 2–3 mV. Basal excitatory synaptic transmission was normal in the dentate gyrus of IRSp53-deficient mice as we observed no significant difference in the selected stimulation intensity, population spike amplitude, and slope of fEPSPs between WT and KO mice (Fig. 4A). Paired-pulse stimulation recorded at the maximum stimulus intensity with intervals of 50, 100, 200, and 500 ms produced paired-pulse facilitation (PPF) of the population spike amplitude, which was significantly more pronounced in IRSp53-deficient animals (Fig. 4B). For inter-pulse intervals of 50 and 200 ms, higher PPF of population spikes amplitude in IRSp53 KO mice was detected by the t test (p < 0.05), and there was a similar tendency for 100- and 500-ms intervals (Fig. 4B).
Subthreshold (i.e. not eliciting a population spike) paired-pulse stimulation with short inter-pulse intervals of 10 and 25 ms produced similar PPFs of the fEPSPs in IRSp53 WT and KO mice (Fig. 4C). Analysis of fEPSPs for longer intervals (50 and 100 ms) further confirmed an increased PPF in IRSp53 KO mice (F(1,12) = 6.834; p = 0.023; two-way analysis of variance; p < 0.05 for 50 ms and p < 0.1 for 100 ms, t test).
Theta-burst stimulation elicited LTP in vivo in both genotypes (Fig. 4D). The levels of potentiation in the slope of fEPSP were: 115 ± 3% in IRSp53+/+ and 121 ± 6% in IRSp53 KO mice. Despite the apparent tendency for potentiation of fEPSPs to be larger in IRSP53 KO mice, there was no significant difference between genotypes in the dentate gyrus.
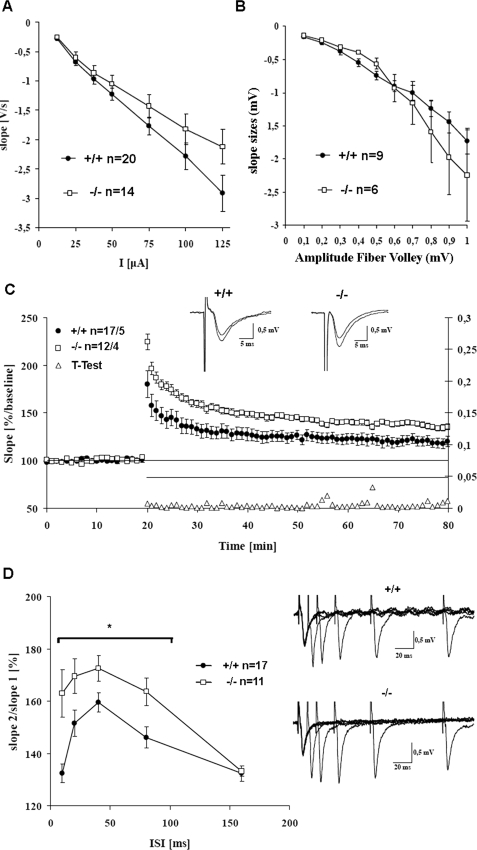
Electrophysiological parameters were also recorded in the CA1 region in the hippocampal slice preparation; again, basal synaptic transmission was unaffected by loss of IRSp53, as detected by fEPSP and fiber volley measurements (Fig. 5, A and B). At higher stimulus intensities and larger fiber volleys, KO mice showed a tendency for larger fEPSP slopes, which were, however, not significantly different from WT animals. Theta-burst stimulation induced LTP in the Schaffer collateral pathway in both genotypes. In this paradigm we observed a stronger potentiation in IRSp53 KO mice, which was significantly different from WT animals at all time points tested, including post-tetanic potentiation (Fig. 5C; p < 0.05, t test). In addition, PPF was significantly increased in KO mice for 10-, 20-, 40-, 80-ms inter-stimulus intervals (Fig. 5D; p < 0.05, t test), as seen before in the dentate gyrus. Thus, collectively, these results showed that IRSp53 KO mice have normal basal synaptic transmission, but increased short-term (PPF in the dentate gyrus and CA1) and long-term (LTP in CA1) synaptic plasticity.
FIGURE 5.
Altered synaptic plasticity in IRSp53 KO mice in the acute hippocampal slice preparation. A, base-line fEPSPs are not significantly changed between WT and KO mice, but KO mice show a tendency to larger fEPSPs at larger stimulation strength. The stimulus electrode was positioned in the CA3 region of the hippocampus and the recording electrode at the CA1 region. B, fiber volley measurements. Responses are largely unchanged in KO animals, with a tendency to larger responses at high stimulus intensities. C, IRSp53-deficient mice exhibit enhanced LTP in the Schaffer collateral pathway. Theta burst stimulation (3 × 100Hz) was applied 20 min after stable base-line recordings. Data points are averaged over six sweeps, and error bars represent S.E. The left abscissa shows the initial slope of the fEPSPs in comparison to base-line values (100%); the right abscissa shows the p values for all t tests performed every minute. KO mice show a significantly higher potentiation starting with post-tetanic potentiation right after theta burst stimulation application. Insets show original fEPSPs during base-line recording and 55 min after theta burst stimulation application. D, PPF is enhanced in KO mice. PPF was measured for the 10, 20, 40, 80, and 160 ms inter-stimulus interval (ISI). Data are plotted as slope2/slope1 against the inter-stimulus interval. The increase in PPF is significant for the 10–80-ms inter-stimulus interval (p < 0.05, t test). The right side shows the original fEPSP sweeps for WT and KO mice.
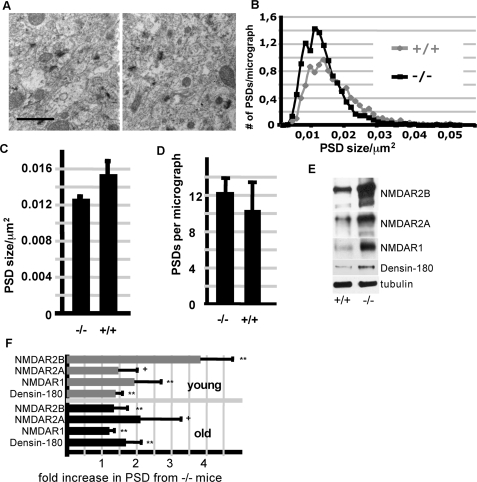
As IRSp53 is strongly enriched in the postsynaptic density, we analyzed whether its loss did result in changes in PSD size or composition. Electron micrographs of the hippocampus showed a slight but significant decrease in the size of the PSDs in KO mice, whereas the density of PSDs was slightly, but not significantly increased (Fig. 6, A and B). In related experiments in cultured neurons from WT and KO mice, we observed that formation of postsynaptic (i.e. PSD-95 positive) clusters is delayed in KO neurons (supplemental Fig. S4).
FIGURE 6.
Altered PSD size and composition in IRSp53-deficient brains. A, hippocampal PSDs of WT (left) and IRSp53-deficient mice (right) were analyzed by electron microscopy; all PSDs identified in at least 20 micrographs per animal (n = 3 animals per genotype; >1200 PSDs analyzed per genotype) were scored for the area covered (scale bar, 0.5 μm). Frequency distribution (B) and statistical analysis (C) indicate that PSDs in WT animals are on average 17% (S.D. ± 14) larger than their counterparts in IRSp53-deficient animals (n = 3; p < 0.05). The total number of PSDs (related to a micrograph covering an area of 8.1 × 10 μm) is slightly but not significantly increased in KO animals (D). E, Western blot of PSD preparations derived from brains of young (2 weeks) WT and KO mice. Shown are representative blots for those four proteins which exhibit an increase in IRSp53-deficient animals. F, quantitative analysis of the data shown in E for young animals, and similar data sets obtained for adult (2 months) animals. Each blot signal was normalized to the signal obtained with anti-tubulin from the same blot. Data obtained from KO animals are then presented as the -fold increase compared with the corresponding tubulin-normalized signal obtained from WT mice (**, p < 0.01; *, p < 0.05; +, p < 0.1; t test n = 3; ±S.D.)
To start unraveling the molecular changes occurring in the PSD of IRSp53-deficient mice, we then compared the levels in the PSD of more than 30 proteins, including most known interaction partners of IRSp53 and Shank proteins. PSDs were prepared from adult (2 months old) as well as from young (2 weeks old) mice. Whereas the abundance of most proteins studied was not changed (supplemental Table S1), we found an increase in Densin-180, which, like IRSp53, interacts with Shank proteins (17). More importantly, we observed an increase in NMDA receptor subunits in KO animals, which was particularly evident in young animals but detectable also in adult mice. Both NMDAR1 and NMDAR2a/b subunit levels were increased (Fig. 6, C and D). No changes were found in the AMPA type GluR1 subunit or the associated stargazin (supplemental Table S1). As induction of LTP depends on the activation of NMDA receptors, the observed increase in NMDA receptors indicates a molecular explanation for the increased LTP in the CA1 region of KO mice.
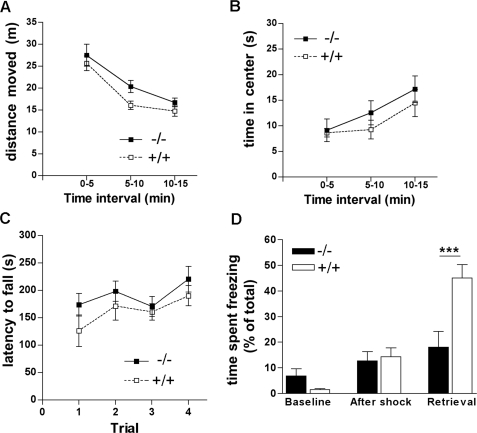
Given the high expression levels of IRSp53 in brain regions involved in cognition (e.g. hippocampus), reward (ventral striatum) and motor control (dorsal striatum, cerebellum), we expected behavioral deficits in KO mice and performed several behavioral assays. The open field test scores motor activity and exploratory behavior of mice; in addition, as the center of the open field area is an anxiogenic stimulus for mice, the test is used to measure anxiety. Here we did not observe any differences between KO mice and their WT littermates in the distance moved (Fig. 7A) and time spent in the center (Fig. 7B) (effect of genotype: F1,17 = 0.9, p > 0.05 for distance moved and F1,17 = 2.4, p > 0.05 for time spent in the center; effect of the interaction between genotype and time interval: F2,34 = 0.3, p > 0.05 for distance moved and F2,34 = 0.8, p > 0.05 for time spent in the center). Both genotypes showed short term familiarization to the open field as indicated by the decrease in distance moved and increase in time spent in the center over the 15-min duration of the test (effect of time interval: F2,34 = 8.3, p < 0.01 for distance moved and F2,34 = 52.6, p < 0.001 for time spent in the center) (Fig. 7, A and B). Similarly, the accelerated Rotarod test indicated that motor coordination of KO mice was unchanged (Fig. 7C). There was no effect of genotype (F1,18 = 1.2, p > 0.05) nor of the interaction between genotype and trials (F3,54 = 0.7, p > 0.05) on the latency to fall from the rotating rod. Both genotypes equally well improved their performances as indicated by the increase in latency to fall over trials (effect of trials: F3,54 = 6.8, p < 0.001), suggesting that cerebellum-dependent motor learning (22) was normal in KO mice. However, KO mice showed significant deficits in the contextual fear conditioning test that has been shown to measure hippocampus-dependent learning and memory (23). Here mice were introduced into a new environment where they received electrical foot shocks. When reintroduced into the same environment to test memory retrieval on the next day, WT mice responded by freezing for prolonged times. In contrast, KO mice spent less time freezing as compared with WT mice during retrieval (p < 0.001, post-hoc comparison after a significant effect of the interaction between genotype and trial: F2,36 = 11.3, p < 0.001), indicating that they did not learn or memorize the association between the foot shocks and the context. No difference was detected between genotypes during the 2 min before and after mice received the first foot shock (Fig. 7D), suggesting that anxiety toward the context and pain sensitivity, respectively, were unchanged in KO mice.
FIGURE 7.
Behavioral analysis. A and B, open field test. Animals (n = 10 per genotype) were placed into the open field box, and the total distance moved (A) as well as the time spent in the center of the box (B) were recorded for three subsequent 5-min intervals. C, Rotarod test. Animals were placed on an accelerated rotating corrugated rod; during four trials (trial 1–3 on the first day; trial 4 on the next day) lasting 300 s each, the latency of mice until they fall from the rod was recorded. D, contextual fear-conditioning test. Mice were placed into a box with a metal grid floor (context); electro-shocks were applied, whereas the time that mice spent motionless (freezing) was continuously monitored during the 2 min before (Base line) and 2 min after the first foot shock (After shock). On the next day the mice were returned to the same box without applying any shock, and the time spent freezing was again monitored for 3 min (Retrieval). Note the significant difference between WT (+/+) and KO (-/-) mice during retrieval, indicating that KO mice do not learn or memorize the association between the foot shocks and the context (***, p < 0.0001, Newman-Keuls post-hoc after two-way analysis of variance for repeated measurements).
DISCUSSION
IRSp53 has been primarily described as a regulator of cellular morphology. Both the N-terminal IMD (IRSp53/missing in metastasis homology domain) and the SH3 domain of IRSp53 may be involved in generating cellular protrusions; whereas the IMD induces filopodial actin bundles and aids in pushing the plasma membrane outwards (10), the SH3 domain recruits actin regulatory proteins such as Eps8, Mena, or WAVE2 in a small G-protein-dependent manner. As brain regions such as the hippocampus are among those tissues with the highest expression levels of IRSp53 (Ref. 13; see also Fig. 1E), we expected to observe changes in dendritic morphology in brains of KO mice. However, our data suggest that in neuronal cells morphological effects of IRSp53 are limited to in vitro conditions. Branching of dendrites is affected by the loss of IRSp53 in cultured neurons but not in the hippocampus of young and adult mice in vivo. Previous work (11) has also demonstrated a role for IRSp53 in the generation of dendritic spines, which is again not mirrored in our measurements in both the dentate gyrus and CA1 region of the hippocampus. This discrepancy may be due to a compensatory effect by other Rho GTPase effectors. A literature survey identifies more than 50 proteins known to regulate spine actin, and most of these work within Rho GTPase controlled signaling pathways, similar to IRSp53. Cultured neuron systems are likely to be more vulnerable to experimental manipulations such that effects of individual actin regulators will be detected more easily. In addition, we reasoned that IRSp53 modulates actin dynamics and, by inference, cell shape, only via some but not all interaction partners. We compared the effects of Eps8, which is particularly relevant as a binding partner of IRSp53 in the hippocampus, and Shank proteins, which interact with IRSp53 specifically in the PSD (12, 13). We have described before that Eps8 and IRSp53 act synergistically to form cellular protrusions (5). Our data indicate that at postsynaptic sites Shank proteins and eventually also synaptopodin are likely to block any effect of IRSp53 on actin dynamics. Interestingly, synaptopodin has been described to block filopodia induction by the Cdc42·IRSp53·Mena complex in kidney podocytes (24) and has been linked to synaptic plasticity (25). In contrast to Mena, Eps8, and WAVE2, Shank proteins as well as synaptopodin lack further actin regulatory modules such as actin capping (Eps8) or Arp2/3 recruiting (WAVE2) domains. In addition, we show here that Shank1 and Shank3 PPP motifs antagonize the bundling activity induced by the respective Eps8-PPP motif, thus interfering with both pathways by which IRSp53 could induce cellular protrusions. Instead, Shank proteins are deeply entrenched within the PSD and together with PSD-95 are likely to determine a postsynaptic role for IRSp53. Shank expression increases strongly during development, suggesting that in mature neurons the effect of Shank proteins is dominant over that of other IRSp53-interacting proteins.
Consistently, our electrophysiological data provide evidence for a synaptic function of IRSp53. The electrophysiological measurements in the hippocampus indicate that IRSp53 is required to limit the extent of paired-pulse facilitation and LTP. One likely explanation for the increase in CA1 LTP observed in IRSp53-deficient mice may be found in the increased incorporation of NMDA receptor subunits into the PSD of these animals, allowing for a stronger induction of LTP. Both IRSp53 and NMDAR2 subunits bind to PDZ1 and PDZ2 of PSD-95 (11, 12, 26); thus, the lack of IRSp53 might allow for enhanced anchoring of NMDA receptors. Another aspect which might be relevant for the increase in LTP is that, in the absence of IRSp53 binding to PSD-95, more “slots” may become available for AMPA receptor-stargazin complexes (also interacting with PSD-95 PDZ domains) to be inserted into the PSD once LTP has been initiated. The number of IRSp53 molecules in a PSD is estimated to about one-fifth that of the number PSD-95-like scaffolds (including PSD-93, SAP102; Ref. 27). This number is likely to increase upon activation of NMDA receptors, as it was shown that IRSp53 is translocated toward the PSD from diffuse dendritic stores after treatment of neurons with NMDA (28). Thus, the lack of IRSp53 may quantitatively affect the number of “open” binding sites for PDZ domain ligands. Consistent with this possibility, several studies have indicated that the limiting factor for AMPA receptor anchoring at synapses is the presence of MAGUK molecules such as PSD-95 (29).
This scenario, however, is unlikely to account for the increase in paired-pulse facilitation, which was apparent both in the dentate gyrus and CA1 synapses in KO mice. PPF is generally regarded as a presynaptic phenomenon, and in keeping with this notion some IRSp53 has been observed presynaptically (supplemental Fig. S5; Ref. (11)). However, it has recently been demonstrated that reduction of postsynaptic PSD-95 levels may also lead to an increase in paired pulse ratio in a retrograde manner (30), indicating that the postsynaptic density might also regulate the presynaptic side. This regulation may occur either via the release of retrograde messenger or through adhesion molecules that span the synaptic cleft. Thus, loss of IRSp53 and loss of PSD-95 both result in increased PPF and CA1 LTP, suggesting that both proteins are not only tightly linked but also contribute in similar ways to synaptic plasticity.
The synaptic role of IRSp53 is also consistent with its putative role as effector of the Rho GTPases, Rac and Cdc42, whose implication in synaptic plasticity has recently emerged. For instance, a Rac1/c-Jun NH2-terminal kinase-dependent signaling pathway was shown to regulate the availability of PSD-95 at synapses during LTP and long term depression (31). Additionally, deficits in LTP have been recorded in mice lacking p21-activated kinase 3 (32), another effector protein downstream of Rac and Cdc42.
Consistent with alterations in CA1 LTP, we also observed behavioral deficits in KO mice. In particular, KO mice were apparently unable to remember the cage in which they previously had experienced electrical foot shocks and, therefore, did not exhibit motionless, fearful behavior (i.e. freezing) when they were reintroduced into this context. We used this test, as it relies at least partially on hippocampus function (23, 33). Other behavioral parameters such as motor coordination (measured in the accelerated Rotarod assay) and exploratory behavior (open field test) were unchanged in KO mice when compared with WT mice, suggesting that IRSp53 deficiency has a specific effect on cognitive function. It may be related to the fact that LTP is facilitated by exploration of a novel environment (34), like the conditioning chamber. The interplay between this influence of novel environment on LTP and exaggerated LTP in IRSp53 KO mice may lead to saturated LTP in specific synapses encoding conditioned context and also to LTP at non-related synapses that are eventually activated in the conditioned context. This would lead to a more “fuzzy” representation of the conditioned environment in KO mice as compared with WT mice. This idea is in line with findings of Xu et al. (35) who showed that the sparsely distributed potentiation of selected synapses and not the level of LTP by itself is important during exploration and learning. Similarly to IRSp53 KO, there is an inverse relationship between LTP and hippocampus-dependent learning in mice lacking PSD-95 or LIM kinase-1, which have enhanced LTP in CA1 and impaired spatial learning in the Morris water maze (36, 37). On the other hand, in many other cases changes in LTP and learning are positively correlated (for a review, see Ref. 38). Thus, additional factors, like maintenance of late LTP and plasticity in synapses beyond CA1 region, may also contribute to impairment of cognitive function in IRSp53 KO mice.
On the molecular level, further work will address how Rac or Cdc42 contribute to the regulation of synaptic plasticity by IRSp53. Active Cdc42 induces membrane localization (5) and interaction of IRSp53 with shank (12). Activation by small G-proteins might, therefore, be the initial stimulus, which leads to incorporation of IRSp53 into the PSD, where it could then act to limit long term potentiation of synaptic transmission.
Supplementary Material
Acknowledgments
We thank Irm Hermans-Borgmeyer and The Center for Molecular Neurobiology Hamburg transgenic service facility for blastocyst injection, Hans-Hinrich Hönck for excellent technical assistance, Daniel Prischl for help with PSD analysis, Dietmar Kuhl and Ora Ohana (Berlin) for access and advice to their Neurolucida setup, and Melitta Schachner for access to behavioral and electrophysiological setups.
This work was supported by Deutsche Forschungsgemeinschaft Kr1321 and FOR885 (to H.-J. K.), Di 702/4-1,2,3 (to A. Dityatev), and SFB497/B8 (to T. M. B.), by Associazione Italiana Ricerca sul Cancro and Progetti di Ricerca di Interesse Nazionale 2007 (to G. S.), and by Fondazione Italiana Ricerca sul Cancro (to A. Disanza).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
Footnotes
The abbreviations used are: IRSp53; insulin receptor substrate of 53 kDa; fEPSP, field excitatory postsynaptic potential; LTP, long term potentiation; PSD, postsynaptic density; PSD-95, PSD protein of 95 kDa; PDZ, PSD-95/discs large/ZO-1; PPF, paired-pulse facilitation; SH3, Src homology; NMDA, N-methyl-d-aspartate; PBS, phosphate-buffered saline.
References
- 1.Govek, E. E., Newey, S. E., and Van Aelst, L. (2005) Genes Dev. 19 1-49 [DOI] [PubMed] [Google Scholar]
- 2.Tashiro, A., Minden, A., and Yuste, R. (2000) Cereb. Cortex. 10 927-938 [DOI] [PubMed] [Google Scholar]
- 3.Miki, H., Yamaguchi, H., Suetsugu, S., and Takenawa, T. (2000) Nature 408 732-735 [DOI] [PubMed] [Google Scholar]
- 4.Krugmann, S., Jordens, I., Gevaert, K., Driessens, M., Vandekerckhove, J., and Hall, A. (2001) Curr. Biol. 11 1645-1655 [DOI] [PubMed] [Google Scholar]
- 5.Disanza, A., Mantoani, S., Hertzog, M., Gerboth, S., Frittoli, E., Steffen, A., Berhoerster, K., Kreienkamp, H. J., Milanesi, F., Di Fiore, P. P., Ciliberto, A., Stradal, T. E., and Scita, G. (2006) Nat. Cell Biol. 8 1337-1347 [DOI] [PubMed] [Google Scholar]
- 6.Lim, K. B., Bu, W., Goh, W. I., Koh, E., Ong, S. H., Pawson, T., Sudhaharan, T., and Ahmed, S. (2008) J. Biol. Chem. 283 20454-20472 [DOI] [PubMed] [Google Scholar]
- 7.Yamagishi, A., Masuda, M., Ohki, T., Onishi, H., and Mochizuki, N. (2004) J. Biol. Chem. 279 14929-14936 [DOI] [PubMed] [Google Scholar]
- 8.Millard, T. H., Bompard, G., Heung, M. Y., Dafforn, T. R., Scott, D. J., Machesky, L. M., and Futterer, K. (2005) EMBO J. 24 240-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suetsugu, S., Kurisu, S., Oikawa, T., Yamazaki, D., Oda, A., and Takenawa, T. (2006) J. Cell Biol. 173 571-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattila, P. K., Pykalainen, A., Saarikangas, J., Paavilainen, V. O., Vihinen, H., Jokitalo, E., and Lappalainen, P. (2007) J. Cell Biol. 176 953-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, J., Ko, J., Racz, B., Burette, A., Lee, J. R., Kim, S., Na, M., Lee, H. W., Kim, K., Weinberg, R. J., and Kim, E. (2005) J. Neurosci. 25 869-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soltau, M., Richter, D., and Kreienkamp, H. J. (2002) Mol. Cell. Neurosci. 21 575-583 [DOI] [PubMed] [Google Scholar]
- 13.Bockmann, J., Kreutz, M. R., Gundelfinger, E. D., and Bockers, T. M. (2002) J. Neurochem. 83 1013-1017 [DOI] [PubMed] [Google Scholar]
- 14.Soltau, M., Berhorster, K., Kindler, S., Buck, F., Richter, D., and Kreienkamp, H. J. (2004) J. Neurochem. 90 659-665 [DOI] [PubMed] [Google Scholar]
- 15.Glaser, E. M., and Van der Loos, H. (1981) J. Neurosci. Methods 4 117-125 [DOI] [PubMed] [Google Scholar]
- 16.Khelfaoui, M., Denis, C., van Galen, E., de Bock, F., Schmitt, A., Houbron, C., Morice, E., Giros, B., Ramakers, G., Fagni, L., Chelly, J., Nosten-Bertrand, M., and Billuart, P. (2007) J. Neurosci. 27 9439-9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quitsch, A., Berhorster, K., Liew, C. W., Richter, D., and Kreienkamp, H. J. (2005) J. Neurosci. 25 479-487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoenica, L., Senkov, O., Gerardy-Schahn, R., Weinhold, B., Schachner, M., and Dityatev, A. (2006) Eur. J. Neurosci. 23 2255-2264 [DOI] [PubMed] [Google Scholar]
- 19.Anderson, W. W., and Collingridge, G. L. (2001) J. Neurosci. Methods 108 71-83 [DOI] [PubMed] [Google Scholar]
- 20.Freitag, S., Schachner, M., and Morellini, F. (2003) Behav. Brain Res. 145 189-207 [DOI] [PubMed] [Google Scholar]
- 21.Thomas, E. A., Foye, P. E., Alvarez, C. E., Usui, H., and Sutcliffe, J. G. (2001) Neurosci. Lett. 309 145-148 [DOI] [PubMed] [Google Scholar]
- 22.Nolan, M. F., Malleret, G., Lee, K. H., Gibbs, E., Dudman, J. T., Santoro, B., Yin, D., Thompson, R. F., Siegelbaum, S. A., Kandel, E. R., and Morozov, A. (2003) Cell 115 551-564 [DOI] [PubMed] [Google Scholar]
- 23.Bast T., Zhang W. N., and Feldon J. (2003) Hippocampus 13 657-675 [DOI] [PubMed] [Google Scholar]
- 24.Yanagida-Asanuma, E., Asanuma, K., Kim, K., Donnelly, M., Young Choi, H., Hyung Chang, J., Suetsugu, S., Tomino, Y., Takenawa, T., Faul, C., and Mundel, P. (2007) Am. J. Pathol. 171 415-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deller, T., Korte, M., Chabanis, S., Drakew, A., Schwegler, H., Stefani, G. G., Zuniga, A., Schwarz, K., Bonhoeffer, T., Zeller, R., Frotscher, M., and Mundel, P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10494-1049920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornau, H. C., Schenker, L. T., Kennedy, M. B., and Seeburg, P. H. (1995) Science 269 1737-1740 [DOI] [PubMed] [Google Scholar]
- 27.Cheng, D., Hoogenraad, C. C., Rush, J., Ramm, E., Schlager, M. A., Duong, D. M., Xu, P., Wijayawardana, S. R., Hanfelt, J., Nakagawa, T., Sheng, M., and Peng, J. (2006) Mol. Cell Proteomics 5 1158-1170 [DOI] [PubMed] [Google Scholar]
- 28.Hori, K., Yasuda, H., Konno, D., Maruoka, H., Tsumoto, T., and Sobue, K. (2005) J. Neurosci. 25 2670-2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elias, G. M., Funke, L., Stein, V., Grant, S. G., Bredt, D. S., and Nicoll, R. A. (2006) Neuron 52 307-320 [DOI] [PubMed] [Google Scholar]
- 30.Futai, K., Kim, M. J., Hashikawa, T., Scheiffele, P., Sheng, M., and Hayashi, Y. (2007) Nat. Neurosci. 10 186-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, M. J., Futai, K., Jo, J., Hayashi, Y., Cho, K., and Sheng, M. (2007) Neuron 56 488-502 [DOI] [PubMed] [Google Scholar]
- 32.Meng, J., Meng, Y., Hanna, A., Janus, C., and Jia, Z. (2005) J. Neurosci. 25 6641-6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders, M. J., Wiltgen, B. J., and Fanselow, M. S. (2003) Eur. J. Pharmacol. 463 217-223 [DOI] [PubMed] [Google Scholar]
- 34.Kemp, A., and Manahan-Vaughan, D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8192-8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, L., Anwyl, R., and Rowan, M. J. (1998) Nature 394 891-894 [DOI] [PubMed] [Google Scholar]
- 36.Migaud, M., Charlesworth, P., Dempster, M., Webster, L. C., Watabe, A. M., Makhinson, M., He, Y., Ramsay, M. F., Morris, R. G., Morrison, J. H., O'Dell, T. J., and Grant, S. G. (1998) Nature 396 433-439 [DOI] [PubMed] [Google Scholar]
- 37.Meng, Y., Zhang, Y., Tregoubov, V., Janus, C., Cruz, L., Jackson, M., Lu, W. Y., MacDonald, J. F., Wang, J. Y., Falls, D. L., and Jia, Z. (2002) Neuron 35 121-133 [DOI] [PubMed] [Google Scholar]
- 38.Lynch, M. A. (2004) Physiol. Rev. 84 87-136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.